Abstract
MicroRNAs (miRNAs) have emerged as pivotal regulators of the development and progression of gastric cancer. Studies have shown that miR-154 is a novel cancer-associated miRNA involved in various cancers. However, the role of miR-154 in gastric cancer remains unknown. Here we aimed to investigate the biological function and the potential molecular mechanism of miR-154 in gastric cancer. We found that miR-154 was significantly downregulated in gastric cancer tissues and cell lines. The overexpression of miR-154 significantly repressed the growth and invasion of gastric cancer cells. Bioinformatics analysis and Dual-Luciferase Reporter Assay data showed that miR-154 directly targeted the 3′-untranslated region of Dishevelled–Axin domain containing 1 (DIXDC1). Real-time quantitative polymerase chain reaction and Western blot analyses showed that miR-154 overexpression inhibited DIXDC1 expression. An inverse correlation of miR-154 and DIXDC1 was also demonstrated in gastric cancer specimens. Overexpression of miR-154 also significantly suppressed the activation of WNT signaling. Moreover, restoration of DIXDC1 expression significantly reversed the inhibitory effect of miR-154 overexpression on the cell proliferation, invasion, and WNT signaling in gastric cancer cells. Overall, these results suggest that miR-154 inhibits gastric cancer cell growth and invasion by targeting DIXDC1 and could serve as a potential therapeutic target for the treatment of gastric cancer.
Key words: Dishevelled–Axin domain containing 1 (DIXDC1), Gastric cancer, miR-154, Wingless-related integration site (WNT)
INTRODUCTION
Gastric cancer derived from gastric mucosa epithelial cells is a prevalent digestive cancer and the second leading cause of cancer-associated deaths worldwide1. Despite advances in cancer treatment, the survival rate and prognosis of patients with advanced gastric cancer have not been significantly improved2,3. The inhibition of malignant growth, local advanced invasion, and metastasis of gastric cancer remains a challenge4. Multiple factors, including environmental and genetic factors, have been proposed as having a role in the development and progression of gastric cancer5–7. However, the precise molecular mechanism of gastric cancer development and progression remains poorly understood. Therefore, it is essential to gain a better understanding of the molecular pathogenesis underlying the development of gastric cancer. Such research will help with the development of novel therapeutic strategies.
In recent years, microRNAs (miRNAs), a new class of noncoding RNAs with 19–25 nucleotides, have emerged as critical regulators for gene expression8. miRNAs negatively modulate gene expression by binding to the 3′-untranslated region (3′-UTR) of target mRNA, inducing mRNA degradation and translational inhibition9,10. Increasing studies have shown that dysregulation of miRNAs is involved in various pathological pathways, regulating numerous biological processes, including cell growth, apoptosis, and differentiation8,11,12. Dysregulation of miRNAs is common in the tumorigenesis and metastasis of gastric cancer, and these dysregulated miRNAs could serve as potential diagnostic, prognostic, and therapeutic targets13,14. Targeting gastric cancer-associated miRNAs may provide potential therapeutic options for gastric cancer.
Dishevelled–Axin domain containing 1 (DIXDC1) is a novel Disheveled–Axin domain containing protein that functions as a positive regulator of wingless-related integration site (WNT) signaling15,16. A growing body of evidence has suggested an oncogenic role for DIXDC1 in many types of cancers, including non-small cell lung cancer17, pancreatic cancer18, prostate cancer19, and glioma20. High levels of DIXDC1 are detected in gastric cancer tissues, and this is correlated with histological intestinal type, lymph node metastasis, and depth of tumor invasion21. DIXDC1 is colocalized with β-catenin in gastric cancer tissues and is involved in the promotion of gastric cancer cell invasion and metastasis by activation of WNT signaling21,22. DIXDC1 may serve as a potential target for the development of novel therapies for gastric cancer. However, the precise molecular mechanism of DIXDC in the progression of gastric cancer requires further study.
Recent studies have reported that miR-154 is a novel cancer-related miRNA involved in various cancers, including breast cancer23, prostate cancer24, glioma25, and hepatocellular carcinoma26. However, whether miR-154 is involved in gastric cancer is unknown. In this study, we aimed to investigate the potential role of miR-154 in gastric cancer. Here we found that miR-154 expression was significantly downregulated in gastric cancer tissues and cell lines. Overexpression of miR-154 impeded the growth and invasion of gastric cancer cells. DIXDC1 was identified as a potential target gene of miR-154. miR-154 inhibits the expression of DIXDC1 in gastric cancer cells. An inverse correlation between miR-154 and DIXDC1 was also detected in gastric cancer specimens. Moreover, miR-154 suppressed the activation of WNT signaling by targeting DIXDC1. The restoration of DIXDC1 significantly reversed the inhibitory effect of miR-154 on gastric cancer cell growth and invasion. Overall, our results suggest that miR-154 inhibits gastric cancer cell growth and invasion by the downregulation of DIXDC1, providing a potential therapeutic target for gastric cancer.
MATERIALS AND METHODS
Tissue Samples
Paired cancer tissues and the matched normal adjacent tissues were collected from 16 patients histologically diagnosed with gastric cancer in Qingdao Jiaozhou City Central Hospital (Shandong, P.R. China). No patients had undergone any radiotherapy or chemotherapy before surgical resection. The resected tissues were stored in liquid nitrogen at −80°C until RNA isolation. All patients enrolled in this study provided informed consent regarding the donation of clinical tissues for research purposes. The experimental protocols were approved by the Institutional Human Experiment and Ethic Committee of Qingdao Jiaozhou City Central Hospital and were performed according to the Helsinki Declaration.
Cell Lines
Human gastric cancer cell lines MGC-803, MKN-87, MKN-45, and AGS and the normal human gastric epithelial cell line GES-1 were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, P.R. China). Cells were grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS) plus 1% streptomycin/penicillin mix (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Cell cultures were maintained at 37°C in a humidified atmosphere of 5% CO2.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was isolated using a miRNeasy Mini Kit (Qiagen, Hilden, Germany). The cDNA was synthesized using miScript II RT Kit (Qiagen) according to the manufacturer’s instructions. PCRs were assayed by SYBR Green Real-Time PCR (Applied Biosystems, Foster City, CA, USA) on an ABI 7900 Real-Time PCR System (Applied Biosystems). U6 small nuclear RNA was used as internal control for normalization of miR-154 expression. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as internal control for normalization of DIXDC1, cyclin D1, and c-myc. Relative expression was evaluated using the 2−ΔΔCt method.
Transient Transfection
Oligonucleotides, miR-154 mimics, and negative control (NC) mimics were purchased from GenePharma (Shanghai, P.R. China). A Bioinformatics method was utilized to identify likely binding partners of miR-154. DIXDC1 was deemed a likely candidate, and its association was explored further. The open reading frame of DIXDC1 was inserted into the pcDNA3.1 vector (Invitrogen). Transient transfection of oligonucleotides or vectors was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. The transfection efficiency was assessed by qPCR or Western blot after 48 h of culture.
Cell Counting Kit-8 (CCK-8) Assay
Cells were seeded into a 96-well plate at 1 × 104 cells per well and cultured overnight. Cells were transfected with miR-154 mimics and incubated for 48 h. A volume of 10 μl of CCK-8 solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated at 37°C for 2 h. Cell proliferation was determined by the measurement of the optical density (OD) value at 490 nm using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Colony Formation Assay
Cells transfected with miR-154 mimics for 48 h were resuspended in RPMI-1640 containing 10% FBS and 0.3% agarose and layered in six-well plates containing RPMI-1640 containing 10% FBS and 0.6% agarose at 1,000 cells per well. Cells were cultured at 37°C for 14 days. Colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma-Aldrich). The colonies were observed and counted under an optical microscope.
Cell Invasion Assay
Cell invasion was detected with Transwell chambers (BD Biosciences, San Jose, CA, USA). The Transwell upper chamber was precoated with Matrigel (BD Biosciences). Gastric cancer cells (1 × 105) suspended into 200 μl of serum-free medium were plated into the upper chambers, and the bottom chambers were filled with 500 μl of complete medium containing 10% FBS to induce cells invading through the membrane. Cells were cultured at 37°C for 24 h, and cells that had invaded across the Transwell membrane were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich). The number of invaded cells was counted under an optical microscope.
Dual-Luciferase Reporter Assay
The wild-type (wt) or mutant (mt) DIXDC1 3′-UTR containing the putative binding site of miR-154 was cloned into the pmirGLO reporter luciferase vector (Promega, Madison, WI, USA). Cells were seeded into 24-well plates and cotransfected with miR-154 mimics and pmirGLO 3′-UTR vector using Lipofectamine 2000 (Invitrogen). After 48 h of culture, the relative luciferase activity was measured by the Dual-Luciferase Reporter Assay (Promega) according to the manufacturer’s instruction.
Western Blot Analysis
Proteins from cell samples were extracted by lysis buffer containing Cocktail protease inhibitor (Sigma-Aldrich). The concentration was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, Haimen, P.R. China). An equivalent amount of proteins was electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gels and then transferred onto a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA, USA) using a wet-transferring method. The membrane was blocked with 5% skimmed milk at room temperature for 1 h followed by incubation with primary antibodies: anti-DIXDC1 and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. Then the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (Abcam, Cambridge, UK) at room temperature for 1 h. Labeled bands were visualized using an enhanced chemiluminescence kit (Pierce, Rockford, IL, USA). Band intensity was analyzed by Image-Pro Plus 6.0 software.
WNT Activity Assay
WNT activity was determined by TOPFlash-dependent luciferase reporter assay. Briefly, cells were seeded into 24-well plates and cotransfected with miR-154 mimics, TOPFlash vector, and Renilla luciferase vector (Promega) and incubated for 48 h. Relative luciferase reporter assay was determined using a Dual-Luciferase Reporter Assay (Promega).
Data Analysis
All data were presented as means ± standard deviation. Statistical analyses were performed using Student’s t-test or a one-way analysis of variance with Bonferroni post hoc test with SPSS 15.0 software (SPSS Inc., Chicago, IL, USA). Correction analysis was conducted using Spearman’s correlation methods. A value of p < 0.05 was regarded as statistically significant.
RESULTS
Decreased Expression of miR-154 in Gastric Cancer Tissues and Cell Lines
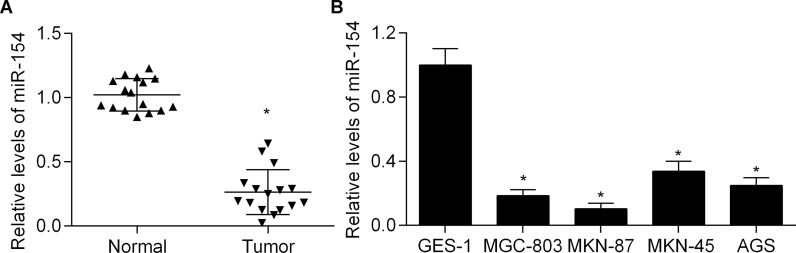
To investigate the potential role of miR-154 in gastric cancer, we first examined the expression status of miR-154 in gastric cancer tissues by qPCR. We found that gastric cancer tissues had significantly decreased expression of miR-154 compared with adjacent normal tissues (Fig. 1A). We then detected the expression status of miR-154 in gastric cancer cell lines. We found that miR-154 expression was frequently downregulated in gastric cancer cells compared with gastric epithelial cells (Fig. 1B). These results suggest that miR-154 might play an important role in the development of gastric cancer.
Figure 1.
Decreased expression of miR-154 in gastric cancer. (A) Quantitative real-time polymerase chain reaction (qPCR) detection of miR-154 expression in gastric cancer tissues and adjacent normal tissues. *p < 0.05 versus normal tissues. (B) Expression levels of miR-154 in the normal human gastric epithelial cell line GES-1 and four human gastric cancer cell lines (MGC-803, MKN-87, MKN-45, and AGS) were detected by qPCR. *p < 0.05 versus GES-1.
Overexpression of miR-154 Inhibits the Growth and Invasion of Gastric Cancers
To investigate the exact biological function of miR-154 in gastric cancer, we performed gain-of-function experiments using MGC-803 and MKN-87 cells by transfection of miR-154 mimics. The qPCR analysis showed that transfection of miR-154 mimics significantly increased the expression of miR-154 in MGC-803 and MKN-87 cells (Fig. 2A). The CCK-8 assay showed that miR-154 overexpression significantly inhibited the proliferation of gastric cancer cells (Fig. 2B). Colony formation showed that miR-154 expression markedly reduced the colony formation capability of gastric cancer cells (Fig. 2C). Moreover, the Transwell invasion assay showed that cell invasion of gastric cancer cells was also markedly suppressed by miR-154 overexpression (Fig. 2D). These results suggest a tumor-suppressive role of miR-154 that exerts an inhibitory effect on gastric cancer cell proliferation and invasion.
Figure 2.
miR-154 inhibits gastric cancer cell proliferation and invasion. MGC-803 and MKN-87 cells were transiently transfected with miR-154 mimics or negative control (NC) mimics for 48 h and subjected to subsequent assays. (A) High expression of miR-154 in miR-154 mimic-transfected cells was detected by qPCR. (B) Cell counting kit-8 (CCK-8) assay was used to detect the effect of miR-154 overexpression on cell proliferation. (C) The effect of miR-154 overexpression on colony formation capability was detected by colony formation assay. (D) The effect of miR-154 overexpression on cell invasion was assessed by Transwell invasion assay. *p < 0.05 versus NC mimics.
DIXDC1 Is a Potential Downstream Target Gene of miR-154
To investigate the molecular mechanism of miR-154 function in gastric cancer, we searched the potential downstream target genes of miR-154 using a bioinformatics method. DIXDC1, an oncogene in gastric cancer21,22, was predicted as a potential target gene of miR-154. As shown in Figure 3A, DIXDC1 3′-UTR contained the putative binding site for miR-154. To detect whether miR-154 directly binds to the putative binding site in DIXDC1 3′-UTR, we performed Dual-Luciferase Reporter Assays. The wt or the mt DIXDC1 3′-UTR containing the putative binding site of miR-154 was cloned into the pmirGLO reporter luciferase vector. We found that overexpression of miR-154 significantly reduced the luciferase activity in cells transfected with luciferase vector containing wt DIXDC1 3′-UTR, while miR-154 overexpression showed no significant effect on cells transfected with luciferase vector containing mt DIXDC1 3′-UTR (Fig. 3B). Moreover, we performed qPCR and Western blot analysis to detect the precise effect of miR-154 on DIXDC1 expression. We found that both the mRNA and protein expression of DIXDC1 was significantly decreased by miR-154 overexpression in gastric cancer cells (Fig. 4A and B). Therefore, our results suggest that DIXDC1 is a direct target gene of miR-154 in gastric cancer cells.
Figure 3.
miR-154 binds to the Dishevelled–Axin domain containing 1 (DIXDC1) 3′-untranslated region (3′-UTR). (A) Schematic of the miR-154 putative binding site in wild type (wt) DIXDC1 3′-UTR and mutant (mt) DIXDC1. (B) Dual-Luciferase Reporter Assay was performed using MGC-803 and MKN-87 cells transfected with miR-154 mimics and pmirGLO reporter luciferase vector containing wt or mt DIXDC1 3′-UTR. *p < 0.05 versus NC mimics.
Figure 4.
miR-154 inhibits DIXDC1 expression. MGC-803 and MKN-87 cells were transfected with miR-154 mimics or NC mimics for 48 h. The mRNA (A) and protein (B) expression of DIXDC1 was detected by qPCR and Western blot, respectively. *p < 0.05 versus NC mimics. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control.
DIXDC1 Is Inversely Correlated With miR-154 Expression in Gastric Cancer Specimens
To further confirm the targeted relationship between miR-154 and DIXDC1, we detected the expression correlation of miR-154 and DIXDC1 expression in gastric cancer specimens. The qPCR analysis showed that DIXDC1 mRNA expression levels were significantly upregulated in gastric cancer tissues compared with adjacent normal tissues (Fig. 5A). Correlation analysis showed that miR-154 expression was inversely correlated with DIXDC1 mRNA expression in gastric cancer specimens (Fig. 5B). These results further confirmed the targeted relationship between miR-154 and DIXDC1 expression in gastric cancer tissues.
Figure 5.
DIXDC1 is inversely correlated with miR-154 expression in gastric cancer specimens. (A) The relative mRNA expression of DIXDC1 in gastric cancer and adjacent normal tissues was detected by qPCR analysis. **p < 0.01 versus normal tissues. (B) An inverse correlation between miR-154 and DIXDC1 expression in gastric cancer tissues was detected by Spearman’s correlation. r = −0.8047; p < 0.001.
Overexpression of miR-154 Inhibits the Activation of WNT Signaling
DIXDC1 has been suggested as a positive regulator of WNT signaling15,16. Considering the regulatory effect of miR-154 on DIXDC1 expression, we detected the effect of miR-154 overexpression on WNT signaling. We found that miR-154 overexpression significantly repressed the activation of WNT signaling (Fig. 6A). Moreover, the mRNA expression levels of cyclin D1 (Fig. 6B) and c-myc (Fig. 6C) were markedly decreased by miR-154 overexpression. These results indicate that miR-154 regulates WNT signaling in gastric cancer.
Figure 6.
Overexpression of miR-154 inhibits the activation of wingless-related integration site (WNT) signaling. (A) The relative activity of WNT signaling was detected by TOPFlash-dependent luciferase reporter assay. MGC-803 and MKN-87 cells were cotransfected with miR-154 mimics, TOPFlash vector, and Renilla luciferase vector for 48 h. qPCR detection of cyclin D1 (B) and c-myc (C) mRNA expression in MGC-803 and MKN-87 in cells transfected with miR-154 mimics or NC mimics for 48 h. *p < 0.05 versus NC mimics.
Restoration of DIXDC1 Expression Reverses the Effect of miR-154
To investigate whether miR-154 functions through DIXDC1, we performed rescue experiments by restoration of DIXDC1. We found that transfection of the pcDNA3.1/DIXDC1 expression vector significantly restored the protein expression of DIXDC1 in miR-154 mimic-transfected cells (Fig. 7A). To detect whether miR-154 regulates WNT signaling through targeting DIXDC1, we detected the effect of DIXDC1 overexpression on WNT signaling. We found that overexpression of DIXDC1 significantly reversed the inhibitory effect of miR-154 on WNT signaling (Fig. 7B). Moreover, the inhibitory effect of miR-154 overexpression on cell proliferation (Fig. 7C) and invasion (Fig. 7D) was partially reversed by restoring DIXDC1 expression. Overall, these data suggest that miR-154 inhibits the cell proliferation and invasion of gastric cancer cells partially through targeting DIXDC1.
Figure 7.
Restoration of DIXDC1 reverses the effect of miR-154. MGC-803 and MKN-87 cells were transfected with miR-154 mimics and pcDNA3.1/DIXDC1 expression vector (without 3′-UTR) and incubated for 48 h. (A) Restoration of DIXDC1 expression was confirmed by Western blot analysis. (B) The relative activity of WNT signaling was detected by TOPFlash-dependent luciferase reporter assay. (C) Cell proliferation was measured by the CCK-8 assay. (D) Cell invasion assay was performed using Transwell chambers. *p < 0.05 versus NC mimics; &p < 0.05 versus miR-154 mimics.
DISCUSSION
Accumulating studies have shown that targeting specific miRNA effectively inhibits the tumorigenesis of gastric cancer27,28. miRNA-based therapy may be a promising strategy for the treatment of gastric cancer. However, the precise role of miRNAs in gastric cancer remains largely unknown. In this study, we identify miR-154 as a novel miRNA involved in the progression of gastric cancer. Decreased expression of miR-154 was observed in gastric cancer tissues and cell lines. We showed that miR-154 inhibits the growth and invasion of gastric cancer cells through targeting DIXDC1. Our findings suggest that miR-154 may serve as a potential target for the development of miRNA-based therapy.
Various studies have reported that miR-154 is located at chromosome 14q32 and is involved in regulating various biological processes, including pulmonary fibrosis, cardiac fibrosis, and osteogenic differentiation29–32. The role of miR-154 in tumorigenesis has been widely studied. miR-154 is reported to be decreased in prostate cancer and functions as a tumor suppressor by targeting cyclin D233, high-mobility group adenine–thymine (AT)-hook 234, and E2F transcription factor 524. A recent study also reported that miR-154 targets the E2F transcription factor 5 to inhibit the growth and invasion of breast cancer cells23. Decreased expression of miR-154 is correlated with advanced clinical stage and lymph node metastasis of colorectal cancer patients35. Overexpression of miR-154 inhibits the growth and motility of colorectal cancer cells by targeting toll-like receptor 236. miR-154 was found to inhibit cell growth of non-small cell lung cancer cells in vitro and in vivo37. Moreover, miR-154 inhibited the migration and invasion of non-small cell lung cancer cells by inhibiting zinc finger E-box binding homeobox 238. Pang et al. reported that miR-154 inhibited the tumor progression of hepatocellular carcinoma cells by targeting zinc finger E-box-binding homeobox 226. All of the above studies suggest a tumor-suppressive role for miR-154 in tumorigenesis. However, a recent study revealed that suppression of miR-154 inhibited the proliferation and migration of glioma stem cells through the promotion of phosphoribosyl pyrophosphate synthetase 1, indicating an oncogenic role of miR-154 in glioma39. In this study, we observed a significant decrease in miR-154 in gastric cancer tissues and cell lines. Our results demonstrated that overexpression of miR-154 inhibited the growth and proliferation of gastric cancer cells. Our study supports a tumor-suppressive role for miR-154. However, the exact role of miR-154 in tumorigenesis requires further investigation.
DIXDC1 has emerged as a critical regulator for tumorigenesis. DIXDC1 is found overexpressed in non-small cell lung cancer tissues and is positively correlated with lymph node metastasis and advanced tumor node metastasis stage17. Moreover, DIXDC1 promotes the migration and invasion of non-small cell lung cancer cells through the activation of the phosphatidylinositol 3 kinase/AKT/activator protein 1 (PI3K/AKT/AP-1) pathway17. Wang et al. reported that overexpression of DIXDC1 upregulated colon cancer cell proliferation by the activation of the PI3K/AKT pathway40. Furthermore, DIXDC1 promotes cell proliferation and the drug resistance of non-Hodgkin’s lymphoma cells through the activation of the PI3K/AKT pathway41. High levels of DIXDC1 in pancreatic cancer patients are correlated with poor overall survival18. Knockdown of DIXDC1 suppresses the growth of pancreatic cancer cells18. Moreover, the silencing of DIXDC1 also impedes the proliferation and metastasis of prostate cancer19 and glioma cells20. In gastric cancer, DIXDC1 is correlated with histological intestinal type, lymph node metastasis, and tumor invasion depth42. Furthermore, gastric cancer patients with high levels of DIXDC1 have poor disease-specific survival42. Also, DIXDC1 is colocalized with β-catenin in gastric cancer42, indicating the involvement of DIXDC1 in regulating WNT signaling in gastric cancer. A recent study shows that DIXDC1 promotes the nuclear accumulation of β-catenin and the activation of WNT signaling by which DIXDC1 upregulates the invasion and metastasis of gastric cancer cells22. These findings suggest that DIXDC1 may serve as a potential prognosis biomarker and therapeutic target. Here we demonstrate that DIXDC1 is a direct target gene of miR-154. Our data show that inhibition of DIXDC1 by miR-154 overexpression significantly suppressed cell proliferation, invasion, and WNT signaling in gastric cancer. Moreover, low levels of miR-154 were inversely correlated with high expression of DIXDC1 in gastric cancer specimens. These data indicate that the dysregulation of miR-154 may contribute to the high expression of DIXDC1 during the progression of gastric cancer. Therefore, the miR-154/DIXDC1 axis might play an important role in gastric cancer.
Recent evidence has revealed that DIXDC1 can be targeted and regulated by miR-582-3p/5p43 and miR-127119, indicating that DIXDC1 undergoes miRNA-mediated epigenetic regulation. Suppression of DIXDC1 by miR-1271 significantly represses the proliferation and invasion of prostate cancer cells19. These studies suggest that miRNAs that target DIXDC1 may function as a potential tool for the inhibition of DIXDC1 expression. In this study, we identified miR-154 as a novel miRNA that targets and regulates DIXDC1 expression. Our results suggest that miR-154 may serve as a potential target for the development of miRNA-based therapy for DIXDC1-associated cancers.
Overall, our results reveal a tumor-suppressive role of miR-154 in gastric cancer. Overexpression of miR-154 impedes the growth and invasion of gastric cancer cells through the downregulation of DIXDC1. These findings indicate that miR-154/DIXDC1 may play a critical role in the development and progression of gastric cancer. Inhibition of DIXDC1 by miR-154 may represent a novel therapeutic strategy for the treatment of gastric cancer.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Bickenbach K, Strong VE. Comparisons of gastric cancer treatments: East vs. West. J Gastric Cancer 2012;12:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22:2403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cunningham D, Starling N, Rao S, Iveson T, Nicolson M, Coxon F, Middleton G, Daniel F, Oates J, Norman AR. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2008;358:36–46. [DOI] [PubMed] [Google Scholar]
- 5. Hou IC, Amarnani S, Chong MT, Bishayee A. Green tea and the risk of gastric cancer: Epidemiological evidence. World J Gastroenterol. 2013;19:3713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berger H, Marques MS, Zietlow R, Meyer TF, Machado JC, Figueiredo C. Gastric cancer pathogenesis. Helicobacter 2016;21 Suppl 1:34–8. [DOI] [PubMed] [Google Scholar]
- 7. Skierucha M, Milne AN, Offerhaus GJ, Polkowski WP, Maciejewski R, Sitarz R. Molecular alterations in gastric cancer with special reference to the early-onset subtype. World J Gastroenterol. 2016;22:2460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 9. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. [DOI] [PubMed] [Google Scholar]
- 10. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466:835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Volny O, Kasickova L, Coufalova D, Cimflova P, Novak J. microRNAs in cerebrovascular disease. Adv Exp Med Biol. 2015;888:155–95. [DOI] [PubMed] [Google Scholar]
- 12. McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–8. [DOI] [PubMed] [Google Scholar]
- 13. Jiang C, Chen X, Alattar M, Wei J, Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22:291–301. [DOI] [PubMed] [Google Scholar]
- 14. Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential diagnostic, prognostic and therapeutic targets of microRNAs in human gastric cancer. Int J Mol Sci. 2016;17:945–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiomi K, Uchida H, Keino-Masu K, Masu M. Ccd1, a novel protein with a DIX domain, is a positive regulator in the Wnt signaling during zebrafish neural patterning. Curr Biol. 2003;13:73–7. [DOI] [PubMed] [Google Scholar]
- 16. Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–92. [DOI] [PubMed] [Google Scholar]
- 17. Xu Z, Liu D, Fan C, Luan L, Zhang X, Wang E. DIXDC1 increases the invasion and migration ability of non-small-cell lung cancer cells via the PI3K-AKT/AP-1 pathway. Mol Carcinog. 2014;53:917–25. [DOI] [PubMed] [Google Scholar]
- 18. Li X, Xiao Y, Fan S, Xiao M, Wang X, Zhu X, Chen X, Li C, Zong G, Zhou G, Wan C. Overexpression of DIXDC1 correlates with enhanced cell growth and poor prognosis in human pancreatic ductal adenocarcinoma. Hum Pathol. 2016;57:182–92. [DOI] [PubMed] [Google Scholar]
- 19. Zhong J, Liu Y, Xu Q, Yu J, Zhang M. Inhibition of DIXDC1 by microRNA-1271 suppresses the proliferation and invasion of prostate cancer cells. Biochem Biophys Res Commun. 2017;484:794–800. [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Shen C, Shi J, Shen J, Chen W, Sun J, Fan S, Bei Y, Xu P, Chang H, Jiang R, Hua L, Ji B, Huang Q. Knockdown of DIXDC1 inhibits the proliferation and migration of human glioma cells. Cell Mol Neurobiol. 2016;37:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J, Zhang Y, Zhang N, Wang C, Herrler T, Li Q. An updated review of mechanotransduction in skin disorders: Transcriptional regulators, ion channels, and microRNAs. Cell Mol Life Sci. 2015;72:2091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan C, Qiao F, Wei P, Chi Y, Wang W, Ni S, Wang Q, Chen T, Sheng W, Du X, Wang L. DIXDC1 activates the Wnt signaling pathway and promotes gastric cancer cell invasion and metastasis. Mol Carcinog. 2016;55:397–408. [DOI] [PubMed] [Google Scholar]
- 23. Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8:2620–30. [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Y, Zhu C, Ma L, Shao P, Qin C, Li P, Cao Q, Ju X, Cheng G, Zhu Q, Gu X, Hua L. miRNA-154-5p inhibits proliferation, migration and invasion by targeting E2F5 in prostate cancer cell lines. Urol Int. 2017;98:102–10. [DOI] [PubMed] [Google Scholar]
- 25. Zhao D, Wang R, Fang J, Ji X, Li J, Chen X, Sun G, Wang Z, Liu W, Wang Y, Cheng G, Zhen H, Sun C, Fei Z. MiR-154 functions as a tumor suppressor in glioblastoma by targeting Wnt5a. Mol Neurobiol. 2017;54:2823–30. [DOI] [PubMed] [Google Scholar]
- 26. Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–9. [DOI] [PubMed] [Google Scholar]
- 27. Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, Zhang B, Wu WK, Cheng AS, Yu J, To KF, Kang W. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer 2017;16:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu B, Chen X, Li J, Gu Q, Zhu Z, Li C, Su L, Liu B. microRNA-29c inhibits cell proliferation by targeting NASP in human gastric cancer. BMC Cancer 2017;17:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun LY, Bie ZD, Zhang CH, Li H, Li LD, Yang J. MiR-154 directly suppresses DKK2 to activate Wnt signaling pathway and enhance activation of cardiac fibroblasts. Cell Biol Int. 2016;40:1271–9. [DOI] [PubMed] [Google Scholar]
- 30. Bernardo BC, Nguyen SS, Gao XM, Tham YK, Ooi JY, Patterson NL, Kiriazis H, Su Y, Thomas CJ, Lin RC, Du XJ, McMullen JR. Inhibition of miR-154 protects against cardiac dysfunction and fibrosis in a mouse model of pressure overload. Sci Rep. 2016;6:22442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J, Hu C, Han L, Liu L, Jing W, Tang W, Tian W, Long J. MiR-154-5p regulates osteogenic differentiation of adipose-derived mesenchymal stem cells under tensile stress through the Wnt/PCP pathway by targeting Wnt11. Bone 2015;78:130–41. [DOI] [PubMed] [Google Scholar]
- 32. Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, McMillan M, Kahn M, Kaminski N. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H, Cao Q, Tao L, Meng X, Ju X, Qin C, Li J, Yin C. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32:31.e9–31.e16. [DOI] [PubMed] [Google Scholar]
- 34. Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, Wang M, Zhang Z, Qin C, Hua L, Yin C, Shao P. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69–75. [DOI] [PubMed] [Google Scholar]
- 35. Kai Y, Qiang C, Xinxin P, Miaomiao Z, Kuailu L. Decreased miR-154 expression and its clinical significance in human colorectal cancer. World J Surg Oncol. 2015;13:195–9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Xin C, Zhang H, Liu Z. miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem. 2014;387:271–7. [DOI] [PubMed] [Google Scholar]
- 37. Lin X, Yang Z, Zhang P, Shao G. miR-154 suppresses non-small cell lung cancer growth in vitro and in vivo. Oncol Rep. 2015;33:3053–60. [DOI] [PubMed] [Google Scholar]
- 38. Lin X, Yang Z, Zhang P, Liu Y, Shao G. miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett. 2016;12:301–6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Yang L, Yan Z, Wang Y, Ma W, Li C. Down-expression of miR-154 suppresses tumourigenesis in CD133(+) glioblastoma stem cells. Cell Biochem Funct. 2016;34:404–13. [DOI] [PubMed] [Google Scholar]
- 40. Wang L, Cao XX, Chen Q, Zhu TF, Zhu HG, Zheng L. DIXDC1 targets p21 and cyclin D1 via PI3K pathway activation to promote colon cancer cell proliferation. Cancer Sci. 2009;100:1801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ouyang Y, Zhong F, Wang Q, Ding L, Zhang P, Chen L, Wang Y, Cheng C. DIXDC1 promotes tumor proliferation and cell adhesion mediated drug resistance (CAM-DR) via enhancing p-Akt in non-Hodgkin’s lymphomas. Leuk Res. 2016;50:104–11. [DOI] [PubMed] [Google Scholar]
- 42. Wang L, Tan C, Qiao F, Wang W, Jiang X, Lian P, Chang B, Sheng W. Upregulated expression of DIXDC1 in intestinal-type gastric carcinoma: Co-localization with beta-catenin and correlation with poor prognosis. Cancer Cell Int. 2015;15:120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uchino K, Takeshita F, Takahashi RU, Kosaka N, Fujiwara K, Naruoka H, Sonoke S, Yano J, Sasaki H, Nozawa S, Yoshiike M, Kitajima K, Chikaraishi T, Ochiya T. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol Ther. 2013;21:610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]