Abstract
MicroRNAs (miRNAs) play important roles in the carcinogenesis of cervical cancer. However, the expression and underlying mechanisms of miRNA in cervical cancer progression remain unclear. In the present study, our data showed that the expression of miR-138-5p was significantly downregulated in cervical cancer tissues, and decreased expression of miR-138-5p was correlated with advanced FIGO stage, poor differentiation, lymph node metastasis, and poor overall survival of cervical cancer patients. Function assays showed that overexpression of miR-138-5p reduced cervical cancer cell proliferation, arrested cells in the G0/G1 phase, and induced cell apoptosis in vitro. Remarkably, SIRT1 was confirmed as a direct target of miR-138-5p in cervical cancer, and miR-138-5p exerted the reduced tumor functions by suppressing SIRT1 expression. Moreover, we further identified that lncRNA H19 could act as a molecular sponge of miR-138-5p in cervical cancer progression. Taken together, these results suggested that miR-138-5p could suppress cervical cancer cell progression by targeting SIRT1.
Key words: miR-138-5p, Long noncoding RNA (lncRNA) H19, SIRT1, Cervical cancer
INTRODUCTION
Cervical cancer is one of the most common gynecological cancers worldwide. According to statistics, there are over 300,000 deaths annually, and more than 85% of these deaths occur in developing countries1,2. Despite great progress in cervical cancer treatments, including surgical, chemotherapy, and radiotherapy in recent years, the clinical outcomes of cervical cancer remain poor3. Therefore, it is critical to identify new biomarkers and therapeutic targets to improve cervical cancer diagnosis and treatment.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs (ncRNAs) of 20–22 nucleotides that play important regulatory roles in biological processes through targeting mRNAs for cleavage and translational repression4. Through negatively mediating their target genes, miRNAs are involved in a variety of biological processes, such as cell proliferation, apoptosis, and differentiation5. Recently, miRNAs represent a promising new family of targets in the current era of molecular therapies in oncology6. For example, Song et al. showed that miR-630 controls cell growth and metastasis in lung cancer by targeting LIM domain only 3 (LMO3)7. Yang et al. showed that miR-506 inhibited cell growth and metastasis via flotilin 1 (FLOT1), but its expression is reduced in clear cell renal cell carcinoma8. Wang et al. showed that miR-98 targets collagen triple helix repeat containing 1 to reduce cell proliferation, migration, and invasion in hepatocellular carcinoma9. However, the expression and functions of miR-138-5p in cervical cancer are still unclear.
In the present study, our data showed that miR-138-5p was decreased in cervical cancer tissues. This decreased expression of miR-138-5p correlated with advanced International Federation of Gynecology and Obstetrics (FIGO) stage, poor differentiation, lymph nodes metastasis, and poor overall survival of cervical cancer patients. In function experiments, we showed that miR-138-5p reduced cervical cancer cell proliferation, arrested cells in the G0/G1 phase, and suppressed silent mating type information regulation 2 homolog 1 (SIRT1) expression to induce cell apoptosis in vitro. In addition, our data indicated that long noncoding RNA (lncRNA) H19 could act as a molecular sponge of miR-138-5p in cervical cancer progression.
MATERIALS AND METHODS
Clinical Specimens
A total of 56 pairs of cervical cancer and adjacent normal tissues were collected from cervical cancer patients who received treatment in Zhengzhou People’s Hospital (Zhengzhou, P.R. China) from January 2010 to December March 2011. None of the patients received any chemotherapy or radiation before surgery. Histopathological diagnoses were based on the World Health Organization (WHO) classification, and clinical stages were in accordance with the FIGO criteria. Written informed consent was obtained from all participants, and the study was approved by the Board and Ethics Committee of Zhengzhou People’s Hospital. The tissues were immediately frozen in liquid nitrogen following surgery and stored at −80°C until use.
Cell Culture and Transfection
Human cervical cancer cell lines (HeLa and SiHa) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin in a cell culture humidified incubator at 37°C with 5% CO2. miR-138-5p mimics and miR-negative control (NC) were purchased from GeneCopoeia Co. Ltd. (Rockville, MD, USA). pcDNA-SIRT1 and H19 small interfering RNA (siRNA) were purchased from Applied Biosystems (Foster City, CA, USA). Transfection was performed in a six-well plate when cell confluence attained 70%–90% according to the Lipofectamine 2000 (Invitrogen) transfection manual.
Quantitative Real-Time Reverse Transcriptase PCR (qRT-PCR)
Total RNA was extracted from tumor samples and cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (500 ng) was reverse transcribed to complementary DNA using PrimeScript™ RT Master Mix (Takara, Dalian, P.R. China). The qRT-PCR was performed in a total reaction volume of 20 ml using SYBR Green qPCR Master Mix (Takara) in the ABI PRISM 7900HT Sequence Detection System. The U6 short nuclear RNA (snRNA) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The relative fold expressions were calculated with the 2−ΔΔCT method. All the qRT-PCRs were run in triplicate.
Western Blotting
Cells were collected and lysed using radioimmunoprecipitation assay (RIPA) protein extraction reagent (Beyotime, Shanghai, P.R. China) supplemented with a protease inhibitor cocktail (Roche, Carlsbad, CA, USA). Then protein concentrations were determined by a Bio-Rad assay kit (Carlsbad, CA, USA). Next, the protein was boiled after adding equal amounts of loading buffer. Protein samples were loaded onto the sodium dodecyl sulfate (SDS)-polyacrylamide gel, electrophoresed, and then transferred onto 0.22-μm nitrocellulose membrane before blocking and probing with primary antibodies at 4°C for 12 h. Then the membranes were subsequently incubated with the corresponding horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. The signals from Western blotting were quantified by densitometry, and the statistical analyses were conducted from three independent experiments.
Cell Proliferation Assay
The proliferation of cervical cancer cells was determined using the Cell Counting Kit-8 (CCK-8; Dojindo, Osaka, Japan) according to the manufacturer’s instructions. Cells were seeded at a density of 3 × 103 cells per well in 96-well plates. CCK-8 (10 μl) was then added to each well at 24, 48, 72, and 96 h and incubated for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm using a Model 680 microplate reader (Bio-Rad).
Flow Cytometry Analysis
For cell apoptosis analysis, cells were harvested and resuspended in staining buffer at a concentration of 1 × 106 cells/ml. The cells were stained with annexin V and propidium iodide (PI) staining using the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions. Apoptotic cells were measured using a BD FACSCanto™ Flow Cytometer (BD Biosciences).
For cell cycle analysis, cells were harvested by trypsin in the logarithmic growth phase and fixed in 75% ethanol at 4°C overnight. Then cells were resuspended in phosphate-buffered saline (PBS) and incubated with BD PharMingen™ PI/RNase staining buffer (BD Biosciences) at room temperature for 30 min in the dark. Finally, cell cycle distribution and DNA content were analyzed using a BD FACSCanto™ Flow Cytometer (BD Biosciences).
Target Gene Identification
The bioinformatics online software program TargetScan (http://www.targetscan.org) was utilized to determine likely target genes for miR-138-5p. One likely target was SIRT1. Starbase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php) was used to identify complementary lncRNA binding partners for miR-138-5p.
Luciferase Reporter Assay
The fragment from H19 containing the predicted miR-138-5p binding site was amplified by PCR and then cloned into a pmirGlO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to form the reporter vector H19-wild type (H19-Wt). To mutate the putative binding site of miR-138-5p in the H19, the sequence of the putative binding site was replaced and named as H19-mutated-type (H19-Mut). Similarly, the fragment from SIRT1 3′-UTR (untranslated region) sequences were amplified by PCR and cloned into pmiR-RB-Report™ vector (RiboBio, Guangzhou, P.R. China) to form the reporter vector SIRT1-Wt. To mutate the putative binding site of miR-138-5p in the 3′-UTR-containing vector, the sequence of putative binding site was replaced and named as SIRT1-Mut. Then the vectors and miR-138-5p mimics were cotransfected into HeLa cells, and the dual-luciferase reporter assay system (Promega) was used for testing the luciferase activity.
Statistical Analysis
The data were expressed as mean ± standard deviation (SD) and analyzed by SPSS17.0. The difference between two groups was evaluated using two-tailed Student’s t-test or one-way ANOVA. Overall survival was defined by Kaplan–Meier and analyzed by log-rank test. Person correlation analysis was used to evaluate the relationship between miR-138-5p and H19. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-138-5p Was Downregulated in Cervical Cancer
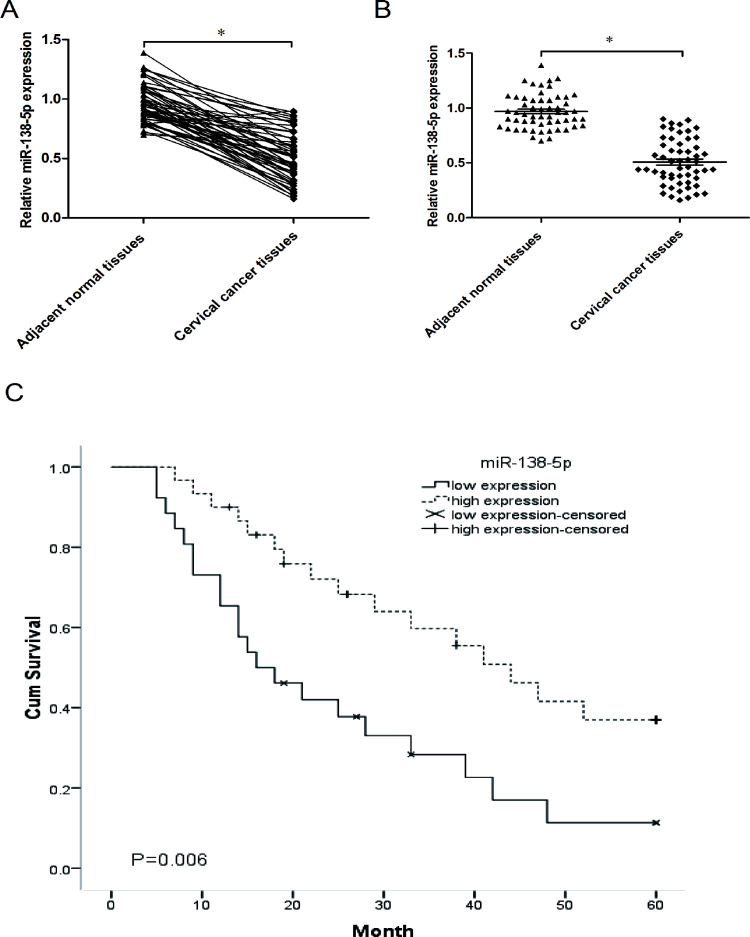
To determine whether miR-138-5p was dysregulated in cervical cancer, we explored miR-138-5p expression in cervical cancer tissues and pair-matched adjacent normal tissues. Using qRT-PCR, we showed that miR-138-5p was significantly decreased in cervical cancer tissues compared with the adjacent normal tissues (p < 0.05) (Fig. 1A and B). The aberrant expression level of miR-138-5p in the cancer tissues indicated that miR-138-5p might play an important role in the development and progression of cervical cancer.
Figure 1.
MicroRNA-138-5p (miR-138-5p) is downregulated in cervical cancer tissues and predicts poor overall survival in cervical cancer patients. (A, B) Expression of miR-138-5p was decreased in cervical cancer tissues compared to adjacent normal tissues as determined by quantitative real-time reverse transcriptase PCR (qRT-PCR). (C) Kaplan–Meier analysis showed that cervical cancer patients with low miR-138-5p expression (<median) have shorter overall survival compared to patients with high miR-138-5p expression (>median). *p < 0.05.
Correlation Between miR-138-5p Expression and Clinical Characteristics and Prognosis
To assess whether miR-138-5p expression was correlated with clinical pathological features and prognosis of cervical cancer, the 56 cervical cancer patients were classified into two groups according to the median of miR-138-5p in tumor tissues: the high miR-138-5p group (n = 28, >median value) and the low miR-138-5p group (n = 28, ≤median value). Correlation analysis showed that low miR-138-5p expression was significantly associated with advanced FIGO stage, poor differentiation, and lymph nodes metastasis (Table 1). However, the expression of miR-138-5p was not related to other clinical parameters, such as age, tumor size, histology, and vascular invasion. Furthermore, Kaplan–Meier analysis showed that patients with low miR-138-5p expression had poor overall survival from cervical cancer compared to patients with high miR-138-5p expression (p < 0.05) (Fig. 1C).
Table 1.
Association of MicroRNA-138-5p (miR-138-5p) Expression With Clinicopathological Features in Cervical Cancer
| Parameters | Total | miR-138-5p Expression | p Value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | 0.420 | |||
| <50 | 25 | 14 | 11 | |
| ≥50 | 31 | 14 | 17 | |
| Tumor size (cm) | 0.415 | |||
| <4 | 23 | 10 | 13 | |
| ≥4 | 33 | 18 | 15 | |
| Histology | 0.397 | |||
| Squamous | 37 | 17 | 20 | |
| Adenocarcinoma | 19 | 11 | 8 | |
| FIGO stage | 0.016 | |||
| Ib–IIa | 29 | 19 | 10 | |
| IIb–IIIa | 27 | 9 | 18 | |
| Differentiation | 0.026 | |||
| Well + moderate | 36 | 22 | 14 | |
| Poor | 20 | 6 | 14 | |
| Lymph nodes metastasis | 0.002 | |||
| No | 35 | 23 | 12 | |
| Yes | 21 | 5 | 16 | |
| Vascular invasion | 0.131 | |||
| No | 41 | 23 | 18 | |
| Yes | 15 | 5 | 10 | |
FIGO, International Federation of Gynecology and Obstetrics.
miR-138-5p Suppressed Cervical Cancer Cell Proliferation by Affecting Cell Apoptosis and Cell Cycle
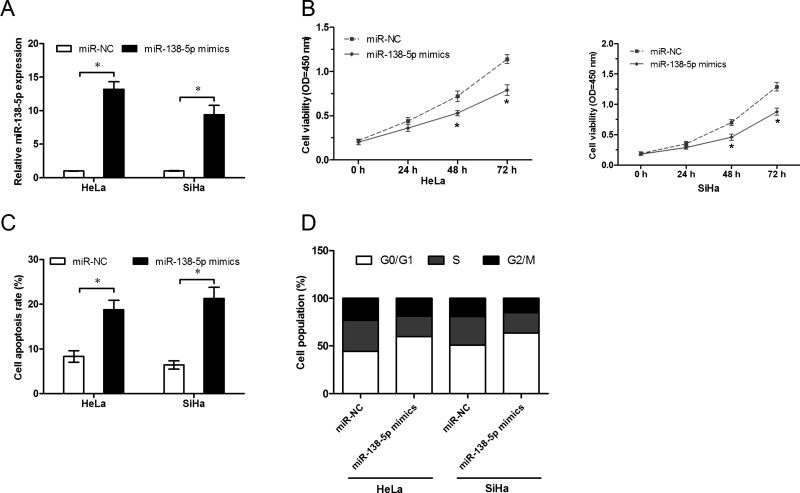
To explore the cellular functions of miR-138-5p in cervical cancer, miR-138-5p mimics were transfected into HeLa and SiHa cells, and the overexpression efficiency was determined by qRT-PCR (p < 0.05) (Fig. 2A). To explore whether miR-138-5p affects the growth of cervical cancer cells, cell proliferation was detected using CCK-8 assays. We found that overexpression of miR-138-5p significantly suppressed the proliferation of cervical cancer cells compared with the miR-NC group (p < 0.05) (Fig. 2B). To investigate whether cell apoptosis and cell cycle contributed to the growth of cervical cancer cells in vitro, we analyzed the effects of miR-138-5p on cell apoptosis and cell cycle of HeLa and SiHa cells. Flow cytometry assay revealed that the apoptosis of HeLa and SiHa cells was greatly enhanced after overexpression of miR-138-5p (p < 0.05) (Fig. 2C). Furthermore, the percentage of cells in the G0/G1 phase significantly increased with miR-138-5p overexpression in HeLa and SiHa cells (p < 0.05) (Fig. 2D). These results indicated that miR-138-5p suppressed cell proliferation by regulating both cell apoptosis and the cell cycle.
Figure 2.
The role of miR-138-5p in regulating the proliferative abilities of cervical cancer cells in vitro. (A) qRT-PCR was used to determine the expression of miR-138-5p in HeLa and SiHa cells transfected with miR-138-5p mimics and miR-negative control (NC). (B) Cell proliferation was measured by cell counting kit-8 (CCK-8) assay in HeLa and SiHa cells transfected with miR-138-5p mimics and miR-NC at indicated time. (C) Flow cytometry showed that the miR-138-5p mimics induced cell apoptosis using fluorescein isothiocyanate (FITC)-annexin V and propidium iodide (PI) staining. (D) Flow cytometry showed that miR-138-5p mimics arrested cervical cancer cell in the G0/G1 phase. *p < 0.01.
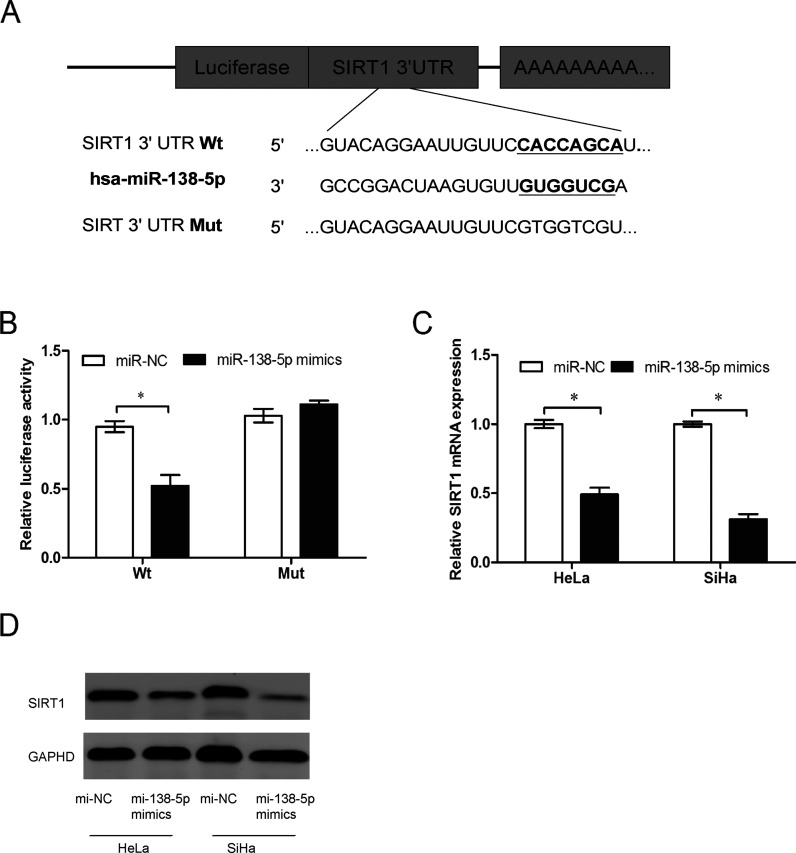
SIRT1 Was a Direct Target of miR-138-5p in Cervical Cancer
Using the bioinformatics software TargetScan for target gene prediction, SIRT1 was identified as one of the potential targets of miR-138-5p (Fig. 3A). The target sequences of SIRT1 3′-UTR Wt or the mutant sequences (Mut) were cloned into the luciferase reporter. After cotransfection of the reporters with miR-138-5p mimics or miR-NC into HeLa cells, the luciferase activity was recorded. Our data showed that miR-138-5p induced a remarkable decrease in the firefly luciferase activity when compared with the miR-NC group. However, the same effect could not be observed in the Mut group (p < 0.05) (Fig. 3B). Additionally, we investigated the regulatory effect of miR-138-5p on SIRT1 by qRT-PCR and Western blotting in HeLa and SiHa cells. We found that the ectopic expression of miR-138-5p suppressed both SIRT1 mRNA and protein levels in HeLa and SiHa cells using qRT-PCR and Western blot (p < 0.05) (Fig. 3C and D). Taken together, it was suggested that miR-138-5p directly targeted SIRT1 in cervical cancer cells.
Figure 3.
Suppressed silent mating type information regulation 2 homolog 1 (SIRT1) is the direct target of miR-138-5p in cervical cancer. (A) The potential miR-138-5p binding sites of the SIRT1 3′-untranslated region (UTR) and the mutated sequences. (B) Luciferase activity assay showed that miR-138-5p suppressed wild-type (Wt) SIRT1 3′-UTR luciferase activity, while it had no effect on mutated (Mut) SIRT1 3′-UTR luciferase activity compared to negative control in HeLa cells. (C) qRT-PCR analysis showed that the relative mRNA levels of SIRT1 were decreased after miR-138-5p mimic transfection in HeLa and SiHa cells. (D) Western blot analysis showed that SIRT1 protein levels were significantly reduced after transfection with miR-138-5p mimics. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *p < 0.05.
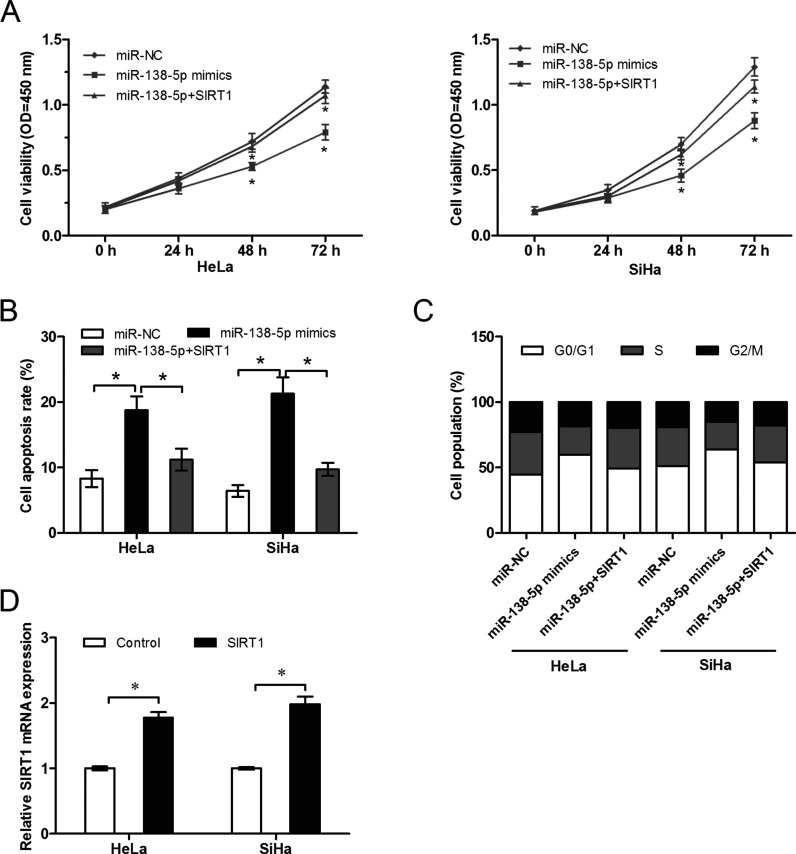
SIRT1 Overexpression Attenuated the Suppressive Effect of miR-138-5p
We further investigated whether overexpression of SIRT1 could reverse the suppressive effect of miR-138-5p. CCK-8 assay (p < 0.05) (Fig. 4A), cell apoptosis assay (p < 0.05) (Fig. 4B), and cell cycle assay (p < 0.05) (Fig. 4C) showed that overexpression of SIRT1 significantly reversed the suppressive effect of miR-138-5p on HeLa and SiHa cells. The effect of pcDNA-SIRT1 was confirmed by qRT-PCR (p < 0.05) (Fig. 4D).
Figure 4.
Overexpression of SIRT1 attenuates the effect of miR-138-5p. (A) HeLa and SiHa cells were cotransfected with miR-138-5p mimics and pcDNA-SIRT1 or the vector. CCK-8 assay was used to measure the proliferation. Cell apoptosis (B) and cell cycle (C) assays of HeLa and SiHa cells cotransfected with miR-138-5p mimics and pcDNA-SIRT1 or the vector are shown. (D) SIRT1 expression was detected by qRT-PCR in HeLa and SiHa cells transfected with miR-138-5p mimics and pcDNA-SIRT1 or the vector. *p < 0.05.
lncRNA H19 Serves as an Endogenous Sponge of miR-138-5p in Cervical Cancer
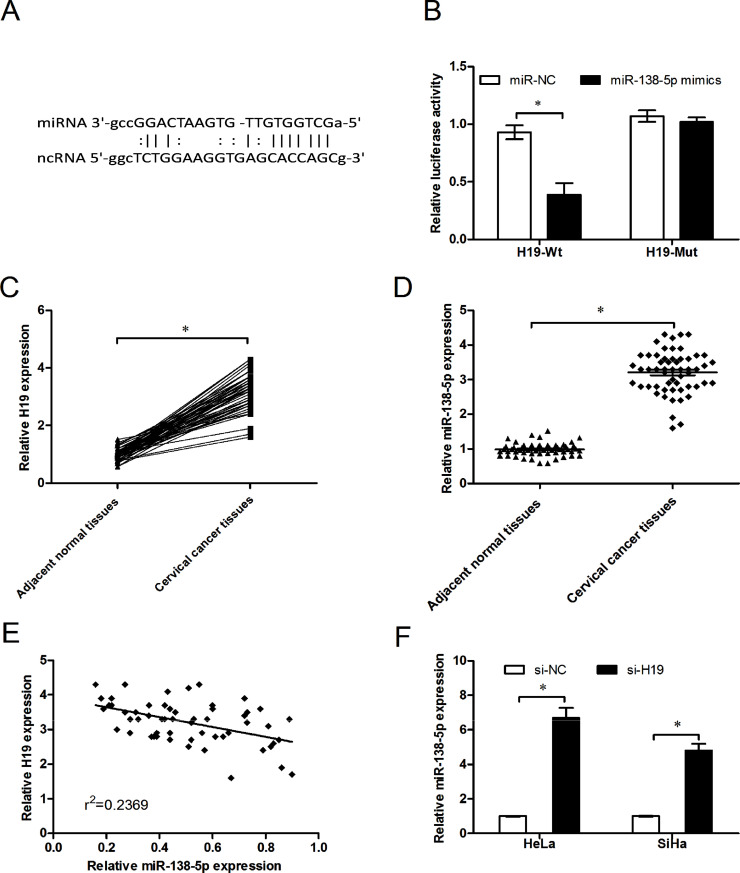
Recent studies have shown that lncRNAs acted as an endogenous sponge of miRNAs. We used Starbase v.2.0 to predict that lncRNA H19 had a complementary sequence for miR-138-5p (Fig. 5A). Dual-luciferase reporter assay was performed to explore whether H19 was a functional target of miR-138-5p. Our data showed that the luciferase activity was significantly decreased by the cotransfection of miR-138-5p mimics and H19-Wt rather than the cotransfection of miR-NC and H19-Wt, whereas cotransfection of miR-138-5p mimics and H19-Mut did not change the luciferase activity (Fig. 5B). Furthermore, we found that the expression of H19 was upregulated in cervical cancer tissues compared with adjacent normal tissues (p < 0.05) (Fig. 5C and D). Moreover, a negative relationship was identified between miR-138-5p and H19 expression (r 2 = 0.2369) (Fig. 5E). In addition, we silenced H19 expression by siRNA in HeLa and SiHa cells. qRT-PCR results showed that H19 knockdown increased miR-138-5p expression in cervical cancer cells (p < 0.05) (Fig. 5F). These findings might explain why miR-138-5p expression was lower in cervical cancer.
Figure 5.
Long noncoding RNA (lncRNA) H19 negatively regulates miR-138-5p expression in cervical cancer. (A) The predicted miR-138-5p binding sites on lncRNA H19. (B) Dual-luciferase reporter assay showed that miR-138-5p mimics reduced the intensity of fluorescence in HeLa cells transfected with H19-Wt but had no effect on the H19-Mut vector. (C, D) Expression of lncRNA H19 was increased in cervical cancer tissues compared to adjacent normal tissues as determined by qRT-PCR. (E) A negative association was verified between H19 and miR-138-5p in a cohort with 58 cervical cancer patients (r2 = −0.2369). (F) Deceased expression of H19 upregulated miR-138-5p expression in HeLa and SiHa cells. *p < 0.05.
DISCUSSION
Cervical cancer is one of the most common gynecological cancers worldwide. Although widespread implementation of screening programs in recent years has decreased the incidence and mortality of this cancer, it continues to be a major public health problem, specifically in advanced cases10. Thus, understanding the molecular mechanism of cervical cancer is urgently needed for the development of effective therapeutic strategies.
miRNAs are regulated in several diseases including cancers, where they have been characterized as oncogenes, tumor suppressors, or components of regulatory pathways that are critical for tumorigenesis6. miR-138-5p is commonly dysregulated in diverse cancers and is involved in various biological processes, including proliferation, migration, invasion, angiogenesis, and metabolism by targeting multiple mRNAs. For example, Yang et al. showed that miR-138-5p suppressed cell proliferation and invasion by targeting survivin in bladder cancer cells11. Zhao et al. found that miR-138-5p acted as a tumor suppressor in colorectal cancer by targeting programmed death ligand 1 (PD-L1)12. Yu et al. revealed that miR-138-5p reduced pancreatic cancer cell growth in a forkhead box protein C1 (FOXC1)-dependent fashion13.
In this study, for the first time, we determined that the expression of miR-138-5p was downregulated in cervical cancer tissues. Decreased expression of miR-138-5p was associated with advanced clinical features and poor overall survival in cervical cancer patients. The data suggested that the decreased expression of miR-138-5p was an unfavorable factor and might lead to the disorder of cell growth in cervical cancer. To confirm this hypothesis, in vitro function assays were designed to explore the biological roles of miR-138-5p in cervical cancer. Our data showed that miR-138-5p mimics significantly reduced cervical cancer cell proliferation, induced cell apoptosis, and arrested cell in the G0/G1 phase in vitro. Our results were consistent with the previous reports in other cancers14.
SIRT1, the human homolog of Sir2, is a member of the sirtuins family15. SIRT1 is a nicotinamide adenine dinucleotide 1 (NAD1)-dependent class III histone deacetylase (HDAC), which plays an important role in the regulation of critical biological processes such as metabolism, oncogenesis, and cancer progression16,17. Increasing studies showed that SIRT1 contributed to cell growth, drug resistance, invasion, metastasis, and recurrence18,19. In the present study, bioinformatics software indicated that SIRT1 was a potential target of miR-138-5p. Luciferase reporter assay showed that miR-138-5p significantly suppressed the luciferase activity of the Wt 3′-UTR but not the Mut 3′-UTR of SIRT1 in HeLa cells. Moreover, we found that the ectopic expression of miR-138-5p suppressed the SIRT1 mRNA and protein level in both HeLa and SiHa cells. In addition, our data showed that SIRT1 overexpression attenuated the suppressive effect of miR-138-5p. The data suggested that SIRT1 was a direct target of miR-138-5p in cervical cancer.
Recent studies showed that deregulation of lncRNAs profoundly influenced the expression of miRNAs20. For example, Li et al. found that through the action of miR-140-5p, the lncRNA maternally expressed 3 (MEG3) reduced adipogenesis and promoted osteogenesis of human adipose-derived mesenchymal stem cells21. Sun et al. revealed that in human nasopharyngeal carcinoma, lncRNA LOC100129148 targets miR-539-5p to function as an oncogene22. In our study, bioinformatics showed that lncRNA H19 had a complementary sequence of miR-138-5p. H19 expression was upregulated in cervical cancer tissues and has a negative relationship with miR-138-5p expression in cervical cancer. In addition, we showed that decreased expression of H19 significantly increased miR-138-5p in cervical cancer cells. These data indicated that lncRNA H19 might serve as an endogenous sponge of miR-138-5p in the progression of cervical cancer.
In conclusion, the present study identified that decreased expression of miR-138-5p in cervical cancer tissues was related to malignant clinical features and poor prognosis. Function assays showed that miR-138-5p could act as a novel inhibitor for tumor growth in cervical cancer. The anticancer functions of miR-138-5p might be due to the inhibition of SIRT1. Together, we indicated that miR-138-5p could become a novel prognostic biomarker and potential therapeutic target in the treatment of cervical cancer.
ACKNOWLEDGMENTS
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, Solomon D, Wentzensen N, Lawson HW. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol. 2013;121(4):829–46. [DOI] [PubMed] [Google Scholar]
- 3. Munoz N, Bravo LE. Epidemiology of cervical cancer in Colombia. Salud Publica Mex. 2014;56(5):431–9. [DOI] [PubMed] [Google Scholar]
- 4. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79. [DOI] [PubMed] [Google Scholar]
- 5. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9. [DOI] [PubMed] [Google Scholar]
- 7. Song YF, Hong JF, Liu DL, Lin QA, Lan XP, Lai GX. miR-630 targets LMO3 to regulate cell growth and metastasis in lung cancer. Am J Transl Res. 2015;7(7):1271–9. [PMC free article] [PubMed] [Google Scholar]
- 8. Yang FQ, Zhang HM, Chen SJ, Yan Y, Zheng JH. MiR-506 Is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PLoS One 2015;10(3):e0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang CY, Zhang JJ, Hua L, Yao KH, Chen JT, Ren XQ. MicroRNA-98 suppresses cell proliferation, migration and invasion by targeting collagen triple helix repeat containing 1 in hepatocellular carcinoma. Mol Med Rep. 2016;13(3):2639–44. [DOI] [PubMed] [Google Scholar]
- 10. Liu W, Gao G, Hu X, Wang Y, Schwarz JK, Chen JJ, Grigsby PW, Wang X. Activation of miR-9 by human papillomavirus in cervical cancer. Oncotarget 2014;5(22):11620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, Fang F, Guo H, Zhang C. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer 2016;15(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, Liu R, Tang A, Li X, Liu F, Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016;7(29):45370–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X, Deng Y, Jiang J, Sun C. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol. 2015;38(3):173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu C, Wang M, Chen M, Huang Y, Jiang J. Upregulation of microRNA-138-5p inhibits pancreatic cancer cell migration and increases chemotherapy sensitivity. Mol Med Rep. 2015;12(4):5135–40. [DOI] [PubMed] [Google Scholar]
- 15. Davenport AM, Huber FM, Hoelz A. Structural and functional analysis of human SIRT1. J Mol Biol. 2014;426(3):526–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25(3):138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang ZY, Hong D, Nam SH, Kim JM, Paik YH, Joh JW, Kwon CH, Park JB, Choi GS, Jang KY, Park CK, Kim SJ. SIRT1 regulates oncogenesis via a mutant p53-dependent pathway in hepatocellular carcinoma. J Hepatol. 2015;62(1):121–30. [DOI] [PubMed] [Google Scholar]
- 18. Côté CD, Rasmussen BA, Duca FA, Zadeh-Tahmasebi M, Baur JA, Daljeet M, Breen DM, Filippi BM, Lam TK. Resveratrol activates duodenal Sirt1 to reverse insulin resistance in rats through a neuronal network. Nat Med. 2015;21(5):498–505. [DOI] [PubMed] [Google Scholar]
- 19. Zhang N, Xie T, Xian M, Wang Y-J, Li H-Y, Ying M-D, Ye Z-M. SIRT1 promotes metastasis of human osteosarcoma cells. Oncotarget 2016;7(48):79654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505(7483):344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z, Jin C, Chen S, Zheng Y, Huang Y, Jia L, Ge W, Zhou Y. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem. 2017;433(1–2):51–60. [DOI] [PubMed] [Google Scholar]
- 22. Sun K-Y, Peng T, Chen Z, Song P, Zhou X-H. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging (Albany NY) 2017;9(3):999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]