Abstract
Renal cell carcinoma (RCC) accounts for approximately 2%–3% of human malignancies and is the most aggressive among urologic tumors. Biological heterogeneity, drug resistance, and chemotherapy side effects are the biggest obstacles to the effective treatment of RCC. The NF-κB transcription factor is one of several molecules identified to be responsible for the aggressive phenotype of this tumor. In the past decade, several studies have demonstrated the activation of NF-κB in RCC, and many have implicated NF-κB1 (p50) as an important molecule in tumor progression and metastasis. In the present study, a lentivirus was used to deliver shRNA targeting NF-κB1 into mouse RCC (Renca) cells. It was determined that the knockdown of the NF-κB1 gene led to a reduction in cell proliferation and late apoptosis/necrosis in vitro. Flow cytometry analysis demonstrated G2/M arrest in the cells. In addition, immunoblotting analysis revealed a significant increase in cyclin B1 and Bax. In vivo experiments showed that Renca-shRNA-NF-κB1 cells have significantly diminished tumorigenicity. Moreover, immunohistochemical analysis revealed an increase in necrotic areas of Renca-shRNA-NF-κB1 tumors. Thus, this study indicates that downregulation of NF-κB1 can suppress RCC tumorigenesis by inducing late apoptosis/necrosis. Therefore, NF-κB1 may be a potential therapeutic target for RCC.
Key words: Renal cell carcinoma (RCC), Proliferation, Short hairpin RNA (shRNA), Nuclear factor κ-light-chain-enhancer of activated B cells 1 (NF-κB1)
INTRODUCTION
Renal cell carcinoma (RCC) is the most common and aggressive urological neoplasia. Among urological cancers, RCC has the second highest mortality rate and accounts for approximately 4% of human malignancies1. Clear cell RCC (ccRCC) is the most frequent histological subtype and accounts for 80%–90% of all RCC cases2. The biological heterogeneity, drug resistance, and side effects of chemotherapy are the biggest obstacles to the effective treatment of RCC. Several molecules have been identified as responsible for the aggressive phenotype of this tumor, one being the transcription factor nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)3. NF-κB is the collective name for the transcription factors of the Rel family. In mammals, five members of this family are known: RelA (p65), RelB, c-Rel, NF-κB1 (p105/p50), and NF-κB2 (p100/p52). The functional role of NF-κB1 in RCC has been investigated in recent years but is far from being fully elucidated4–7. The Nf-kB1 gene encodes the p105 protein, which undergoes cleavage to remove the carboxy-terminal portion by the 26S proteasome, generating the p50 protein that is capable of binding to DNA but lacks transcriptional activity. For this reason, the p50 homodimers are considered inhibitors of transcription. However, these dimers can recruit a coactivator, B-cell CLL/lymphoma 3 (Bcl-3), and thereby promote the activation of transcription8,9. The NF-κB1 and RelA (p50/p65) heterodimer, the first NF-κB protein discovered, is more abundant and regulates the expression of more genes than other heterodimers and homodimers.
Oya et al. were the first to identify the activation of NF-κB in renal adenocarcinoma cell lines in 2001. In this work, supershift gel experiments showed that p50 is the subunit involved in tumorigenesis. In addition, they demonstrated that cell lines susceptible to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis showed low levels of NF-κB activation, whereas apoptosis-resistant strains showed high activation10. In a later study, the same group investigated the expression of NF-κB in 45 primary renal tumor samples. The highest levels of NF-κB activation were found in the more advanced stage samples. Two independent groups, Meteoglu et al. and Djordjević et al., analyzed histological samples from patients with metastatic RCC and found that an increase in p50 activity was correlated with the expression of angiogenesis and apoptosis markers [epithelial growth factor receptor (EGFR), vascular endothelial growth factor (VEGF), Bcl-2, and tumor protein p53 (p53)]6,7. An and colleagues studied the effects of NF-κB blockade on the induction of apoptosis in two RCC lines (R11 and 444RCC). The authors showed that inhibition of NF-κB was not sufficient to induce apoptosis, but its blockade was necessary for the induction of apoptosis by the drug bortezomib5. The impact of NF-κB inhibition on renal adenocarcinoma was also evaluated by Morais et al. In vitro and in vivo assays showed that when RCC cell lines were treated with the drug pyrrolidine dicarbamate (PDTC), NF-κB expression was inhibited, and consequently, cellular viability and proliferation decreased11.
In a recent work, we demonstrated that endostatin treatment resulted in a significant reduction of p50 expression. Immunoprecipitation analysis confirmed the presence of the p50/Bcl-3 complex in nuclear extracts from metastatic lung cancer cells, suggesting an important role of p50 dimers in the transcriptional regulation of genes in ccRCC12. In this study, the effect of Nf-kB1 knockdown on the proliferation and tumor growth of RCC cells was evaluated.
MATERIALS AND METHODS
Cell Culture
The mouse RCC cell line (Renca) was obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% of fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (all obtained from Gibco, Grand Island, NY, USA) in a humidified incubator supplemented with 5% CO2 at 37°C.
Lentivirus Production
Short hairpin RNA (shRNA) targeting NF-κB1 mRNA and the empty vector-transfected control cells (Renca-Mock) were obtained from Mission® shRNA Lentiviral Plasmids (Sigma-Aldrich, St. Louis, MO, USA). After transformation into Escherichia coli DH5α (Subcloning Efficiency DH5-α Competent Cells; Invitrogen, Carlsbad, CA, USA), a two-plasmid system was used to pack the virus particles (pSPAX2-ψ PSI and pCMV-VsVg; Addgene, Cambridge, MA, USA) and the lentiviral particles were cotransfected into HEK 293T cells with constructed lentiviral interference vectors using Lipofectamine® 2000 (transfection reagent; Invitrogen). The concentrated virus supernatant was collected after 48 h.
Infection of Renca Cells With Lentivirus
Renca cells were seeded in six-well plates at 5 × 104 cells per plate in medium containing lentivirus with NF-κB1–shRNA or scrambled-shRNA and polybrene (hexadimethrine bromide; Sigma-Aldrich). After 6 h, the medium with 10% FBS was refreshed, and after 48 h, puromycin (Invitrogen) was added to select the cells. The cells were divided into three groups: Renca-wild type (WT) (cells without viral transfection), Renca-Mock (empty vector-transfected control cells), and Renca-shRNA-NF-κB1 (cells transfected with lentivirus NF-κB1-shRNA).
RNA Extraction and Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Total cellular RNA was extracted with TRIzol® (Life Technologies, Carlsbad, CA, USA). Next, cDNA was synthesized with QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol using 2 μg of RNA, and the cDNA was stored at −20°C. Absolute SYBR Green qPCR Mix® (Invitrogen) was used according to the manufacturer’s instructions. The reactions were carried out in a 10-μl volume reaction system. The PCR conditions consisted of 40 cycles, with a 15-s denaturation at 95°C, a 1-h annealing at 60°C, and a 1-min elongation step at 72°C. Target gene expression levels were normalized to tubulin mRNA levels. The primers used were as follows: NF-κB1, 5′-GGCTACACCGAAGCAATTGAA-3′ (forward) and 5′-CAGCGAGTGGGCCTGAGA-3′ (reverse); tubulin, 5′-CGCATCTCGGAGCAGTTCA-3′ (forward) and 5′-CGTGTACCAGTGCAGGAAAGC-3′ (reverse). Relative quantification of gene expression was performed with the 2−ΔΔCT method13. The PCR was carried out using the ABI-Prism 7000 quantitative PCR instrument (Applied Biosystems Company, Foster City, CA, USA).
Protein Extraction and Western Blot Analysis
Protein was extracted from cells using CelLytic™ M (Sigma-Aldrich). After centrifugation, the supernatant was collected, and a protease inhibitor cocktail powder (Sigma-Aldrich) was added. The protein concentration was determined using the BCA method, and the samples were stored at −80°C until use. Protein was loaded at 50 μg per lane and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinyl difluoride (PVDF) membranes GE Hybond-P, and blocked in 5% skim milk Tris-buffered saline at room temperature for 1 h. Membranes were incubated overnight at 4°C with the primary antibodies anti-NF-κB1 p105/p50, anti-β-actin and anti-Bcl-2-associated X protein (Bax; Abcam, Milton, Cambridge, UK), anti-cyclin B1 (Cell Signaling Technology, Danvers, MA, USA), anti-cyclin-dependent kinase inhibitor 1A (p21; Bioss), anti-Bcl-2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Millipore, Boston, MA, USA), all diluted 1:500. Goat or mouse horseradish peroxidase (HRP)-conjugated secondary antibodies, diluted 1:1,000, were added, and the membranes were incubated for 2 h at room temperature. The bands on the Western blot were detected with the SuperSignal® West Pico Chemiluminescent Substrate Kit (Thermo Scientific, Carlsbad, CA, USA). Images were taken with the Uvitec Cambridge Alliance 4.7 equipment.
Proliferation Assay: Doubling Time Analysis
Renca cells were seeded (5 × 103 cells per well) in 24-well plates. After 24 h, the medium was replaced with serum-free medium for cell synchronization, and after 24 h the medium was replaced again with medium containing 10% FBS. During the 4 days of culture, one quadruplicate was trypsinized and a cell count was performed. The number of viable cells was assessed using the trypan blue dye exclusion method. The experiment was carried out in triplicate. The doubling time was determined using the formula (ΔT = 1/K): K = 3.32 × [(logvf − logvi)/Tf − Ti].
Cell Cycle Analysis
Renca cells were treated with trypsin, centrifuged at 700 × g for 5 min, and fixed with prechilled 75% ethanol for 24 h at 4°C. The cells were stained with 50 μg/ml propidium iodide (PI) and 0.3 mg/ml ribonuclease A (Sigma-Aldrich) for 20 min at room temperature. The cell cycle distribution was then analyzed by flow cytometry with the Attune® Acoustic Focusing Cytometer (Life Technologies). The proportion of cells at each stage was analyzed using the multicycle DNA content and FlowJo cell analysis software. Each experiment was performed in triplicate.
Apoptosis Analysis
Cells were analyzed for apoptosis and necrosis with the Annexin V-fluorescein isothiocyanate (FITC) kit (Life Technologies) according to the manufacturer’s instructions. Fluorescence-activated cell sorting (FACS) was used to detect the binding of annexin V-fluorescein and determine the exclusion or inclusion of PI.
In Vivo Tumorigenicity Assay
The BALB/c female mice were obtained from the Nuclear and Energy Research Institute (IPEN/CNEN, São Paulo, Brazil). Renca cells were injected subcutaneously into the flanks of 8- to 10-week-old BALB/c female mice (n = 5/group). The tumor volume was calculated using the formula V = largest measure × (smaller measure)2 × 0.5. The mice were euthanized 17 days after injection, and the tumors were collected, excised, fixed in methacarn, and routinely processed for paraffin embedding. All animals were cared for in accordance with the standards of the institute, under a protocol approved by the Animal Experimentation Ethics Committee (No. of Process: 162/15). Histological analysis was performed on 4-μm sections stained with hematoxylin and eosin (H&E).
Immunohistochemistry (IHC)
Immunohistochemical labeling for Ki-67 (Santa Cruz Biotechnology) was performed. Briefly, paraffin-embedded tissue sections were deparaffinized with graduated xylene and alcohol and immersed in a citrate solution (pH 6.0). The tissues were incubated with methanol and hydrogen peroxide (3%) 1:1, followed by the primary antibody, diluted in 1:1,000, at 4°C overnight and the biotinylated secondary antibody (Vector Laboratories Inc., Burlingame, CA, USA) at 37°C for 30 min. After washing, 3,3′-diaminobenzidine tetrahydrochloride (DAB) solution was used to visualize the staining. The stained slides were evaluated by standard light microscopy (Eclipse E600; Nikon, Tokyo, Japan).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). The results are presented as the mean ± standard deviation (SD). Statistically significant differences between groups for each assay were analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni’s test. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
In all experiments, Renca-WT and nontarget shRNA-transduced cells (Renca-Mock) were used as controls. Nf-kB1 mRNA production was evaluated by qRT-PCR, demonstrating lower expression levels in the Renca-shRNA-NF-κB1 cells, with an mRNA level reduction of 70.0 ± 0.12% (p < 0.01) compared to the Renca-Mock cells (Fig. 1A). The protein expression of p105 and p50 in Renca-shRNA-NF-κB1 cells was reduced by 84.0 ± 0.01% (p < 0.01) and 77.0 ± 0.08% (p < 0.001), respectively, compared to the Renca-Mock cells (Fig. 1B and C).
Figure 1.
Stable knockdown of nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB1) in mouse renal cell carcinoma cell line (Renca) cells. Renca cells were transfected with short hairpin (shRNA)-NF-κB1. (A) mRNA expression was reduced by 70% (**p < 0.01). (B) Western blot analysis. (C) The optical density values of the corresponding bands were quantified using ImageJ software, with β-actin serving as an internal control. The results show a reduction in p105 (**p < 0.01) and p50 (***p < 0.001) at the protein level. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Bonferroni’s test.
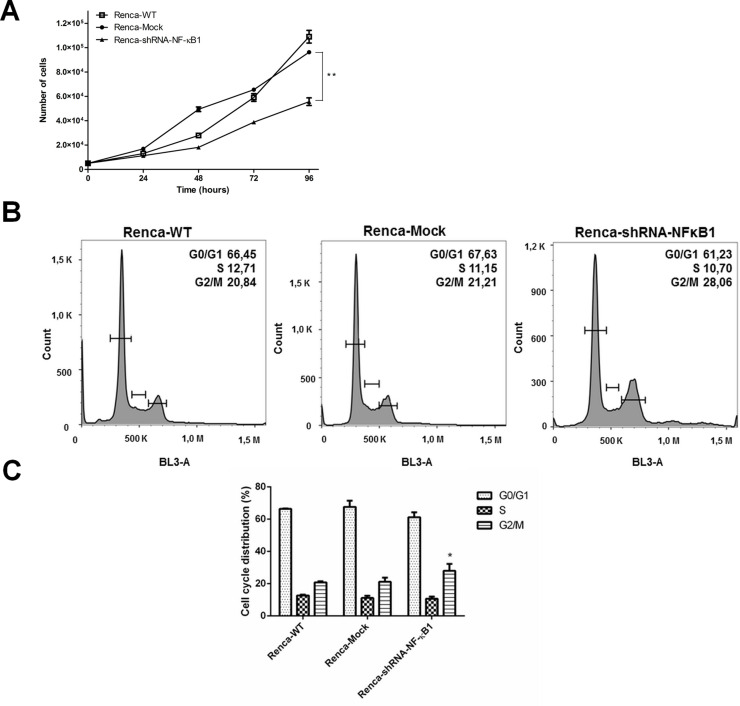
To examine whether NF-κB knockdown affected Renca cell growth, cellular proliferation was evaluated at different time points (24, 48, 72, and 96 h). The results demonstrated that cell growth significantly decreased in the Nf-kB1 knockdown Renca cells at 48, 72, and 96 h compared to the Renca-Mock cells (Fig. 2A). The doubling cell time, in hours, for the Renca-WT cells was 21.4 h, for the Renca-Mock cells was 21.8 h, and for the Renca-shRNA-NF-κB1 cells was 27.0 h (p < 0.01 vs. Renca-Mock cells).
Figure 2.
NF-κB knockdown significantly elongates the doubling time in Renca cells and leads to G2/M arrest. (A) The proliferation of Renca-shRNA-NF-κB1 cells was markedly inhibited in a time-dependent manner (**p < 0.01). (B) Cell cycle distribution. (C) Cell cycle analysis showed a significant increase in the G2/M population of Renca-shRNA-NF-κB1 cells (*p < 0.05 vs. Renca-Mock). The results represent the mean ± SD of three experiments. Statistical analysis was performed using one-way ANOVA followed by Bonferroni’s test.
To further confirm the impact of NF-κB1 knockdown on cell growth, the cell cycle distribution in the population was analyzed by flow cytometry. The results demonstrated that the number of Renca-shRNA-NF-κB1 cells in the G2/M phase was significantly increased (p < 0.05 vs. Renca-Mock) (Fig. 2B and C).
Renca cells were harvested and analyzed for apoptosis and necrosis using the annexin V/PI flow cytometry assay. As shown in Figure 3, the percentage of late apoptotic/necrotic cells in the Renca-WT and Renca-Mock groups was 6.17 ± 0.72% and 5.44 ± 0.63%, respectively, which was significantly lower compared to the Renca-shRNA-NF-κB1 group (8.96 ± 3.07%, p < 0.05), indicating that the knockdown of the Nf-kB1 gene leads to a significant increase in the baseline apoptosis levels of Renca cells.
Figure 3.
Knockdown of NF-κB1 promotes late apoptosis/necrosis in the Renca cell line. (A) Diagrams of the flow cytometry of Renca cells after label with annexin V-fluorescein isothiocyanate (FITC) (AnexV) and propidium iodide (PI) showing the following: Q1, dead cells; Q2, late apoptosis; Q3, early apoptosis; and Q4, normal cells. (B) Flow cytometric analysis of Renca cells. Data are expressed as the mean ± SD of three independent experiments. *p < 0.05. Statistical analysis was performed using one-way ANOVA followed by Bonferroni’s test.
To elucidate the molecular mechanism by which NF-κB1 regulates the proliferation of RCC cells, the expression of p21, cyclin B1, Bax, and Bcl-2, which are involved in the progression of cell cycle and cell death, was evaluated. Western blot analysis revealed that knockdown of NF-κB1 increased the expression of p21, cyclin B1, and Bax compared to the control group, while Bcl-2 expression decreased subtly (Fig. 4). The Bax/Bcl-2 ratio was increased in the Renca-shRNA-NF-κB1 cells 5.64 ± 0.33-fold compared with the control group Renca-Mock (p < 0.05), indicating that both Bax and Bcl-2 are active in the response of Renca cells to apoptosis and that knockdown of NF-κB1 deregulates the ratio between these proteins.
Figure 4.
Effect of NF-κB1 knockdown on the expression of cell cycle regulators. (A) The protein expression levels of cyclin-dependent kinase inhibitor 1A (p21), cyclin B1, B-cell CLL/lymphoma 2 (Bcl-2), and Bcl-2-associated X protein (Bax) were detected by Western blot. (B) Quantitative analysis of the protein levels using ImageJ 1.6.0_24 software and normalized to glyceraldehyde 3-phosphate (GAPDH). (C) Increase in the Bax/Bcl-2 ratio of the protein levels in knockdown for NF-κB1 cells. The values shown represent the mean ± SD. *p < 0.05, **p < 0.01 versus Renca-Mock. Statistical analysis was performed using one-way ANOVA followed by Bonferroni’s test.
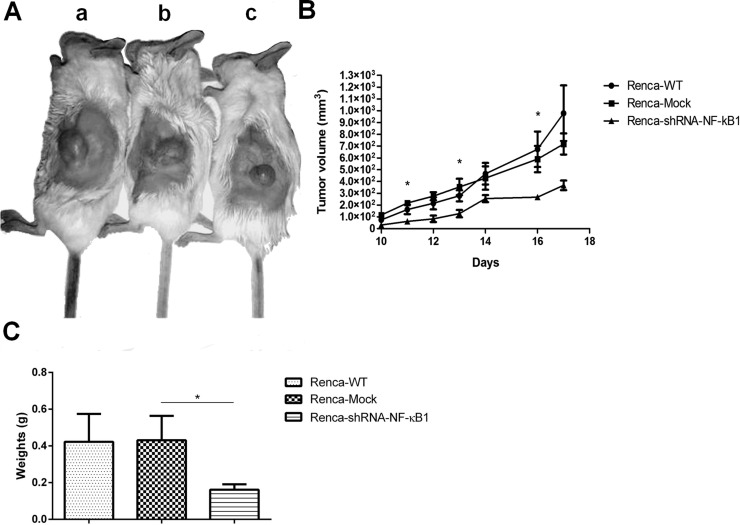
The in vivo tumorigenesis assay revealed that tumor growth was significantly slower in BALB/c mice inoculated with the Renca-shRNA-NF-κB1 cells compared to the controls (Fig. 5A). The mean tumor volume for the Renca-shRNA-NF-κB1 group was 367.51 ± 40.57 mm3, for the Renca-WT group was 979.29 ± 235.47 mm3, and for the Renca-Mock group was 718.19 ± 89.37 mm3. Throughout the experiment, a significant difference was found on days 11, 13, and 16 (p < 0.05 vs. Renca-Mock) (Fig. 5B). The mean tumor weight of the resected tumors is shown in Figure 5C. The tumor weight was significantly reduced in the NF-κB1 shRNA knockdown xenograft tumors (p < 0.05 vs. Renca-Mock).
Figure 5.
Effects of NF-κB1 knockdown in Renca cells. (A) BALB/c mice were subcutaneously inoculated with Renca cells transfected with Renca-wild type (WT) (a) and Renca-Mock (b) or shRNA-NF-κB1 (c). The image is representative of tumors formed. (B) Growth curves of tumor volumes. The graph is representative of tumor growth 17 days after inoculation. (C) Graph showing the mean tumor weight. All data are presented as the mean ± SD (n = 5) and represent three independent experiments; *p < 0.05 versus Renca-Mock. Statistical analysis was performed using one-way ANOVA followed by Bonferroni’s test.
Immunohistochemical analysis of the xenograft tissues was performed to confirm the effect of NF-κB1 knockdown in Renca cells on proliferation (Fig. 6). The results showed that the necrotic area in the Renca-shRNA-NF-κB1 samples was significantly increased compared to the Renca-Mock tumors (p < 0.001). Ki-67 labeling was significantly reduced in the Renca-shRNA-NF-κB1 group compared to the other two groups (p < 0.05).
Figure 6.
Histological and morphometric analysis of paraffin-embedded tumor sections in different groups. (A) Tumor sections were subjected to hematoxylin and eosin (H&E) staining and immunohistochemical staining for anti-Ki-67. (B) Quantitative analysis was achieved by morphometry. Tumor tissue in the Renca-shRNA-NF-κB1 group showed a significant increase in the necrotic area (***p < 0.001). Moreover, Ki-67 expression was slightly but significantly reduced (*p < 0.05). Statistical analysis was performed using one-way ANOVA followed by Bonferroni’s test.
DISCUSSION
In the past decade, several studies have correlated the activation of NF-κB in RCC with angiogenesis, cell proliferation, resistance to chemotherapy, and resistance to apoptosis14–17. Here we showed that NF-κB1 may constitute a therapeutic target for RCC. qRT-PCR and Western blot results indicate that NF-κB1 expression at both the mRNA and protein levels was significantly decreased in Renca-shRNA-NF-κB1 cells. Knocking down the expression of NF-κB1 results in a time-dependent inhibition of cell proliferation. Recently, Chen and collaborators demonstrated that the NF-κB1 polymorphism (rs4648068) is associated with cell proliferation and motility in gastric cancer. They found that the GG genotype leads to higher levels of p105/p50 and increased cell proliferation and motility compared to the AA genotype18.
To verify the causal relationship between low NF-κB1 levels and the inhibition of cellular proliferation, the cell cycle distribution was analyzed. The Renca-shRNA-NF-κB1 cell line exhibited an increased number of cells in the G2/M phase, accompanied by a decreased number of cell number in the S and G0/G1 phases. Moreover, these cells were analyzed by the annexin V/PI flow cytometry method, and an increase in late apoptotic/necrotic cells was discovered. These data are in agreement with the previous findings of Yu and collaborators that described the participation of p50 dimers in cell proliferation-related and antiapoptotic signaling pathways19.
To explore the mechanism underlying late apoptosis/necrosis in Renca cells that is mediated by NF-κB1 downregulation, we employed Western blot analysis and demonstrated a significant increase in cyclin B1 and Bax, whereas the expression of p21 was slightly upregulated. The accumulation of cyclin B1 in the G2/M phase with an increase in apoptosis has been previously reported20. The inability of Renca-shRNA-NF-κB1 cells to exit the G2 stage may be explained by assuming that the active cyclin B1–cell division cycle 2 (cdc2) complexes are confined to the cytoplasmic compartment21.
A syngeneic animal model was used to assess the tumorigenicity of Renca-shRNA-NF-κB1 cells, and our data showed that NF-κB1 silencing markedly inhibited tumor growth. H&E staining of Renca-shRNA-NF-κB1 tumor sections uncovered large necrotic areas compared to the control groups. These data are consistent with the results of the annexin V/PI assay as well as with the increased levels of Bax, since Bax regulates primary cellular necrosis through an apoptosis-independent mechanism22.
Immunohistochemical staining of Renca cells for Ki-67, a cell proliferation marker of several malignancies, including ccRCC, was consistent with the in vitro findings23.
In conclusion, knockdown of the NF-κB1 gene efficiently reduced Renca cell growth by inducing G2/M arrest and late apoptosis/necrosis. It may be useful as a pathological biomarker as well as a chemotherapy target in RCC.
ACKNOWLEDGMENTS
We would like to thank Ms. Evelin Caroline da Silva for her valuable technical support on this project. This study was supported by FAPESP (Process No. 2014/19265-7).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Quan Z, He Y, Luo C, Xia Y, Zhao Y, Liu N, Wu X. Interleukin 6 induces cell proliferation of clear cell renal cell carcinoma by suppressing hepaCAM via the STAT3-dependent up-regulation of DNMT1 or DNMT3b. Cell Signal. 2017;32:48–58. [DOI] [PubMed] [Google Scholar]
- 2. Zhang F, Ma X, Li H, Guo G, Li P, Li H, Gu L, Li X, Chen L, Zhang X. The predictive and prognostic values of serum amino acid levels for clear cell renal cell carcinoma. Urol Oncol. 2017;35(6):392–400. [DOI] [PubMed] [Google Scholar]
- 3. Morais C, Gobe G, Johnson DW, Healy H. The emerging role of nuclear factor kappa B in renal cell carcinoma. Int J Biochem Cell B 2011;43:1537–49. [DOI] [PubMed] [Google Scholar]
- 4. Steiner T, Junker U, Henzgen B, Nuske K, Durum SK, Schubert J. Interferon–alpha suppresses the antiapoptotic effect of NF–kB and sensitizes renal cell carcinoma cells in vitro to chemotherapeutic drugs. Eur Urol. 2001;39:478–83. [DOI] [PubMed] [Google Scholar]
- 5. An J, Sun Y, Fisher M, Rettig MB. Maximal apoptosis of renal cell carcinoma by the proteasome inhibitor bortezomib is nuclear factor-κB dependent. Mol Cancer Ther. 2004;3:727–36. [PubMed] [Google Scholar]
- 6. Meteoglu I, Erdogdu IH, Meydan N, Erkus M, Barutca S. NF-kappaB expression correlates with apoptosis and angiogenesis in clear cell renal cell carcinoma tissues. J Exp Clin Cancer Res. 2008;27:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Djordjević G, Matusan-Ilijas K, Sinozić E, Damante G, Fabbro D, Grahovac B, Lucin K, Jonjić N. Relationship between vascular endothelial growth factor and nuclear factor-kappaB in renal cell tumors. Croat Med J. 2008;49:608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karin M. NF-κB as a critical link between inflammation and cancer. CSH Perspect Biol. 2009;1:a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Napetschnig J, Wu H. Molecular basis of NF-κB signaling. Annu Rev Biophys. 2013;42:443–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oya M, Ohtsubo M, Takayanagi A, Tachibana M, Shimizu N, Murai M. Constitutive activation of nuclear factor-kappa B prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene 2001;20:3888. [DOI] [PubMed] [Google Scholar]
- 11. Morais C, Healy H, Johnson DW, Gobe G. Inhibition of nuclear factor kappa B attenuates tumour progression in an animal model of renal cell carcinoma. Nephrol Dial Transplant. 2010;25:1462–74. [DOI] [PubMed] [Google Scholar]
- 12. de Souza Braga M, da Silva Paiva KB, Foguer K, Chaves KCB, de Sá Lima L, Scavone C, Bellini MH. Involvement of the NF-κB/p50/Bcl-3 complex in response to antiangiogenic therapy in a mouse model of metastatic renal cell carcinoma. Biomed Pharmacother. 2014;68:873–9. [DOI] [PubMed] [Google Scholar]
- 13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 14. Oya M, Takayanagi A, Horiguchi A, Mizuno R, Ohtsubo M, Marumo K, Shimizu N, Murai M. Increased nuclear factor-kappa B activation is related to the tumor development of renal cell carcinoma. Carcinogenesis 2003;24:377–84. [DOI] [PubMed] [Google Scholar]
- 15. Riemann K, Becker L, Struwe H, Nückel H, Dührsen U, Alakus H, Winde G, Neuhäuser M, Rübben H, Schmitz KJ, Wohlschlaeger J, Schmid KW, Siffert W. No association of the NFKB1 insertion/deletion promoter polymorphism with survival in colorectal and renal cell carcinoma as well as disease progression in B-cell chronic lymphocytic leukemia. Pharmacogenet Genom. 2006;16:783–8. [DOI] [PubMed] [Google Scholar]
- 16. Matušan-Ilijaš K, Damante G, Fabbro D, Dorđević G, Hadžisejdić I, Grahovac M, Avirović M, Grahovac B, Jonjić N, Lučin K. EGFR expression is linked to osteopontin and Nf-κB signaling in clear cell renal cell carcinoma. Clin Transl Oncol. 2013;15:65–71. [DOI] [PubMed] [Google Scholar]
- 17. Sourbier C, Danilin S, Lindner V, Steger J, Rothhut S, Meyer N, Jacqmin D, Helwig JJ, Lang H, Massfelder T. Targeting the nuclear factor-kB rescue pathway has promising future in human renal cell carcinoma therapy. Cancer Res. 2007;67:11668–76. [DOI] [PubMed] [Google Scholar]
- 18. Chen Y, Lu R, Zheng H, Xiao R, Feng J, Wang H, Gao X, Guo L. The NFKB1 polymorphism (rs4648068) is associated with the cell proliferation and motility in gastric cancer. BMC Gastroenterol. 2015;15:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu Y, Wan Y, Huang C. The biological functions of NF-κB1 (p50) and its potential as an anti-cancer target. Curr Cancer Drug Targets 2009;9:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindqvist A, van Zon W, Rosenthal CK, Wolthuis RM. Cyclin B1–Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zaffaroni N, De Marco C, Villa R, Riboldi S, Daidone MG, Double JA. Cell growth inhibition, G2M cell cycle arrest and apoptosis induced by the imidazoacridinone C1311 in human tumour cell lines. Eur J Cancer 2001;37:1953–62. [DOI] [PubMed] [Google Scholar]
- 22. Whelan RS, Konstantinidis K, Wei AC, Chen Y, Reyna DE, Jha S, Yang Y, Calvert JW, Lindsten T, Thompson CB, Crow MT, Gavathiotis E, Dorn GW II, O’Rourke B, Kitsis RN. Bax regulates primary necrosis through mitochondrial dynamics. Proc Natl Acad Sci USA 2012;109:6566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tollefson MK, Thompson RH, Sheinin Y, Lohse CM, Cheville JC, Leibovich BC, Kwon ED. Ki-67 and coagulative tumor necrosis are independent predictors of poor outcome for patients with clear cell renal cell carcinoma and not surrogates for each other. Cancer 2007;110:783–90. [DOI] [PubMed] [Google Scholar]