Abstract
As documented in numerous studies, microRNAs (miRNAs) play key roles in various biological processes associated with melanoma occurrence and development. In this study, we found that miRNA-342 (miR-342) was significantly downregulated in melanoma tissues and cell lines. Additionally, the ectopic expression of miR-342 prohibited the cell proliferation and invasion of melanoma. Moreover, zinc-finger E-box-binding homeobox 1 (ZEB1) was identified as a direct target gene of miR-342 in melanoma. Similar with the results induced by miR-342 overexpression, ZEB1 knockdown attenuated cell proliferation and invasion in melanoma. Furthermore, the restoration of ZEB1 expression reversed the suppressive effects of miR-342 on the proliferation and invasion of melanoma cells. These findings suggest that miR-342 may play tumor-suppressing roles in melanoma, at least partially, by directly inhibiting ZEB1 expression. Therefore, miR-342 may be developed as a potential candidate for the treatment of patients with this aggressive type of cancer.
Key words: Melanoma, MicroRNA-342 (miR342), Proliferation, Invasion, Zinc-finger E-box-binding homeobox 1 (ZEB1)
INTRODUCTION
Melanoma is an aggressive form of skin cancer that develops from melanocytes1. This condition is the fifth most common cancer in males and the seventh most common cancer in females around the world2. Melanoma is characterized by high invasion, high metastasis, and high mortality3. In the last 20 years, the morbidity of melanoma globally has increased compared with those of other malignancies4. Patients who acquire melanoma at an early stage could be treated with surgical management; however, patients at an advance stage respond poorly to current treatments5. Despite the advancements in treatments, including surgery and radiotherapy, the therapeutic outcome of patients with melanoma remains unsatisfactory, presenting a 5-year survival rate of less than 15%4. Numerous studies have recently demonstrated that aberrantly expressed oncogenes and tumor suppressors are involved in the initiation and progression of melanoma7,8. However, the detailed molecular mechanism underlying the pathogenesis of melanoma remains largely unknown. Therefore, full understanding of the mechanisms underlying melanoma formation and progression is urgently needed, and novel therapeutic strategies to treat this disease must be developed.
MicroRNAs (miRNAs) are a type of endogenous, non-protein-coding, short RNAs of 19 to 22 nucleotides in length9. miRNAs serve as important regulators of gene expression through direct interaction with seed sequences in the 3′-untranslated regions (3′-UTRs) of their target genes, thus resulting in mRNA degradation or translational inhibition10. To date, more than 1,400 miRNAs have been validated in the human genome and are estimated to regulate the expression of approximately 30% of all human genes11. Altered miRNA expression has been identified in a variety of human malignancies, including melanoma12. miRNAs may participate in carcinogenesis and progression by regulating a wide range of biological processes, including cell proliferation, cell cycle, apoptosis, migration, invasion, epithelial–mesenchymal transition, metastasis, and angiogenesis13–15. Dysregulated miRNAs in human cancer can act as tumor suppressors or oncogenes depending on the nature of their target genes16. Accordingly, considerable effort is needed to elucidate the expression level, roles, and underlying mechanism of cancer-related miRNAs, and the outcome may be particularly useful for providing potential therapeutic targets in the treatment of patients with cancer.
The deregulation of miR-342 has been reported in numerous cancer types17–20. However, the expression pattern and biological roles of miR-342 in melanoma remain largely unknown. In this study, we detected the expression level and investigated the roles of miR-342 in the progression of melanoma as well as the underlying molecular mechanism.
MATERIALS AND METHODS
Clinical Sample Collection
A total of 27 paired melanoma tissues and matched adjacent normal tissues were acquired from patients with melanoma who underwent surgical resection in the Hubei Provincial Hospital of Traditional Chinese Medicine (Hubei, P.R. China) between February 2014 and August 2016. None of the patients had been treated with cutaneous radiotherapy or chemotherapy prior to surgery. All tissue samples were immediately frozen and stored in liquid nitrogen following surgical resection. This study was approved by the Ethics Committee of Hubei Provincial Hospital of Traditional Chinese Medicine, and all patients who participated in this research provided written informed consent.
Cell Culture and Transfection
Human epidermal melanocytes (HEMs) were purchased from ScienCell Research Laboratories, Inc. (San Diego, CA, USA) and cultured in melanocyte medium (ScienCell Research Laboratories, Inc.) according to the manufacturer’s instructions. Four human melanoma cell lines (A375, A2058, SK-MEL-28, and HT144) were obtained from the American Type Culture Collection (Manassas, VA, USA). All melanoma cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and maintained at 37°C in a humidified incubator with a mixture of 5% CO2.
For functional experiments, miR-342 mimic, miRNA negative control mimic (miR-NC), small interfering RNA (siRNA) targeting zinc-finger E-box-binding homeobox 1 (ZEB1 siRNA), and negative control siRNA (NC siRNA) were purchased from Guangzhou RiboBio Co., Ltd (Guangzhou, P.R. China). ZEB1 overexpression plasmid pCMV-ZEB1 and empty plasmid pCMV were synthesized by Guangzhou GeneCopoeia Co., Ltd (Guangzhou, P.R. China). Cells were plated into six-well plates at a density of 6 × 105 cells/well 24 h prior to transfection. Cell transfection was conducted using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer’s protocols.
RNA Isolation and Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to extract total RNA from tissues or cells as recommended by the manufacturer’s protocol. To analyze miR-342 expression, reverse transcription was performed with a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). Subsequent qPCR was carried out on an ABI 7500 thermocycler (Applied Biosystems) using a TaqMan MicroRNA PCR Kit (Applied Biosystems). For detection of ZEB1 mRNA expression, a PrimeScript Reverse Transcription PCR Kit (TaKaRa, Shiga, Japan) was used to convert RNA into cDNA. Afterward, the SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, P.R. China) was used to perform qPCR following the manufacturer’s instructions. U6 and β-action were employed as an internal reference for miR-342 and ZEB1 mRNA, respectively. Relative gene expression was analyzed by the 2−ΔΔCq method21.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
MTT assay was applied to measure cell proliferation. Cells were collected at 24 h posttransfection and seeded into 96-well plates with a density of 3,000 cells per well. Then the transfected cells were cultured at 37°C with 5% CO2 for 0, 24, 48, or 72 h. A total of 10 μl of MTT solution (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added into each well and incubated for an additional 4 h at 37°C. Then the supernatant containing MTT solution was removed, and 150 μl of dimethyl sulfoxide (Sigma) was added into each well. After incubation at 37°C for 20 min, optical density (OD) was determined at a wavelength of 490 nm using a SpectraMax M5 microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Each assay was independently conducted in triplicate.
Cell Invasion Assay
Matrigel (BD Bioscience, San Jose, CA, USA)-coated Transwell chambers (Corning Incorporated, Corning, NY, USA) with 8-μm pore filter inserts were used to evaluate cell invasion capacity. Following 48 h of transfection, the transfected cells were digested and suspended in DMEM without FBS. A total of 5 × 104 transfected cells were seeded into the upper chamber. The lower chambers were filled with DMEM containing 20% FBS to serve as a chemoattractant. Then the chambers were incubated at 37°C with 5% CO2 for 24 h. Noninvaded cells that did not pass through the 8-μm pore in the top chamber were removed using a cotton swab. The invaded cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet, and washed with phosphate buffer solution. After drying, the number of invaded cells was observed and counted under an Olympus BX51/61 microscope (magnification: 200×; Olympus Corporation, Tokyo, Japan) on five randomly selected fields.
Target Prediction
The putative targets of miR-342 were searched for using TargetScan (www.targetscan.org) and miRBase (http://www.mirbase.org/).
Dual-Luciferase Reporter Assay
Luciferase reporter plasmids containing wild-type or mutant-binding sequences in the 3′-UTR of ZEB1 were chemically synthesized and confirmed by Shanghai GenePharma Co., Ltd (Shanghai, P.R. China) and named as pGL3-ZEB1-3′-UTR WT and pGL3-ZEB1-3′-UTR MUT, respectively. Cells were seeded into 24-well plates and cotransfected with the luciferase reporter plasmid along with miR-342 mimic or miR-NC using Lipofectamine™ 2000 in accordance with the manufacturer’s protocols. After culture at 37°C for 48 h, the activities of Renilla luciferase and firefly luciferase were detected using the Dual Luciferase® Reporter Assay System (Promega Corporation) in accordance with the manufacturer’s instructions. Renilla luciferase activity was used to normalize firefly luciferase activity.
Western Blot Analysis
Ice-cold radioimmunoprecipitation assay buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was adopted to isolate total protein from tissues or cells. The total protein concentration was examined using a BCA assay kit (Santa Cruz Biotechnology). Equal quantities of total protein were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA), and then blocked in TBS/0.1% Tween (TBST) containing 5% milk at room temperature for 2 h. The membranes were incubated at 4°C overnight with primary antibodies: mouse anti-human ZEB1 monoclonal antibody (sc-81428; 1:1,000 dilution; Santa Cruz Biotechnology) or mouse anti-human GAPDH monoclonal antibody (sc-137179; 1:1,000 dilution; Santa Cruz Biotechnology). After being washed with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (sc-2005; 1:5,000 dilution; Santa Cruz Biotechnology) at room temperature for 2 h. Finally, an enhanced chemiluminescence reagent (Bio-Rad Laboratories, Hercules, CA, USA) was used to detect protein signals. GAPDH was used as internal control.
Statistical Analysis
All data were expressed as mean ± standard deviation. Statistical analysis was performed using SPSS 18.0 software (SPSS, Chicago, IL, USA) with a paired t-test for two groups and one-way analysis of variance with Student–Newman–Keuls post hoc test for three or more groups. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
miR-342 Is Downregulated in Melanoma Tissues and Cell Lines
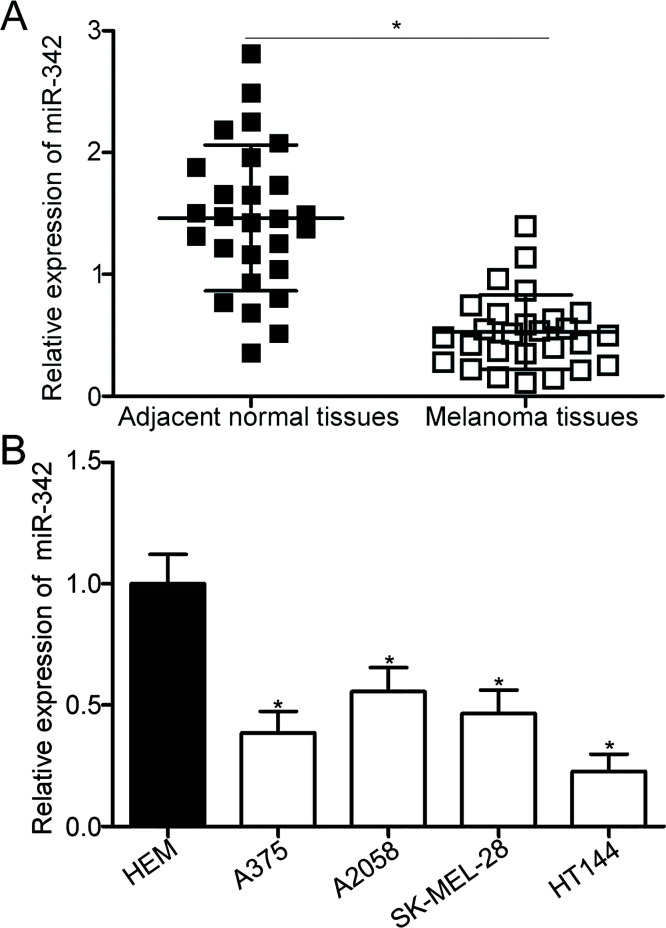
For investigating the role of miR-342 in melanoma, the expression levels of miR-342 in 27 paired melanoma tissues and matched adjacent normal tissues were detected using RT-qPCR. The expression level of miR-342 was downregulated in melanoma tissues compared with adjacent normal tissues (p < 0.05) (Fig. 1A). In addition, miR-342 expression was determined in four melanoma cell lines and HEMs. The results of the RT-qPCR analysis revealed that the expression level of miR-342 was lower in all four melanoma cell lines compared with those in HEMs (p < 0.05) (Fig. 1B). These results suggest that the downregulation of miR-342 may be associated with melanoma progression.
Figure 1.
Expression level of microRNA-342 (miR-342) in melanoma tissues and cell lines. (A) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis was used to detect miR-342 expression in 27 paired melanoma tissues and matched adjacent normal tissues. *p < 0.05 compared with adjacent normal tissues. (B) miR-342 expression in four melanoma cell lines (A375, A2058, SK-MEL-28, and HT144) and human epidermal melanocytes (HEMs) was determined using RT-qPCR. *p < 0.05 compared with HEM.
miR-342 Overexpression Prohibits Cell Proliferation and Invasion in Melanoma
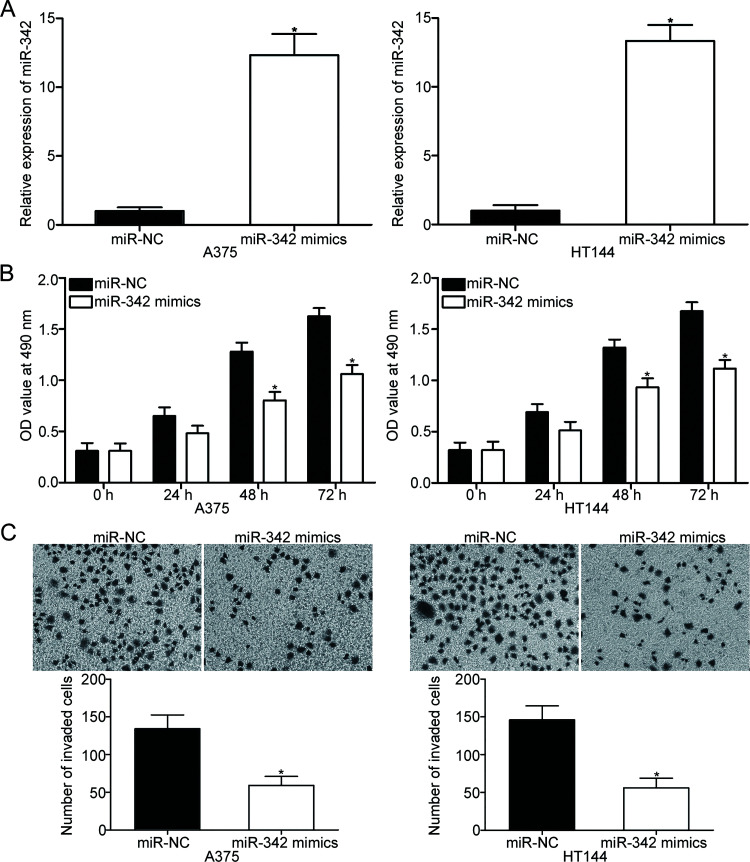
To investigate the biological functions of miR-342 in melanoma, we transfected miR-342 mimic into the A375 and HT144 cells, both of which expressed relatively low miR-342 levels. RT-qPCR analysis demonstrated that A375 and HT144 cells after transfection with miR-342 mimic showed significantly higher miR-342 expression compared with cells transfected with miR-NC (p < 0.05) (Fig. 2A). The effect of miR-342 overexpression on melanoma cell proliferation was examined using MTT assay. As shown in Figure 2B, the induced expression of miR-342 inhibited the proliferative capacity of A375 and HT144 cells (p < 0.05) (Fig. 2B). Cell invasion assay was used to characterize the effect of miR-342 on melanoma cell invasion. The number of invaded A375 and HT144 cells transfected with miR-342 mimic was significantly fewer than that from the miR-NC group (p < 0.05) (Fig. 2C). Accordingly, these results suggest that miR-342 may play tumor-suppressive roles in melanoma cell proliferation and invasion.
Figure 2.
Effect of miR-342 overexpression on the proliferation and invasion of A375 and HT144 cells. (A) A375 and HT144 cells were transfected with miR-342 mimic or miR-negative control (NC). RT-qPCR was performed to measure miR-342 expression after 48 h of transfection. *p < 0.05 compared with miR-NC. (B) Cell proliferation was evaluated by MTT assay in A375 and HT144 cells transfected with miR-342 mimic or miR-NC. *p < 0.05 compared with miR-NC. (C) Cell invasion assay was applied to analyze cell invasion capacity in A375 and HT144 cells following transfection with miR-342 mimic or miR-NC. *p < 0.05 compared with miR-NC.
ZEB1 Is a Novel Target Gene of miR-342 in Melanoma
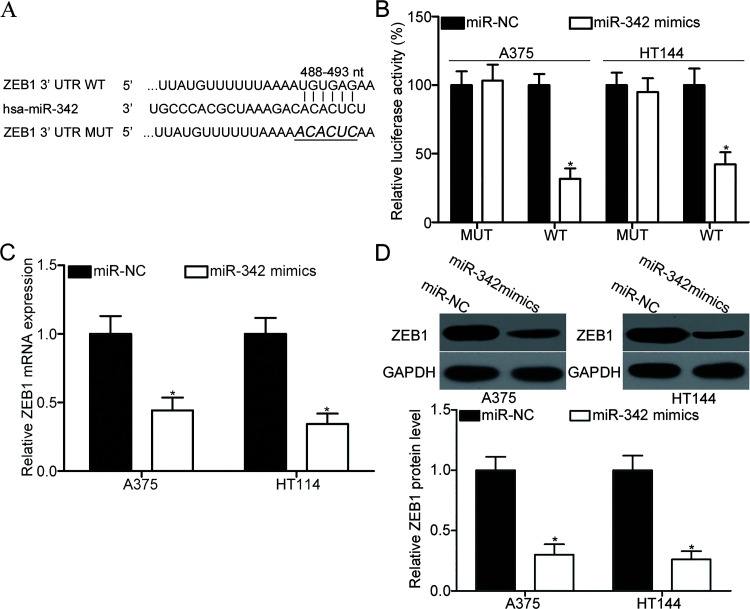
To explore the mechanism underlying the action of miR-342 in melanoma, we predicted the potential target genes of miR-342 through bioinformatics analysis. As illustrated in Figure 3A, the binding sequences of miR-342 matched the 3′-UTR of ZEB1, suggesting that ZEB1 was a candidate target of miR-342. Dual-luciferase reporter assay was performed to confirm whether miR-342 could directly target the 3′-UTR of ZEB1. The luciferase reporter plasmid containing the WT or MUT binding sequences for miR-342 in the 3′-UTR of ZEB1 was introduced into A375 and HT144 cells along with miR-342 mimic or miR-NC. The forced expression of miR-342 decreased the luciferase activity in A375 and HT144 cells with the WT plasmid (p < 0.05), although no significant change was observed in cells transfected with the MUT plasmid (Fig. 3B). For detecting whether miR-342 can affect ZEB1 expression in melanoma cells, RT-qPCR and Western blot analysis were employed to quantify ZEB1 mRNA and protein expression in A375 and HT144 cells after transfection with miR-342 mimic or miR-NC. As indicated in Figure 3C and D, the upregulation of miR-342 decreased ZEB1 expression in A375 and HT144 cells at the mRNA (p < 0.05) and protein (p < 0.05) levels. These findings collectively demonstrate that ZEB1 is a direct target of miR-342 in melanoma.
Figure 3.
Zinc-finger E-box-binding homeobox 1 (ZEB1) is a direct target of miR-342 in melanoma. (A) Putative wild type (WT) or mutant (MUT) seed-matching sites for miR-342 in the 3′-untranslated region (3′-UTR) of ZEB1. (B) pGL3-ZEB1-3′-UTR WT or pGL3-ZEB1-3′-UTR MUT along with miR-342 mimic or miR-NC was transfected into A375 and HT144 cells. After transfection for 48 h, cells were subjected to dual-luciferase reporter assay for the detection of luciferase activity. *p < 0.05 compared with miR-NC. A375 and HT144 cells were transfected with miR-342 mimic or miR-NC. RT-qPCR and Western blot analysis were applied to analyze ZEB1 mRNA (C) and protein (D) expression, respectively. *p < 0.05 compared with miR-NC.
ZEB1 Knockdown Exhibits Similar Effects With miR-342 Overexpression in Melanoma
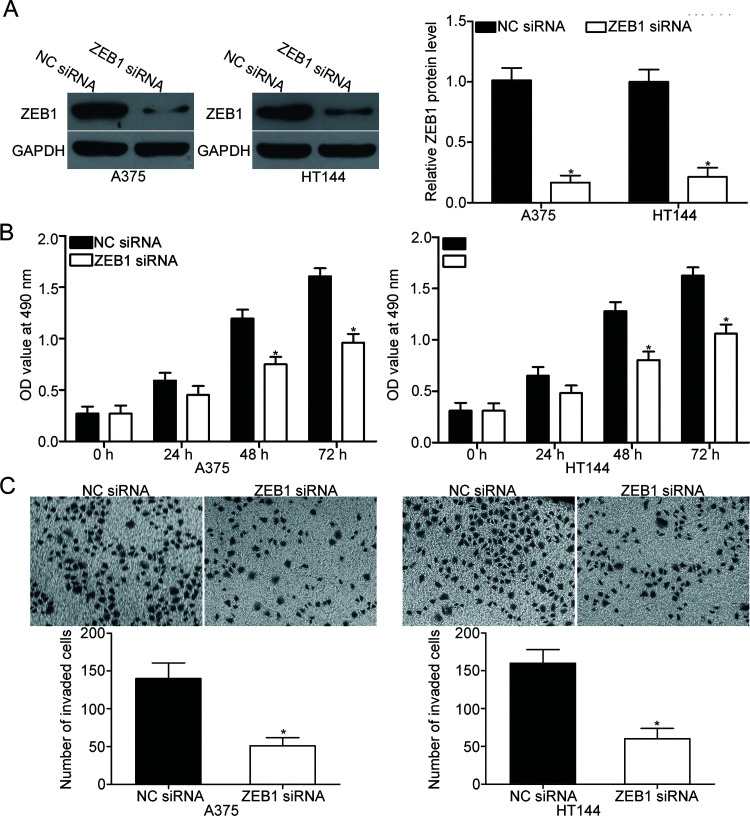
ZEB1 siRNA or NC siRNA was transfected into A375 and HT144 cells to investigate the functions of the ZEB1 in melanoma. After transfection, Western blot analysis demonstrated that ZEB1 protein expression was markedly downregulated in A375 and HT144 cells after transfection with ZEB1 siRNA compared with that in cells transfected with NC siRNA (p < 0.05) (Fig. 4A). Subsequent MTT and cell invasion assays revealed that ZEB1 knockdown suppressed the proliferation (p < 0.05) (Fig. 4B) and invasion (p < 0.05) (Fig. 4C) of A375 and HT144 cells; these findings were similar to those induced by miR-342 overexpression. These results suggest that ZEB1 is a functional target of miR-342 in melanoma.
Figure 4.
ZEB1 knockdown inhibits A375 and HT144 cell proliferation and invasion. (A) ZEB1 small interfering RNA (siRNA) or NC siRNA was transfected into A375 and HT144 cells. At 72 h posttransfection, the knockdown of ZEB1 was confirmed by Western blot analysis. *p < 0.05 compared with NC siRNA. MTT (B) and cell invasion (C) assays were employed to investigate the effects of ZEB1 knockdown on the proliferation and invasion of A375 and HT144 cells. *p < 0.05 compared with NC siRNA.
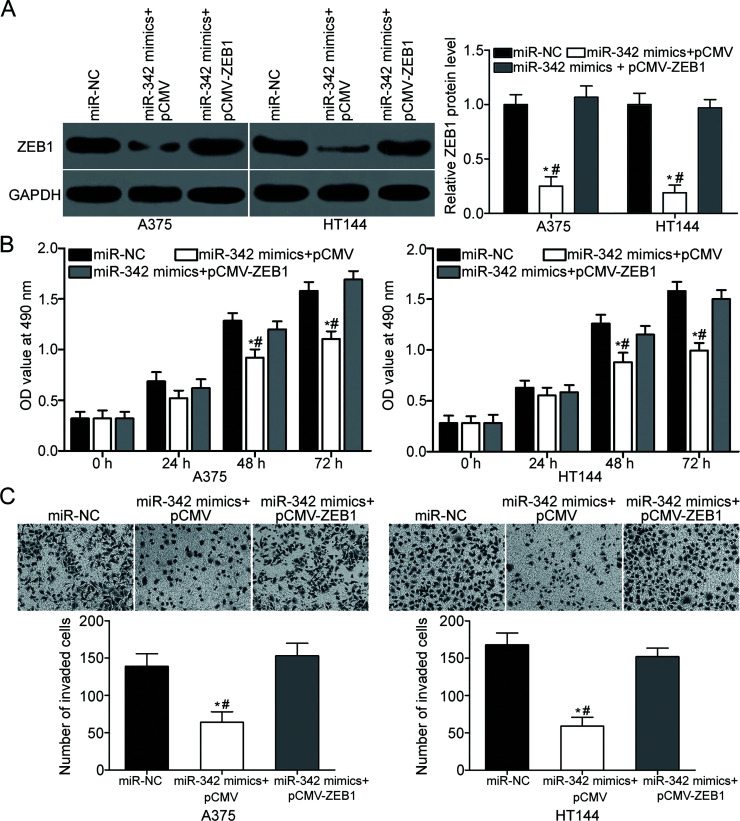
Restoration of ZEB1 Expression Reverses the Inhibitory Effects of miR-342 on the Proliferation and Invasion of Melanoma Cells
Considering that ZEB1 was identified as a direct target of miR-342, we speculated that restoration of ZEB1 expression could rescue the biological phenotypes caused by the upregulation of miR-342 in melanoma. For clarifying this speculation, miR-342 was transfected into A375 and HT144 cells along with ZEB1 overexpression plasmid pCMV-ZEB1 or empty plasmid pCMV. ZEB1 protein expression was confirmed by Western blot analysis, and the downregulation of ZEB1 caused by miR-342 overexpression was recovered in A375 and HT144 cells after cotransfection with pCMV-ZEB1 (p < 0.05) (Fig. 5A). Moreover, functional experiments indicated that the ectopic expression of ZEB1 reversed the inhibitory effects of miR-342 overexpression on proliferation (p < 0.05) (Fig. 5B) and invasion (p < 0.05) (Fig. 5C) in A375 and HT144 cells. Our findings suggest that the tumor-suppressing roles of miR-342 in melanoma were at least partially related to the downregulation of ZEB1.
Figure 5.
ZEB1 can rescue the effect of miR-342 overexpression in melanoma. (A) A375 and HT144 cells were cotransfected with miR-342 mimic and pCMV or pCMV-ZEB1. Western blot analysis was conducted to determine ZEB1 protein expression at 72 h posttransfection. *p < 0.05 compared with miR-NC. #p < 0.05 compared with miR-342 mimic + pCMV-ZEB1. (B) MTT assay was used to detect cell proliferation in A375 and HT144 cells cotransfected with miR-342 mimic and pCMV or pCMV-ZEB1. *p < 0.05 compared with miR-NC. #p < 0.05 compared with miR-342 mimic + pCMV-ZEB1. (C) Cell invasion capacity in A375 and HT144 cells cotransfected with miR-342 mimic and pCMV or pCMV-ZEB1 was evaluated using cell invasion assay. *p < 0.05 compared with miR-NC. #p < 0.05 compared with miR-342 mimic + pCMV-ZEB1.
DISCUSSION
As documented in numerous studies, miRNAs play key roles in various biological processes associated with melanoma occurrence and development22–24. Therefore, further investigation on melanoma-related miRNAs may facilitate the identification of novel diagnostic and therapeutic targets in patients with this malignancy. In this study, we found that miR-342 expression was frequently downregulated in melanoma tissues and cell lines. The ectopic expression of miR-342 inhibited melanoma cell proliferation and invasion in vitro. The results of the bioinformatics analysis, dual-luciferase reporter assay, RT-qPCR, and Western blot analysis demonstrated that ZEB1 was a direct target of miR-342 in melanoma. Moreover, ZEB1 knockdown reduced the cell proliferation and invasion of melanoma, and these findings were similar to those induced by miR-342 overexpression. Furthermore, restored ZEB1 expression rescued the inhibitory effects of miR-342 on the proliferation and invasion of melanoma cells. These results suggest that the miR-342 may exhibit potential as a novel therapeutic target for the treatment of patients with melanoma.
Dysregulation of miR-342 has been implicated in several types of human cancers. For example, miR-342 was underexpressed in hepatocellular carcinoma tissues and cell lines. Low miR-342 expression was associated with histologic grade and tumor TNM stage. Patients with hepatocellular carcinoma and low miR-342 expression level showed poorer overall survival than those with high miR-342 level. Univariate analysis identified miR-342 as an independent prognosis biomarker for patients with hepatocellular carcinoma17. miR-342 was downregulated in colorectal cancer18. Patients with colon cancer and high miR-342 level showed shorter survival time than patients with low miR-34219. The expression level of miR-342 decreased in glioma tissues, and this downregulation was strongly correlated with WHO grades and KPS scores of glioma patients20. miR-342 downregulation was also observed in non-small cell lung cancer25, cervical cancer26, and acute myeloid leukemia27. These findings suggest that the expression level of miR-342 may be a promising biomarker for the diagnosis and prognosis of these specific types of human cancers.
miR-342 reportedly plays important roles in the initiation and progression of multiple human cancers. For instance, Zhao et al. found that restoring the expression of miR-342 induced significant suppression of cell proliferation in hepatocellular carcinoma28. Wang et al. reported that the upregulation of miR-342 reduces the proliferation and invasion of colorectal cancer cells, induces G0/G1 cell cycle arrest in vitro, and decreases tumor growth and lung metastasis in vivo18. Lu et al. demonstrated that miR-342 reexpression represses glioma proliferation and invasion and increases apoptosis in glioma20. Zhang et al. showed that the resumption of miR-342 expression inhibits osteosarcoma cell growth and metastasis29. Xie et al. revealed that miR-342 overexpression attenuates cell proliferation and invasion in vitro and tumor growth in vivo25. Li et al. illustrated that enforced expression of miR-342 prohibits the growth and motility of cervical cancer cells26. These studies suggest that miR342 may perform tumor-suppressive roles in various human cancers and may be developed as a therapeutic target for cancer treatments.
Several direct targets of miR-342, including p21 (cyclin-dependent kinase inhibitor 1A)-activated kinase 4 (PAK4)20 in glioma, Ras-related RAP-2B (RAP2B)25 in non-small cell lung cancer, forkhead box M1 (FOXM1)26 in cervical cancer, nuclear factor κ light chain enhancer of activated B cells inhibitor γ (IKK-γ)28, transforming growth factor-β (TGF-β)-activated kinase 1-binding protein 2 (TAB2)28 and TAB328 in hepatocellular carcinoma, and DNA methyl transferase 1 (DNMT1)18, FOXM130, and FOXQ130 in colorectal cancer, have been previously validated. In our current study, ZEB1, a member of the zinc finger family31, was identified as a direct target of miR-342 in melanoma. ZEB1 was found to be aberrantly upregulated in numerous cancer types, such as ovarian cancer32, glioblastoma33, lung cancer34, colorectal cancer35, and gastric cancer36. ZEB1 was also previously reported to be upregulated in melanoma tissues. ZEB1 may contribute to the occurrence and progression of melanoma through the regulation of cell proliferation, colony formation, migration, invasiveness, epithelial–mesenchymal transition, and tumorigenicity37,38. Given the important roles of ZEB1 in melanoma, ZEB1 may be considered as a novel promising therapeutic opportunity for treating this aggressive cancer.
In conclusion, the results of this study provided sufficient evidence suggesting that miR-342 may serve as a tumor suppressor in melanoma, at least in part, by regulation of ZEB1. The miR-342/ZEB1 axis may provide novel insights into the formation and progression of melanoma and could be explored as a novel potential therapeutic target for the treatment of this malignancy.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. J Invest Dermatol. 2009;129:1666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trotter SC, Sroa N, Winkelmann RR, Olencki T, Bechtel M. A global review of melanoma follow-up guidelines. J Clin Aesthet Dermatol. 2013;6:18–26. [PMC free article] [PubMed] [Google Scholar]
- 3. Tarhini AA. Immunotherapy of melanoma. Curr Mol Pharmacol. 2016;9:196–207. [DOI] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 5. Brozyna AA, Jochymski C, Janjetovic Z, Jozwicki W, Tuckey RC, Slominski AT. CYP24A1 expression inversely correlates with melanoma progression: Clinico-pathological studies. Int J Mol Sci. 2014;15:19000–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 7. Park WY, Hong BJ, Lee J, Choi C, Kim MY. H3K27 Demethylase JMJD3 employs the NF-kappaB and BMP signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Res. 2016;76:161–70. [DOI] [PubMed] [Google Scholar]
- 8. Bang W, Jeon YJ, Cho JH, Lee RH, Park SM, Shin JC, Choi NJ, Choi YH, Cho JJ, Seo JM, Lee SY, Shim JH, Chae JI. Beta-lapachone suppresses the proliferation of human malignant melanoma cells by targeting specificity protein 1. Oncol Rep. 2016;35:1109–16. [DOI] [PubMed] [Google Scholar]
- 9. Farrokhnia F, Aplin JD, Westwood M, Forbes K. MicroRNA regulation of mitogenic signaling networks in the human placenta. J Biol Chem. 2014;289:30404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 11. Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA 2008;105:14879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W, Zhang J, Xu H, Dai J, Zhang X. The negative regulation of miR-149-5p in melanoma cell survival and apoptosis by targeting LRIG2. Am J Transl Res. 2017;9:4331–40. [PMC free article] [PubMed] [Google Scholar]
- 13. Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–7. [DOI] [PubMed] [Google Scholar]
- 14. Zagryazhskaya A, Zhivotovsky B. miRNAs in lung cancer: A link to aging. Ageing Res Rev. 2014;17:54–67. [DOI] [PubMed] [Google Scholar]
- 15. Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J Mol Med. (Berl) 2011;89:445–57. [DOI] [PubMed] [Google Scholar]
- 16. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–8. [DOI] [PubMed] [Google Scholar]
- 17. Gao Y, Zhang SG, Wang ZH, Liao JC. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Eur Rev Med Pharmacol Sci. 2017;21:2098–102. [PubMed] [Google Scholar]
- 18. Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu R, Pan Z, Kang T, Huang W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis 2011;32:1033–42. [DOI] [PubMed] [Google Scholar]
- 19. Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 20. Lu X, Wang H, Su Z, Cai L, Li W. MicroRNA-342 inhibits the progression of glioma by directly targeting PAK4. Oncol Rep. 2017;38:1240–50. [DOI] [PubMed] [Google Scholar]
- 21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 22. Mimeault M, Batra SK. Novel biomarkers and therapeutic targets for optimizing the therapeutic management of melanomas. World J Clin Oncol. 2012;3:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qiu H, Yuan S, Lu X. miR-186 suppressed CYLD expression and promoted cell proliferation in human melanoma. Oncol Lett. 2016;12:2301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao XH, Chen M, Wang Y, Cui PG, Liu SB, Xu ZY. MicroRNA-21 regulates the ERK/NF-kappaB signaling pathway to affect the proliferation, migration, and apoptosis of human melanoma A375 cells by targeting SPRY1, PDCD4, and PTEN. Mol Carcinog. 2017;56:886–94. [DOI] [PubMed] [Google Scholar]
- 25. Xie X, Liu H, Wang M, Ding F, Xiao H, Hu F, Hu R, Mei J. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36:5031–8. [DOI] [PubMed] [Google Scholar]
- 26. Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298–307. [DOI] [PubMed] [Google Scholar]
- 27. Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani R, Berri F, Vanhamme L, Burny A, Martiat P, Rouas R, Badran B. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013;11:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-kappaB pathway. Biochem Biophys Res Commun. 2015;457:370–7. [DOI] [PubMed] [Google Scholar]
- 29. Zhang S, Liu L, Lv Z, Li Q, Gong W, Wu H. MicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1). Oncol Res. 2017;25:1505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 are promising prognostic biomarkers and novel targets of tumor-suppressive miR-342 in human colorectal cancer. Clin Cancer Res. 2016;22:4947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol. 2012;105:830–4. [DOI] [PubMed] [Google Scholar]
- 32. Li X, Huang R, Li RH, Trope CG, Nesland JM, Suo Z. Expression of zinc finger E-box-binding homeobox factor 1 in epithelial ovarian cancer: A clinicopathological analysis of 238 patients. Mol Clin Oncol. 2016;4:18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Devers KG, Yachnis AT, Kupper MD, Neal D, Nabilsi NH, Kladde MP, Suslov O, Brabletz S, Brabletz T, Reynolds BA, Steindler DA. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5:1196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, Girard L, Behrens C, Wistuba II, Gazdar AF, Hayward NK, Minna JD. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Z, Sun B, Qi L, Li H, Gao J, Leng X. Zinc finger E-box binding homeobox 1 promotes vasculogenic mimicry in colorectal cancer through induction of epithelial-to-mesenchymal transition. Cancer Sci. 2012;103:813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jia B, Liu H, Kong Q, Li B. Overexpression of ZEB1 associated with metastasis and invasion in patients with gastric carcinoma. Mol Cell Biochem. 2012;366:223–9. [DOI] [PubMed] [Google Scholar]
- 37. Dou J, He X, Liu Y, Wang Y, Zhao F, Wang X, Chen D, Shi F, Wang J. Effect of downregulation of ZEB1 on vimentin expression, tumour migration and tumourigenicity of melanoma B16F10 cells and CSCs. Cell Biol Int. 2014;38:452–61. [DOI] [PubMed] [Google Scholar]
- 38. Wels C, Joshi S, Koefinger P, Bergler H, Schaider H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial–mesenchymal transition-like phenotype in melanoma. J Invest Dermatol. 2011;131:1877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]