Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that are involved in human carcinogenesis and progression. miR-204 has been reported to be a tumor suppressor in several cancer types. However, the function and underlying molecular mechanism of miR-204 in cervical cancer (CC) are still unclear. In the present study, the expression level of miR-204 was measured using the qRT-PCR method in 30 paired CC clinical samples and in 6 CC cell lines. We found that the expression of miR-204 was significantly downregulated in CC tissues and cell lines compared to normal cervical tissues and cell line. miR-204 was overexpressed by transfection with the miR-204 mimic in HeLa and C33A cell lines in the following experiments. The results showed that overexpression of miR-204 dramatically suppressed cell proliferation, migration, and invasion, caused cell cycle arrest at the G0/G1 phase, promoted cell apoptosis in vitro, and inhibited tumor growth in vivo. Western blot results indicated that overexpressing miR-204 decreased the expressions of CDK2, cyclin E, MMP2, MMP9, Bcl2, whereas it enhanced Bax expression and suppressed the activation of the PI3K/AKT signaling pathways in CC cells. Ephrin type B receptor 2 (EphB2) was identified as a direct target of miR-204 in CC cells according to bioinformatics analysis and luciferase reporter assay. Furthermore, knockdown of EphB2 mimicked the inhibitory effect of miR-204 on the proliferation, invasion, and migration of CC cells. These findings suggested that miR-204 might serve as a tumor suppressor in the development of CC by directly targeting EphB2.
Key words: MicroRNA, Cervical cancer (CC), Cell proliferation, Metastasis, Ephrin type-B receptor 2 (EphB2)
INTRODUCTION
Cervical cancer (CC) is one of the most common gynecologic cancers and causes major cancer-related deaths in females worldwide, with an estimate of more than 500,000 new cases diagnosed every year1. Although previous studies have demonstrated that the initiation and progression of CC are associated with the persistent infection with high-risk human papillomavirus (HPV), growing evidence suggests that invasive CC is a complex and multistep pathology that involves a range of normal factors and cervical epithelium deterioration2. Surgery and radiotherapy are the major treatment strategies for CC in the clinic; however, the 5-year survival rates for advanced CC patients remain unsatisfactory, and about 40% of patients develop lymph node recurrence and distant metastasis3. Recent investigations have shown that the molecular therapeutic targets possess the potential value to improve the prognosis of metastasis in carcinogenesis4. Therefore, the molecular mechanisms underlying the genetics and biology of cervical carcinogenesis need to be fully understood.
MicroRNAs (miRNAs) are a class of endogenous, noncoding regulatory RNAs 21–25 nucleotides (nt) in length that regulate gene expression at the posttranscriptional level through binding to the 3′-untranslated regions (3′-UTRs) of specific target genes5. Because bioinformatics prediction shows that a single gene can be modulated by many RNA transcripts, accumulating evidence has illustrated that miRNAs are involved in multiple biological processes including cell proliferation, differentiation, apoptosis, migration, and development6. Recent studies have demonstrated that miRNAs are dysregulated in various types of human cancers and act as oncogenes or tumor suppressors. miR-204 embedded in intron 6 of the transient receptor potential melastain 3 gene has been identified to play a crucial role in metabolism and cardiovascular disease7. Recently, the expression of miR-204 was reported to be downregulated in various human cancers, and its expression is associated with the poor prognosis of cancer patients8. However, the exact role of miR-204 in CC progression has not yet been thoroughly investigated.
Therefore, in the present study, the function and mechanism of miR-204 were explored in the progression of CC. Our data revealed that miR-204 was dramatically downregulated in CC tissue and cell lines. Biological function analysis showed that ectopic expression of miR-204 suppressed CC cell proliferation, invasion, and migration, caused cell cycle arrest at the G0/G1 stage, and promoted apoptosis. Moreover, the 3′-UTR of ephrin type B receptor 2 (EphB2) was identified as a direct target of miR-204 in CC, and silencing EphB2 obtained the similar inhibitory effect of miR-204 mimic on CC development.
MATERIALS AND METHODS
Tumor Specimens
A total of 30 cases of CC tissue samples and adjacent normal cervical epithelial tissues were collected from newly diagnosed CC patients who underwent surgery between September 2012 and November 2015 in the Shaanxi Nuclear Industry 215 Hospital (Shaanxi, P.R. China). Written consent was obtained from all patients. None of the patients received radiotherapy or chemotherapy treatment prior to surgery. The protocol of the present study was approved by the Ethics Committee of Shaanxi Nuclear Industry 215 Hospital and was in accordance with the Helsinki Declaration. The fresh specimens were immediately frozen in liquid nitrogen for further investigation.
Cell Culture and Transfection
Human CC cell lines (HeLa, C33A, SiHa, MS751, HCC94, and CaSki) and a normal human cervix epithelial cell line (Ect1/E6E7) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere of 5% CO2.
miRNA and siRNA Transfection
To upregulate miR-204 expression or downregulate EphB2 expression in the Hela and C33A cell lines, the miR-204 mimic and corresponding negative control (miR-204 NC) and EphB2 siRNA and corresponding negative control (NC siRNA) were designed and synthesized by GenePharma Co., Ltd. (Shanghai, P.R. China). HeLa and C33A cell lines were transfected with miRNA or siRNA using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
RNA Extraction and Quantitative RT-PCR
The total RNA was isolated from cell lines and tissues using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Reverse transcription was carried out using PrimeScript™ RT reagent kit (Takara, Dalian, P.R. China). Then miR-204 expression levels were quantified with SYBR Green Master Mix (Invitrogen) on an ABI Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, New York, NY, USA). U6 was used as an internal control for miR-204. The relative quantification of miRNAs was calculated using the comparative 2−ΔΔCt method.
Western Blot Analysis
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Nanjing, P.R. China) according to the manufacturer’s instructions. The concentration of protein was measured using a bicinchoninic acid (BCA) assay kit (Beyotime) according to the manufacturer’s instructions. A total of 20 μg of protein was separated by 12% SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% nonfat milk for 1 h at room temperature and then were incubated with primary antibodies against EphB2, CDK2, cyclin E, Bcl2, Bax, MMP2 (matrix metalloproteinase 2), MMP9, p-AKT, t-AKT, and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. After rinsing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology) at room temperature for 2 h. The blots were detected using an enhanced chemiluminescence (ECL) detection system (Amersham Biosciences Corp., Piscataway, NJ, USA) according to the manufacturer’s instructions. The results were analyzed using the ImageJ program (NIH, Bethesda, MD, USA). GAPDH was used as internal control.
Cell Proliferation Assay
Cell proliferation was assessed using the CCK-8 reagent (Dojindo, Tokyo, Japan) following the manufacturer’s protocol. Briefly, HeLa and C33A cells were seeded into 96-well plates at a density of 2 × 103 each well and then transfected with miR-204 mimic or NC for 96 h at 37°C. Cell proliferation was measured every 24 h; CCK-8 solution was added to each well and incubated for an additional 4 h at 37°C. The optical density (OD) values of cells were measured at 490 nm using a microplate reader (Bio-Rad, Baltimore, MD, USA).
Colony Formation Assay
HeLa and C33A cells transfected with the miR-204 mimic or NC were seeded in a six-well plate at a density of 1 × 103 cells each well and cultured for 14 days. The culture medium was replaced at 3-day intervals. Colonies were fixed with methanol and stained with crystal violet for 15 min. The number of colonies (>50 cells) was calculated under an invert microscope (Olympus, Tokyo, Japan).
Migration and Invasion Assay
Migration and invasion assays were performed using Transwell chambers (8-μm pore size; Costar, Corning, NY, USA). Transfected cells (5 × 104 and 5 × 103) in 100 μl of FBS-free DMEM medium were added to the upper chambers precoated with Matrigel or without Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), respectively, for invasion and migration assays. DMEM medium (500 μl) containing 10% FBS was added to the lower chambers to serve as the chemoattractant. After incubation at 37°C for 36 h, cells remaining on the upper chambers were carefully removed using cotton swabs. The invaded and migrated cells were stained with 0.1% crystal violet at room temperature. The migrated/invaded cells were counted at five randomly selected fields under an inverted microscope (Olympus).
Cell Cycle Assay
Cell cycle assay was performed using flow cytometry. Briefly, HeLa and C33A cells were transfected with miR-204 or NC for 36 h, and then the transfected cells were washed with ice-cold PBS and fixed in ice-cold 70% ethanol at 4°C for 1 h. After rinsing, the cells were treated with RNase A (Sigma-Aldrich, Carlsbad, CA, USA) and stained with propidium iodide (PI; Sigma-Aldrich) for 30 min at 37°C in the dark. Cell cycle distribution was analyzed by FACScalibur flow cytometry (BD Biosciences, Glencoe, MA, USA).
Cell Apoptosis Analysis
HeLa and C33A cells were transfected with miR-204 or NC for 48 h, and then transfected cells were collected and double stained with annexin V-FITC and PI using Annexin-V-FITC Apoptosis Detection Kit (Invitrogen) according to the manufacturer’s protocols. Cell apoptosis was quantitatively analyzed by a FACScalibur flow cytometer (BD Biosciences) and CellQuest software (BD Biosciences).
Bioinformatics Analysis and Luciferase Report Assay
Bioinformatics analysis was performed using microRNA.org (http://www.microrna.org/microrna/home.do) to predict the potential targets of miR-204. The wild-type (WT) or mutant (MU) EphB2 3′-UTR binding site of miR-204 was cloned into the pGL3 luciferase vector (Invitrogen). HeLa and C33A cells were seeded into 24-well plates and were cotransfected with pGL3-EphB2-3′UTR WT or pGL3-EphB2-3′UTR MU, and miR-204 mimic or NC using Lipofectamine 2000 following the manufacturer’s instructions. After transfection for 48 h, luciferase activities were detected using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Renilla luciferase activity was used for normalization.
In Vivo Experiments
An in vivo model of CC was established using female BALB/c nude mice (5 weeks old, n = 20). All mice were obtained from the Laboratory Animal Center of The Fourth Military Medical University and housed under specific pathogen-free conditions. The animal protocols were approved by the Animal Care and Use Committee of Shaanxi Nuclear Industry 215 Hospital. HeLa cells (1 × 107) transfected with the miR-204 mimic or NC suspended in PBS were subcutaneously injected into the flank of each mouse (n = 10 for each group). Tumor weight was assessed using calipers every week and measured according to the formula: volume (mm3) = 0.5 × length × width2. After 5 weeks, all mice were euthanized, and tumor tissues were isolated and weighed. The miR-204 level in tumor tissues was calculated by the qRT-PCR method as described above.
Statistical Analysis
All data were analyzed using GraphPad Prism version 5 (GraphPad Software, La Jolla, CA, USA) and were expressed as mean ± standard deviation (SD). The differences between two groups were analyzed using the unpaired two-tailed Student’s t-test. Each experiment was done at least three independent times. A value of p < 0.05 was considered a statistically significant difference.
RESULTS
miR-204 Was Downregulated in Human CC Tissues and Cell Lines
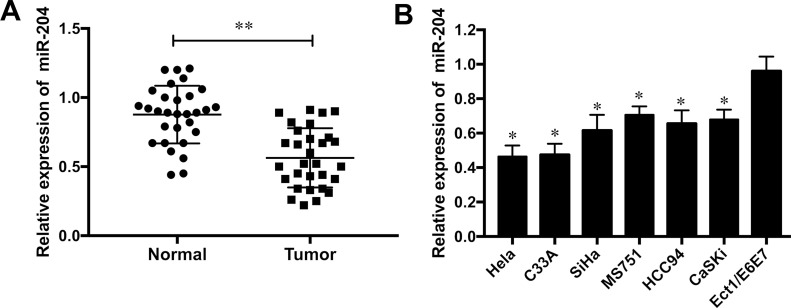
To measure the role of miR-204 on CC, the expression levels of miR-204 were validated in CC tissue samples and adjacent normal tissues by qRT-PCR assays. The results showed that the expression of miR-204 was noticeably downregulated in human CC tissues when compared to that in adjacent normal tissues (p < 0.01) (Fig. 1A). Furthermore, the expression levels of miR-204 in CC cell lines HeLa, C33A, SiHa, MS751, HCC94, and CaSKi were significantly decreased compared to the normal human cervix epithelial cell line Ect1/E6E7 (all p < 0.05) (Fig. 1B). HeLa and C33A cell lines were used for subsequent investigations as the downregulation of miR-204 was more obvious than in the other cell lines.
Figure 1.
miR-204 was downregulated in human cervical cancer (CC) tissues and cell lines. (A) The expression level of miR-204 was detected in 30 CC specimens and paired adjacent normal tissues by qRT-PCR. (B) The expression level of miR-204 was detected in six CC cell lines (HeLa, C33A, SiHa, MS751, HCC94, and CaSki) and normal human cervix epithelial cell line (Ect1/E6E7). *p < 0.05, **p < 0.01 versus the control group.
Overexpression of miR-204 Inhibited Cell Proliferation and Colony Formation in CC Cells
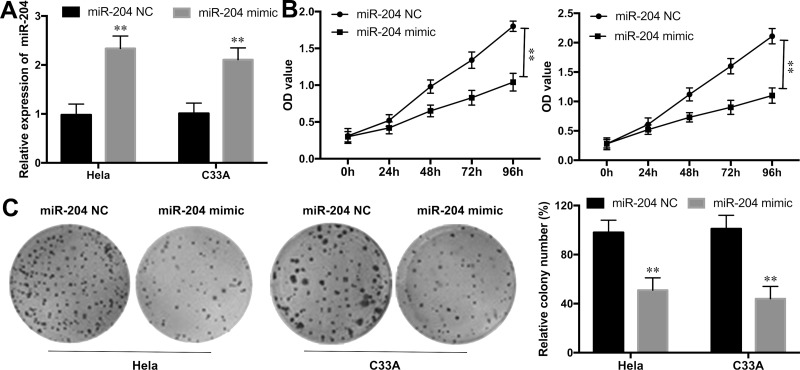
To determine the biological role of miR-204 overexpression on CC progression in vitro, miR-204 mimics or miR-204 NC was transfected into HeLa and C33A cell lines. The efficiency of the transfection was validated by qRT-PCR (p < 0.01) (Fig. 2A). Then CCK-8 assay was performed to explore the effect of miR-204 overexpression on CC cell proliferation. The results revealed that ectopic expression of miR-204 markedly inhibited cell growth in HeLa and C33A cell lines compared to that in NC groups during the 96-h detection (p < 0.01) (Fig. 2B). Moreover, cell colony formation assay also revealed that the cell growth ability in HeLa and C33A cell lines with miR-204 overexpression was significantly inhibited in comparison to NC groups (p < 0.01) (Fig. 2C).
Figure 2.
Overexpression of miR-204 inhibited cell proliferation and colony formation in CC cells. (A) The expression levels of miR-204 in Hela and C33A cell lines with the miR-204 mimic or NC transfection was determined by qRT-PCR. (B) The cell proliferation rate in the Hela and C33A cell lines with the miR-204 mimic or NC transfection was measured by the CCK-8 assay. (C) The colony number of Hela and C33A cell lines with miR-204 mimic or NC transfection was detected by cell colony formation assay. **p < 0.01 versus the NC group.
Overexpression of miR-204 Promoted CC Cell Apoptosis and Cell Cycle at the G0/G1 Phase
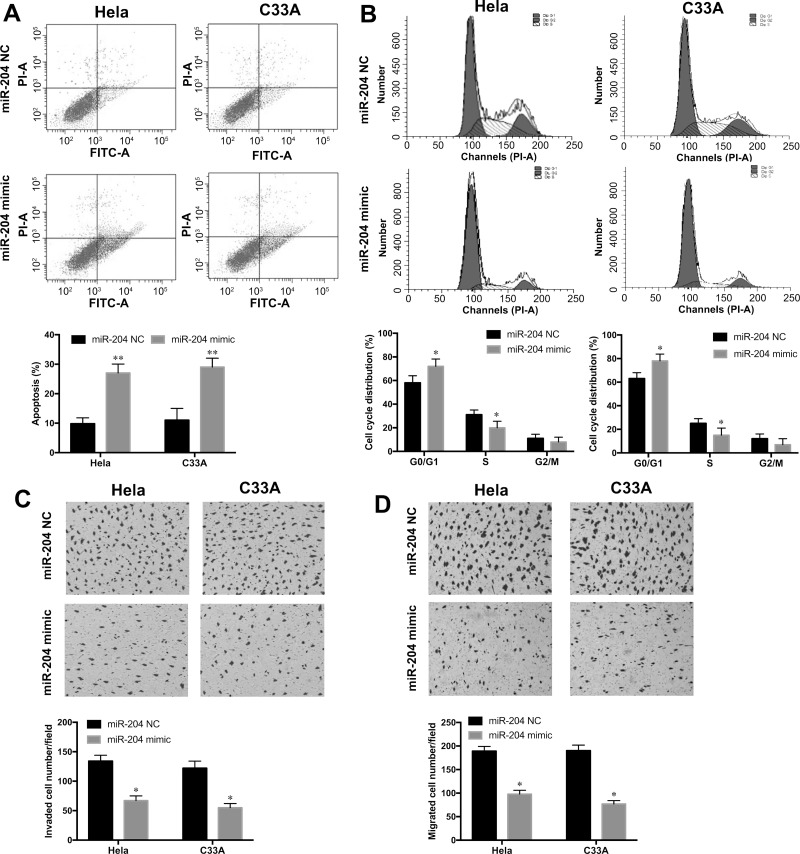
Cell apoptosis and cell cycle distribution were examined by annexin V/FITC staining and PI staining, respectively, in HeLa and C33A cell lines with miR-204 mimic or NC transfection. The annexin V/FITC staining assay showed that cell apoptosis rates in the HeLa and C33A cell lines with miR-204 mimic transfection were remarkably increased when compared to the NC groups (p < 0.01) (Fig. 3A). Furthermore, we also found that overexpression of miR-204 significantly increased the percentage of G0/G1 phase cells, whereas it reduced the percentage of S phase cells in HeLa and C33A cell lines compared with those in the NC groups (p < 0.05) (Fig. 3B). Thus, these results indicated that miR-204 overexpression efficiently induced cell apoptosis and cell cycle arrest at the G0/G1 phase in CC cells.
Figure 3.
Overexpression of miR-204 promoted CC cell apoptosis and cell cycle at the G0/G1 phase and inhibited cell invasion and migration. (A) Cell apoptosis rate in HeLa and C33A cell lines with the miR-204 mimic or NC transfection was determined by annexin V/FITC staining. (B) Cell cycle distribution in HeLa and C33A cell lines with the miR-204 mimic or NC transfection was examined by propidium iodide (PI) staining. Cell invasion ability (C) and migration ability (D) were measured in Hela and C33A cell lines with the miR-204 mimic or NC transfection by Transwell assays. *p < 0.05, **p < 0.01 versus the NC group.
Overexpression of miR-204 Suppressed Migration and Invasion of CC Cells
To further investigate the influence of miR-204 overexpression on CC cell metastasis, Transwell assays were performed in HeLa and C33A cell lines with the miR-204 mimic or NC transfection. The results showed that ectopic expression of miR-204 remarkably suppressed the invasive abilities of HeLa and C33A cells compared to the NC groups (p < 0.05) (Fig. 3C). Furthermore, the migration abilities of HeLa and C33A cells were also significantly inhibited with miR-204 mimic transfection when compared with those in the NC groups (p < 0.05) (Fig. 3D). These results suggested that overexpression of miR-204 contributed to the inhibitory ability of metastasis in CC cells.
The Effects of miR-204 Overexpression on the Activation of the PI3K/AKT Signaling Pathway
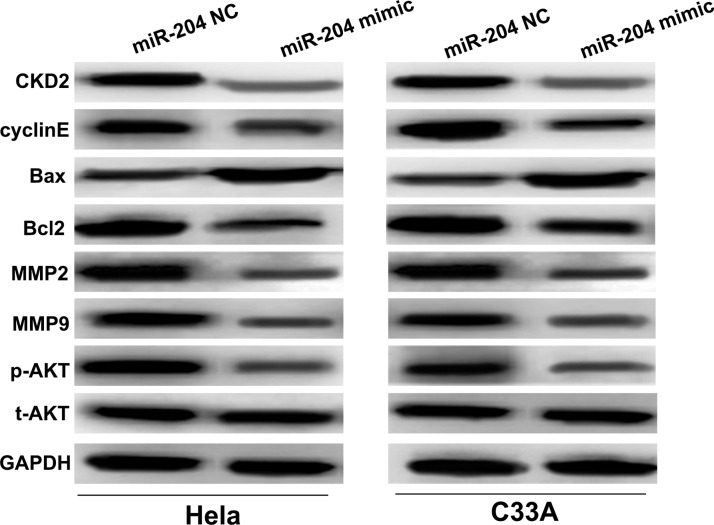
Given that the proliferation, cell cycle, invasion, and migration were inhibited, and the apoptosis was promoted, in HeLa and C33A cells with miR-204 overexpression, the expression levels of crucial cancer-related signaling pathway PI3K/AKT and its key regulatory components in CC cells were evaluated by Western blot. As shown in Figure 4, compared to NC groups, the expression levels of two cell cycle regulators CDK2 and cyclin E were significantly reduced in HeLa and C33A cell lines with miR-204 mimic transfection. Overexpression of miR-204 could remarkably increase Bax expression levels, whereas it decreased Bcl2 expression levels in the HeLa and C33A cell lines. The metastasis-related proteins MMP2 and MMP9 were both dramatically downregulated in HeLa and C33A cell lines with miR-204 overexpression. Furthermore, the expression levels of phosphorylated (p)-AKT was significantly downregulated in miR-204 mimic transfection cells compared to NC groups. However, no change was found on the total (t)-AKT expression among different groups. These findings suggested that miR-204 might regulate CC progression through PI3K/AKT signaling and its downstream key regulators.
Figure 4.
miR-204 overexpression inhibited the activation of the PI3K/AKT signaling pathway and altered its downstream regulators. The protein expressions of CDK2, cyclin E, Bax, Bcl2, MMP2, MMP9, p-AKT, and t-AKT in the HeLa and C33A cell lines with the miR-204 mimic or NC transfection were determined by Western blot.
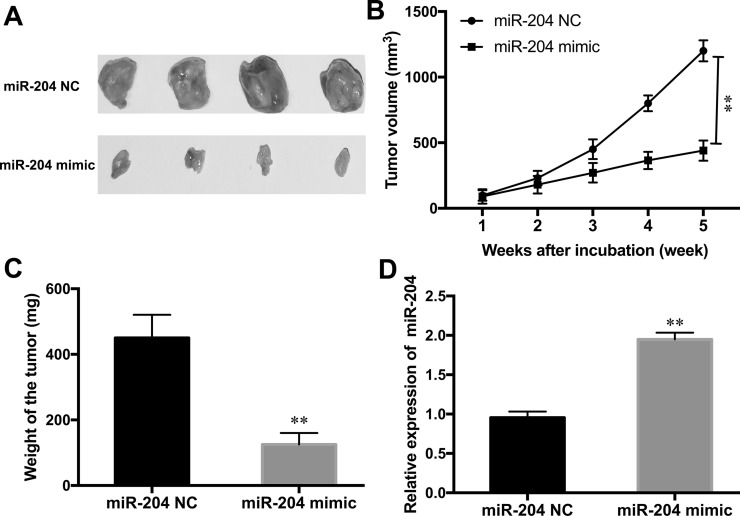
Overexpression of miR-204 Inhibited CC Tumor Growth In Vivo
Because miR-204 plays a vital role on CC cell proliferation, metastasis, and apoptosis in vitro, a xenograft was established in nude mice to investigate the effect of miR-204 in vivo. The results indicated that overexpression of miR-204 significantly inhibited tumor growth by suppressing the tumor volume and tumor weight in the xenograft (Fig. 5A–C). Moreover, qRT-PCR detection of tumor tissues showed that miR-204 expression was noticeably upregulated in the miR-204 mimic-transfected cell injection group when compared to the NC group (p < 0.01) (Fig. 5D). These data suggested that overexpression of miR-204 inhibited tumor development of CC in vivo.
Figure 5.
Overexpression of miR-204 inhibited CC tumor growth in vivo. (A) Representative images of CC isolated from nude mice injected with the miR-204 mimic or NC transfection HeLa cells. Tumor volume (B) and tumor weight (C) were measured in nude mice treated with the miR-204 mimic or NC. (D) The expression level of miR-204 in tumor xenograft from nude mice was determined by qRT-PCR. **p < 0.01 versus the NC group.
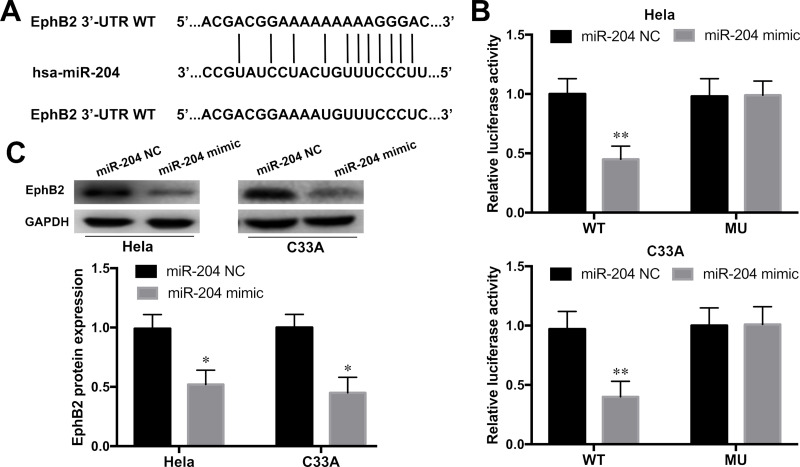
EphB2 Was a Direct Target of miR-204
To understand the molecular mechanism underlying miR-204 overexpression on the suppression of CC development, the 3′-UTR of target mRNAs for miR-204 was predicted by bioinformatics analysis (microRNA.org). As shown in Figure 6A, the 3′-UTR of EphB2 was predicted as a potential binding site for miR-204. To examine whether EphB2 was a candidate target of miR-204, luciferase report assay was performed. The results revealed that the relative luciferase activities of the HeLa and C33A cell lines were significantly inhibited with pGL3-EphB2-3′UTR WT and miR-204 mimic cotransfection (p < 0.01) (Fig. 6B). The luciferase activities of HeLa and C33A cell lines with pGL3-EphB2-3′UTR MU and miR-204 mimic cotransfection showed no effect. Furthermore, the effect of miR-204 on the expression of EphB2 was further measured in HeLa and C33A cell lines by Western blot. As shown in Figure 6C, miR-204 overexpression significantly inhibited the expression of EphB2 in HeLa and C33A cell lines when compared to the NC group (p < 0.05). These findings suggested that EphB2 was a direct target of miR-204 in CC cells.
Figure 6.
Ephrin type B receptor 2 (EphB2) was a direct target of miR-204. (A) The potential binding site in 3′-UTR EphB2 of miR-204 was predicted by microRNA.org. (B) Relative luciferase activity was measured in Hela and C33A cell lines cotransfected with pGL3-EphB2-3′UTR WT or MU reporter plasmids and the miR-204 mimic or NC by luciferase report assay. (C) The protein expression levels of EphB2 in HeLa and C33A cell lines with the miR-204 mimic or NC transfection were determined by Western blot. *p < 0.05, **p < 0.01 versus the NC group.
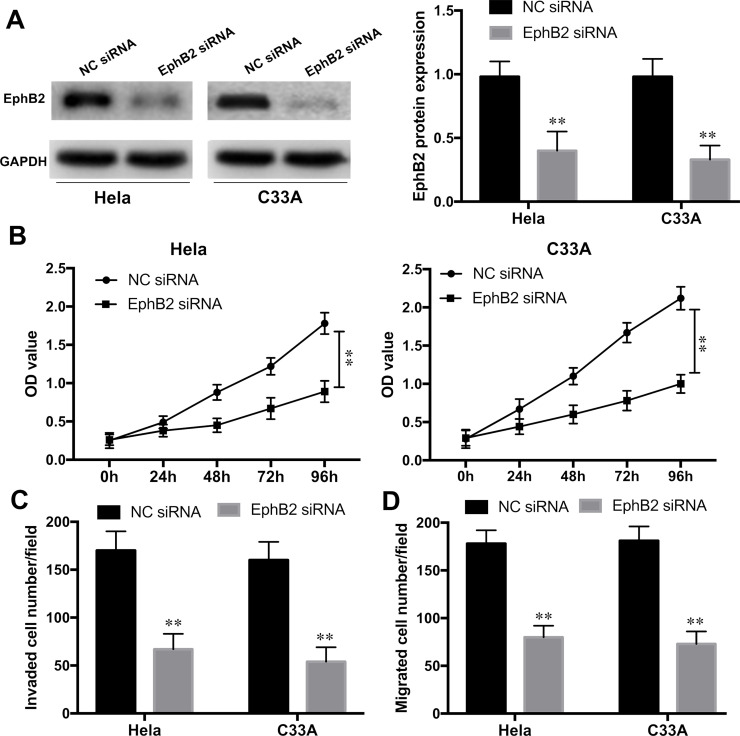
EphB2 Knockdown Inhibited Proliferation and Metastasis of CC Cells
To further evaluate whether EphB2 was involved in miR-204-mediated proliferation and metastasis of CC cells, EphB2 expression was knocked down in HeLa and C33A cell lines by EphB2 siRNA transfection. Western blot assay showed that EphB2 expression was significantly silenced by EphB2 siRNA when compared with that in the NC group (p < 0.01) (Fig. 7A). Furthermore, the CCK-8 assay revealed that the cell growth rates in HeLa and C33A cells were significantly inhibited with EphB2 knockdown compared to NC siRNA groups (p < 0.01) (Fig. 7B). Transwell assays also showed that the invaded and migrated cells in the HeLa and C33A cell lines were remarkably suppressed with EphB2 siRNA transfection (all p < 0.01) (Fig. 7C and D). These data suggested that EphB2 silencing could mimic the inhibitory effect of miR-204 overexpression on CC proliferation and metastasis.
Figure 7.
EphB2 knockdown inhibited proliferation and metastasis of CC cells. (A) The efficiency of EphB2 knockdown in HeLa and C33A cell lines with EphB2 siRNA or NC siRNA transfection was detected by Western blot. (B) The cell proliferation of HeLa and C33A cell lines with EphB2 siRNA or siRNA NC transfection was examined by the CCK-8 assay. Cell invasion (C) and migration (D) abilities of HeLa and C33A cell lines with EphB2 siRNA or siRNA NC transfection were determined by Transwell assays. **p < 0.01 versus the NC group.
DISCUSSION
Cervical carcinoma remains the second most common cancer killer in women following breast cancer and seriously threatens the health of females in developing countries, especially in P.R. China9. The efficiencies of conventional therapeutic approaches are limited in metastatic CC patients. Thus, it is urgent to elucidate the underlying molecular mechanisms of CC progression and explore efficient strategies to overcome this disorder. Growing evidence indicates that interference of the key components of signaling pathways or tumor regulatory factors by targeted therapies has received increased attention10. In order to keep the balance of cell proliferation in normal cellular conditions, cell proliferation is promoted by pro-oncogenes and inhibited by counterpart tumor suppressors. Malignant tumor proliferation occurs once mutations increase pro-oncogene activity or inactivate tumor suppressors11. The abnormal expression of miRNAs acting as either oncogenes or suppressors is associated with progression and carcinogenesis of multiple cancers12. Thus, a better understanding of the functions of miRNAs on CC development might improve the therapeutic options for refractory CC treatment.
Previous research has illustrated the tumor suppressor role of miR-204 in the development of some types of malignant tumors. For instance, downregulation of miR-204 has been verified to be closely related with lymphatic metastasis and poor prognosis in human nasopharyngeal carcinoma13. Sun et al. suggested that miR-204 inhibits invasion and epithelial–mesenchymal transition in esophageal cancer14. Wu et al. demonstrated that downregulation of miR-204 is associated with proliferation and invasion of human retinoblastoma cells15. Xia et al. found that miR-204 expression is decreased in non-small cell lung cancer tissues, and miR-204 silencing promoted cell invasion and proliferation in vitro16. Yin et al. also illustrated that miR-204 inhibited proliferation and invasion and enhanced chemotherapeutic sensitivity of colorectal cancer cells17. Thus, in this study, we supposed that the aberrant miR-204 might play an essential role in CC cell proliferation and invasion.
Consistent with previous findings, the present data revealed that the expression level of miR-204 was downregulated in CC tissues compared with adjacent normal tissues. Furthermore, miR-204 was also expressed lower in six CC cell lines than in the normal cell line. CCK-8 and colony formation assays showed that overexpression of miR-204 could dramatically inhibit the cell proliferation and colony formation abilities in CC cells. Furthermore, the cell cycle distribution assay suggested that restoration of miR-204 expression caused cell cycle arrest at the G0/G1 phase, and annexin V/FITC staining assay revealed an increased apoptosis rate in CC cells with miR-204 overexpression. Transwell assays also indicated that ectopic expression of miR-204 could significantly suppress cell invasion and migration capacities in CC cells. In addition, a tumor xenograft of CC was established to investigate the role of miR-204 in vivo. Overexpression of miR-204 markedly suppressed tumor growth and resulted in reduced tumor weight and volume in the xenograft. These findings strongly suggested that miR-204 acted as a tumor suppressor during CC development.
The bioinformatics analysis prediction revealed that the 3′-UTR of EphB2 was a potential binding site for miR-204, which was further verified by dual-luciferase reporter and Western blot assays in CC cells. EphB2 belongs to the membrane-bound ephrin (Eph) family of receptor tyrosine kinases (RTKs), which contains a cytoplasmic kinase domain, a hydrophobic transmembrane domain, and an N-terminal domain involved in ligand binding18. In humans, there are 10 members of the EphA receptor (EphA1–10) and 6 members of the EphB receptor (EphB1–6)19. Eph receptors and their corresponding ligands are expressed at the cell membrane of adjacent cells, which makes signal receiving between receptor cells and ligand cells more efficient. A number of cellular biological activities are mediated by Eph receptors and their ligands, including angiogenesis, cell proliferation, motility, and cell attachment20,21. Recent reports have demonstrated that EphB2 plays an essential role in various human cancer progressions. For instance, a high expression of EphB2 predicts poor overall survival and high mortality rate in lung adenocarcinoma patients22. Restoration of EphB2 expression induces autophagy, apoptosis, and invasion in human breast cancer cells23. Knockdown of EphB2 expression inhibits proliferation, migration, and invasion of cutaneous squamous cell carcinoma24. Overexpression of EphB2 promotes cancer progression by inducing epithelial–mesenchymal transition in CC25. In our study, we found that EphB2 knockdown by siRNA significantly reduced cell proliferation, migration, and invasion of CC cells, which could phenocopy the inhibitory effect of miR-204 overexpression on CC progression. Thus, these results strengthened the evidence that miR-204 regulated the development of CC cells via binding to EphB2.
It has been shown that the abnormal expression of EphB2 affects its major downstream PI3K/AKT signaling pathway, which is the key molecular mechanism of tumorigenesis and development26,27. Thus, in the present study, we also ascertained whether miR-204 overexpression affected the activation of the PI3K/AKT pathway in CC cells. Our results showed that overexpression of miR-204 inhibited p-AKT expression. Furthermore, the downstream components of the PI3K/AKT pathway were also investigated in this study. For instance, CDK2/cyclin E complexes regulate the G1-to-S phase transition and play an important role in G1 phase progression28. Bcl2 inhibits apoptosis, whereas Bax stimulates mitochondrial damage and promotes apoptosis29. MMP2 and MMP9 are two major members of the MMP family and play essential roles in cell invasion and metastasis30. In this study, overexpression of miR-204 reduced the expressions of CDK2, cyclin E, MMP2, MMP9, and Bcl2 and increased the expression of Bax. However, the precise mechanisms still need further explanation. Taken together, the detections of downstream components of PI3K/AKT demonstrated that overexpression of miR-204 might suppress cell growth, cell cycle transition, invasion, and migration and promote apoptosis in CC by targeting EphB2 through the PI3K/AKT and its downstream signaling pathways.
In conclusion, this study demonstrated that miR-204 served as a novel tumor suppressor miRNA in the proliferation and metastasis of CC progression. miR-204 exhibited its inhibitory effect on the proliferation, apoptosis, migration, and invasion of CC cells via directly targeting EphB2. The miR-204/EphB2 axis might provide a novel insight in understanding the mechanisms underlying CC development and progression and thereby improve therapeutic outcome for patients with advanced CC.
ACKNOWLEDGMENTS
The authors would like to thank Shaanxi Nuclear Industry 215 Hospital for providing the human cervical tissue specimens.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: From pathogenesis to therapy. Tumour Biol. 2014;35(6):5083–93. [DOI] [PubMed] [Google Scholar]
- 3. Smith RA, Brooks D, Cokkinides V, Saslow D, Brawley OW. Cancer screening in the United States, 2013: A review of current American Cancer Society guidelines, current issues in cancer screening, and new guidance on cervical cancer screening and lung cancer screening. CA Cancer J Clin. 2013;63(2):88–105. [DOI] [PubMed] [Google Scholar]
- 4. Manzo-Merino J, Contreras-Paredes A, Vazquez-Ulloa E, Rocha-Zavaleta L, Fuentes-Gonzalez AM, Lizano M. The role of signaling pathways in cervical cancer and molecular therapeutic targets. Arch Med Res. 2014;45(7):525–39. [DOI] [PubMed] [Google Scholar]
- 5. Li J, Tan S, Kooger R, Zhang C, Zhang Y. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev. 2014;43(2):506–17. [DOI] [PubMed] [Google Scholar]
- 6. Li Z, Rana TM. Therapeutic targeting of microRNAs: Current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38. [DOI] [PubMed] [Google Scholar]
- 7. Hall DP, Cost NG, Hegde S, Kellner E, Mikhaylova O, Stratton Y, Ehmer B, Abplanalp WA, Pandey R, Biesiada J, Harteneck C, Plas DR, Meller J, Czyzyk-Krzeska MF. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell 2014;26(5):738–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37(9):11667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi JF, Canfell K, Lew JB, Qiao YL. The burden of cervical cancer in China: Synthesis of the evidence. Int J Cancer 2012;130(3):641–52. [DOI] [PubMed] [Google Scholar]
- 10. Diaz-Padilla I, Monk BJ, Mackay HJ, Oaknin A. Treatment of metastatic cervical cancer: Future directions involving targeted agents. Crit Rev Oncol Hematol. 2013;85(3):303–14. [DOI] [PubMed] [Google Scholar]
- 11. Qiu B, Simon MC. Oncogenes strike a balance between cellular growth and homeostasis. Semin Cell Dev Biol. 2015;43(1):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seven M, Karatas OF, Duz MB, Ozen M. The role of miRNAs in cancer: From pathogenesis to therapeutic implications. Future Oncol. 2014;10(6):1027–48. [DOI] [PubMed] [Google Scholar]
- 13. Ma L, Deng X, Wu M, Zhang G, Huang J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014;588(9):1562–70. [DOI] [PubMed] [Google Scholar]
- 14. Sun Y, Yu X, Bai Q. miR-204 inhibits invasion and epithelial-mesenchymal transition by targeting FOXM1 in esophageal cancer. Int J Clin Exp Pathol. 2015;8(10):12775–83. [PMC free article] [PubMed] [Google Scholar]
- 15. Wu X, Zeng Y, Wu S, Zhong J, Wang Y, Xu J. MiR-204, down-regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting cyclinD2 and MMP-9. FEBS Lett. 2015;589(5):645–50. [DOI] [PubMed] [Google Scholar]
- 16. Xia Y, Zhu Y, Ma T, Pan C, Wang J, He Z, Li Z, Qi X, Chen Y. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588(20):3703–12. [DOI] [PubMed] [Google Scholar]
- 17. Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, Song M, Bian Z, Wang Q, Ni S, Hu Y, Mao Y, Zhou L, Wang Y, Yu J, Du X, Hua D, Huang Z. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20(23):6187–99. [DOI] [PubMed] [Google Scholar]
- 18. Yu M, Liang W, Wen S, Zhao T, Zhu MX, Li HH, Long Q, Wang M, Cheng X, Liao YH, Yuan J. EphB2 contributes to human naive B-cell activation and is regulated by miR-185. FASEB J. 2014;28(8):3609–17. [DOI] [PubMed] [Google Scholar]
- 19. Khansaard W, Techasen A, Namwat N, Namwat N, Yongvanit P, Khuntikeo N, Puapairoj A, Loilome W. Increased EphB2 expression predicts cholangiocarcinoma metastasis. Tumour Biol. 2014;35(10):10031–41. [DOI] [PubMed] [Google Scholar]
- 20. Schaupp A, Sabet O, Dudanova I, Ponserre M, Bastiaens P, Klein R. The composition of EphB2 clusters determines the strength in the cellular repulsion response. J Cell Biol. 2014;204(3):409–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin S, Wang B, Getsios S. Eph/ephrin signaling in epidermal differentiation and disease. Semin Cell Dev Biol. 2012;23(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao C, Wang A, Lu F, Chen H, Fu P, Zhao X. Overexpression of junctional adhesion molecule-A and EphB2 predicts poor survival in lung adenocarcinoma patients. Tumour Biol. 2017;39(2):1010428317691000. [DOI] [PubMed] [Google Scholar]
- 23. Husa AM, Magic Z, Larsson M, Fornander T, Perez-Tenorio G. EPH/ephrin profile and EPHB2 expression predicts patient survival in breast cancer. Oncotarget 2016;7(16):21362–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Farshchian M, Nissinen L, Siljamaki E, Riihila P, Toriseva M, Kivisaari A, Ala-Aho R, Kallajoki M, Verajankorva E, Honkanen HK, Heljasvaara R, Pihlajaniemi T, Grenman R, Peltonen J, Peltonen S, Kahari VM. EphB2 promotes progression of cutaneous squamous cell carcinoma. J Invest Dermatol. 2015;135(7):1882–92. [DOI] [PubMed] [Google Scholar]
- 25. Gao Q, Liu W, Cai J, Li M, Gao Y, Lin W, Li Z. EphB2 promotes cervical cancer progression by inducing epithelial-mesenchymal transition. Hum Pathol. 2014;45(2):372–81. [DOI] [PubMed] [Google Scholar]
- 26. Jiang J, Wang ZH, Qu M, Gao D, Liu XP, Zhu LQ, Wang JZ. Stimulation of EphB2 attenuates tau phosphorylation through PI3K/Akt-mediated inactivation of glycogen synthase kinase-3beta. Sci Rep. 2015;5(1):11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu J, Li Z, Su Q, Zhao J, Ma J. TRIM29 promotes progression of thyroid carcinoma via activating P13K/AKT signaling pathway. Oncol Rep. 2017;37(3):1555–64. [DOI] [PubMed] [Google Scholar]
- 28. Zhao Z, Liu J, Wang C, Wang Y, Jiang Y, Guo M. MicroRNA-25 regulates small cell lung cancer cell development and cell cycle through cyclin E2. Int J Clin Exp Pathol. 2014;7(11):7726–34. [PMC free article] [PubMed] [Google Scholar]
- 29. Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014(1):150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan J, Wang X, Li L. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol. 2015;47(2):690–700. [DOI] [PubMed] [Google Scholar]