Abstract
Recent studies have suggested that the dysregulation of microRNAs (miRNAs) plays a critical role in the progression of human cancers, including gastric cancer (GC). miR-143 had been reported to function as a tumor suppressor in GC. However, the exact molecular mechanism of how miR-143 participates in GC progression remains to be determined. In this present study, we revealed that the expression of miR-143 was significantly downregulated in human GC tissues and cell lines compared with normal tissues and a normal gastric epithelium cell line. In addition, upregulation of the expression of miR-143 in a GC cell line inhibited cell proliferation and induced cell cycle arrested in the G0/G1 phase. Furthermore, GATA6 was identified as a direct target of miR-143 in GC using the luciferase reporter assay. Upregulation of miR-143 inhibited the expression of GATA6 in GC cell lines. Moreover, the overexpression of GATA6 could attenuate the effect of miR-143 on cell proliferation in the GC cell lines. Collectively, these data indicated that miR-143 plays a tumor suppressor role partly through regulating the expression of GATA6 in GC. Therefore, targeting miR-143 may be a novel therapeutic method for GC.
Key words: miR-143, Gastric cancer (GC), GATA-binding factor 6 (GATA6), Cell proliferation, Cell cycle
INTRODUCTION
Gastric cancer (GC) is the leading cause of cancer-related deaths worldwide, which accounts for about 700,000 deaths annually1. China is a high-risk area for digestive cancers2 and was estimated to suffer 498,000 GC deaths in 20153. The 5-year overall survival rate of patients with GC is about 10–30% because most patients are diagnosed at advanced disease stages4. Therefore, there is an urgent need to identify specific genetic alterations in GC with the aim to validate novel potential therapeutic targets.
MicroRNAs (miRNAs) are endogenously expressed 19–24 nt noncoding RNAs that control protein expression mainly through binding to the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs)5. Increasing studies have demonstrated that miRNAs play important roles in human cancer development and progression6–9. miR-143, located at chromosome 5q33, has been reported to be involved in multiple biological processes including cell proliferation and migration through targeting genes including Bcl-27, COX-28, MAPK79, ERK510, and ELK111. Previous studies have suggested that miR-143 was downregulated in several human cancers including osteosarcoma9, breast cancer10, non-small cell lung cancer12, colorectal cancer13, and prostate cancer14. miR-143 expression has also been reported to be downregulated in human GC tissues and cell lines15. Zhuang et al. found that miR-143 was involved with the cisplatin resistance of GC cells via targeting IGF1R and Bcl-216. In addition, Wu et al. showed that miR-143 could suppress GC cell growth and induce apoptosis by targeting COX-28. Recently, Zhang et al. found that miR-143 also could inhibit GC cell proliferation and invasion by targeting DNMT3A17. However, to date, the biological function and underlying mechanism of miR-143 in GC progression remain unclear, and therefore further efforts are needed to fully elucidate the regulatory role of miR-143 in GC.
In our present study, we measured the expression level of miR-143 in GC tissues and cell lines and explored its function and underlying mechanisms in the progression of GC. Our results revealed that miR-143 expression was significantly downregulated in GC tissues and cell lines, and the overexpression of miR-143 inhibited cell proliferation and cell cycle progression in GC. Further, bioinformatics and luciferase reporter assay confirmed that GATA-binding factor 6 (GATA6) was a target gene of miR-143 in GC. Thus, our results presented in this study indicated that miR-143 functions as a tumor suppressor by targeting GATA6 in GC. miR-143 may be a potential target for treating GC in the future.
MATERIALS AND METHODS
Participants
A total of 46 paired GC tissues and corresponding noncancerous tissues were collected from GC patients who underwent treatment in the Department of Clinical Oncology, The First Affiliated Hospital of Zhengzhou University between October 2009 and April 2011. The patients’ clinicopathological features were collected. Each patient gave written informed consent for research purposes. The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Cell Culture
The human GC cell lines HGC-27, AGS, SGC-7901, and BGC-823 and a normal gastric epithelium cell line, GES-1, were obtained from the Cell Resource Center, Institute of Biochemistry and Cell Biology at the Chinese Academy of Science. These cells were grown in RPMI-1640 medium supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA). The cells were incubated in a humidified atmosphere at 37°C containing 5% CO2.
Cell Transfection
The miR-143 mimic, miR-143 inhibitor, and negative control miRNA (miR-NC) were designed and synthesized by GenePharma (Shanghai, P.R. China). The GATA6 open reading frame (ORF) expression construct was purchased from GenScript (Nanjing, P.R. China). The small interfering RNA (siRNA) targeting GATA6 (si-GATA6) and NC siRNA were designed and synthesized by GenePharma. The cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s recommendations.
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-qPCR) Analysis
For RT-qPCR analysis, total RNA was extracted from the GC tissues and cell lines using the TRIzol reagent (Invitrogen), and miRNAs were obtained using the miRneasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The cDNA was synthesized using the SYBR Premix Ex Taq Kit (Takara, Dalian, P.R. China) according to the manufacturer’s protocols. The TaqMan miRNA RT-qPCR Kit (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the expression level of miR-143. U6 RNA was used as an internal control for miR-143. The SYBR Green PCR Master Mix Kit (Applied Biosystems) was used to determine the expression level of GATA6. GAPDH was used as internal control for GATA6. RT-qPCR was carried out with ABI Prism 7500 Sequence Detection System (Applied Biosystems). The primers used in this study were as follows: GATA6, 5′-CACACGCTGACAGTGCTGG-3′ (forward) and 5′-TACAGGGCGATACAAAGCAGGAGAA-3′ (reverse); miR-143, 5′-ACACTCGAGCTGGGGCTTCTCCTGGCTCTCC-3′ (forward) and 5′-TGGTGTGGTGGAGTCG-3′ (reverse); U6 RNA, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-ACGCTTCACGAATTTGCGT-3′ (reverse); GAPDH, 5′-ACGGGAAGCTCACTGGCATGG-3′ (forward) and 5′-GGTCCACCACCCTGTTGCTGTA-3′ (reverse). The relative expression was calculated using the 2−ΔΔCt method.
Western Blot Analysis
Total protein was extracted from tissues and cells using radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma-Aldrich, St. Louis, MO, USA). The protein concentration was determined by the BCA assay kit (Beyotime, Haimen, Jiangsu, P.R. China). The same amount of protein (50 μg) was separated using 10% SDS-PAGE gel, and then the protein was transfected to a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with fat-free milk and incubated with antibody against GATA6 (1:1,000; ab106066; Abcam, Cambridge, MA, USA) and GAPDH (1:1,000; ab8245; Abcam) at 4°C overnight. Finally, the membrane was incubated with HRP-conjugated rabbit anti-mouse IgG (1:5,000; ab6728; Abcam). The protein was detected with the enhanced chemiluminescence method (Beyotime). Protein signals were quantified using Quantity One Software (Bio-Rad, Hercules, CA, USA).
Cell Proliferation Analysis by Cell Counting Kit-8 (CCK-8) Assay
The cell proliferation rate was measured using the CCK-8 assay (Beyotime). Briefly, cells transfected with the miRNAs or GATA6 expression construct were seeded into a 96-well plate at a density of 3 × 103 cells/well. At indicated time points (0, 1, 2, and 3 days), 10 μl of the CCK-8 solution was added to each well and cultured for another 2 h. Finally, the absorbance was measured at 450 nm using a microplate reader (Multiskan MK3; Thermo Fisher Scientific, Inc.).
Cell Cycle Analysis by Flow Cytometry
The cell cycle distribution was assessed using flow cytometry. For cell cycle analysis, the cells were fixed with 70% ethanol at 4°C overnight. The cells were then stained with 50 mg/ml propidium iodide (PI; Sigma-Aldrich) in the presence of RNase for 30 min at 37°C. The cell cycle phase proportion was determined using the ModFit software (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Luciferase Reporter Assay
The mutant (Mut) and wild-type (WT) 3′-UTR of GATA6 that contain the putative target region of miR-143 were designed and synthesized by GenePharma and cloned into the psiCHECK™-2 Dual Luciferase miRNA target expression vector (Promega, Madison, WI, USA). The cells were cotransfected with Mut or WT 3′-UTR of GATA6 constructs and miR-143 mimic or miR-NC using Lipofectamine 2000 (Invitrogen). The luciferase and Renilla signals were measured using the Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol after 48 h of transfection.
Statistical Analysis
Data were presented as mean ± standard deviation. Statistical analysis was carried out using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Student’s t-test and one-way ANOVA were used to determine the significance of differences between two groups or among multiple groups, respectively. Pearson’s correlation analysis was used to evaluate the correlation of miR-143 with GATA6. Chi-square test was used to analyze the associations between miR-143 expression and the clinicopathological features. Differences with a value of p < 0.05 were considered statistically significant.
RESULTS
miR-143 Was Downregulated in GC Tissues and a Number of GC Cell Lines
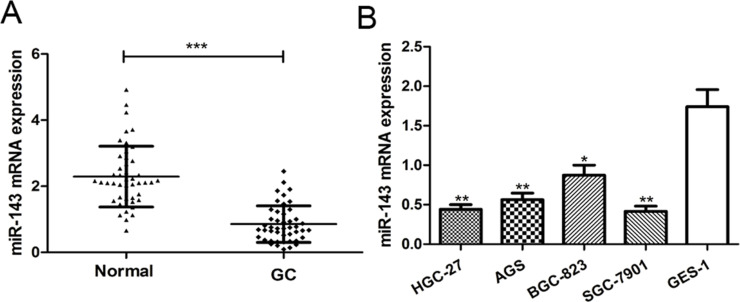
To investigate the potential significance of miR-143 in GC progression, we first examined miR-143 expression in GC tissues and corresponding noncancerous tissues using RT-qPCR. We found the expression of miR-143 was significantly decreased in tumor tissues compared to the corresponding noncancerous tissues (p < 0.001) (Fig. 1A). We examined the expression of miR-143 in four human GC cell lines (HGC-27, AGS, SGC-7901, and BGC-823) and one normal gastric epithelium cell line (GES-1). The results showed that miR-143 expression was significantly reduced in all four GC cell lines compared with the normal gastric epithelium cell line GES-1 (p < 0.01) (Fig. 1B). Among the four GC cell lines selected in this study, the expression of miR-143 was lowest in the SGC-7901 cell line and was highest in the BGC-823 cell line (Fig. 1B). Therefore, these two cell lines were selected for further experiments. Collectively, these findings indicated that miR-143 may play a critical role in GC progression.
Figure 1.
MicroRNA-143 (miR-143) expression is downregulated in gastric cancer (GC) tissues and cell lines. (A) Quantitative real-time reverse transcriptase polymerase chain reaction (RT-qPCR) was used to analyze miR-143 expression in 46 paired GC tissues and adjacent noncancerous tissues. (B) RT-qPCR was used to analyze miR-143 expression in four GC cell lines (HGC-27, AGS, SGC-7901, and BGC-823) and a normal gastric epithelium cell line (GES-1). *p < 0.05, **p < 0.01, ***p < 0.001.
miR-143 Was Correlated With the Clinicopathological Features of GC Patients
To investigate the clinical significance of miR-143 expression in GC, we collected the clinicopathological features of the 46 enrolled patients and analyzed the correlation between miR-143 expression and the clinicopathological features. The 75th percentile of the miR-143 expression level in the 46 patients was used as a cutoff value, and these patients were divided into two groups (high miR-143 expression group, n = 12; low miR-143 expression group, n = 34). We also investigated the correlations between miR-143 expression and other clinicopathological features. We found that low expression of miR-143 was significantly correlated with tumor stage (p = 0.012), tumor size (p = 0.010), and differentiation (p = 0.017), but no significant correlation was observed with age and gender (p > 0.05)
Increasing Expression of miR-143 Can Inhibit the Cell Proliferation of GC Cell Lines
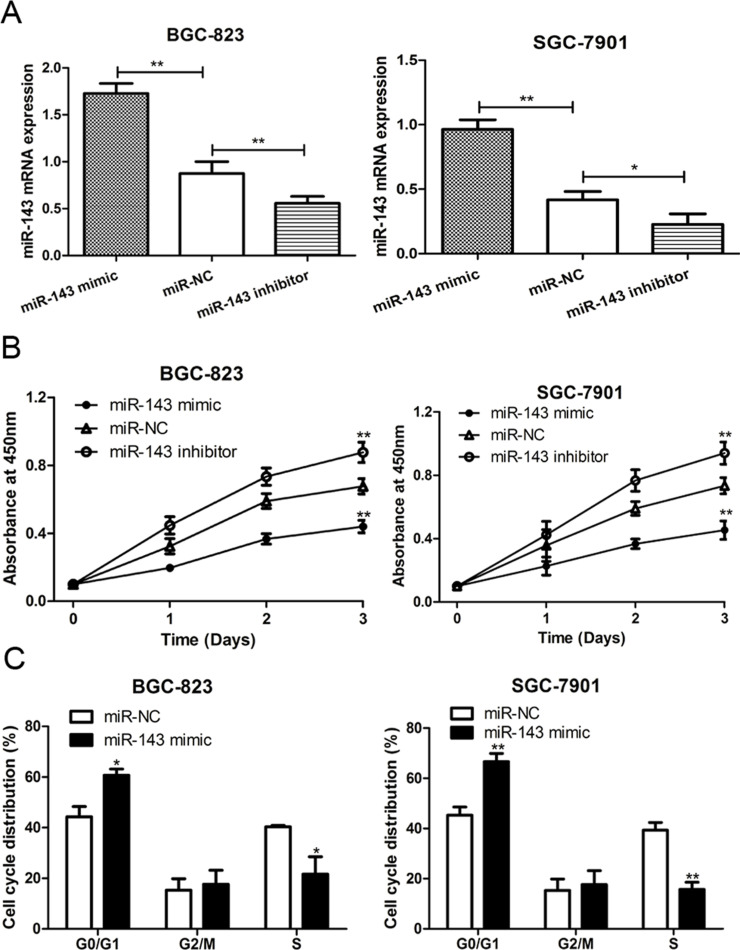
To explore the biological function of miR-143 in GC progression, the SGC-7901 and BGC-823 cell lines were transfected with the miR-143 mimic, miR-143 inhibitor, and miR-NC. RT-qPCR was performed to measure the relative expression posttransfection. The results presented in Figure 2A indicated that the transfection of the miR-143 mimic could significantly upregulate the expression of miR-143, whereas the miR-143 inhibitor could reduce the miR-143 expression. Additionally, CCK-8 assay was performed to measure the relative cell viability. As shown in Figure 2B, increased expression of miR-143 led to an obvious reduction in the cell proliferation rate of both SGC-7901 and BGC-823 cell lines. Conversely, decreased expression of miR-143 led to an increase in the cell proliferation rate in both cell lines. These results suggest that increasing expression of miR-143 may repress GC cell proliferation ability.
Figure 2.
miR-143 inhibited cell proliferation and cell cycle progression in GC. (A) RT-qPCR was used to analyze miR-143 expression in SGC-7901 and BGC-823 cell lines after miR-143 mimic, miR-143 inhibitor, or miR-NC (negative control) transfection. (B) Cell counting kit-8 (CCK-8) assay was used to analyze cell proliferation rate in SGC-7901 and BGC-823 cell lines after miR-143 mimic, miR-143 inhibitor, or miR-NC transfection. (C) Flow cytometry was used to analyze cell cycle distribution in SGC-7901 and BGC-823 cell lines after miR-143 mimic, miR-143 inhibitor, or miR-NC transfection. *p < 0.05, **p < 0.01.
Increasing Expression of miR-143 Can Induce Cell Cycle Arrest in the G0/G1 Phase
To further understand the regulatory role of miR-143 in cell proliferation, we analyzed the effect of miR-143 expression on cell cycle progression by flow cytometry. As shown in Figure 2C, we found that the G0/G1 phase cell proportion was increased, but the S phase cell proportion was decreased when cells were transfected with the miR-143 mimic, compared with miR-NC group in both SGC-7901 and BGC-823 cell lines. Thus, these results suggested that increasing expression of miR-143 suppressed cell cycle progression in GC.
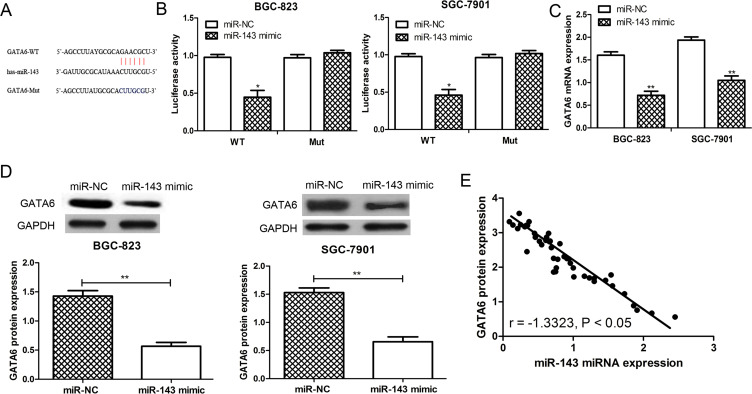
GATA6 Was a Direct Target of miR-143 in GC
To identify potential target genes of miR-143 in GC, we applied computational algorithms to predict the mRNA targets of miRNAs by TargetScan. As a result, the oncogene GATA6 was found to contain the putative binding sequence for miR-143 and might be a potential target for miR-143 (Fig. 3A). The dual-luciferase reporter assay was performed to validate whether GATA6 could be regulated by miR-143. Not surprisingly, miR-143 had no significant effect on luciferase activity in the Mut GATA6 3′-UTR construct, but produced a significant reduction in luciferase activity in the WT GATA6 3′-UTR construct compared with the control (p < 0.05) (Fig. 3B). Furthermore, the expression of GATA6 in the miR-143 mimic- or miR-NC-transfected SGC-7901 and BGC-823 cell lines was examined. The results from the RT-qPCR showed that the expression of GATA6 was downregulated by miR-143 in these two cell lines (p < 0.01) (Fig. 3C). The results from the Western blot analysis further confirmed that the expression of GATA6 was reduced by miR-143 overexpression in the SGC-7901 and BGC-823 cell lines (p < 0.01) (Fig. 3D). Furthermore, the analysis of the expression of miR-143 and GATA6 in tissues revealed that GATA6 expression was inversely correlated with the expression of miR-143 (p < 0.05) (Fig. 3E). These results suggested that GATA6 was a direct target of miR-143 in GC.
Figure 3.
GATA-binding factor 6 (GATA6) was a direct target of miR-143 in GC. (A) GATA6 was predicted as a potential target of miR-143 by TargetScan online software. (B) The relative luciferase activity in BGC-823 or SGC-7901 cells was detected after the wild-type (WT) or mutant (Mut) GATA6 3′-untranslated region (3′-UTR) genes were cotransfected with miR-NC or miR-143 mimic. (C) RT-qPCR. (D) Western blot was used to analyze GATA6 expression in SGC-7901 and BGC-823 cell lines after miR-143 mimic or miR-NC transfection. (E) The expression of GATA6 was inversely correlated with the expression of miR-143 in GC. *p < 0.05, **p < 0.01.
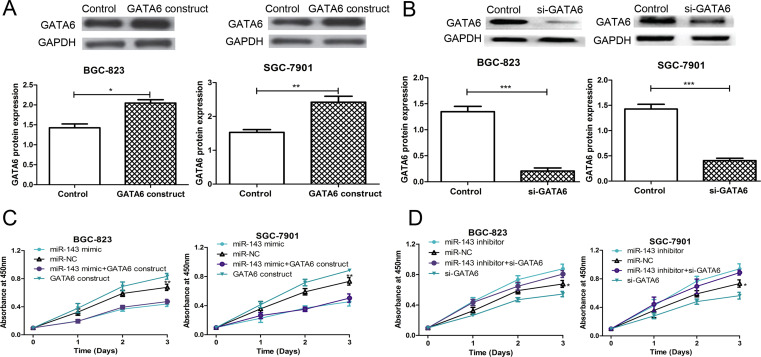
GATA6 Was Involved in miR-143-Mediated Suppression of Cell Proliferation
To further determine the association between miR-143 and GATA6 in regulating GC cell proliferation, rescue experiments were performed by cotransfecting with the miR-143 mimic or miR-143 inhibitor and the GATA6 ORF expression construct or si-GATA6 into SGC-7901 and BGC-823 cell lines. The results demonstrated that the protein expression of GATA6 was elevated when the GATA6 ORF expression construct was transfected into the cell lines (Fig. 4A). On the contrary, the protein expression of GATA6 was reduced when si-GATA6 was transfected into the cell lines (Fig. 4B). However, the cell proliferation rate of the SGC-7901 and BGC-823 cell lines was partially inhibited by cotransfecting with the miR-143 mimic and GATA6 ORF expression construct compared with the GATA6 ORF expression construct-transfected groups (Fig. 4C). We also observed that the cell proliferation rate of the SGC-7901 and BGC-823 cell lines was partially restored by cotransfecting with the miR-143 inhibitor and si-GATA6 compared with the si-GATA6-transfected groups (Fig. 4D). Taken together, these results illustrated that GATA6 was involved in miR-143-mediated suppression of cell proliferation.
Figure 4.
GATA6 was involved in miR-143-mediated cell proliferation suppression. (A) Western blot was used to analyze GATA6 expression in SGC-7901 and BGC-823 cell lines after GATA6 open reading frame (ORF) expression construct or empty vector control transfection. (B) Western blot was used to analyze GATA6 expression in SGC-7901 and BGC-823 cell lines after si-GATA6 or control siRNA transfection. (C) CCK-8 assay was used to analyze the cell proliferation rate in SGC-7901 and BGC-823 cell lines after miR-143 mimic, miR-NC, or GATA6 ORF expression construct transfection. (D) CCK-8 assay was used to analyze the cell proliferation rate in SGC-7901 and BGC-823 cell lines after miR-143 inhibitor, miR-NC, or si-GATA6 transfection. *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
GC is the fourth most common human malignant disease and the second most frequent cause of cancer-related deaths worldwide, with particularly high frequencies in East Asia1. Despite many efforts that have been made, the prognosis of GC is still unsatisfactory18,19. Thus, exploring the underlying mechanism of GC and finding new treatment targets are essential for improving the survival rate of GC patients.
miRNAs are important factors in tumorigenesis and have been the subject of research in many types of cancers20,21. Accumulating evidence has suggested that miR-143 functions as a tumor suppressor in multiple human cancers including GC, which involves many biological processes including tumor initiation and progression7–15. However, the precise functional role of miR-143 in GC is not yet known. In this study, we examined the expression pattern of miR-143 in GC and investigated the molecular mechanism of miR-143 in GC progression. We found that miR-143 was downregulated in GC tumor tissues compared with adjacent nontumor tissues. A low expression level of miR-143 was also observed in four GC cell lines compared with normal gastric epithelium cell line. Furthermore, the low expression of miR-143 was correlated with several clinicopathological features including tumor size, tumor stage, and differentiation through analyzing the clinical data collected from the enrolled patients. Our results indicated that miR-143 might play an important role in GC.
Previous studies indicated that the abnormal expression of miR-143 is associated with cancer cell proliferation and apoptosis in human cancers7–10. Therefore, in vitro functional assays were also performed to investigate the role of miR-143 in GC progression. The CCK-8 assay revealed that upregulation of miR-143 could suppress cell proliferation in GC cell lines, whereas downregulation of miR-143 could promote GC cell proliferation. Furthermore, by flow cytometry analysis, we found that upregulation of miR-143 could suppress cell cycle progression through arresting the cell cycle in the G0/G1 phase. Based on these findings, we demonstrated that miR-143 functioned as a tumor suppressor in GC. However, the underlying mechanism of miR-143 in cell proliferation and cell cycle regulation in GC is still unclear.
Several studies have demonstrated that miRNAs function as an oncogene or tumor suppressor gene through regulating the expression of their downstream target genes in tumors7–10. Therefore, in this study, the online software TargetScan was used to predict the targets of miR-143 in GC and, as a result, GATA6 was selected for further studies. We found that the overexpression of miR-143 could downregulate the expression of GATA6, suggesting that GATA6 was directly involved in miR-143-mediated regulation in GC. Further experiments demonstrated that the overexpression of GATA6 could partially counter the effect of miR-143 overexpression on cell proliferation, which validated that GATA6 was a direct target of miR-143 in GC. Therefore, a novel epigenetic pathway was identified in which miR-143 may suppress the progression of GC by targeting GATA6.
In conclusion, our data demonstrated that miR-143 was downregulated in GC, and restoration of its expression inhibits cell proliferation and induces cell cycle arrest at the G0/G1 phase in GC cell lines. The growth-inhibitory activity of miR-143 in GC was linked to downregulation of GATA6. Thus, miR-143 may serve as a potential target for treatment of GC.
REFERENCES
- 1. Gigek CO, Chen ES, Calcagno DQ, Wisnieski F, Burbano RR, Smith MA. Epigenetic mechanisms in gastric cancer. Epigenomics 2012;4(3):279–94. [DOI] [PubMed] [Google Scholar]
- 2. Li MM, Wan X, Wang YH, Sun YY, Yang GH, Wang L. Time trends of esophageal and gastric cancer mortality in China, 1991–2009: An age-period-cohort analysis. Sci Rep. 2017;7(1):6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. [DOI] [PubMed] [Google Scholar]
- 4. Hartgrink HH, Jansen EP, van Grieken NC, Van de Velde CJ. Gastric cancer. Lancet 2099;374(9688):477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008;455(7209):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu X, Li Z. The role of miRNAs in cutaneous squamous cell carcinoma. J Cell Mol Med. 2016;20(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang CQ, Huang JY, Ma PY, Yu GF. MicroRNA-143 acts as a suppressor of hemangioma growth by targeting Bcl-2. Gene 2017;628:211–7. [DOI] [PubMed] [Google Scholar]
- 8. Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. microRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19(43):7758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong XC, Lv B, Li YS, Cheng QH, Su C, Yin GY. Mir-143 regulates the proliferation and migration of osteosarcoma cells through targeting MAPK7. Arch Biochem Biophys. 2017;630:47–53. [DOI] [PubMed] [Google Scholar]
- 10. Zhou LL, Dong JL, Huang G, Sun ZL, Wu J. MicroRNA-143 inhibits cell growth by targeting ERK5 and MAP3K7 in breast cancer. Braz J Med Biol Res. 2017;50(8):e5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009;460(7256):705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang HB, Sun LC, Ling L, Cong LH, Lian R. miR-143 suppresses the proliferation of NSCLC cells by inhibiting the epidermal growth factor receptor. Exp Ther Med. 2016;12(3):1795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai JW, Xue HZ, Zhang C. Down-regulation of microRNA-143 is associated with colorectal cancer progression. Eur Rev Med Pharmacol Sci. 2016;20(22):4682–7. [PubMed] [Google Scholar]
- 14. Zhou P, Chen WG, Li XW. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am J Cancer Res. 2015;5(6):2056–63. [PMC free article] [PubMed] [Google Scholar]
- 15. Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009;77(1):12–21. [DOI] [PubMed] [Google Scholar]
- 16. Zhuang M, Shi Q, Zhang XW, Ding YB, Shan LQ, Shan X, Qian JQ, Zhou X, Huang ZB, Zhu W, Ding Y, Cheng W, Liu P, Shu Y. Involvement of miR-143 in cisplatin resistance on gastric cancer cells via targeting IGF1R and BCL2. Tumor Biol. 2015;36(4):2737–45. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Q, Feng Y, Liu P, Yang J. Mir-143 inhibits cell proliferation and invasion by targeting DNAMT3 in gastric cancer. Tumor Biol. 2017;39(7):1–8. [DOI] [PubMed] [Google Scholar]
- 18. Badgwell B, Das P, Ajani J. Treatment of localized gastric and gastroesophageal adenocarcinoma: The role of accurate staging and preoperative therapy. J Hematol Oncol. 2017;10(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abozeid M, Rosato A, Sommaggio R. Immunotherapeutic strategies for gastric carcinoma: A review of preclinical and clinical recent development. Biomed Res Int. 2017;2017:5791262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan Y, Wang R, Guan W, Qiao M, Wang L. Role of microRNAs in cancer associated fibroblasts of gastric cancer. Pathol Res Pract. 2017;213(7):730–6. [DOI] [PubMed] [Google Scholar]
- 21. Liu X, Liu X, Wu Y, Wu Q, Wang Q, Yang Z, Li L. MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget 2017;8(19):32370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]