Abstract
MicroRNAs (miRNAs) play important roles in several human cancers. Although miR-188 has been suggested to function as a tumor repressor in cancers, its precise role in glioma and the molecular mechanism remain unknown. In the present study, we investigated the effect of miR-188 on glioma and explored its relevant mechanisms. We found that the expression of miR-188 is dramatically downregulated in glioma tissues and cell lines. Subsequent investigation revealed that miR-188 expression was inversely correlated with β-catenin expression in glioma tissue samples. Using a luciferase reporter assay, β-catenin was determined to be a direct target of miR-188. Overexpression of miR-188 reduced β-catenin expression at both the mRNA and protein levels, and inhibition of miR-188 increased β-catenin expression. Moreover, we found that overexpression of miR-188 suppressed glioma cell proliferation and cell cycle G1–S transition, whereas inhibition of miR-188 promoted glioma cell proliferation. Importantly, silencing β-catenin recapitulated the cellular and molecular effects seen upon miR-188 overexpression, which included inhibiting glioma cell proliferation and G1–S transition. Taken together, our results demonstrated that miR-188 inhibits glioma cell proliferation by targeting β-catenin, representing an effective therapeutic strategy for glioma.
Key words: Glioma, MicroRNA-188 (miR-188), β-Catenin, Proliferation, Cell cycle
INTRODUCTION
Gliomas are the most common type of primary cancers arising from glial cells in the central nervous system, accounting for 40%–50% of all central nervous system tumors1. Gliomas can be divided into astrocytomas, anaplastic astrocytomas, glioblastomas, oligodendrogliomas, glioblastoma multiforme, ependymomas, and medulloblastomas2. The incidence rate of gliomas has gradually increased and accounts for 1.9% of total cancer incidences in the world3,4. Despite advances in surgery, radiotherapy, chemotherapy, and multimodality therapy, glioma-related mortality remains high because it is a complex disease. The carcinogenesis and progression of glioma involve many genetic and environmental factors, in multistep processes5–7. Accumulating evidence shows there are a series of molecular events underlying the tumorigenesis and progression of gliomas, which remain unclear. Therefore, it is of vital clinical significance to understand the molecular mechanisms of glioma, which could identify novel drug targets and develop therapeutic strategies for patients with gliomas.
MicroRNAs (miRNAs) are a class of endogenous, single-stranded, small noncoding, highly conserved RNA molecules with a length of 18–24 nucleotides, which regulate the gene expression at a posttranscriptional level through binding to the 3′-untranslated region (3′-UTR) of the respective target’s mRNA, leading to mRNA degradation and/or suppression of translation8–11. Accumulating studies have shown that miRNAs play key roles in a variety of biological processes. In particular, more and more types of miRNAs have been shown to involve tumorigenesis and development, including the regulation of cancer cell survival, proliferation, cycles, differentiation, apoptosis, metabolism, invasion, and migration12–18. According to their functions, miRNAs may serve as oncogenes or antioncogenes, making them potential therapeutic targets in tumor treatment16,19. In human gliomas, altered miRNA expression and biogenesis have been demonstrated to play an important role in cancer-related signaling pathways, which are associated with a range of tumor characteristics including gliomagenesis, proliferation, apoptosis, invasion, and malignancy. According to previous studies, we found that downregulation of miR-188 acts as an antioncogene in several forms of cancers, such as oral carcinoma, nasopharyngeal carcinoma, and prostate carcinoma20–22. However, the molecular mechanisms underlying the role of miR-188 in gliomas remain unclear.
In the present study, we investigated the role and molecular mechanisms of miR-188 in gliomas. We found that the expression of miR-188 was remarkably downregulated in glioma tissues and correlated with clinicopathological characteristics. Furthermore, miR-188 potently inhibited glioma cell proliferation and cell cycle progression. More importantly, we provide evidence, for the first time, that β-catenin is a direct and functional target of miR-188. Our data suggest that miR-188 may be a potential therapeutic target in glioma therapy.
MATERIALS AND METHODS
Preparation of Human Tissue Samples
Human glioma samples (81) were collected from patients who were diagnosed at The First Affiliated Hospital, Xi’an Jiaotong University (Xi’an, Shaanxi, P.R. China). Normal brain samples (26) were obtained from people with traumatic brain injury, for whom a partial resection of brain tissues was conducted in order to reduce the intracranial pressure. We obtained informed consent from each patient before specimen collection. The samples were stored at −80°C. The experiments were approved by the Ethics Committee of Xi’an Jiaotong University Health Science Center.
Cell Culture
Human glioma cell lines H4, U87, SNB19, and LN229 and primary normal human astrocytes (NHAs) were purchased from the Cell Bank (Shanghai Genechem Co., Ltd., Shanghai, P.R. China). These cells were cultivated in Roswell Park Memorial Institute (RPMI)-l640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% (v/v) fetal bovine serum (Gibco BRL, Grand Island, NY, USA) and were incubated at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from human tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. SYBR Premix Ex Taq II Kit and PrimeScript RT Reagent Kit (TaKaRa Biotechnology Co., Ltd., Dalian, P.R. China) were used for the detection of miR-188 expression and β-catenin mRNA expression. qRT-PCR was conducted using the iCycler iQ Multicolor qRT-PCR Detection System (Bio-Rad, Hercules, CA, USA). The results were normalized to RNU6B (U6) gene or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression, as recommended by the manufacturer. The primer sequences were as follows: miR-188 reverse-transcribed primer, 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCCTCCA-3′; miR-188, 5′-ATCCAGTGCGTGTCGTG-3′ (forward) and 5′-TGCTCATCCCTTGCATGG-3′ (reverse); U6 reverse-transcribed primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); β-catenin, 5′-GTGTGGCGACATATGCAGCT-3′ (forward) and 5′-CAAGATCAGCAGTCTCATTC-3′ (reverse); GAPDH, 5′-GAAGGTGAAGGTCGGAGTCA-3′ (forward) and 5′-TTGAGGTCAATGAAGGGGTC-3′ (reverse). All reactions were performed in triplicate.
Dual-Luciferase Assay
The 3′-UTR of human β-catenin mRNA was constructed with synthetic oligonucleotides and cloned in the pmirGLO Dual-Luciferase miRNA target expression vector (Promega, Madison, WI, USA). The pmirGLO-β-catenin-3′-UTR vector was cotransfected with miR-188 into HEK293T cells using the pmirGLO vector as their control. These cells were collected and lysed 24 h after transfection. The Dual-Luciferase Reporter Assay System (Promega) was used to examine reporter activity according to the manufacturer’s protocol.
Expression Vector Construction
Hsa-miR-188 precursor expression vector (named miR-188) and control vector (named miR-Ctrl) were constructed with synthetic oligonucleotides and interpolated into pcDNA6.2-GW/EmGFPmiR vector according to the manufacturer’s instructions. Transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
miR-188 Inhibitor Synthesis, siRNA Synthesis, and Transfection
Interfering RNA oligonucleotides were used as miR-188 inhibitors (named anti-miR-188) and purchased from GenePharma (Shanghai, P.R. China). The sequence of anti-miR-188 was 5′-CCCUCCACCAUGCAAGGGAUG-3′. Scramble siRNA was used as negative control (named anti-miR-Ctrl), and the sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. RNA oligonucleotides were transfected into glioma LN229 cells with Lipofectamine 2000. siRNA was used for human β-catenin gene silencing. Human β-catenin siRNA-1 (sense: 5′-GGCUACUGUUGGAUUGAUUTT-3′, antisense: 5′-AAUCAAUCCAACAGUAGCCTT-3′), siRNA-2 (sense: 5′-GUCAACGUCUUGUUCAGAATT-3′, antisense: 5′-UUCUGAACAAGACGUUGACTT-3′), and negative control-siRNA (NC-siRNA; sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′) were chemically synthesized by GenePharma Corporation. Lipofectamine 2000 was used to transfect siRNA. The siRNAs were diluted to 60 nM in the plated cells for further experimental procedures.
MTT Assay
The LN229 cells (5,000 cells/well in 200 μl of RPMI-l640 medium) were seeded into 96-well plates and incubated for 1 day. The cells were treated with miR-Ctrl, miR-188, anti-miR-Ctrl, anti-miR-188, NC-siRNA (60 nM), siRNA-1, or siRNA-2 for 1, 2, and 3 days, respectively. Cell viability was measured using MTT assay FLUOstar OPTIMA (BMG Labtechnologies, Offenburg, Germany) at a wavelength of 490 nm. Each experiment contained four replicates and was repeated at least three times.
Cell Cycle Analysis
The LN229 cells were cultured in six-well plates in triplicate and treated by miR-Ctrl, miR-188, anti-miR-Ctrl, anti-miR-188, NC-siRNA, siRNA-1, or siRNA-2 for 2 days. The cells were harvested and washed in phosphate-buffered saline (PBS) and fixed in ice-cold ethanol overnight at 4°C. The fixed cells were washed in PBS and stained with 50 μg/ml propidium iodide (PI) containing 50 μg/ml RNase A (DNase free) for 20 min at room temperature. The cells were examined by fluorescence-activated cell sorting (BD Biosciences, San Diego, CA, USA). The cell cycle populations were determined by ModFit software.
Cell Apoptosis Analysis
The cells were seeded into six-well plates in triplicate and treated for 2 days. Cell apoptosis analysis was performed with Annexin-V Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Invitrogen) according to the manufacturer’s instructions. Then the stained cells were examined using a flow cytometer (BD Biosciences). Quantification of apoptosis was determined by ModFit software.
Western Blot Analysis
Total protein was extracted using a radioimmunoprecipitation assay (RIPA) lysis buffer (Wolsen, Xi’an, P.R. China) from cells or tissue samples, separated in 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and electrophoretically transferred to nitrocellulose membrane (Roche, Basel, Switzerland). Then the membrane was incubated with primary antibodies overnight at 4°C. The primary monoclonal antibodies included rabbit monoclonal anti-β-catenin (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit monoclonal anti-c-Myc (1:1,000; Santa Cruz Biotechnology), mouse monoclonal anti-cyclin D1 (1:1,000; Santa Cruz Biotechnology), and mouse monoclonal anti-GAPDH (1:2,000; Santa Cruz Biotechnology). The membranes were incubated with enhanced chemiluminescence (ECL; Pierce, Rockford, IL, USA) for chemiluminescence detection. The blots were scanned, and the band density was measured with Quantity One imaging software.
Statistical Analysis
The data were presented as mean ± SEM from at least three separate experiments. Statistical analysis was performed with SPSS 21.0 software (Abbott Laboratories, Chicago, IL, USA). Student’s t-test was used to analyze the difference between two groups. Chi-square test was employed to examine the relationships between miR-188 expression and clinicopathologic characteristics, and Pearson’s correlation analysis was calculated to estimate the correlation between miR-188 and β-catenin in glioma tissues. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-188 Is Frequently Reduced in Human Glioma Tissues and Glioma Cell Lines
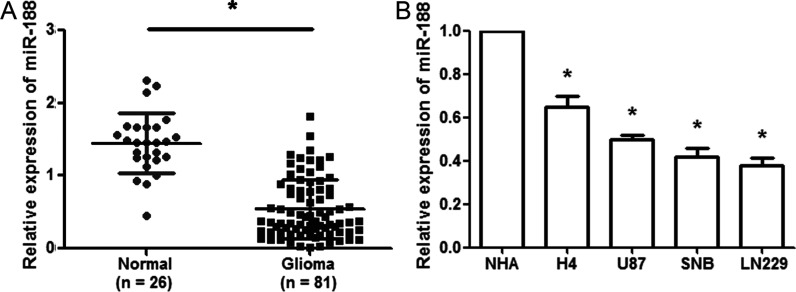
To explore the role of miR-188 in glioma, we performed real-time PCR to investigate its expression in 81 primary glioma samples and 26 normal brain tissues and cell lines. The real-time PCR assays showed that miR-188 expression was significantly lower in glioma tissues than in normal brain tissues (p < 0.01) (Fig. 1A). We chose the median value of miR-188 expression as a cutoff point (median value = 0.516): glioma tissues with a relative expression exceeding the median value were deemed to have high miR-188 expression with all other glioma tissues considered to have low miR-188 expression. Among 81 patients with gliomas, 53 (65.43%) were placed in the low miR-188 expression group and 28 (34.57%) were placed in the high miR-188 expression. Subsequently, we investigated the correlation of miR-188 expression with clinicopathological characteristics, including age, sex, tumor size, Karnofsky performance status (KPS) score, tumor location, and WHO grade. A significant correlation was observed between low miR-188 expression level and advanced WHO pathological grade of glioma (p < 0.01), high KPS (p < 0.01), as well as tumor size (p < 0.05). However, miR-188 expression level was not significantly associated with age, sex, or tumor location. In addition, miR-188 expression in glioma cell lines (H4, U87, SNB, and LN229) was downregulated compared with NHA cells (p < 0.01) (Fig. 1B). These data suggested that miR-188 might be a useful biomarker for malignant status of glioma.
Figure 1.
MicroRNA-188 (miR-188) is downregulated in glioma tissues and cell lines. (A) The qualitative real-time reverse transcription polymerase chain reaction (qRT-PCR) assay revealed that miR-188 expression was significantly decreased in glioma tissues (n = 81) compared with normal brain tissues (n = 26). (B) qRT-PCR was used to analyze miR-188 expression in the glioma cell lines H4, U87, SNB, and LN229 and the normal human astrocyte (NHA). *p < 0.01.
β-Catenin Is a Direct Target of miR-188
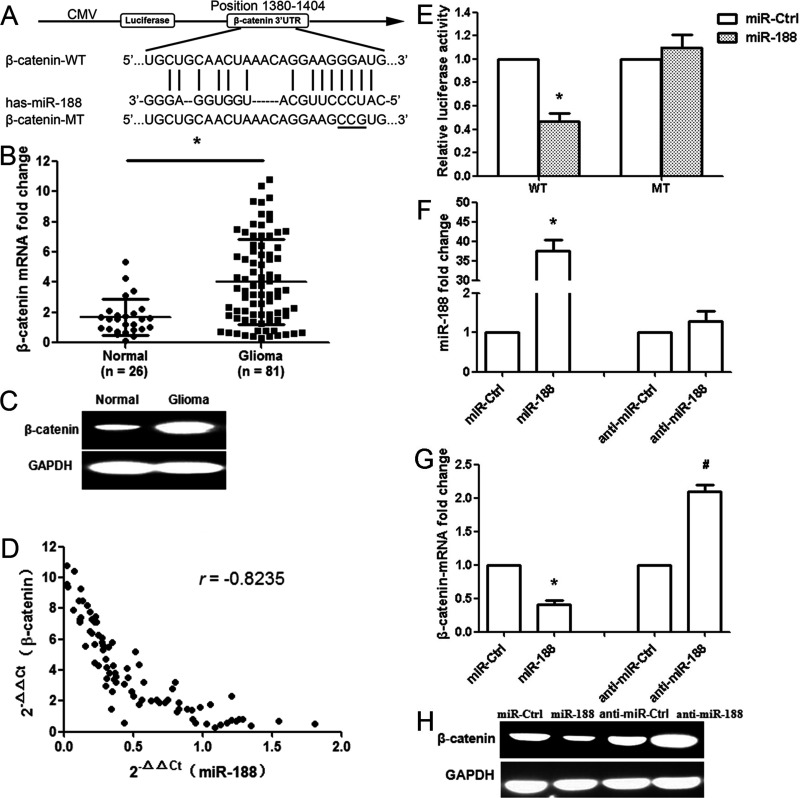
We used bioinformatic databases (PicTar and miRanda) to identify a large number of potential target genes of miR-188. Among these candidates, β-catenin was selected for further analysis. A binding site of miR-188 was observed in the 3′-UTR of β-catenin mRNA ranging from nucleotide 1380 to 1404 bp (Fig. 2A). Next, we examined β-catenin expression at the mRNA and protein levels. The results showed that the expression of β-catenin significantly increased at both levels in glioma tissues compared with the normal tissues (p < 0.01) (Fig. 2B and C). The effect of miR-188 on β-catenin was assessed based on the data obtained from the qRT-PCR. A significant inverse correlation was identified between β-catenin and miR-188 (n = 81, r = −0.8235, p < 0.001, Pearson’s correlation) (Fig. 2D). To validate the hypothesis that β-catenin might be a target of miR-188, a dual-luciferase reporter system containing wild type (WT) and mutant (MT) 3′-UTR of β-catenin was used. HEK293T cells were cotransfected with reporter plasmids and pre-miR-188 or pmirGLO control vector (miR-Ctrl). As a result, pre-miR-188/WT-β-catenin-UTR-transfected cells showed a significant reduction of luciferase activity (p < 0.01), and pre-miR-188/MT-β-catenin-UTR-transfected cells restored the relative luciferase activity (Fig. 2E), indicating that miR-188 may inhibit β-catenin expression by its binding sequences at the 3′-UTR. Because miR-188 expression was the lowest in LN229 cells, this cell line was used in subsequent experiments. The miR-188 expression was upregulated in transfected LN229 cells with pre-miR-188 vector compared with that in transfected cells with control vector (p < 0.01) (Fig. 2F). Overexpression of miR-188 significantly downregulated the mRNA and protein expression of β-catenin. Inhibition of miR-188 promoted the expression of β-catenin at both the mRNA and protein levels (p < 0.01) (Fig. 2G and H). These results indicate that miR-188 directly recognizes the 3′-UTR of β-catenin mRNA and inhibits β-catenin expression.
Figure 2.
miR-188 directly targets the β-catenin gene. (A) Bioinformatics predicted interactions of miR-188 and their binding sites at the 3′-untranslated region (3′-UTR) of β-catenin (PicTar and miRanda). CMV, cytomegalovirus; MT, mutant; WT, wild type. (B) β-Catenin mRNA expression in glioma (n = 81) and normal brain tissues (n = 26). (C) β-Catenin protein level was measured by Western blot. Relative optical density was normalized to glyceralde 3-phosphate dehydrogenase (GAPDH) to indicate protein content. (D) miR-188 and β-catenin levels were inversely correlated. 2−ΔΔCt values of miR-188 and β-catenin were subjected to a Pearson’s correlation analysis (n = 81, r = −0.8235, p < 0.001, Pearson’s correlation). (E) The luciferase reporter plasmid containing wild- or mutant-type β-catenin 3′-UTR was cotransfected into HEK293T cells in combination with miR-188 or miR-Ctrl. Luciferase activity was examined by the dual-luciferase assay. (F) The miR-188 expression was determined in glioma LN229 cells after miR-188 overexpression or anti-miR-188 treatment. (G) The β-catenin mRNA was determined after miR-188 overexpression or anti-miR-188 treatment. (H) The expression of β-catenin protein was analyzed by Western blot. GAPDH was used as control. *p < 0.01, compared with the miR-Ctrl group; #p < 0.01, compared with the anti-miR-Ctrl group, n = 3.
Overexpression of miR-188 Inhibits the Proliferation of Glioma LN229 Cells by Suppressing the β-Catenin Signaling Pathway
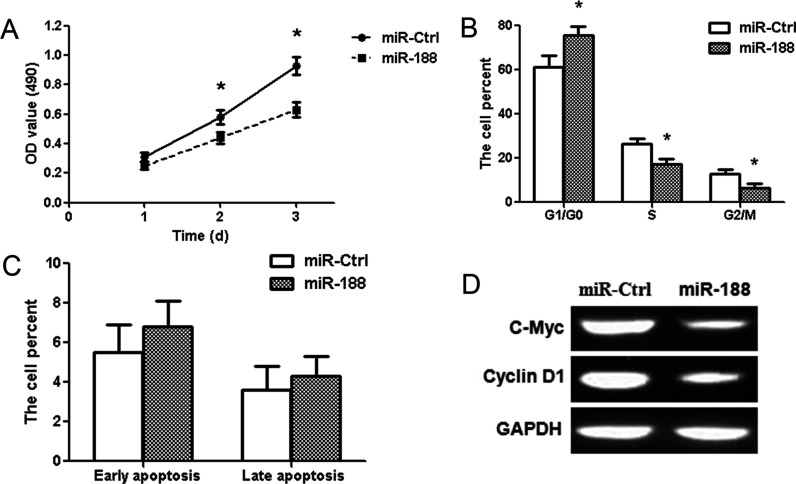
To explore the role of miR-188 in glioma, MTT assay, cell cycle analysis, and cell apoptosis analysis were adopted. The results showed that the transient overexpression of miR-188 led to the inhibition of the proliferation of LN229 cells at 2 and 3 days after transfection (p < 0.01) (Fig. 3A). Because the cell cycles are involved in the regulation of cell proliferation, we analyzed the processes with a flow cytometer. Our results revealed that cell cycles were arrested significantly at the G1/G0 stage in the miR-188 group (p < 0.01) (Fig. 3B). In addition, there was no significant difference among the proportion of early apoptosis and late apoptosis in the different treatment groups (Fig. 3C). To further investigate the possible molecular mechanisms of miR-188-induced cell proliferation repression, we examined the downstream pathway regulators of β-catenin by Western blot after transfection with the pre-miR-188 and miR-Ctrl vector. The results showed that miR-188 can reduce the expression of c-Myc and cyclin D1 proteins (Fig. 3D). These results suggest that miR-188 may suppress glioma cell proliferation and cell cycle by regulating β-catenin signaling pathway.
Figure 3.
Overexpression of miR-188 suppresses proliferation in glioma LN229 cells. (A) MTT assay showed that miR-188 overexpression reduced the cell activity at 2 and 3 days. (B) Cell cycle was detected in LN229 cells 2 days after transfection by propidium iodide (PI) staining flow cytometry. Histogram represented the percentage of cells in the G0/G1, S, and G2/M phases. (C) Cell apoptosis was visualized using annexin V/PI staining. The data showed the percentages of early apoptosis and late apoptosis. (D) The expressions of c-Myc and cyclin D1 proteins were measured by Western blot in the miR-188 group. *p < 0.01, compared with the miR-Ctrl group, n = 3.
Inhibition of miR-188 Promotes Proliferation in Glioma LN229 Cells via Regulating β-Catenin Signaling Pathway
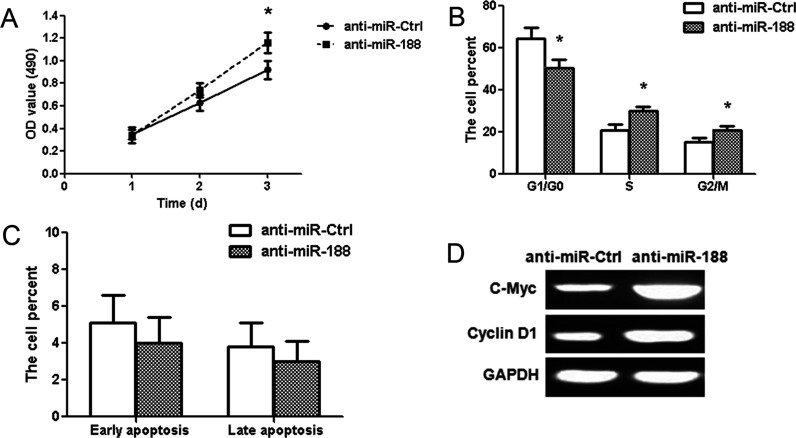
Human glioma LN229 cells were transfected with miR-188 antisense oligonucleotides (anti-miR-188). The MTT assay showed that anti-miR-188 promoted the proliferation of glioma LN229 cells (p < 0.01) (Fig. 4A). The number of S and G2/M phase cells significantly increased in the anti-miR-188 group compared with the anti-miR-Ctrl group; however, the number of G1/G0 phase cells remarkably decreased (p < 0.01) (Fig. 4B). There was no significant difference among the proportion of early and late apoptosis in the different treatment groups (Fig. 4C). c-Myc and cyclin D1 protein expressions upregulated in transfected cells with anti-miR-188 (Fig. 4D).
Figure 4.
Inhibition of miR-188 promotes cell proliferation in glioma LN229 cells. (A) MTT assay showed that anti-miR-188 increased the cell activity at 3 days. (B) Cell cycle analysis showed the percentage of cells in the G1/G0, S, and G2/M phases. S and G2/M phase cells significantly increased in the anti-miR-188 group. (C) The data showed the percentage of early and late apoptosis in the anti-miR-188 group. (D) c-Myc and cyclin D1 protein expressions were examined in the anti-miR-188 group. *p < 0.01, compared with the anti-miR-Ctrl group, n = 3.
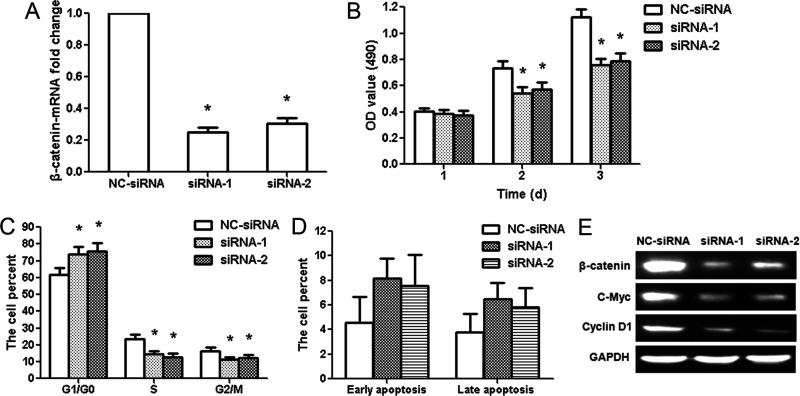
Knockdown of β-Catenin Suppresses Glioma LN229 Cell Proliferation
miR-188 regulated cell proliferation and cell cycle in glioma cells, and β-catenin was validated as a direct target of miR-188. Thus, we knocked down β-catenin expression in glioma LN229 cells by RNA interference (RNAi) to confirm its involvement in the antitumor effects of miR-188. β-Catenin mRNA expression was specifically knocked down by siRNA-1 or siRNA-2 (p < 0.01) (Fig. 5A). Silencing of β-catenin significantly decreased the cell activity at 2 and 3 days after transfection (p < 0.01) (Fig. 5B). The cell cycles were arrested significantly at the G1/G0 phases in the siRNA-1 or siRNA-2 group. The number of S and G2/M phase cells significantly decreased in the siRNA-1 or siRNA-2 group (p < 0.01) (Fig. 5C). There was no significant change among the proportion of early and late apoptosis in different groups (Fig. 5D). These were similar to those of miR-188 overexpression, indicating a similar effect of β-catenin knockdown and miR-188 overexpression. In addition, we analyzed knockdown efficiency of β-catenin siRNA at the protein level. The protein expression of β-catenin decreased significantly in the siRNA-1 or siRNA-2 group compared to the NC-siRNA group; c-Myc and cyclin D1 protein expressions were also reduced (Fig. 5E).
Figure 5.
β-Catenin small interfering RNAs (siRNAs) inhibit the proliferation of glioma LN229 cells. (A) qRT-PCR results showed the knockdown efficiency of β-catenin siRNAs in LN229 cells. (B) MTT assay showed that β-catenin siRNAs decreased the activity of LN229 cells at 2 and 3 days. (C) Flow cytometry analysis showed the percentage of cells in the G1/G0, S, and G2/M phases. G1/G0 phase cells increased after β-catenin siRNA treatment, whereas S and G2/M phase cells decreased. (D) The data showed the percentage of early and late apoptosis after β-catenin siRNA treatment. (E) β-Catenin, c-Myc, and cyclin D1 protein expressions were examined after β-catenin siRNA treatment. *p < 0.01, compared with the NC-siRNA group, n = 3.
DISCUSSION
In the last decade, growing evidence has confirmed the important roles of miRNAs in glioma23,24. Although the miRNA signatures have been well characterized in glioma, the role of dysregulated miRNAs on glioma progression and development remains largely unknown. Identifying miRNAs and elucidating their precise biological functions in glioma will aid the search for novel targets for diagnosis and therapy of the disease. It is reported that miR-188 regulates the progression of several human cancers, including oral carcinoma, nasopharyngeal carcinoma, prostate carcinoma, and hepatocellular carcinoma20–22,25. During the course of our study, Ding and colleagues reported that miR-188 may act as a tumor suppressor in glioma by directly targeting the insulin-like growth factor 2-binding protein 2 (IGF2BP2) gene26. However, the clinical significance and function of miR-188 in glioma were not entirely clear. In this study, we further verify that miR-188 expression is frequently downregulated in both glioma tissues and cell lines. The clinicopathological significance of miR-188 expression was also analyzed. We found that low miR-188 levels are significantly associated with advanced WHO pathological grade of glioma, high KPS, and tumor size. No significant correlation is identified between miR-188 expression and other clinicopathological characteristics. Our experiment demonstrates that miR-188 suppresses glioma cell proliferation by blocking G1–S transition in vitro. These findings indicate that miR-188 plays an important role in glioma development and progression.
In this study, we provide the first evidence that miR-188 suppresses glioma cell proliferation by targeting a novel miR-188 target, β-catenin. β-Catenin was postulated to be a target of miR-188 using two different databases. As indicated on reporter assaying, miR-188 repressed the construct with the β-catenin 3′-UTR. Overexpression of miR-188 suppressed β-catenin 3′-UTR luciferase activity, and this effect was abolished by mutation of the miR-188 seed binding site; there is an inverse correlation between miR-188 and β-catenin expression in glioma tissues; and miR-188 overexpression repressed expression of the β-catenin mRNA and protein in glioma cell. These results indicate that miR-188 may play the role of a negative regulator or tumor suppressor for the cell growth partly mediated by repressing β-catenin expression. The wingless-related integration site (Wnt)/β-catenin pathway is an evolutionarily conserved signaling pathway, which is important in initiating and regulating a diverse range of biological process, including embryogenesis, carcinogenesis, calcium homeostasis, cell proliferation, apoptosis, cell polarity, and so on27–31. For example, activation of the Wnt/β-catenin signaling pathway can promote the proliferation of embryonic, skin, intestinal, and neural stem cells, and induce the self-renewal and differentiation of stem-like cells32–37. Our results showed that knockdown of β-catenin suppressed glioma cell proliferation. There are a number of downstream target proteins in the Wnt/β-catenin signaling pathway, including c-Myc and cyclin D138. The results demonstrated that miR-188 may have specific effects on the Wnt/β-catenin signal pathway by targeting β-catenin. To investigate these effects of miR-188 on the Wnt/β-catenin pathway, c-Myc and cyclin D1 protein expression levels were measured using Western blot analysis. The results showed that miR-188 overexpression may reduce the expression of c-Myc and cyclin D1, and miR-188 inhibition may promote their expression. In brief, miR-188 suppresses glioma cell proliferation by directly targeting β-catenin.
The first gap (G1) phase of the cell cycle is a crucial stage when cells respond to environmental signals to determine their cell fate such as cell survival, proliferation, and cellular senescence. Important cell cycle regulators include cyclin A-cyclin-dependent kinase 2 (CDK2) and cyclin D-CDK4/6 protein kinase complexes, which regulate the cellular progression through the G1/G0-to-S phase of the cell cycle39. CDK4 and CDK6 are stably expressed in a cell cycle-independent manner, but the expression of cyclins (D1, D2, and D3) fluctuates during the cell cycle, indicating that different D-type cyclins play important roles in the regulation of cell cycle variations40. After the extracellular mitogenic stimulation, D-cyclins can lead to the release of the E2F transcription factors and drive cell entry into the S phase of the cell cycle. It was reported that cyclin D1 is involved in human tumorigenesis41. In this experiment, we demonstrate that miR-188 overexpression and β-catenin siRNA may suppress the expression of cyclin D1 and induce G1 phase cell cycle arrest, while anti-miR-188 may promote the expression of cyclin D1 and lead more cells into the S and G2/M phases. These results suggest that miR-188 may inhibit the expression of cyclin D1 and induce G1 phase cell cycle arrest by targeting β-catenin.
In conclusion, our study reports a tumor suppressor role for miR-188 in glioma. We find that miR-188 is downregulated in glioma and demonstrates that miR-188 suppresses glioma cell proliferation, decreases cyclin D1 expression, and induces G1 phase cell cycle arrest by targeting β-catenin. These findings add to our understanding of the molecular pathogenesis of glioma and provide an effective therapeutic strategy for glioma.
ACKNOWLEDGMENTS
This study was supported by Xi’an City’s Science and Technology Projects (No. 2017115SF/YX009).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Hollon T, Hervey-Jumper SL, Sagher O, Orringer DA. Advances in the surgical management of low-grade glioma. Semin Radiat Oncol. 2015;25:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang C, Bao Z, Zhang W, Jiang T. Progress on molecular biomarkers and classification of malignant gliomas. Front Med. 2013;7:150–6. [DOI] [PubMed] [Google Scholar]
- 3. Huang J, Chen H, Wei Q, Zhang Z, Zhong Z, Xu Y. Downregulation of LKB1 promotes tumor progression and predicts unfavorable prognosis inpatients with glioma. Oncol Lett. 2017;13:1688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo E, Liu H, Liu X. Overexpression of SCUBE2 inhibits proliferation, migration, and invasion in glioma cells. Oncol Res. 2017;25:437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhattacharya K, Maiti S, Mandal C. PTEN negatively regulates mTORC2 formation and signaling in grade IV glioma via Rictor hyperphosphorylation at Thr1135 and direct the mode of action of an mTORC1/2 inhibitor. Oncogenesis 2016;5:e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duran CL, Lee DW, Jung JU, Ravi S, Pogue CB, Toussaint LG, Bayless KJ, Sitcheran R. NIK regulates MT1-MMP activity and promotes glioma cell invasion independently of the canonical NF-κB pathway. Oncogenesis 2016;5:e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenall SA, Lim YC, Mitchell CB, Ensbey KS, Stringer BW, Wilding AL, O’Neill GM, McDonald KL, Gough DJ, Day BW, Johns TG. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis 2017;6:e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cano A, Gomez R. Mir-661: A key factor in embryo-maternal dialog with potential clinical application to predict implantation outcome? EBioMedicine 2015;2:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Danilenko M, Studzinski GP. Keep harm at bay: Oxidative phosphorylation induces Nrf2-driven antioxidant response via ERK5/MEF2/miR-23a signaling to Keap-1. EBioMedicine 2016;3:4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gallo A, Jang SI, Ong HL, Perez P, Tandon M, Ambudkar I, Illei G, Alevizos I. Targeting the Ca(2+) sensor STIM1 by exosomal transfer of Ebv-miR-BART13-3p is associated with Sjögren’s syndrome. EBioMedicine 2016;10:216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao LY, Tong DD, Xue M, Ma HL, Liu SY, Yang J, Liu YX, Guo B, Ni L, Liu LY, Qin YN, Wang LM, Zhao XG, Huang C. MeCP2, a target of miR-638, facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Oncogenesis 2017;6:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Li C, Li H, Song Y, Zhao Y, Zhai L, Wang H, Zhong R, Tang H, Zhu D. miR-378 activates the pyruvate-pep futile cycle and enhances lipolysis to ameliorate obesity in mice. EBioMedicine 2016;5:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao Y, Zhang L, Wei M, Jiang X, Jia D. MicroRNA-409-3p represses glioma cell invasion and proliferation by targeting high-mobility group nucleosome-binding domain 5. Oncol Res. 2017;25:1097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang J, Yang J, Lei Y, Gao H, Wei T, Luo L, Zhang F, Chen H, Zeng Q, Guo L. An ANCCA/PRO2000-miR-520a-E2F2 regulatory loop as a driving force for the development of hepatocellular carcinoma. Oncogenesis 2016;5:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu L, Wang Y, Bai R, Yang K, Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis 2016;5:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan Z, Cui H, Yu H, Ji Q, Kang L, Han B, Wang J, Dong Q, Li Y, Yan Z, Yan X, Zhang X, Lin Z, Hu Y, Jiao S. MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis 2016;5:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q, Navitskaya S, Chakravarthy H, Huang C, Kady N, Lydic TA, Chen YE, Yin KJ, Powell FL, Martin PM, Grant MB, Busik JV. Dual anti-inflammatory and anti-angiogenic action of miR-15a in diabetic retinopathy. EBioMedicine 2016;11:138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu J, Lin H, Li G, Sun Y, Chen J, Shi L, Cai X, Chang C. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine 2016;12:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J, Wei J, Wu Y, Wang Z, Guo Y, Lee P, Li X. Metformin induces ER stress-dependent apoptosis through miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis 2015;4:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37:4105–13. [DOI] [PubMed] [Google Scholar]
- 21. Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, Huang N, Xie W, Xu N, Zhang Y. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo J, Wang Y, Xu Y. miR-188-5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget 2015;6:6092–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hummel R, Maurer J, Haier J. MicroRNAs in brain tumors: A new diagnostic and therapeutic perspective? Mol Neurobiol. 2011;44:223–34. [DOI] [PubMed] [Google Scholar]
- 24. Palumbo S, Miracco C, Pirtoli L, Comincini S. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol. 2014;229:277–86. [DOI] [PubMed] [Google Scholar]
- 25. Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–85. [DOI] [PubMed] [Google Scholar]
- 26. Ding L, Wang L, Guo F. microRNA-188 acts as a tumour suppressor in glioma by directly targeting the IGF2BP2 gene. Mol Med Rep. 2017;16(5):7124–30. [DOI] [PubMed] [Google Scholar]
- 27. Zhao L, Liu Y, Tong D, Qin Y, Yang J, Xue M, Du N, Liu L, Guo B, Hou N, Han J, Liu S, Liu N, Zhao X, Wang L, Chen Y, Huang C. MeCP2 promotes gastric cancer progression through regulating FOXF1/Wnt5a/β-Catenin and MYOD1/Caspase-3 signaling pathways. EBioMedicine 2017;16:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohtsuka M, Ling H, Ivan C, Pichler M, Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X, Chen M, Vidhu F, Bartholomeusz GA, Toiyama Y, Kusunoki M, Doki Y, Mori M, Song S, Gunther JR, Krishnan S, Slaby O, Goel A, Ajani JA, Radovich M, Calin GA. H19 noncoding RNA, an independent prognostic factor, regulates essential Rb-E2F and CDK8-β-catenin signaling in colorectal cancer. EBioMedicine 2016;13:113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su YJ, Chang YW, Lin WH, Liang CL, Lee JL. An aberrant nuclear localization of E-cadherin is a potent inhibitor of Wnt/β-catenin-elicited promotion of the cancer stem cell phenotype. Oncogenesis 2015;4:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lefèvre L, Omeiri H, Drougat L, Hantel C, Giraud M, Val P, Rodriguez S, Perlemoine K, Blugeon C, Beuschlein F, de Reyniès A, Rizk-Rabin M, Bertherat J, Ragazzon B. Combined transcriptome studies identify AFF3 as a mediator of the oncogenic effects of β-catenin in adrenocortical carcinoma. Oncogenesis 2015;4:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mohammed MK, Shao C, Wang J, Wei Q, Wang X, Collier Z, Tang S, Liu H, Zhang F, Huang J, Guo D, Lu M, Liu F, Liu J, Ma C, Shi LL, Athiviraham A, He TC, Lee MJ. Wnt/β-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumori-genesis and cancer chemoresistance. Genes Dis. 2016;3:11–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee DK, Nathan Grantham R, Trachte AL, Mannion JD, Wilson CL. Activation of the canonical Wnt/beta-catenin pathway enhances monocyte adhesion to endothelial cells. Biochem Biophys Res Commun. 2006;347:109–16. [DOI] [PubMed] [Google Scholar]
- 33. Sinnberg T, Makino E, Krueger MA, Velic A, Macek B, Rothbauer U, Groll N, Pötz O, Czemmel S, Niessner H, Meier F, Ikenberg K, Garbe C, Schittek B. A nexus consisting of beta-catenin and Stat3 attenuates BRAF inhibitor efficacy and mediates acquired resistance to vemurafenib. EBioMedicine 2016;8:132–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon JH, Eun JW, Choi WS, Kim O, Nam SW, Lee JY, Park WS. NKX6.3 is a transcription factor for Wnt/β-catenin and Rho-GTPase signaling-related genes to suppress gastric cancer progression. EBioMedicine 2016;9:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osawa Y, Oboki K, Imamura J, Kojika E, Hayashi Y, Hishima T, Saibara T, Shibasaki F, Kohara M, Kimura K. Inhibition of cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB)-binding protein (CBP)/β-catenin reduces liver fibrosis in mice. EBioMedicine 2015;2:1751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moor AE, Anderle P, Cantù C, Rodriguez P, Wiedemann N, Baruthio F, Deka J, André S, Valenta T, Moor MB, Győrffy B, Barras D, Delorenzi M, Basler K, Aguet M. BCL9/9L-β-catenin signaling is associated with poor outcome in colorectal cancer. EBioMedicine 2015;2:1932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Q, Yang D, Zong H, Zhu L, Wang L, Wang X, Zhu X, Song X, Wang J. Growth-induced stress enhances epithelial-mesenchymal transition induced by IL-6 in clear cell renal cell carcinoma via the Akt/GSK-3β/β-catenin signaling pathway. Oncogenesis 2017;6:e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao J, Yan XR, Liu T, Han XB, Yu JJ, Liu SH, Wang LB. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH 1 via the Wnt/β-catenin signaling pathway in colorectal cancer. Oncol Lett. 2017;14:3795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao LY, Zhang J, Guo B, Yang J, Han J, Zhao XG, Wang XF, Liu LY, Li ZF, Song TS, Huang C. MECP2 promotes cell proliferation by activation ERK1/2 and inhibiting p38 activity in human hepatocellular carcinoma HEPG2 cells. Cell Mol Biol. (Noisy-le-grand) 2013;Suppl 59:OL1876–81. [PubMed] [Google Scholar]
- 40. Zhao LY, Yao Y, Han J, Yang J, Wang XF, Tong DD, Song TS, Huang C, Shao Y. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci. 2014;59:1743–53. [DOI] [PubMed] [Google Scholar]
- 41. Chu X, Zhang T, Wang J, Li M, Zhang X, Tu J, Sun S, Chen X, Lu F. Alternative splicing variants of human Fbx4 disturb cyclin D1 proteolysis in human cancer. Biochem Biophys Res Commun. 2014;447:158–64. [DOI] [PubMed] [Google Scholar]