Abstract
Epithelial ovarian cancer (EOC) is the one of most common gynecological malignant tumors with high mortality. A series of long noncoding RNAs (lncRNAs) have been validated to play a vital role in EOC tumorigenesis. Colon cancer-associated transcript 2 (CCAT2) has been verified as an oncogenic lncRNA in multiple tumors; however, the role of CCAT2 in EOC genesis is still unclear. The purpose of the present study was to probe the function of CCAT2 on EOC. Preliminary experiments found that CCAT2 expression was significantly upregulated in EOC tissues and cell lines compared to noncancerous tissue and cells. CCAT2 knockdown induced by interfering oligonucleotides could inhibit proliferation and promote apoptosis and induce cell cycle arrest at the G0/G1 phase. Bioinformatics analysis predicted that miR-424 targeted CCAT2, which was confirmed by luciferase reporter assay. Moreover, the miR-424 inhibitor rescued the tumorigenesis inhibition induced by CCAT2 knockdown. In summary, our findings illustrate that CCAT2 acts as competing endogenous RNA (ceRNA) or sponge via negatively targeting miR-424, providing a novel diagnostic marker and therapeutic target for EOC.
Key words: Epithelial ovarian cancer (EOC), Long noncoding RNAs (lncRNAs), CCAT2, miR-424
INTRODUCTION
Epithelial ovarian cancer (EOC) is a primary gynecological malignant tumor with third highest morbidity and highest mortality in women worldwide1. The 5-year survival rate of EOC patients is near 45%, which is significantly lower than other gynecological tumors, such as breast cancer (about 90%), endometrial cancer (about 80%), and cervical cancer (about 68%)2. Because there is insufficient effective early detection, the majority of EOC patients are diagnosed at later stages, and recurrence occurs in most EOC patients within 16 months3. Because of the lack of early symptoms, it is always accompanied with extensive metastases in the pelvic and abdominal cavity as the EOC is diagnosed, leading to lower overall survival rate and poor prognosis4. Thus, the in-depth mechanism of pathogenesis for EOC is urgently needed.
Long noncoding RNAs (lncRNAs) are non-protein coding nucleotide sequences with length longer than 200 nt5. Many studies have reported that lncRNAs are dysregulated in cancers, suggesting that the aberrant expression of lncRNAs is associated with tumorigenesis, metastasis, and prognosis in tumorigenesis6. For instance, lncRNA HOTAIR is overexpressed in EOC patients and predicts poor prognosis and promotes tumor metastasis7. Furthermore, the expression of lncRNA RP11-190D6.2 was markedly decreased in EOC tumor tissues compared with normal tissues, suggesting it is positively correlated with WWOX expression8. Colon cancer-associated transcript 2 (CCAT2) was originally identified as an oncogenic lncRNA in colorectal cancer, which has been verified to play an oncogenic molecular role in multiple tumors9. However, the role of CCAT2 in EOC genesis is still unclear.
Similar to lncRNAs, microRNAs (miRNAs) have more widely been recognized in oncogenesis due to their comprehensive distribution and vast previous studies10,11. As well as in EOC, miRNAs are widely identified to contribute to the tumorigenesis and progression of multiple tumors, including proliferation, migration, invasion, and apoptosis12. For instance, miR-127-3p was downregulated and directly regulated the BAG5 gene and acted as a tumor suppressor in EOC13. miR-424 has been shown to inhibit tumor cell migration, invasion, and epithelial–mesenchymal transition by downregulating DCLK1 in ovarian clear cell carcinoma14.
In the present study, we found that CCAT2 was overexpressed in EOC tissue and predicted metastasis and a poor prognosis. Thus, this study was aimed to investigate the potential physiological effect of CCAT2 on EOC tumorigenesis and progression and further validate the miRNA sponge role of CCAT2 for miR-424.
MATERIALS AND METHODS
Tissue Sample Acquirement
The collection of tumor specimens from 31 cases of EOC patients at Huai’an First People’s Hospital, Nanjing Medical University, was used in the present study. Tumor tissues and adjacent nontumor tissues were excised during surgery and immediately stored in −80°C liquid nitrogen for use. No patients experienced radiotherapy and chemotherapy before the operation. The study was approved by the ethics committee and institutional review board of Nanjing Medical University. Informed written consent was signed by all the enrolled patients.
Cell Lines and Culture
EOC cell lines (SKOV3, OMC685, A2780, and HO8910) and a normal ovarian cell line (IOSE386) were purchased from the Shanghai Institute of Cell Biology at the China Academy of Sciences (Shanghai, P.R. China). Cells were cultured in RPMI-1640 (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin at 37°C with 5% CO2 atmosphere.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The extracted RNA was performed to synthesize cDNA using Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Rockford, IL, USA). PCR was performed using GoTaq qPCR Master Mix with SYBR Green (Promega, Madison, WI, USA) according to the manufacturer’s protocol. GAPDH was used as endogenous control. All primers were designed and synthesized by GenePharma (Shanghai, P.R. China) as follows: CCAT2, 5′-CTAGGGTCGTATAGAGAAAACG-3′ (forward) and 5′-AACCATACTCCAATGGTAACAC-3′ (reverse); miR-424, 5′-GCTACGCTAACTGGGTAACCTTG-3′ (forward) and 5′-GTCGAGGGGCATCGGTT-3′ (reverse); and GAPDH, 5′-CGACACTTTATCATGGCTA-3′ (forward) and 5′-TTGTTGCCGATCACTGAAT-3′ (reverse). The level of gene expression was expressed relative to GAPDH and calculated using the 2−ΔΔCt method.
Cell Transfection
Interfering RNAs in the study were obtained from GenePharma Co. Ltd. For the transfection of the miR-424 mimics, inhibitors, and CCAT2 small interfering RNAs (siRNAs) (si-CCAT2), EOC cell lines (2 × 105) were seeded in six-well plates and transfected with 100 nM siRNAs using Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA, USA). The sequences are as follows: miR-424 mimics, 5′-CUUUUUGCGGUCUGGGCUUGC-3′; miR-424 inhibitor, 5′-GCAAGCCCAGACCGCAAAAAG-3′; and si-CCAT2, 5′-AAACUUUAAAGAACACUACUG-3′.
Cell Proliferation Assay
Cell proliferation assay was performed with the cell counting kit-8 (CCK-8; DOJINDO, Japan). The transfected cells were seeded in 96-well plates with a density of 5 × 103 per well. The cells were then added with 10 ml of CCK-8 solution at 37°C for 90 min, and the absorbance was assessed at 450 nm after incubation.
Colony Formation Assay
EOC cells were plated in six-well culture plates at a density of 100 cells per well. After incubation for 14 days at 37°C, cells were fixed with methanol and stained with 0.2% crystal violet and then washed three times with phosphate-buffered saline (PBS). Finally, the visible colonies consisting of >50 cells were manually counted.
Luciferase Reporter Assays
Luciferase assays were performed using the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer’s instructions. Briefly, the full-length 3′-UTR of CCAT2 containing miR-424 binding sites was cloned downstream of the firefly luciferase gene in pGL3 (Invitrogen) to construct pGL3-luc-CCAT2. Cells were seeded into 96-well plates and transfected with pGL3-luc-CCAT2 (50 ng) and Renilla luciferase (5 ng), and miR-424 mimics or NC nucleotides (5 pmol) using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s instructions. After 48 h, luciferase activity was detected using Dual-Glo Luciferase Assay System (Promega). For comparison, the activity was normalized with Renilla luciferase activity. All experiments were performed in triplicate.
Cell Cycle and Apoptosis Analysis
Cell cycle and apoptosis analysis were performed using the cell cycle analysis kit (Lianke, Shanghai, P.R. China) and Annexin-V/Dead Cell Apoptosis Kit (Invitrogen). Briefly, cells were seeded in six-well plates at a density of 4 × 105 per well. Cells were starved in FBS-free medium for 12 h and then treated with cell cycle and apoptosis agents for 48 h. The adherent cells were washed and trypsinized, and centrifuged at 12,000 × g for 5 min. The collected cells were fixed in 75% cold ethanol at −20°C overnight and resuspended in annexin-binding buffer and 5 μl of 7AAD/annexin V/propidium iodide at room temperature for 30 min, and then incubated at room temperature for 15 min in the dark. Fluorescence was analyzed by flow cytometry.
Statistical Analyses
The data are presented as the mean ± SD from at least three independent experiments. Statistical analyses were performed with an analysis of one-way ANOVA or a two-tailed Student’s t-test using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 18.0 (Chicago, IL, USA). The significance level was set as p < 0.05. Survival was analyzed based on Kaplan–Meier curves with the log-rank test analysis.
RESULTS
CCAT2 Expression Was Upregulated in EOC Tissues and Cell Lines
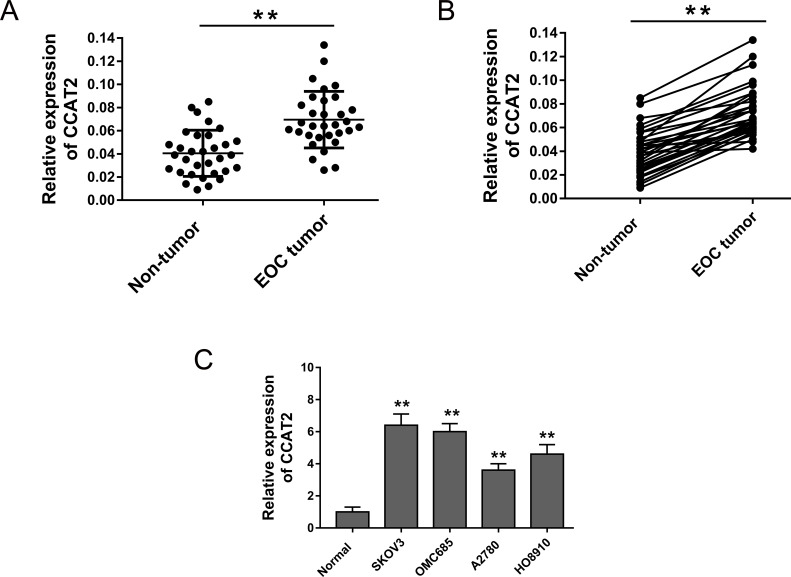
To test the abundance of CCAT2 in EOC tissue, we performed RT-PCR to assess its expression level in EOC tissue and cell lines. Primarily, the results showed that CCAT2 expression was significantly increased in 31 cases of EOC tissues samples (Fig. 1A). CCAT2 expression was upregulated in 90.32% (28/31) of EOC tissues compared with their adjacent nontumor tissues (Fig. 1B). Moreover, the expression of CCAT2 of the EOC cell lines (SKOV3, OMC685, A2780, and HO8910) was significantly higher than that of normal ovarian cell line (IOSE386) (Fig. 1C). Overall, the present study revealed that CCAT2 was markedly overexpressed in EOC tissues and cell lines.
Figure 1.
Colon cancer-associated transcript 2 (CCAT2) expression was upregulated in epithelial ovarian cancer (EOC) tissue samples and cell lines. (A) RT-PCR showed the expression level of CCAT2 in EOC tumor tissues and adjacent nontumor tissues. (B) CCAT2 expression was upregulated in 90.32% (28/31) of EOC tissues compared with pair-matched adjacent noncancerous tissues. (C) Expression of CCAT2 was detected in EOC cell lines (SKOV3, OMC685, A2780, and HO8910) and a normal ovarian cell line (IOSE386). Data are presented as mean ± SD. **p < 0.01 compared with the corresponding control group.
CCAT2 Knockdown Suppressed EOC Cell Line Proliferation and Promoted Apoptosis
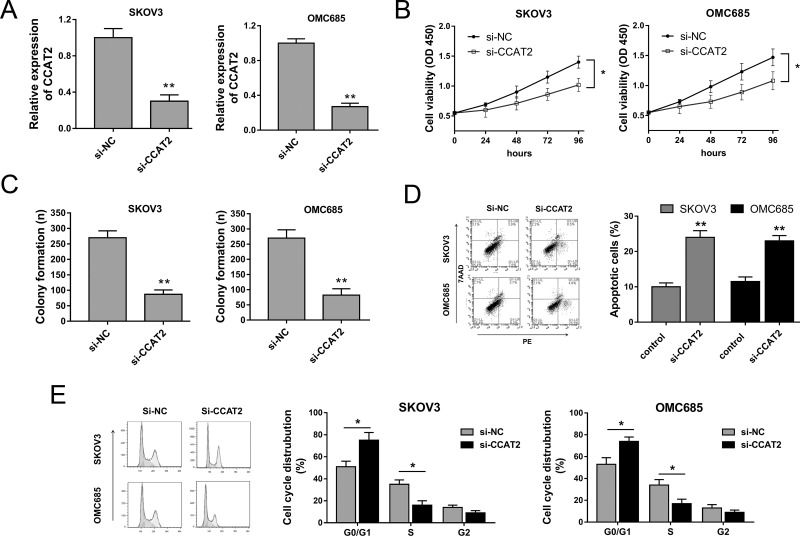
Because CCAT2 was significantly overexpressed in EOC tissue and cell lines, we hypothesized that CCAT2 acted as an oncogenic lncRNA in EOC oncogenesis and played a vital role. Loss-of-function experiments aimed at CCAT2 were performed to verify the hypothesis. To knock down CCAT2 expression and abundance, small interfering nucleotide (si-CCAT2) and control (si-NC) were synthesized and transfected into EOC cell lines (SKOV3 and OMC685), showing the effective interfering efficiency of si-CCAT2 (Fig. 2A). Cell proliferation and colony formation assays showed that CCAT2 knockdown induced by si-CCAT2 could significantly repress the proliferation and colony formation capacity (Fig. 2B and C). Moreover, the flow cytometry assay showed that CCAT2 knockdown could accelerate the apoptosis of SKOV3 and OMC685 cells and induce cell cycle arrest at the G0/G1 phase (Fig. 2D and E). In summary, the results strongly revealed that CCAT2 knockdown could suppress EOC cell lines’ proliferation and promoted apoptosis, suggesting the oncogenic role of CCAT2 in EOC oncogenesis.
Figure 2.
CCAT2 knockdown suppressed EOC cell lines’ proliferation and promoted apoptosis in vitro. (A) Expression level of CCAT2 in EOC cell lines (SKOV3 and OMC685) transfected with small interfering nucleotide (si-CCAT2) and control (si-NC), showing effective interfering efficiency. (B) Cell proliferation ability was detected by the cell counting kit-8 (CCK-8) assay. (C) Cell proliferation ability was detected by colony formation assay. (D) Apoptosis of SKOV3 and OMC685 cells was detected by flow cytometry assay. (E) Cell cycle analysis of SKOV3 and OMC685 detected by flow cytometry revealed the cell cycle arrest at the G0/G1 phase. Data are presented as mean ± SD. *p < 0.05, **p < 0.01 compared with the corresponding control group.
Bioinformatics Analysis Predicted That CCAT2 Was a Target of miR-424
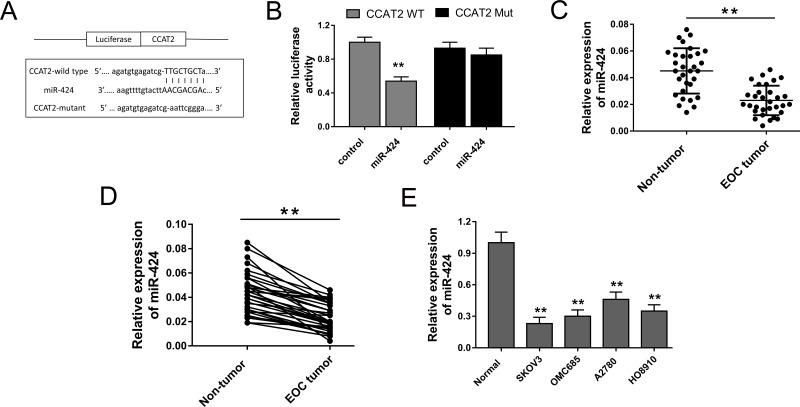
The present study confirmed that CCAT2 was upregulated in EOC tissue, indicating that CCAT2 might function as oncogenic lncRNA in EOC oncogenesis. Admittedly, the major approach that lncRNA exerted physiological function is to act as miRNA “sponge” to recede the target miRNAs’ abundance to indirectly modulate functional miRNAs. Thus, we probed the possible target miRNAs of CCAT2 using bioinformatics analysis. Bioinformatics prediction programs (starBase, http://starbase.sysu.edu.cn) revealed that miR-424 shared complementary binding sites with CCAT2 (Fig. 3A). The luciferase reporter assay verified that miR-424 mimics bound with CCAT2 wild type and decreased luciferase activity (Fig. 3B). RT-PCR was performed to test the miR-424 expression in EOC tissue, and the results showed the lower-expressed miR-424 in EOC tissues compared to the adjacent nontumor tissues (Fig. 3C). miR-424 expression was downregulated in 87.09% (27/31) of EOC tissues compared with their adjacent nontumor tissues (Fig. 3D). Moreover, in EOC cell lines, miR-424 expression levels were significantly suppressed (Fig. 3E). All the results showed that miR-424 was negatively correlated with CCAT2 and could act as target for CCAT2.
Figure 3.
CCAT2 was directly targeted with miR-424. (A) The predicted complementary binding sites of miR-424 and CCAT2. (B) Luciferase reporter assay showed that luciferase activity was decreased in miR-424 mimics binding to CCAT2 wild type. (C) RT-PCR showed lower miR-424 expression in EOC tissues compared to adjacent nontumor tissues. (D) miR-424 expression was downregulated in 87.09% (27/31) of EOC tissues compared with pair-matched adjacent noncancerous tissues. (E) Expression of miR-424 in EOC cell lines. Data are presented as mean ± SD. **p < 0.01 compared with the corresponding control group.
miR-424 Targeted CCAT2 to Modulate Tumorous Progression of EOC Cells
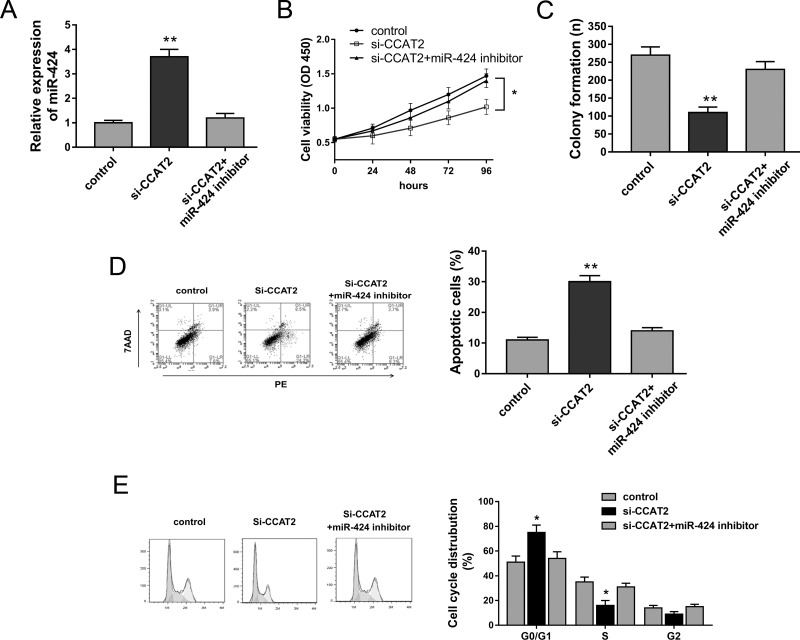
In EOC tumor tissue and cell lines, miR-424 expression was negatively correlated with CCAT2; thus, we hypothesized that miR-424 and CCAT2 exerted inversely regulating roles in EOC cell tumor progression. The existing literature has reported that miR-424 acted as a tumor suppressor in multiple tumorigenesis15,16. Previous experiments had shown that CCAT2 knockdown could suppress EOC cell tumorigenesis; hence, we performed miR-424 knockdown and rescue experiments to verify the competing endogenous role of CCAT2 in SKOV3 cell lines. CCAT2 knockdown induced by si-CCAT2 could increase the miR-424 expression level, which was reversed by the miR-424 inhibitor (Fig. 4A). The CCK-8 assay and colony formation assay showed that the miR-424 inhibitor rescued the inhibition of CCAT2 knockdown on cell proliferation ability in SKOV3 cells (Fig. 4B and C). Furthermore, flow cytometry demonstrated that CCAT2 knockdown accelerated apoptosis and induced cell cycle arrest at the G0/G1 phase, whereas it was reversed by the miR-424 inhibitor (Fig. 4D and E). In summary, a series of rescue experiments revealed that the miR-424 inhibitor could reverse the tumor suppressor role of CCAT2 knockdown, suggesting the targeting binding within CCAT2 and miR-424.
Figure 4.
miR-424 targeted CCAT2 to modulate EOC cells’ proliferation and apoptosis. (A) miR-424 expression in SKOV3 cells transfected with si-CCAT2 and/or miR-424 inhibitor. (B) Cell proliferation of SKOV3 cells was detected by CCK-8. (C) Colony formation assay. (D) Apoptosis of EOC cells was assessed by flow cytometry. (E) Cell cycle analysis showed the cycle distribution. Data are presented as mean ± SD. *p < 0.05, **p < 0.01 compared with the corresponding control group.
DISCUSSION
EOC is the most aggressive subtype of ovarian cancer with a significantly higher recurrence and mortality rate. Recently, lncRNAs have become a research hotspot involving cardiovascular system disease, endocrine system disease, and tumors17. For ovarian cancer, lncRNAs are performed to elucidate the complex mechanisms of tumorigenesis.
CCAT was originally identified as an oncogenic lncRNA in colorectal cancer, which similarly has been identified as oncogenic molecular in other tumors18. In the present study, CCAT2 was significantly upregulated in EOC tissue and cell lines. CCAT2 knockdown suppressed EOC cell lines’ proliferation and promoted apoptosis, suggesting the oncogenic role of CCAT2 in EOC oncogenesis. In addition to CCAT2, several lncRNAs have been identified to modulate carcinogenesis. The lncRNA HOTAIR was increased in EOC tissues compared with corresponding normal tissues, acting as a useful biomarker for the predisposition to EOC and for the early diagnosis of the disease19. Moreover, urothelial cancer-associated 1 (UCA1) was aberrantly upregulated in EOC tissues and cells, acting as a prognostic factor for EOC patients’ overall survival and endogenous sponge by directly binding to miR-485-5p20. Hence, lncRNAs had been recognized for their vital pathological roles in EOC tumorigenesis.
Admittedly, the major approach for lncRNAs to exert physiological function is to act as an miRNA “sponge” to recede target miRNAs’ abundance to indirectly modulate functional miRNAs. Thus, we investigated the possible target miRNAs of CCAT2 using bioinformatics analysis. The results showed that miR-424 shares complementary binding sites with CCAT2 at the 3′-UTR, indicating potential targeting miRNA for CCAT2. It has been validated that miR-424 functions as a tumor suppressor in the carcinogenesis of breast cancer, non-small cell lung cancer, and cervical cancer21–23. In the present study, miR-424 was downregulated in EOC tissues and cell lines, being inversely correlated with that of CCAT2. The luciferase reporter assay revealed internal matching function. Generally, CCAT2 acts as an miR-424 sponge, and the miR-424 inhibitor rescued the tumorigenesis inhibition induced by CCAT2 knockdown. The regulation of miR-424 and CCAT2 is antagonistic, suggesting paired binding. The role that lncRNAs acts as an miRNA sponge is a widespread acting mechanism in multiple types of tumorigenesis24. In hepatocellular carcinoma, lncRNA MALAT1 downregulated miR-146b-5p, which inhibited the phosphorylation of Akt mediated by TRAF6, to promote cancer growth and metastasis25. Thus, miRNA sponge is a vital regulatory mechanism in oncogenesis, integrating miRNA and targeting gene mRNAs and composing a complete regulating loop.
Aberrant expression of lncRNAs might act as diagnostic and therapeutic targets for multiple tumors, as well as EOC. Because lncRNAs exist in the cytoplasm or nuclei, their expression levels could be detected in the early phase of solid tumors. Huang et al. reported that CCAT2 was upregulated in the EOC tissue and was associated with shorter overall survival and disease-free survival, predicting poor prognosis26. lncRNA–mRNA coexpression network analyses are conducted to predict lncRNA expression trends and the potential target genes of lncRNAs27.
In summary, the present study demonstrates the overexpression of lncRNA CCAT2 and indicates the potential correlation with miR-424 in tumorigenesis, revealing the ceRNA mechanism or the miRNA sponge and providing a novel diagnostic marker and therapeutic target for EOC.
ACKNOWLEDGMENTS
This work was supported by The Basic Medical Research Center of Nanjing Medical University and the Department of Gynecology, Huai’an First People’s Hospital.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Scalici JM, Arapovic S, Saks EJ, Atkins KA, Petroni G, Duska LR, Slack-Davis JK. Mesothelium expression of vascular cell adhesion molecule-1 (VCAM-1) is associated with an unfavorable prognosis in epithelial ovarian cancer (EOC). Cancer 2017;123(6):977–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kajiyama H, Mizuno M, Shibata K, Yamamoto E, Kawai M, Nagasaka T, Kikkawa F. Recurrence-predicting prognostic factors for patients with early-stage epithelial ovarian cancer undergoing fertility-sparing surgery: A multi-institutional study. Eur J Obstet Gynecol Reprod Biol. 2014;175:97–102. [DOI] [PubMed] [Google Scholar]
- 3. Bentivegna E, Gouy S, Maulard A, Pautier P, Leary A, Colombo N, Morice P. Fertility-sparing surgery in epithelial ovarian cancer: A systematic review of oncological issues. Ann Oncol. 2016;27(11):1994–2004. [DOI] [PubMed] [Google Scholar]
- 4. Sun S, Cai J, Yang Q, Zhu Y, Zhao S, Wang Z. Prognostic value and implication for chemotherapy treatment of ABCB1 in epithelial ovarian cancer: A meta-analysis. PLoS One 2016;11(11):e0166058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hulstaert E, Brochez L, Volders PJ, Vandesompele J, Mestdagh P. Long non-coding RNAs in cutaneous melanoma: Clinical perspectives. Oncotarget 2017;8(26):43470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Min L, Garbutt C, Tu C, Hornicek F, Duan Z. Potentials of long noncoding RNAs (LncRNAs) in sarcoma: From biomarkers to therapeutic targets. Int J Mol Sci. 2017;18(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134(1):121–8. [DOI] [PubMed] [Google Scholar]
- 8. Tong W, Yang L, Yu Q, Yao J, He A. A new tumor suppressor lncRNA RP11-190D6.2 inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells. Onco Targets Ther. 2017;10:1227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xin Y, Li Z. CCAT2: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;54(3):242–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barciszewska AM. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol. 2016;54(4):369–74. [DOI] [PubMed] [Google Scholar]
- 11. Liang Z, Xi Y. MicroRNAs mediate therapeutic and preventive effects of natural agents in breast cancer. Chin J Nat Med. 2016;14(12):881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halvorsen AR, Kristensen G, Embleton A, Adusei C, Barretina-Ginesta MP, Beale P, Helland A. Evaluation of prognostic and predictive significance of circulating microRNAs in ovarian cancer patients. Dis Markers 2017;54(5):309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bi L, Yang Q, Yuan J, Miao Q, Duan L, Li F, Wang S. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep. 2016;36(5):2563–70. [DOI] [PubMed] [Google Scholar]
- 14. Wu X, Ruan Y, Jiang H, Xu C. MicroRNA-424 inhibits cell migration, invasion, and epithelial mesenchymal transition by downregulating doublecortin-like kinase 1 in ovarian clear cell carcinoma. Int J Biochem Cell Biol. 2017;85:66–74. [DOI] [PubMed] [Google Scholar]
- 15. Li Q, Qiu XM, Li QH, Wang XY, Li L, Xu M, Dong M, Xiao YB. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol Rep. 2015;33(5):2354–60. [DOI] [PubMed] [Google Scholar]
- 16. Zhang D, Shi Z, Li M, Mi J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5:e1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-coding RNAs: A rising biotarget in colorectal cancer. Oncotarget 2017;8(13):22187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan YH, Fang H, Ji CX, Xie H, Xiao B, Zhu XG. Long noncoding RNA CCAT2 can predict metastasis and poor prognosis: A meta-analysis. Clin Chim Acta 2017;466:120–6. [DOI] [PubMed] [Google Scholar]
- 19. Wu H, Shang X, Shi Y, Yang Z, Zhao J, Yang M, Li Y, Xu S. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget 2016;7(27):41047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y, Jiang Y, Wan Y, Zhang L, Qiu J, Zhou S, Cheng W. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. 2016;37(8):10633–41. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Lv Z, Fu J, Wang Z, Fan Z, Lei T. Endogenous microRNA-424 predicts clinical outcome and its inhibition acts as cancer suppressor in human non-small cell lung cancer. Biomed Pharmacother. 2017;89:208–14. [DOI] [PubMed] [Google Scholar]
- 22. Zhou Y, An Q, Guo RX, Qiao YH, Li LX, Zhang XY, Zhao XL. miR424-5p functions as an anti-oncogene in cervical cancer cell growth by targeting KDM5B via the Notch signaling pathway. Life Sci. 2017;171:9–15. [DOI] [PubMed] [Google Scholar]
- 23. Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ, Gorbatenko A, Birnbaum D, Ezhkova E, Cordon-Cardo C, Finetti P, Llobet-Navas D, Silva JM. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayers D, Vandesompele J. Influence of microRNAs and long non-coding RNAs in cancer chemoresistance. Genes (Basel) 2017;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, Bi J, Qu K, Zhang J, Liu C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget 2017;8(17):28683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang S, Qing C, Huang Z, Zhu Y. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J, Lv M, Gu Y, Zhang J, Hua X, Jia G, Xu S, Jia X, Xu P. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. [DOI] [PMC free article] [PubMed] [Google Scholar]