Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide. MicroRNA (miRNA), a class of noncoding single-stranded RNA molecules, is involved in regulating cancer cell proliferation, metastasis, migration, invasion, and apoptosis. We showed that the expression of miR-346 was significantly increased in HCC tissues and cell lines, compared with noncancerous controls, and was associated with poor prognosis. Overexpression of miR-346 promoted proliferation and inhibited apoptosis of SMMC-7721 cells, while knockdown of miR-346 significantly suppressed proliferation and induced apoptosis of HepG2 cells. Then we identified breast cancer metastasis suppressor 1 (BRMS1) as a direct target of miR-346 based on luciferase reporter assays. There was a negative correlation between miR-346 and BRMS1 expression at both the protein and mRNA levels. Furthermore, inhibition of BRMS1 expression reversed the tumor-suppression effects of miR-346 downregulation in HepG2 cells. These results indicate that miR-346 promotes HCC progression by regulating BRMS1 expression.
Key words: MicroRNA-346, Breast cancer metastasis suppressor 1 (BRMS1), Hepatocellular carcinoma (HCC), Proliferation, Migration and invasion
INTRODUCTION
Hepatocellular carcinoma (HCC) is a kind of primary liver cancer with high mortality rate. It is one of the most common malignant tumors in the world, especially in Asia, Africa, and southern Europe1. The incidence of HCC is on the rise, and about 250,000 people die of HCC every year worldwide, with China accounting for about 45% of them. HBV infection, HCV infection, cirrhosis, aflatoxin exposure, and genetic factors are identified as major causes of HCC2,3. At present, the available treatment of HCC includes surgery, transcatheter arterial chemoembolization (TACE), radiofrequency therapy, biological targeted therapy, and so on. Although the clinical treatment of HCC continues to progress, the long-term prognosis of HCC is still dismal. Therefore, there is an urgent need to understand the molecular mechanism of HCC progression to identify new treatment targets.
MicroRNAs (miRNAs) are a class of noncoding single-stranded RNA molecules encoded by an endogenous gene with a length of about 22 nucleotides, which are involved in the regulation of posttranscriptional gene expression in plants and animals. A growing body of evidence suggests that miRNAs play important roles in a variety of cellular processes, such as differentiation, apoptosis, epithelial–mesenchymal transition, and metastasis4–6. miRNA acts as a tumor suppressor or an oncogene by targeting specific genes in a 3′-untranslated region (3′-UTR)-dependent manner. miR-346, a less characterized oncogenic miRNA, has been shown to play an important role in follicular thyroid carcinoma7, rheumatoid arthritis8, cutaneous squamous cell carcinoma9, and cervical cancer10,11. Wang et al. indicated that miR-346 positively regulated human mesenchymal stem cell osteogenic differentiation by targeting glycogen synthase kinase (GSK)-3β and activating the wingless-related integration site (Wnt)/β-catenin pathway12. Moreover, Chen et al. reported that ectopic expression of miR-346 promoted A431 cell proliferation and migration9. However, the role of miR-346 and its mechanisms of action in HCC have not been investigated yet. Our current study is to explore the biological functions of miR-346 in HCC and to investigate the underlying mechanisms of action.
We found that the expression of miR-346 was higher in HCC tissues than in matched normal tissues. Ectopic expression of miR-346 promoted the HCC cell migration and invasion through directly targeting breast cancer metastasis suppressor 1 (BRMS1).
MATERIALS AND METHODS
Ethics Statement
This study was approved by the Ethical Committee of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, Henan, P.R. China), and written informed consent from each participant was obtained before operation according to the system of our hospital.
Tissue Specimens
A total of 40 pairs of surgically resected HCC tissues and the corresponding noncancerous tissues (more than 2 cm from tumor margin and no cancer cell confirmed by pathological examination) were collected from patients during operation in our hospital. All patients were pathologically confirmed and were followed up by medical records or telephone follow-up for 50 months.
Cell Culture and Transfection
Human HCC cell lines Bel-7402, Huh7, HepG2, MHCC-97H, and SMMC-7721 and human immortalized normal liver epithelial cell line LO2 were obtained from the Cell Bank of the Shanghai Institutes for Biological Sciences (Shanghai, P.R. China). All HCC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco). LO2 cells were cultured in RPMI-1640 medium (Gibco) with 10% FBS. Cells were maintained in a humidified incubator containing 5% CO2 at 37°C.
SMMC-7721 and HepG2 cells were plated into 24-well plates and incubated overnight to obtain >70% convergence at the time of transfection. miR-346 mimic, miR-346 inhibitor, and their corresponding control vectors were synthesized from GenePharma (Shanghai, P.R. China). Twenty-nanometer olignonucleotides were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Small interference RNA of BRMS1 (BRMS1-siRNA) and the negative control (NC) RNA oligonucleotides (GenePharma) were transfected into HepG2 cells using similar methods.
RNA Extraction and Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The A260/A280 ratio was measured, and RNA concentration was determined via a NanoDrop ultraviolet spectrophotometer. qRT-PCR was run on the StepOnePlus thermocycler (Applied Biosystems, Foster City, CA, USA). The relative miR-346 and BRMS1 expression levels were determined after normalization to the U6 small nuclear RNA or actin expression based on the comparative cycle threshold (ΔΔCt) method.
Cell Proliferation and Colony Formation Assays
Transfected cells (5 × 103 cells/well) in exponential growth period with complete medium were seeded into 96-well plates, followed by incubation for 24, 48, 72, 96, and 120 h. Afterward, 10 μl of cell counting kit 8 (CCK-8) solution (Dojindo Laboratories, Japan) was added to each well and incubated for 2 h at 37°C. Cell activity was detected by measuring absorbance at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA). All experiments were conducted in triplicate and were repeated three times. For the colony formation assay, transfected cells were seeded in a six-well plate at a density of 1,000 cells per well and cultured for 12 days. The number of colonies was counted after being fixed with 4% paraformaldehyde and stained with crystal violet.
In Vitro Invasion and Migration Assay
We performed the invasion assay using Matrigel (BD Biosciences, San Jose, CA, USA)-precoated Transwell chambers (8.0-μm pore size with polyethylene tetraphthalate membrane) according to the manufacturer’s protocol. Briefly, 1 × 105 transfected cells suspended with serum-free medium were seeded in the upper chamber of the Transwell insert (Corning, Corning, NY, USA), while the lower chamber was added with DMEM and 10% FBS as a chemoattractant. After 24 h of incubation, noninvaded cells in the upper chamber were wiped away, and cells that had invaded to the lower chamber were fixed with 4% paraformaldehyde and stained with crystal violet. At least six separate fields of cells on the filters were photographed and counted. Cell migration assay was similar to the invasion assay, except that Matrigel was not required.
Apoptosis Assay
Cell apoptosis was detected using an Annexin-V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining kit (Roche, Basel, Switzerland). SMMC-7721 and HepG2 cells were harvested 48 h after transfection, resuspended in 500 μl of binding buffer, and incubated with 5 μl of annexin V/FITC and 5 μl of PI at room temperature in the dark for 15 min. Results were obtained using a FACScan flow cytometer (BD Biosciences).
Bioinformatics Analysis
Bioinformatics analysis using the TargetScan and PicTar databases was performed to identify potential 3′-UTR-binding target genes for miR-346. The interaction with a likely candidate was then explored by Dual-Luciferase Reporter assay, and its expression was determined.
Luciferase Reporter Assay
SMMC-7721 cells (4 × 105) were seeded in six-well plates and cotransfected with the reporter constructs together with the miR-346 mimic or NC using Lipofectamine 2000 according to the manufacturer’s recommendation (Invitrogen). Renilla luciferase was used for endogenous normalization. Dual-luciferase activity was determined after 48 h according to the manufacturer’s instructions (Promega, Beijing, P.R. China).
Western Blotting
Transfected cells were harvested and washed twice with PBS (Solarbio, Beijing, P.R. China). Cell lysis buffer (120 μl; Thermo Fisher) was added to each cell and incubated on ice for 30 min. After centrifugation at 4°C, 13,000 rpm, for 10 min, the supernatant was carefully collected to obtain the total protein. Protein concentration was measured using the BCA protein assay kit (Solarbio). Equal amounts of protein (30 μg) were separated by 12% SDS-PAGE (Beyotime) and then transferred to PVDF membranes (Thermo Fisher). Membranes were blocked with Tris-buffered saline (TBS; Solarbio) containing 5% fat-free milk powder (Yili, Beijing, P.R. China) at room temperature for 1 h. Then the membrane was incubated overnight at 4°C with rabbit anti-BRMS1 antibody (Proteintech, Wuhan, Hubei, P.R. China) or mouse anti-actin antibody (Proteintech). The following day, membranes were incubated with secondary antibodies conjugated with horseradish peroxidase against mouse IgG or rabbit IgG (Proteintech) for 1 h at room temperature after washing three times with TBST. Blots were detected with the infrared laser scanning imaging system (CDYSSEY CLx; General Electric Company).
Statistical Analysis
Statistical analyses were performed with SPSS17.0 software (SPSS Statistics, Chicago, IL, USA). All experiments were repeated at least three times. Data are shown as the mean ± standard deviation. Student’s t-test was used to determine the statistical significance. Paired t-test was used for paired samples. A value of p < 0.05 was considered to be statistically significant.
RESULTS
miR-346 Is Upregulated in HCC Cell Lines and Tissues and Is Associated With Poor Prognosis
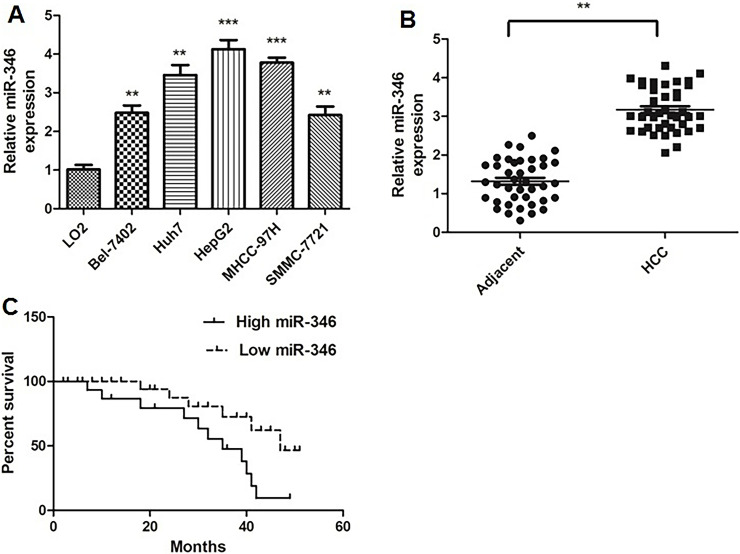
Using qRT-PCR, we found that miR-346 was significantly upregulated in all the five HCC cell lines (Bel-7402, Huh7, HepG2, MHCC-97H, and SMMC-7721) compared with normal LO2 liver cells (Fig. 1A). We then examined miR-346 expression in 40 pairs of HCC tissues and adjacent normal liver tissues. The results showed that miR-346 was upregulated in HCC tissues compared with adjacent normal liver tissues (Fig. 1B).
Figure 1.
MicroRNA-346 (miR-346) expression was upregulated in hepatocellular carcinoma (HCC) cells and tissues and was associated with poor prognosis. (A) miR-346 was measured in HCC cell lines and normal liver cells using quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR). **p < 0.01, ***p < 0.001 compared to LO2 cells. (B) Expression of miR-346 was examined in 40 pairs of HCC tissue specimens and adjacent normal liver tissues using qRT-PCR. **p < 0.01 compared to adjacent tissue. (C) Kaplan–Meier overall survival curves of HCC patients separated into high and low miR-346 expression according to the mean value of miR-346 expression (3.17).
The 40 patients with HCC were further divided into low and high miR-346 groups according to the mean value of the miR-346 expression (3.17). Kaplan–Meier analysis revealed that the high expression of miR-346 was significantly correlated with poorer prognosis of HCC (Fig. 1C). These results indicated that high miR-346 might be associated with HCC progression.
Effect of miR-346 on the Proliferation, Migration, and Invasion of HCC Cells
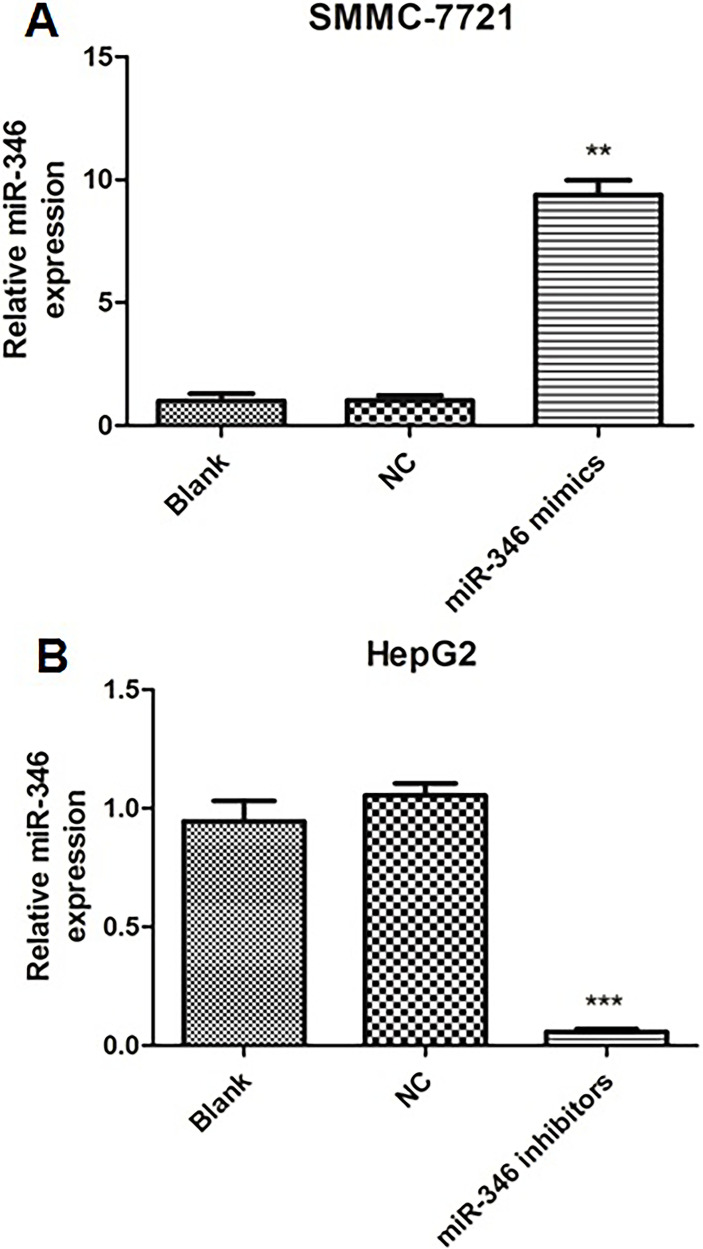
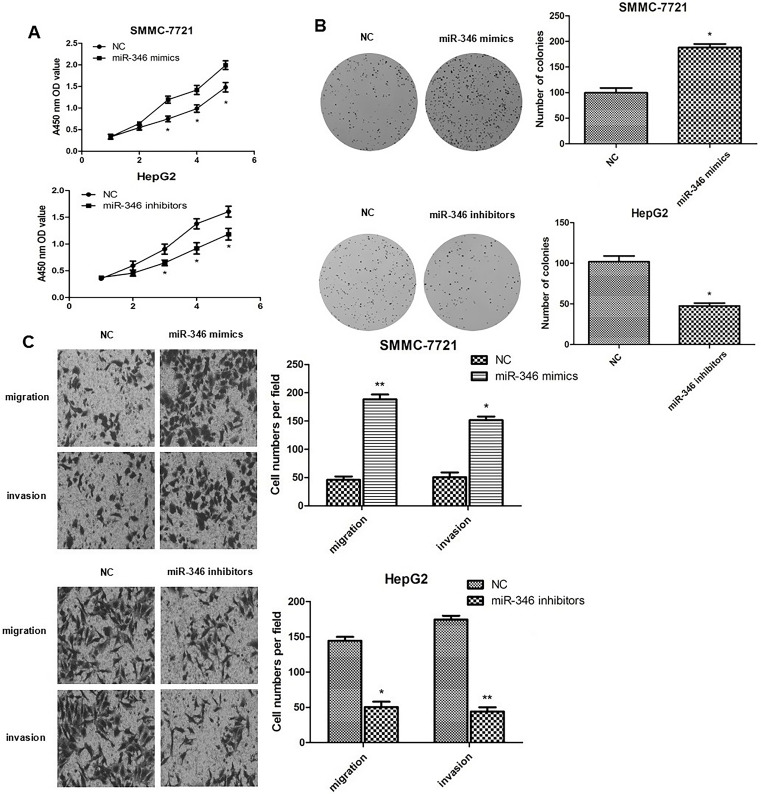
To investigate the biological functions of miR-346, knockdown and overexpression experiments were done. As shown in Figure 1A, HepG2 cells displayed significantly high miR-346 expression level, while SMMC-7721 cells displayed low expression. For comparison, we transfected miR-346 mimics into SMMC-7721 cells and miR-346 inhibitors into HepG2 cells, respectively. Compared to the NC group, the expression level of miR-346 was significantly upregulated in SMMC-7721 cells and remarkably downregulated in HepG2 cells (Fig. 2). CCK-8 assays and colony formation assays were performed in SMMC-7721 and HepG2 cells to evaluate the effect of miR-346 on cell proliferation. Data showed that upregulation of miR-346 promoted SMMC-7721 cell proliferation and increased the number of colonies, whereas downregulation of miR-346 inhibited HepG2 cell proliferation and decreased the number of colonies (Fig. 3A and B). Furthermore, Transwell migration and Matrigel invasion assay were conducted to evaluate the role of miR-346 in tumor cell migration and invasion. The results revealed that overexpression of miR-346 promoted HCC cell migration and invasion, while knockdown of miR-346 suppressed HCC cell migration and invasion (Fig. 3C). Collectively, these results indicated that miR-346 promotes cell proliferation, migration, and invasion in vitro.
Figure 2.
The expression level of miR-346 following overexpressive or inhibitory transfection. The expression level of miR-346 in SMMC-7721 cells (A) and HepG2 cells (B) transfected with blank, negative control (NC), miR-346 mimics, or miR-346 inhibitors was detected using qRT-PCR. **p < 0.01, ***p < 0.001 compared to NC.
Figure 3.
miR-346 promotes HCC cell growth, migration, and invasion. (A) Cell viability was determined by cell counting kit 8 (CCK-8) assays on SMMC-7721 cells and HepG2 cells transfected with miR-346 mimics or inhibitors, respectively. (B) Colonies formed in SMMC-7721 cells and HepG2 cells transfected with miR-346 mimics or inhibitors, respectively. (C) Migration and invasion assays were performed using Transwell migration and Matrigel invasion chambers after transfection of miR-346 mimics or inhibitors in SMMC-7721 cells and HepG2 cells, respectively (magnification: 40×). *p < 0.05, **p < 0.01 compared to NC.
Effect of miR-346 on the Apoptosis of HCC Cells
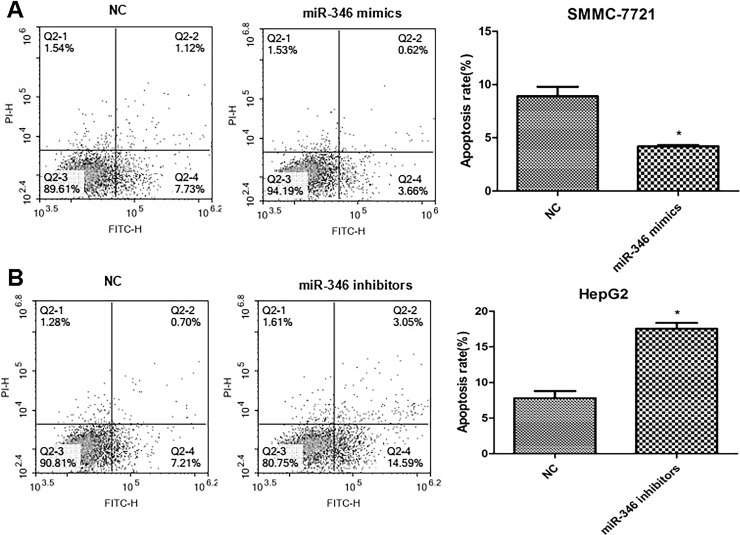
The annexin V-FITC/PI staining method to detect cell apoptosis demonstrated that compared with their respective NC group, upregulation of miR-346 significantly inhibited the apoptotic rate of SMMC-7721 cells, and downregulation of miR-346 enhanced the apoptotic rate of HepG2 cells (Fig. 4). Our results showed that miR-346 can reduce HCC cell apoptosis.
Figure 4.
Effect of miR-346 on the apoptosis of HCC cells. (A) Upregulation of miR-346 inhibited the apoptosis of SMMC-7721 cells. *p < 0.05 compared to NC. (B) Downregulation of miR-346 enhanced the apoptosis of HepG2 cells. *p < 0.05 compared to NC.
BRMS1 Is the Target of miR-346
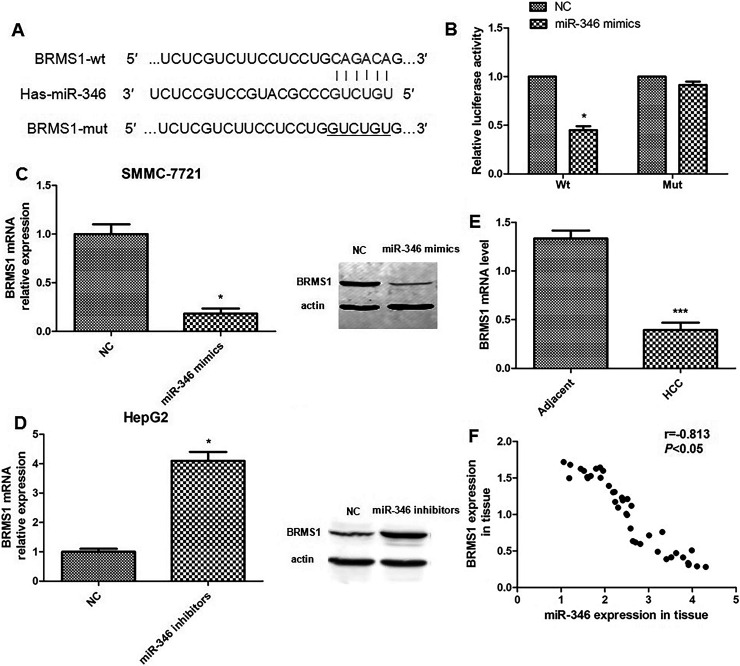
The TargetScan and PicTar databases showed that BRMS1 was one of the predicted miR-346 targets whose 3′-UTR contained putative miR-346 target sites (Fig. 5A). The luciferase reporter assay indicated that the BRMS1-3′-UTR luciferase signal dropped by 50% in the miR-346 mimics group. However, the BRMS1 mut-3′-UTR luciferase activity had no significant difference from the NC group (Fig. 5B). To further determine whether BRMS1 is a target of miR-346, Western blot was performed in miR-346-upregulated SMMC-7721 cells and miR-346-downregulated HepG2 cells. The results indicated that compared with the NC group, the protein level of BRMS1 in the SMMC-7721 cells transfected with miR-346 mimics was significantly decreased, while HepG2 cells transfected with miR-346 inhibitors have the contrary results. The real-time PCR assay gets the same results (Fig. 5C and D).
Figure 5.
Breast cancer metastasis suppressor 1 (BRMS1) is a target of miR-346. (A) Bioinformatic analysis showed that the 3′-untranslated region (3′-UTR) of BRMS1 contained a putative binding site for miR-346. (B) Relative luciferase activities were analyzed in SMMC-7721 cells cotransfected with miR-NC or miR-346 mimics and luciferase reporters carrying wild-type (WT) or mutant (Mut) BRMS1 3′-UTR, respectively. (C) Expression of BRMS1 was detected in SMMC-7721 cells transfected with miR-NC or miR-346 mimics using qRT-PCR and Western blot. (D) Expression of BRMS1 was measured in HepG2 cells transfected with miR-NC or miR-346 inhibitors using qRT-PCR and Western blot. (E) qRT-PCR was conducted to determine the mRNA level of BRMS1 in HCC tissues compared to adjacent normal tissues. (F) A reverse correlation between the miR-346 expression and BRMS1 expression is indicated in 40 HCC tissue samples. *p < 0.05, ***p < 0.001 compared to NC or adjacent, respectively.
We then examined the BRMS1 mRNA in 40 pairs of HCC tissue samples. We found that the average level of BRMS1 expression was lower in HCC tissues than in the adjacent nontumor tissues (Fig. 5E). A significant inverse correlation was observed between BRMS1 and miR-346 in the 40 HCC tissue samples (Fig. 5F).
BRMS1 Reverses the Role of miR-346 on HCC Cell Migration and Invasion
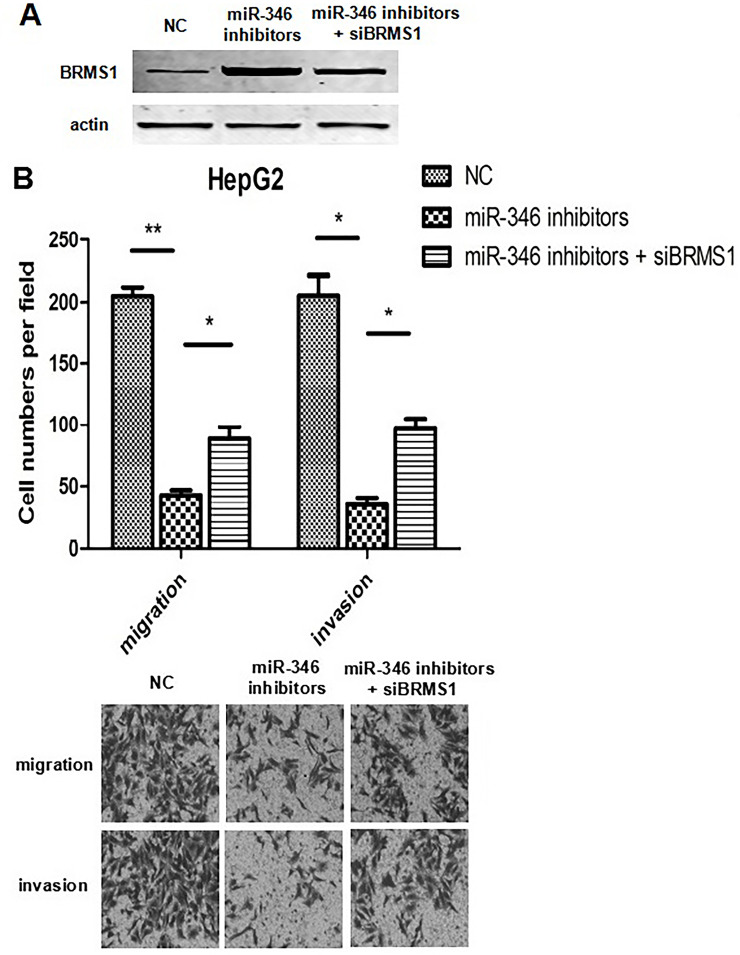
We then tested if miR-346 induces the progression of HCC by targeting BRMS1. We inhibited BRMS1 expression in miR-346-downregulated HepG2 cells. Strikingly, inhibited expression of BRMS1 significantly increased the migration and invasion capacities of HepG2 cells transfected with miR-346 inhibitors (Fig. 6). These results collectively suggested that miR-346 promotes the invasiveness of HCC by targeting BRMS1.
Figure 6.
Inhibiting expression of BRMS1 reverses the effects of miR-346 on cell migration and invasion. (A) Western blot analysis of BRMS1 protein in HepG2 cells transfected with NC or miR-346 inhibitors and/or short interfering BRMS1 miRNA (siBRMS1). (B) Transwell migration and Matrigel invasion assays were performed on each group (magnification: 40×). *p < 0.05, **p < 0.01.
DISCUSSION
miRNAs play a variety of roles in cancer development and progression13. For instance, miR-21 has been reported to promote K-ras-dependent lung tumorigenesis by activating the Ras/MEK/ERK pathway14. The miR-17-92 cluster is upregulated in a wide range of cancer types including lymphoma, lung, breast, stomach, colon, and pancreatic cancers15. In this study, we found that the expression of miR-346 was significantly increased in HCC tissues and cell lines, compared with noncancerous controls. Gain-of-function experiments found that upregulation of miR-346 promoted HCC cell proliferation, migration, and invasion, and inhibited apoptosis, while depletion of miR-346 acquires the opposite results. Our results suggest that miR-346 has oncogenic function in HCC progression by inhibiting cell apoptosis. The oncogenic role of miR-346 has also been described in other studies. miR-346 was found to facilitate human non-small cell lung cancer cell growth and metastasis and suppress cell apoptosis by regulation of the XPC/ERK/E-cadherin pathway16. Yang and his colleagues also reported that miR-346 promotes breast cancer cell proliferation, colony formation, migration, and invasion, reduced apoptosis, and sensitivity to docetaxel17.
Bioinformatics analysis can predict a lot of candidate target genes of specific miRNAs. Here we identified BRMS1 as a direct target of miR-346. BRMS1, which maps to human chromosome 11q13.1-q13.2, is a metastasis suppressor that was first identified in breast cancer18. BRMS1 inhibition of tumor metastasis has also been reported in melanoma, ovarian carcinoma, non-small cell lung cancer, and others19–21. In this study, we found that miR-346 could target BRMS1 3′-UTR, and there was a negative correlation between miR-346 and BRMS1 expression at both the protein and mRNA levels. In HCC tissue samples, we got the same results. To further test whether miR-346 induces the progression of HCC by targeting BRMS1, we silenced BRMS1 expression in miR-346-downexpressing HepG2 cells. As expected, reducing the expression levels of BRMS1 reversed the role of miR-346 on HCC cells. This suggests that miR-346 promotes the migration and invasion of HCC cells by targeting BRMS1.
In conclusion, our study clarifies that upregulation of miR-346 plays a vital role in the progression of HCC via targeting BRMS1. Although the more exact molecular mechanism of how miR-346 regulates BRMS1 expression and how BRMS1 regulates progression of HCC needs further investigation, our study proves that miR-346 and BRMS1 could serve as potential therapeutic targets for HCC.
ACKNOWLEDGMENTS
Zhixian Guo and Jingjing Li made an equal contribution to the study. Jihong Sun obtained the specimens. Lu Sun contributed to the study. Yubing Zhou provided language help. Zujiang Yu conducted the work.
REFERENCES
- 1. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007;132(7):2557–76. [DOI] [PubMed] [Google Scholar]
- 2. Ringelhan M, Heikenwalder M, Protzer U. Direct effects of hepatitis B virus-encoded proteins and chronic infection in liver cancer development. Dig Dis. 2013;31(1):138–51. [DOI] [PubMed] [Google Scholar]
- 3. Thomas DL. Global control of hepatitis C: Where challenge meets opportunity. Nat Med. 2013;19(7):850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeker LT, Bluestone JA. MicroRNA regulation of T-cell differentiation and function. Immunol Rev. 2013;253(1):65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li C, Hashimi SM, Good DA, Cao S, Duan W, Plummer PN, Mellick AS, Wei MQ. Apoptosis and microRNA aberrations in cancer. Clin Exp Pharmacol Physiol. 2012;39(8):739–46. [DOI] [PubMed] [Google Scholar]
- 6. Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31(3–4):653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human microRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91(9):3584–91. [DOI] [PubMed] [Google Scholar]
- 8. Semaan N, Frenzel L, Alsaleh G, Suffert G, Gottenberg JE, Sibilia J, Pfeffer S, Wachsmann D. miR-346 controls release of TNF-alpha protein and stability of its mRNA in rheumatoid arthritis via tristetraprolin stabilization. PLoS One 2011;6(5):e19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen B, Pan W, Lin X, Hu Z, Jin Y, Chen H, Ma G, Qiu Y, Chang L, Hua C, Zou Y, Gao Y, Ying H, Lv D. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol. 2016;37(2):2765–71. [DOI] [PubMed] [Google Scholar]
- 10. Guo J, Lv J, Liu M, Tang H. miR-346 up-regulates argonaute 2 (AGO2) protein expression to augment the activity of other microRNAs (miRNAs) and contributes to cervical cancer cell malignancy. J Biol Chem. 2015;290(51):30342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song G, Wang R, Guo J, Liu X, Wang F, Qi Y, Wan H, Liu M, Li X, Tang H. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci Rep. 2015;5:15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q, Cai J, Cai XH, Chen L. miR-346 regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting the Wnt/beta-catenin pathway. PLoS One 2013;8(9):e72266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010;18(3):282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Lorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M, Previati M, Ambs S, Palumbo T, Garofalo M, Veronese A, Bottoni A, Gasparini P, Harris CC, Visone R, Pekarsky Y, de la Chapelle A, Bloomston M, Dillhoff M, Rassenti LZ, Kipps TJ, Huebner K, Pichiorri F, Lenze D, Cario S, Buendia MA, Pineau P, Dejean A, Zanesi N, Rossi S, Calin GA, Liu CG, Palatini J, Negrini M, Vecchione A, Rosenberg A, Croce CM. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20(5):589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun CC, Li SJ, Yuan ZP, Li DJ. MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging 2016;8(10):2509–24. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ, Ding L, Li J, Chen D, Ma R, Wu JZ, Tang JH. MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene 2017;600:21–8. [DOI] [PubMed] [Google Scholar]
- 18. Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60(11):2764–9. [PubMed] [Google Scholar]
- 19. Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273(2):229–39. [DOI] [PubMed] [Google Scholar]
- 20. Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer 2006;16(2):522–31. [DOI] [PubMed] [Google Scholar]
- 21. Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276(2):196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]