Abstract
Cholangiocarcinoma (CCA) is a highly metastatic tumor, and the majority of patients with CCA have a short survival time because there are no available effective treatments. Hence, a better understanding regarding CCA metastasis may provide an opportunity to improve the strategies for treatment. A comparison study between the highly metastatic cells and their parental cells is an approach to uncover the molecular mechanisms underlying the metastatic process. In the present study, a lung metastatic CCA cell line, KKU-214L5, was established by the in vivo selection of the tail vein-injected mouse model. KKU-214L5 cells possessed mesenchymal spindle-like morphology with higher migration and invasion abilities in vitro than the parental cells (KKU-214). KKU-214L5 also exhibited extremely aggressive lung colonization in the tail vein-injected metastatic model. Epithelial–mesenchymal transition (EMT) was clearly observed in KKU-214L5 cells. Significant downregulation of epithelial markers (ZO-1 and claudin-1), with unique upregulation of E-cadherin and mesenchymal markers (vimentin, β-catenin, and slug), was observed in KKU-214L5. Increasing MMP-2 and MMP-9 activities and CD147 expression reflected the high invasion activity in KKU-214L5 cells. Suppression of vimentin using siRNA significantly decreased the migration and invasion capabilities of KKU-214L5 to almost the basal levels of the parental cells without any change on the expression levels of other EMT markers and the activities of MMPs. These results suggest that vimentin activation is essential to potentiate the metastatic characters of CCA cells, and suppression of vimentin expression could be a potential strategy to improve the treatment of CCA, a highly metastatic cancer.
Key words: Bile duct cancer, Epithelial–mesenchymal transition (EMT), Metastasis, Metalloproteinases (MMPs)
INTRODUCTION
Metastasis is a multistep process that almost always leads to the patient’s death. The cancer cells dissociate from a primary tumor, intravasate, survive in the circulation, attach, extravasate, and then colonize to form a secondary tumor. The biology of a metastatic tumor is different from its origin1. Therefore, the standard treatment which aims to target the primary tumor might not be effective for the metastatic tumor2–4. Cholangiocarcinoma (CCA) is a dismal cancer due to its high metastasis. It is a major health problem in the northeastern part of Thailand. Even though CCA is a rare liver cancer, its incidence is now rising worldwide5,6. Poor patient outcomes caused by metastasis have been reported.7,8 Lymph nodes and lungs are the common metastatic sites of CCA9. The molecular basis underlying CCA metastasis is still limited; hence, an intensive study of the metastatic process of the highly metastatic CCA cells may provide a better understanding of the process, leading to a better and more effective treatment of CCA.
Epithelial–mesenchymal transition (EMT) is an important process for cancer metastasis10,11. It is required to transform cancer cells from being epithelial-like to mesenchymal-like cells and detach from neighboring cells before invading through the extracellular matrix (ECM) and getting into the circulation. The associations of losing epithelial and/or increased mesenchymal properties with metastasis and with shorter survival of CCA patients have been frequently reported12–14. Suppression of epithelial cadherin (E-cadherin), an epithelial marker, promoted migration and invasion of CCA cell lines by disruption of the E-cadherin/β-catenin interaction15. On the other hand, overexpression of the mesenchymal markers, Twist and neural (N)-cadherin, increased the migration and invasion capabilities of transforming growth factor-β (TGF-β)-stimulated CCA cells16.
In this study, to obtain a better understanding of the molecular basis involved in CCA metastasis, a highly metastatic CCA cell line, KKU-214L5, was established by five consecutive rounds of intravenous injections of the parental cells (KKU-214) into nonobese diabetic/severe combined immunodeficient/janus kinase 3 null (NOD/SCID/Jak3null; NOJ) mice17. The metastatic phenotypes and molecular alterations of the EMT markers of KKU-214L5 cells were compared with the parental, KKU-214, cell line. The associations of vimentin with migration and invasion in the highly metastatic cells were elucidated, and the potential of vimentin function and EMT for the transition of CCA to metastatic phenotypes is discussed.
MATERIALS AND METHODS
CCA Cell Line and Cell Culture
The KKU-214 cell line was derived from the primary tumor of a Thai patient with histologically proven tubular CCA as previously described18. The ova of liver fluke, Opisthorchis viverrini, were detected in the tissue, possibly a pathogenic trigger for CCA. The KKU-214 cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank, Osaka, Japan. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and a 1% antibiotic–antimycotic unless otherwise specified in the protocols. Cells were cultured in an incubator with a humidified atmosphere of 5% CO2, 37°C. All cell culture reagents were supplied by Gibco BRL (Grand Island, NY, USA).
Antibodies
Anti-zona occludens 1 (ZO-1; D7D12), anti-claudin-1 (D5H1D), anti-E-cadherin (24E10), anti-β-catenin (D10A8), anti-snail homolog 2 (slug; C19G7), and anti-rabbit immunoglobulin G (IgG) were from Cell Signaling Technology (Danvers, MA, USA). The other antibodies were obtained from various sources: anti-vimentin (V9; Dako, Carpinteria, CA, USA), anti-cytokeratin-19 (CK-19; HPA002465), and anti-β-actin (AC-15) from Sigma-Aldrich (St. Louis, MO, USA); anti-mouse IgG from GE Healthcare (Buckinghamshire, UK); and all secondary fluorescent antibodies from Invitrogen (Carlsbad, CA, USA).
Establishment of a CCA Lung Metastatic Cell Line, KKU-214L5, and an In Vivo Metastatic Assay
Mice were housed and monitored in the animal research facility unit according to the institutional guidelines; food and drink were provided ad libitum on a 12-h light/dark cycle. The experimental protocols were approved by the Institutional Animal Care and Use Committee, Kumamoto University, Japan.
In order to generate the highly metastatic cell line, 5 × 105 KKU-214 cells were injected intravenously via tail veins into the NOD/SCID/Jak3null (NOJ) mice17. Mice were carefully observed and were euthanized when breathing difficulty was observed. CCA cells were isolated from lung nodules and cultured until a homogenous morphology, KKU-214L1, was obtained. The in vivo selection processes were repeated five times to generate KKU-214L5. The metastatic KKU-213L5 cell line was generated in a similar fashion to confirm that any changes in protein expression were due to metastasis rather than being cell specific (data not shown)19.
For the in vivo metastatic assay, KKU-214 and KKU-214L5 cells (5 × 104 cells/mouse) were injected intravenously into Balb/c recombination-activating gene 2 knockout (Rag-2−/−)/Jak3−/− mouse via tail veins (n = 3–4 mice/group)20. Mice were observed and euthanized when signs of stress were shown (31–32 days). Lungs were removed and cryopreserved in optimal cutting temperature (OCT) media (Leica Microsystems, Tokyo, Japan). Immunohistochemistry staining of CK-19 was performed as a standard protocol21. Signals were enhanced using the Vectastain Elite ABC Standard Kit (Vector Laboratories, Burlingame, CA, USA). Detection was performed using the Histofine® diaminobenzidine (DAB) substrate kit (Nichirei Bioscience, Tokyo, Japan). The numbers of metastatic CK-19+ nodules were determined under the microscope (BZ-8100 Biozero fluorescence; Keyence, Osaka, Japan) and were quantified using a BZ-II Analyzer (Keyence). Tumor nodules with diameters of >150 μm were defined as macrometastatic nodules, and smaller nodules were counted as micrometastatic nodules.
For the immunohistochemistry staining of vimentin, the EnVision+ System horseradish peroxidase (HRP)-labeled polymer anti-mouse (Dako) was applied after anti-vimentin incubation, and the signal was developed using DAB (Sigma-Aldrich). Sections were counterstained with hematoxylin and eosin (H&E).
Genotypic Characterization
Genomic DNAs of KKU-214 and KKU-214L5 were extracted using the QIAamp® DNA Micro Kit (QIAGEN, Valencia, CA, USA). The samples were genotyped using the AmpFℓSTR® Identifiler® Plus Polymerase Chain Reaction (PCR) Amplification Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. Short tandem repeat (STR) PCR products were profiled using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems), and the data were analyzed using GeneMapper® ID Software v3.2 (Applied Biosystems). The likelihood ratios and percent probabilities were calculated according to STR loci frequencies of the northeastern Thailand population22.
Cell Proliferation Assay
Cell proliferation was accessed using the cell counting kit-8 (WST-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s recommendations. OD450 was determined at 0, 24, 48, and 72 h after seeding. The growth rates were calculated relative to the OD450 at 0 h. The data were the means ± SD from two independent experiments.
Wound Healing Assay
KKU-214 and KKU-214L5 were cultured as a monolayer until confluent. A scratch wound was performed with a sterile 200-μl pipette tip. Wound images were captured using an Olympus CK40-F100 microscope (Olympus, Tokyo, Japan) with 4× objective lens at 0 and 6 h, and then at 2-h intervals until the wound closed. The migrating distances were analyzed as (distance at time 0 − distance at the indicated time)/distance at time 0.
Migration and Invasion Assays
Migration and invasion assays were determined using the Boyden chamber under similar protocols, except that the upper chambers for the invasion assay were coated overnight with 40 μg of Matrigel (BD Biosciences, San Jose, CA, USA). Cells (2.5 × 105) in a serum-free DMEM were added into the upper chamber of an 8-μm pore size Transwell (Corning Costar, Corning, NY, USA). DMEM with 10% FBS, serving as the chemoattractant, filled the lower chamber. Cells were incubated for a further 12 h. The migrated or invaded cells underneath the filter were fixed with 4% paraformaldehyde and were stained with 0.4% sulforhodamine B. The total stained cells were counted under a light microscope using the 10× objective lens. The results are presented as means ± SD from two independent experiments.
Western Blot
Cells were lysed with lysis buffer containing 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and protease and phosphatase inhibitors (Roche Diagnostics, Mannheim, Germany). Protein amounts were determined using the Bradford assay (Bio-Rad, Hercules, CA, USA). Five to 30 μg of protein was subjected to sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane (GE Healthcare). Detection was performed with the enhanced chemiluminescence (ECL) Prime Western Blotting Detection System (GE Healthcare). The signals were captured and analyzed by ImageQuant™ LAS 4000 mini (GE Healthcare).
Gelatin Zymography
KKU-214L5 and KKU-214 (2.5 × 105 cells) were plated into six-well plates and cultured for 24 h in 10% FBS-DMEM. Then media were replaced with serum-free DMEM, and cells were cultured for a further 24 h. Conditioned media were collected and processed as previously described23.
Immunocytofluorescence Staining
Cells were cultured in 24-well plates for 72 h and then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4, for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 3 min. After washing, cells were blocked with 5% normal horse sera and incubated with the primary antibodies at 4°C overnight. Cells were then incubated with the corresponding IgG-conjugated Alexa Fluor® 488. Finally, nuclei were stained with Hoechst 33342 (Molecular Probes, Eugene, OR, USA), and pictures were taken using an inverted fluorescence microscope (Olympus IX71; Olympus).
Vimentin Knockdown Using Small Interfering RNA (siRNA)
siRNAs against vimentin (si-vimentin) were sense 5′-GCA GGA UGA GAU UCA GAA UdTdT-3′ and antisense 5′-AUU CUG AAU CUC AUC CUG CdTdT-3′ (Nippon Gene, Tokyo, Japan). For scramble control, Silencer Negative Control siRNA#1 was purchased from Ambion (Austin, TX, USA). KKU-214L5 was transiently transfected using Lipofectamine® 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. After transfection, cells were cultured for a further 48 h before being subjected to further experiments.
Statistical Analysis
Data were analyzed using SPSS version 16.0 software (SPSS, Chicago, IL, USA). In vitro quantitative analyses were presented as means ± SD from at least two independent experiments. Statistical comparisons between the experimental groups were calculated using the Student’s t-test. A probability of p < 0.05 was considered as statistically significant.
RESULTS
Establishment of the Highly Metastatic KKU-214L5 Cells and Characterization of Genetic and Morphological Phenotypes
To generate the highly metastatic CCA cells, KKU-214 parental cells (5 × 105 cells/mouse) were injected intravenously via tail veins into the NOD/SCID/Jak3null (NOJ) mice as described in Materials and Methods. After the first injection, CCA cells colonizing the mouse lung were found. The cells from the lung nodules were isolated, cultured, and again injected into new mice with the same protocols via tail veins. This procedure was repeated five times to generate KKU-214L5.
STR is a repetitive DNA composed of repeated units of 2 to 13 nucleotides, typically repeating between 5 and several hundreds of times in a row on the DNA strand. STR DNA profiling analysis is a standard molecular biology method that can be used to identify an individual. A standard of 15 STR loci and amelogenin previously used in the forensic sciences24,25 is now generally applied for authentication of human cell lines. To ensure that the established KKU-214L5 cells were the subline of KKU-214, the genotypic profiles of KKU-214 and KKU-214L5 were determined using STR analysis. As shown in Table 1, the STR DNA profiling of KKU-214 and KKU-214L5 cells were compatible. The likelihood ratio (the reciprocal of the probability of a random match) and the percent probability indicated that the STR profiling of KKU-214 and KKU-214L5 were identical and hence the two cell lines had the same origin.
Table 1.
Short Tandem Repeat Profiles (STR) of KKU-214L5 and KKU-214 Cells
| Loci | STR | |
|---|---|---|
| KKU-214L5 | KKU-214 | |
| D8S1179 | 11, 13 | 11, 13 |
| D21S11 | 29 | 29 |
| D7S820 | 11 | 11 |
| CSF1PO | 9, 13 | 9, 13 |
| D3S1358 | 15 | 15 |
| TH01 | 7 | 7 |
| D13S317 | 8, 12 | 8, 12 |
| D16S539 | 9, 11 | 9, 11 |
| D2S1338 | 22, 24 | 22, 23, 24 |
| D19S433 | 13, 14 | 13, 14 |
| vWA | 19 | 18, 19 |
| TPOX | 8 | 8 |
| D18S51 | 14 | 14 |
| Amelogenin | X | X |
| D5S818 | 9 | 9 |
| FGA | 16 | 16 |
The likelihood ratio = 1.2 × 10−25, and the percent probability = 100%.
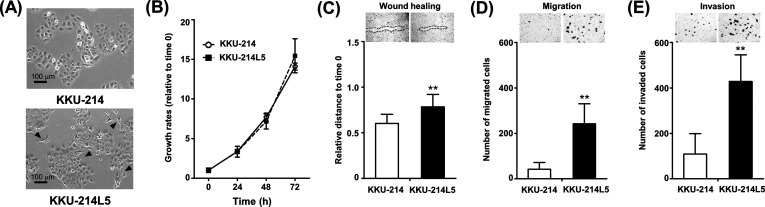
The morphologies of KKU-214L5 and its parental cells (KKU-214) were compared. KKU-214L5 exhibited a spindle-like shape (Fig. 1A, arrowheads), while KKU-214 possessed a polygonal shape. The characteristics related to metastatic phenotypes were next compared. The growth rates of KKU-214 and KKU-214L5 were not significantly different (Fig. 1B); however, the wound healing assay indicated a higher migrating ability of KKU-214L5 than the KKU-214 cells (Fig. 1C). At 14 h after wound scratching, the relative migration distance of KKU-214L5 was 0.78 ± 0.14 and that of KKU-214 was 0.60 ± 0.10 (p < 0.01). KKU-214L5 also had higher migration and invasion abilities than its parental cell line (Fig. 1D). The number of migrated cells of KKU-214L5 (242 ± 88 cells) was 5.7-fold higher than that of the parental cells (42 ± 30 cells) (p < 0.01). In parallel, the number of invaded cells of KKU-214L5 (429 ± 117 cells) was 3.9-fold higher than that of the KKU-214 cells (109 ± 90 cells) (p < 0.01) (Fig. 1E). These in vitro results clearly indicate that KKU-214L5 cells possessed higher migration and invasion abilities than the parental cells.
Figure 1.
Phenotypic characteristics of KKU-214L5 and its parental cells (KKU-214). (A) KKU-214L5 cells exhibited spindle-like shapes (arrows), while KKU-214 cells are polygonal shapes (phase contrast, scale bar: 100 μm). (B) KKU-214L5 cells had no significant growth differences but significantly higher migration and invasion capabilities than the parental KKU-214 cells as shown by the (C) wound healing, (D) migration, and (E) invasion Boyden chamber assays. The data are means ± SD of two independent experiments. **p < 0.01, Student’s t-test.
Highly Metastatic KKU-214L5 Cells Exhibited High Lung Metastasis Ability In Vivo
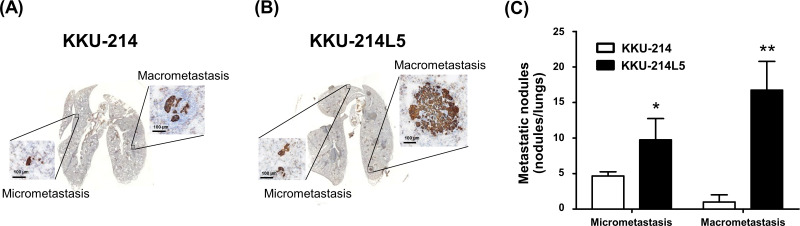
The metastatic ability of KKU-214L5 in vivo was examined using the tail vein metastatic assay. Lungs were collected at 31 days postinjection. The lung metastatic nodules were visualized by CK-19 immunostaining. The CK-19+ cells were counted as micro- and macrometastatic foci. The numbers of metastatic foci from lungs injected with KKU-214L5 were significantly higher than those from lungs injected with KKU-214 (p < 0.05) (Figs. 2A and B). The micrometastatic foci of KKU-214L5-injected mice (10 ± 3) were twofold higher than those of KKU-214-injected mice (5 ± 0.6). The macrometastatic foci of KKU-214L5-injected mice (17 ± 4) were 17-fold higher than those of KKU-214-injected mice (1 ± 1) (Fig. 2C).
Figure 2.
KKU-214L5 exerted a highly metastatic capability in vivo. The metastatic potential of KKU-214L5 was demonstrated in the lung metastatic mouse tail vein injection model. (A, B) KKU-214L5 generated significantly higher numbers of lung colonization of micrometastatic (≤150 μm) and macrometastatic nodules (>150 μm) than KKU-214 cells. (C) The quantitative analysis of the micro- and macrometastatic nodules. Data presented are means ± SD. *p < 0.05, **p < 0.01, Student’s t-test. Scale bar: 100 μm.
Downregulation of Epithelial Markers and Upregulation of Mesenchymal Markers Were the Characteristics of KKU-214L5
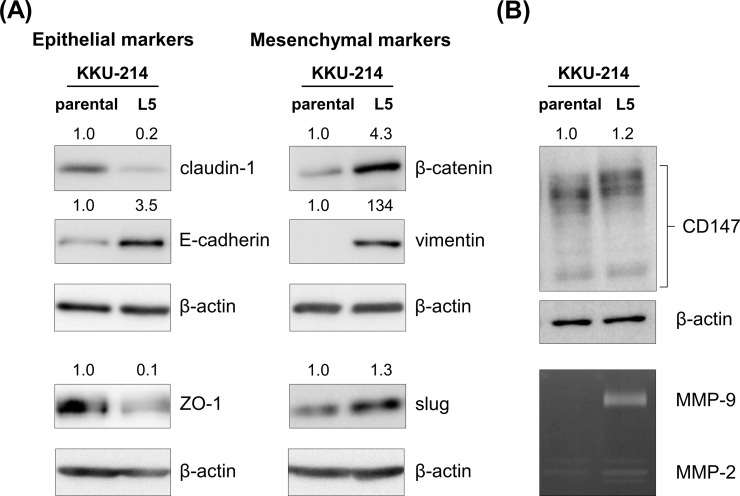
The spindle-like shape with high migration/invasion abilities of KKU-214L5 indicated the acquisition of EMT in the highly metastatic cells. To test this, the expressions of EMT markers were determined using Western blot analysis. The expressions of the EMT markers of KKU-214L5 were compared to those of the KKU-214 cells. As shown in Figure 3A, in KKU-214L5 cells, the protein expressions of the epithelial markers (claudin-1 and ZO-1) were downregulated, whereas the expressions of the mesenchymal markers (β-catenin, vimentin, and slug) were upregulated. In addition, the expression of CD147 or ECM metalloproteinase inducer (EMMPRIN) and the activity of its downstream matrix metalloproteinase enzymes (MMP-2 and MMP-9) were increased in KKU-214L5 cells (Fig. 3B). On the other hand, the protein expression of E-cadherin, an epithelial marker, was high in KKU-214L5 (Fig. 3A), while those of N-cadherin, snail, and zinc finger E-box-binding homeobox 1 (ZEB1) were not detected by Western blotting in either the KKU-214 or KKU-214L5 cells.
Figure 3.
Mesenchymal markers were upregulated in KKU-214L5 cells. Epithelial and mesenchymal markers were determined using Western blotting; β-actin was used as the loading control. (A) KKU-214L5 had downregulation of epithelial markers (claudin-1 and zona occludens 1 [ZO-1]) and upregulation of mesenchymal markers (β-catenin, vimentin, and slug), with an increase in epithelial (E)-cadherin. (B) Cluster of differentiation 147 (CD147) expression and matrix metalloproteinase 2 (MMP-2) and MMP-9 activities were elevated in KKU-214L5. Protein lysates of 5 μg were used for all epithelial–mesenchymal transition (EMT) markers, except ZO-1 and snail homolog 2 (slug) were at 30 μg. The numbers on the top of the Western blot indicate the relative protein band intensities by giving those of KKU-214 as 1. Parental = KKU-214; L5 = KKU-214L5.
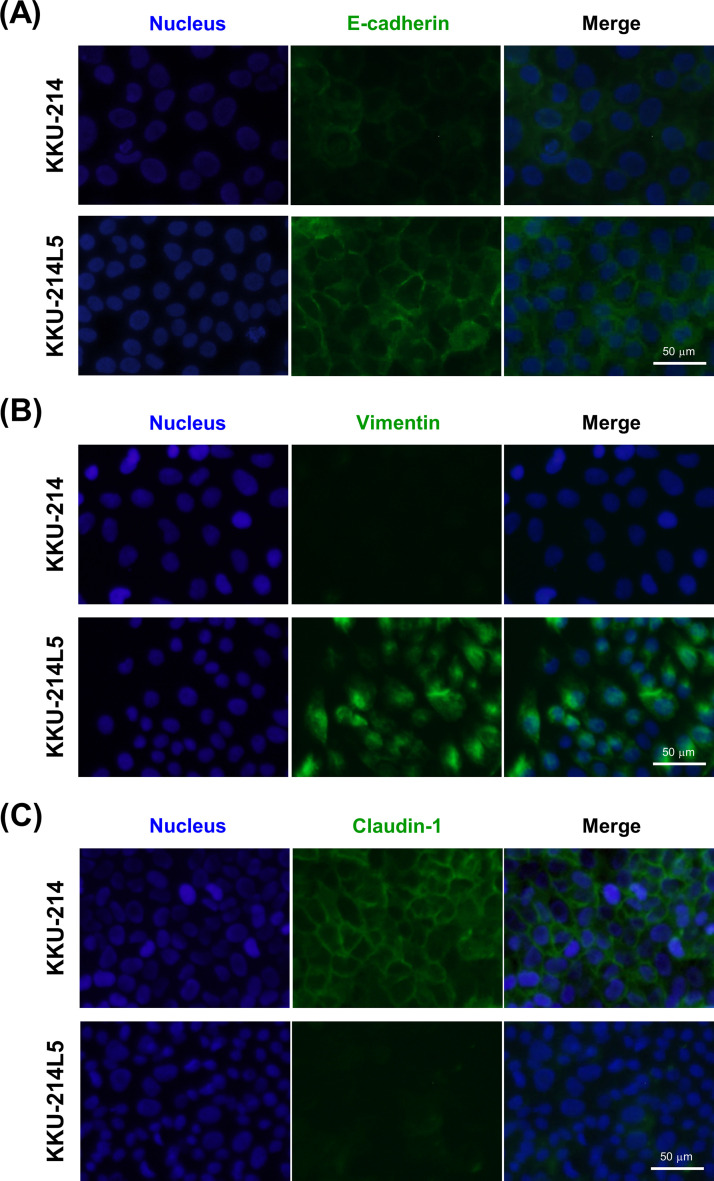
To elucidate the localizations of claudin-1, E-cadherin, and vimentin in KKU-214 and KKU-214L5 cells, immunocytofluorescence staining of these proteins was performed. As shown in Figure 4, the epithelial markers claudin-1 and E-cadherin were detected on the plasma membrane, while vimentin was stained in the cytoplasm. Consistent with the Western blotting, the expression levels of E-cadherin (Fig. 4A) and vimentin (Fig. 4B) were higher in the KKU-214L5 than in the KKU-214 cells, whereas decreased expression of claudin-1 was observed in KKU-214L5 cells (Fig. 4C).
Figure 4.
Immunocytofluorescence staining of EMT markers in KKU-214L5 compared to KKU-214. The protein expression of E-cadherin, vimentin, and claudin-1 were determined using immunocytofluorescence stain with immunoglobulin G (IgG)-conjugated Alexa Fluor® 488 (green). Nuclei were stained with Hoechst 33342 (blue). Overexpression of (A) E-cadherin and (B) vimentin and suppression of (C) claudin-1 was observed in KKU-214L5 cells. Scale bar: 50 μm.
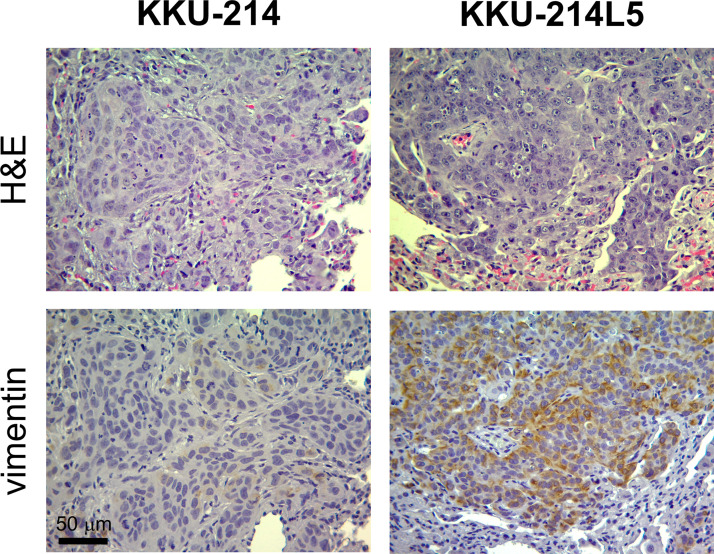
In order to affirm the upregulation of vimentin in KKU-214L5 in vivo, vimentin histochemistry staining was further performed using lung tissues from KKU-214L5 and KKU-214 tail vein-injected mice. Similar to the in vitro results (Figs. 3A and 4B), strongly stained vimentin was observed in the lungs of KKU-214L5-injected mice compared to the KKU-214-injected mice (Fig. 5).
Figure 5.
Vimentin was elevated in KKU-214L5-injected lung tissue. The expression of vimentin was determined using immunohistochemistry. Stronger staining of vimentin was observed in KKU-214L5-injected lung tissue when compared to KKU-214-injected lung tissue. Hematoxylin and eosin (H&E) staining was performed to indicate cancer cells. Scale bar: 50 μm.
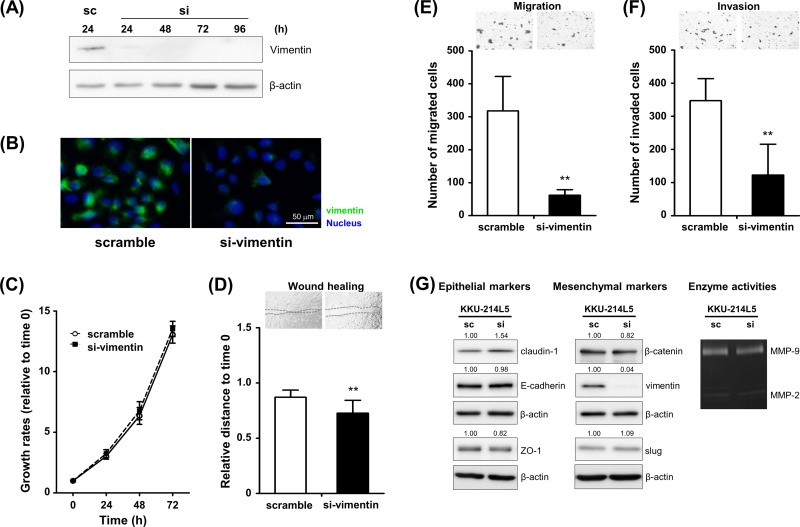
Silencing of Vimentin Expression Decreased the Migration and Invasion Abilities of the Highly Metastatic KKU-214L5 Cells to Almost the Basal Levels of the Parental Cells
Since vimentin expression was remarkably increased in highly metastatic cells, we next clarified whether vimentin is accountable for the metastatic progression observed in KKU-214L5 cells. Transient knockdown of vimentin was performed in KKU-214L5 cells using vimentin siRNA, and the expression levels of vimentin were determined using Western blots. Compared with the scramble control, si-vimentin effectively suppressed vimentin expression from 24 to 96 h (Fig. 6A). Suppression of vimentin expression in vimentin knockdown cells was confirmed using immunocytofluorescence staining (Fig. 6B). The decrease in vimentin expression did not affect the proliferation (Fig. 6C), but suppressed the migration and invasion abilities of KKU-214L5 cells (Fig. 6D–F). The migrating distance of KKU-214L5 cells treated with si-vimentin was significantly reduced (0.73 ± 0.12) to 80% of the control cells (0.87 ± 0.07) (p < 0.01) (Fig. 6D). Similarly, suppression of vimentin reduced the number of migrated KKU-214L5 cells to 20% of the scramble control cells (62 ± 17 cells vs. 318 ± 104 cells) (p < 0.01) (Fig. 6E) and decreased the invasion of KKU-214L5 cells to 35% of the scramble control cells (123 ± 93 cells vs. 347 ± 66 cells) (p < 0.01) (Fig. 6F). Suppression of vimentin, however, did not affect the expression levels of other EMT markers and the activity of MMPs (Fig. 6G). These results strongly suggest that the acquired migration and invasion abilities observed in the highly metastatic KKU-214L5 cells are induced via the vimentin activation.
Figure 6.
Depletion of vimentin suppressed the migration and invasion capacities of KKU-214L5. KKU-214L5 cells were treated with short interfering RNA for vimentin (si-vimentin) or a scrambled sequence (sc), and the expression levels of vimentin were determined using (A) Western blotting 24–96 h posttreatment and (B) immunocytofluorescence stain 48 h posttreatment. Magnification: 200×; scale bar: 50 μm. (C) No significant differences in proliferation were found between si-vimentin-treated KKU-214L5 and the scramble control cells. (D, E) Treatment of si-vimentin significantly suppressed the migration and (F) invasion capacities of KKU-214L5 compared to the scramble control cells 48 h posttreatment. (G) Suppression of vimentin did not alter the expression levels of other EMT markers and the enzyme activities of MMPs. Protein lysates of 10 μg were used for all EMT markers, except ZO-1 and slug were at 30 μg. The numbers indicate the comparison of the band intensities by giving those of scramble control as 1. **p < 0.01. sc = scramble control cells; si = si-vimentin-treated cells.
DISCUSSION
CCA metastasis is an example of the major cause of cancer-related death24. A better understanding of the underlying mechanisms related to metastasis may lead to better and effective therapy. In the present study, a highly metastatic CCA subline, KKU-214L5, was established from the parental cells (KKU-214) as a model for the comparative studies. The highly metastatic subline had a mesenchymal spindle-like shape with a more aggressive phenotype with increased migration and invasion compared to the parental cells. The aggressiveness of KKU-214L5 was demonstrated in the in vivo tail vein-injected metastatic mouse model. The underlying mechanism appears obviously to be due to the transition from epithelial- to mesenchymal-like cells. The particular characteristics observed in KKU-214L5 were the striking overexpressions of a mesenchymal marker, vimentin, and an epithelial marker, E-cadherin. The significant role of vimentin in supporting progression of CCA cells was elucidated. Suppression of vimentin using siRNA significantly reduced the migration and invasion capabilities of KKU-214L5 to levels that were close to the basal level of the parental KKU-214 cells (compare Figs. 1 and 6).
To determine the molecular mechanism underlying the metastatic process, a highly metastatic KKU-214L5 subline was first established using an in vivo tail vein metastasis mouse model. The model introduced cancer cells directly into the blood circulation and was colonized mainly in the lungs25, which have been reported to be the second most common metastatic site in CCA patients9. KKU-214L5 was generated by tail vein intravenous injection of KKU-214 to the NOJ mice, and cancer cells that colonized in the lung tissues were isolated, cultured, and reinjected via the tail vein for four more rounds. The established subline, KKU-214L5, was shown to have a common genetic origin of the parental cells as shown by STR analysis (Table 1). Cellular morphology, however, of KKU-214L5 was observed to be a mesenchymal spindle-like shape (Fig. 1A), which indicates the transition of epithelial to mesenchymal cells.
There are several biological capabilities acquired by cancer cells to achieve metastasis, for example, cell proliferation, migration, and invasion. In this study, cell migration was assessed using wound healing and Boyden chamber assays. The wound healing assay is a laboratory technique used to study lateral migration of cells and cell–cell interaction, whereas the Boyden chamber assay is an experimental tool established for the evaluation of the chemotactic ability of eukaryotic cells and has been modified to determine cell migration according to chemotactic ability. These two migration assays have different principles, have different measurements, and act independently to each other. It is obvious in the present study that KKU-214L5 cells had higher migratory activity than KKU-214 cells: 1.3-fold of migration distance for the wound healing assay and 5.7-fold of migratory cells for the Boyden chamber migration assay. KKU-214L5 also showed higher invasion capacities in the in vitro proliferative activity compared to the parental KKU-214 cells (Fig. 1B–E), indicating the necessity of detecting migration/invasion activities rather than proliferative activity for the metastasis of CCA cells in this model. The higher metastatic potential of KKU-214L5 than KKU-214 was affirmed by the in vivo metastatic mouse lung model (Fig. 2). The elevations of migration and invasion appear to be acquired properties of highly metastatic sublines after the epithelial–mesenchymal-like transition of parental cells, so called EMT.
EMT is a process in which epithelial cells transform into mesenchymal cells. It is involved with physiological development and also relates to cancer progression10,11. During the EMT process, several epithelial markers are downregulated, while mesenchymal markers are upregulated. In the present study, the expressions of epithelial markers (ZO-1 and claudin-1) were decreased, while the mesenchymal proteins β-catenin, vimentin, and slug were highly expressed in the highly metastatic KKU-214L5 subline (Fig. 3A). The suppression of claudin-1 and overexpressions of E-cadherin and vimentin were confirmed by immunocytofluorescence staining (Fig. 4). In addition, the elevations of MMP-2 and MMP-9 activities and CD147 expression (Fig. 3B) may increase the invasion ability of KKU-214L5 cells. CD147 is an integral membrane glycoprotein of an immunoglobulin superfamily and promotes cancer cell invasion through the stimulation of MMP production26,27. Upon activation, MMPs cleave cell surface proteins and degrade ECM components to facilitate the migration of the cells to invade neighboring tissues and the basement membrane28. The transition from epithelial to mesenchymal features may be essential to potentiate the high migration and invasion capacities observed in KKU-214L5 cells.
It is of interest that E-cadherin, a type 1 transmembrane protein that plays essential roles in cell–cell adhesion, was also upregulated in the highly metastatic KKU-214L5 cells as shown by Western blotting and immunocytofluorescence staining (Figs. 3A and 4A). To make sure that this observation was not cell type specific, the expression levels of E-cadherin and vimentin were determined in one more highly metastatic cell, KKU-213L5, that was established in a similar way to KKU-214L519. The Western blot results revealed that KKU-213L5 also had a higher expression of E-cadherin and vimentin than its parental cells (data not shown). The high expression of E-cadherin in the highly metastatic CCA cells denotes the important function of E-cadherin in metastasis. The association of E-cadherin expression with metastasis has been documented in some studies29–34. In the study with the prostate cancer cell lines, upregulation of E-cadherin was associated with the colonization ability in vivo31. The prostate cell line S-DU145, a negative E-cadherin subline, could not form tumors, whereas the T-DU145 subline, which had a strong E-cadherin expression, formed tumors with osteolytic activity. The association of E-cadherin expression and metastatic activity was supported by the observations found in breast29,30,34 and prostate cancer patients32,33 that the metastasized tumors exhibited strong expression of E-cadherin. The regulation of E-cadherin function related to the cellular migration and invasion, however, had not been reported in these metastatic cells. Further studies are needed to clarify the molecular pathway of E-cadherin underlying the positive metastatic process. The molecular mechanisms of E-cadherin in the highly metastatic CCA cells are being explored in a separate study.
KKU-214L5 possesses a striking level of vimentin expression both in vitro and in vivo (Figs. 3A, 4B, and 5), designating the involvement of vimentin with the highly metastatic potential of KKU-214L5. Vimentin is one of the type III intermediate filaments and has been reported as a mesenchymal marker. Vimentin plays a crucial role in cancer metastasis35–39. In CCA, the association of overexpression of vimentin with CCA metastasis has been revealed by immunohistochemistry staining40,41. In addition, upregulation of vimentin together with increased migration and invasion was reported in CCA cells16. The direct association of vimentin function with metastasis in CCA cells, however, has never been investigated. Herein it was demonstrated that suppression of vimentin using si-vimentin had no effect on cell proliferation, but significantly suppressed the migration and invasion of KKU-214L5 cells almost to the basal levels of the parental KKU-214 cells (Fig. 6). These results suggested that vimentin activity might not be necessary for the proliferative ability, but it plays a significant role on the increased migration and invasion capacities of KKU-214L5 cells. In addition, it also implies that the upregulation of migration and invasion via vimentin activation may be important for the regulation of metastatic potential of these CCA cells. The precise molecular mechanisms by which vimentin regulates cell migration and invasion need to be elucidated.
In conclusion, suppression of epithelial markers with increased mesenchymal markers could potentiate the metastatic potential, namely, migration and invasion by CCA cells. Deprivation of vimentin effectively defeated the metastatic activity of highly metastatic KKU-214L5 cells almost to the level of parental cells. Therapy targeting vimentin may be an effective approach to treat the highly metastatic cancer CCA. Moreover, the highly metastatic subline, KKU-214L5, established from this study will be suitable for the study of CCA metastasis, especially on EMT-related functions.
ACKNOWLEDGMENTS
This work was co-supported by TRF Senior Research Scholar Grant and Khon Kaen University (KKU) to S.W. (RTA5780012) and the Invitation Research Grant, Faculty of Medicine, KKU (IN58243). The authors would also like to thank the support given by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission through the Center of Excellence in SHeP-GMS cluster of KKU (M54209), and Japan Student Services Organization to W. Saentaweesuk (JASSO 2015-2016), as well as the RGJ-PhD Program, TRF (PHD/0021/2556) to W. Saentaweesuk and C.W. The authors would like to thank Professor James A. Will at the Publication Clinic, KKU, for the English language presentation of the manuscript, and Dr. R. Kraiklang, Faculty of Public Health, and Dr. P. Dana, Faculty of Medicine, KKU, for their technical assistance on the protein expression analysis.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Klein CA. Selection and adaptation during metastatic cancer progression. Nature 2013;501(7467):365–72. [DOI] [PubMed] [Google Scholar]
- 2. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007;7(11):834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer 2010;10(12):871–7. [DOI] [PubMed] [Google Scholar]
- 4. Klein CA. Framework models of tumor dormancy from patient-derived observations. Curr Opin Genet Dev. 2011;21(1):42–9. [DOI] [PubMed] [Google Scholar]
- 5. Blechacz B, Komuta M, Roskams T, Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2011;8(9):512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Endo I, Gonen M, Yopp AC, Dalal KM, Zhou Q, Klimstra D, D’Angelica M, DeMatteo RP, Fong Y, Schwartz L, Kemeny N, O’Reilly E, Abou-Alfa GK, Shimada H, Blumgart LH, Jarnagin WR. Intrahepatic cholangiocarcinoma: Rising frequency, improved survival, and determinants of outcome after resection. Ann Surg. 2008;248(1):84–96. [DOI] [PubMed] [Google Scholar]
- 7. Blechacz BR, Gores GJ. Cholangiocarcinoma. Clin Liver Dis. 2008;12(1):131–50. [DOI] [PubMed] [Google Scholar]
- 8. de Jong MC, Nathan H, Sotiropoulos GC, Paul A, Alexandrescu S, Marques H, Pulitano C, Barroso E, Clary BM, Aldrighetti L, Ferrone CR, Zhu AX, Bauer TW, Walters DM, Gamblin TC, Nguyen KT, Turley R, Popescu I, Hubert C, Meyer S, Schulick RD, Choti MA, Gigot JF, Mentha G, Pawlik TM. Intrahepatic cholangiocarcinoma: An international multi-institutional analysis of prognostic factors and lymph node assessment. J Clin Oncol. 2011;29(23):3140–5. [DOI] [PubMed] [Google Scholar]
- 9. Goodman ZD, Terracciano LM, Wee A. Tumours and tumour-like lesions of the liver. In: Burt AD, Portmann BC, Ferrell LD, editors. MacSween’s pathology of the liver, 6th ed Edinburgh: Churchill Livingstone; 2012. p. 761–851. [Google Scholar]
- 10. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2(6):442–54. [DOI] [PubMed] [Google Scholar]
- 11. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139(5):871–90. [DOI] [PubMed] [Google Scholar]
- 12. Gu MJ, Choi JH. Epithelial-mesenchymal transition phenotypes are associated with patient survival in intrahepatic cholangiocarcinoma. J Clin Pathol. 2014;67(3):229–34. [DOI] [PubMed] [Google Scholar]
- 13. Ryu HS, Chung JH, Lee K, Shin E, Jing J, Choe G, Kim H, Xu X, Lee HE, Kim DG, Lee H, Jang JJ. Overexpression of epithelial-mesenchymal transition-related markers according to cell dedifferentiation: Clinical implications as an independent predictor of poor prognosis in cholangiocarcinoma. Hum Pathol. 2012;43(12):2360–70. [DOI] [PubMed] [Google Scholar]
- 14. Sato Y, Harada K, Itatsu K, Ikeda H, Kakuda Y, Shimomura S, Shan Ren X, Yoneda N, Sasaki M, Nakanuma Y. Epithelial-mesenchymal transition induced by transforming growth factor-{beta}1/Snail activation aggravates invasive growth of cholangiocarcinoma. Am J Pathol. 2010;177(1):141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Techasen A, Loilome W, Namwat N, Khuntikeo N, Puapairoj A, Jearanaikoon P, Saya H, Yongvanit P. Loss of E-cadherin promotes migration and invasion of cholangiocarcinoma cells and serves as a potential marker of metastasis. Tumor Biol. 2014;35(9):8645–8652. [DOI] [PubMed] [Google Scholar]
- 16. Duangkumpha K, Techasen A, Loilome W, Namwat N, Thanan R, Khuntikeo N, Yongvanit P. BMP-7 blocks the effects of TGF-beta-induced EMT in cholangiocarcinoma. Tumour Biol. 2014;35(10):9667–76. [DOI] [PubMed] [Google Scholar]
- 17. Okada S, Harada H, Ito T, Saito T, Suzu S. Early development of human hematopoietic and acquired immune systems in new born NOD/Scid/Jak3null mice intrahepatic engrafted with cord blood-derived CD34 + cells. Int J Hematol. 2008;88(5):476–82. [DOI] [PubMed] [Google Scholar]
- 18. Sripa B, Leungwattanawanit S, Nitta T, Wongkham C, Bhudhisawasdi V, Puapairoj A, Sripa C, Miwa M. Establishment and characterization of an opisthorchiasis-associated cholangiocarcinoma cell line (KKU-100). World J Gastroenterol. 2005;11(22):3392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uthaisar K, Vaeteewoottacharn K, Seubwai W, Talabnin C, Sawanyawisuth K, Obchoei S, Kraiklang R, Okada S, Wongkham S. Establishment and characterization of a novel human cholangiocarcinoma cell line with high metastatic activity. Oncol Rep. 2016;36(3):1435–46. [DOI] [PubMed] [Google Scholar]
- 20. Ono A, Hattori S, Kariya R, Iwanaga S, Taura M, Harada H, Suzu S, Okada S. Comparative study of human hematopoietic cell engraftment into BALB/c and C57BL/6 strain of rag-2/jak3 double-deficient mice. J Biomed Biotechnol. 2011;2011:539748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaeteewoottacharn K, Kariya R, Dana P, Fujikawa S, Matsuda K, Ohkuma K, Kudo E, Kraiklang R, Wongkham C, Wongkham S, Okada S. Inhibition of carbonic anhydrase potentiates bevacizumab treatment in cholangiocarcinoma. Tumour Biol. 2016;37(7):9023–35. [DOI] [PubMed] [Google Scholar]
- 22. Shotivaranon J, Chirachariyavej T, Leetrakool N, Rerkamnuaychoke B. DNA database of populations from different parts in the Kingdom of Thailand. Forensic Sci Int Genet. 2009;4(1):e37–8. [DOI] [PubMed] [Google Scholar]
- 23. Toth M, Fridman R. Assessment of gelatinases (MMP-2 and MMP-9) by gelatin zymography. Methods Mol Med. 2001;57:163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta GP, Massague J. Cancer metastasis: Building a framework. Cell 2006;127(4):679–95. [DOI] [PubMed] [Google Scholar]
- 25. Pearson HB, Pouliot N. Modeling metastasis in vivo. Madame Curie Bioscience Database [Internet]. Texas: Landes Bioscience; 2000. [Google Scholar]
- 26. Kanekura T, Chen X. CD147/basigin promotes progression of malignant melanoma and other cancers. J Dermatol Sci. 2010;57(3):149–54. [DOI] [PubMed] [Google Scholar]
- 27. Nabeshima K, Iwasaki H, Koga K, Hojo H, Suzumiya J, Kikuchi M. Emmprin (basigin/CD147): Matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol Int. 2006;56(7):359–67. [DOI] [PubMed] [Google Scholar]
- 28. Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: A tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3(3):207–14. [DOI] [PubMed] [Google Scholar]
- 29. Bukholm IK, Nesland JM, Borresen-Dale AL. Re-expression of E-cadherin, alpha-catenin and beta-catenin, but not of gamma-catenin, in metastatic tissue from breast cancer patients. J Pathol. 2000;190(1):15–9. [DOI] [PubMed] [Google Scholar]
- 30. Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003;5(6):R217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Putzke AP, Ventura AP, Bailey AM, Akture C, Opoku-Ansah J, Celiktas M, Hwang MS, Darling DS, Coleman IM, Nelson PS, Nguyen HM, Corey E, Tewari M, Morrissey C, Vessella RL, Knudsen BS. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179(1):400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rubin MA, Mucci NR, Figurski J, Fecko A, Pienta KJ, Day ML. E-cadherin expression in prostate cancer: A broad survey using high-density tissue microarray technology. Human Pathol. 2001;32(7):690–7. [DOI] [PubMed] [Google Scholar]
- 33. Saha B, Arase A, Imam SS, Tsao-Wei D, Naritoku WY, Groshen S, Jones LW, Imam SA. Overexpression of E-cadherin and beta-catenin proteins in metastatic prostate cancer cells in bone. Prostate 2008;68(1):78–84. [DOI] [PubMed] [Google Scholar]
- 34. Saha B, Chaiwun B, Imam SS, Tsao-Wei DD, Groshen S, Naritoku WY, Imam SA. Overexpression of E-cadherin protein in metastatic breast cancer cells in bone. Anticancer Res. 2007;27(6B):3903–8. [PubMed] [Google Scholar]
- 35. Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, Foidart JM. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 2003;63(10):2658–64. [PubMed] [Google Scholar]
- 36. Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, Wu MC, Huang JF, Zeng WF, Sham JS, Yang M, Guan XY. Association of vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene 2004;23(1):298–302. [DOI] [PubMed] [Google Scholar]
- 37. Jin H, Morohashi S, Sato F, Kudo Y, Akasaka H, Tsutsumi S, Ogasawara H, Miyamoto K, Wajima N, Kawasaki H, Hakamada K, Kijima H. Vimentin expression of esophageal squamous cell carcinoma and its aggressive potential for lymph node metastasis. Biomed Res. 2010;31(2):105–12. [DOI] [PubMed] [Google Scholar]
- 38. Lang SH, Hyde C, Reid IN, Hitchcock IS, Hart CA, Bryden AA, Villette JM, Stower MJ, Maitland NJ. Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate 2002;52(4):253–63. [DOI] [PubMed] [Google Scholar]
- 39. McInroy L, Maatta A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun. 2007;360(1):109–14. [DOI] [PubMed] [Google Scholar]
- 40. Korita PV, Wakai T, Ajioka Y, Inoue M, Takamura M, Shirai Y, Hatakeyama K. Aberrant expression of vimentin correlates with dedifferentiation and poor prognosis in patients with intrahepatic cholangiocarcinoma. Anticancer Res. 2010;30(6):2279–85. [PubMed] [Google Scholar]
- 41. Mao X, Chen D, Wu J, Li J, Zhou H, Wu Y, Duan X. Differential expression of fascin, E-cadherin and vimentin: Proteins associated with survival of cholangiocarcinoma patients. Am J Med Sci. 2013;346(4):261–8. [DOI] [PubMed] [Google Scholar]