Abstract
Cholangiocarcinoma (CCA) is one of the most malignant adenocarcinomas arising from bile duct epithelial cells. However, the molecular mechanism regulating CCA development and progression still needs to be investigated. Here we found that miR-365 was downregulated in CCA tissues compared with adjacent normal tissues. By functional experiments, we found that overexpression of miR-365 significantly inhibited CCA cell proliferation and promoted cellular apoptosis in vitro. Furthermore, administration with miR-365 markedly suppressed the growth of tumor tissues in vivo. Mechanistically, we identified E2F2 as the target gene of miR-365 in CCA cells. We found that overexpression significantly inhibited the expression of E2F2 in CCA cells, and there was an inverse correlation between the expression levels of E2F2 and miR-365 in CCA tissues. We also found that E2F2 was highly expressed in CCA tissues and cell lines. Restoration of E2F2 in miR-365-overexpressing CCA cells promoted cell viability and reduced cellular apoptosis in CCA. Collectively, our study demonstrated the essential role of miR-365 and its functional mechanism in CCA cells, which provided a new insight on the design of therapeutic targets for CCA treatment.
Key words: Cholangiocarcinoma (CCA), miR-365, Proliferation, Apoptosis, E2F transcription factor 2 (E2F2)
INTRODUCTION
Cholangiocarcinoma (CCA), one of the most invasive and metastatic cancers, gives rise to large numbers of cancer-related deaths every year around the world1. The incidence and motility are markedly increasing each year worldwide2. Nowadays, the main effective curative method seems to be surgical excision for early stage patients with CCA3. Unfortunately, the recurrence rate and clinical outcomes of CCA patients are very high and poor, respectively, because of tumor invasion and metastasis4. Thus, determination of the molecular mechanism regulating the carcinogenesis, progression, and metastasis of CCA is urgently required.
In recent decades, the research of oncogenes and microRNAs (miRNAs) has been helpful to further discover the pathogenesis of CCA and new molecular targeted therapy. miRNAs are a group of small (19 to 25 nts) endogenous noncoding RNA molecules, which act as gene regulators by binding to partially complementary target sites in mRNA 3′-untranslated regions (3′-UTRs) that results in the degradation of the target mRNAs or translational repression of the encoded proteins5. miRNAs have been demonstrated to be involved in various biological processes, such as proliferation, apoptosis, and differentiation6–8. Dysregulation of miRNAs is closely related with human diseases, especially tumorigenesis. For instance, a previous study reported that miR-144 was reduced in CCA tissues; miR-144 may be an essential suppressor of CCA cell proliferation and invasion through targeting lissencephaly 1 protein (LIS1; platelet-activating factor acetylhydrolase 1b regulatory subunit 1)9. Wang et al. reported that downregulation of miR-138 enhanced the proliferation, migration, and invasion of CCA cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-910. Liu et al. indicated that miR-122 overexpression inhibited the proliferation and invasion, and stimulated apoptosis of CCA cells11. Therefore, understanding the mechanism by which miRNAs participate in CCA progression will benefit the development of effectively curative methods for CCA.
miR-365 has been reported to inhibit tumor growth in a diversity of human cancers. For example, miR-365 induces hepatocellular carcinoma cell apoptosis12. Wang et al. showed that miR-365 inhibits ovarian cancer progression13. Additionally, Bai et al. reported that miR-365 inhibits growth, invasion, and metastasis of malignant melanoma14. However, the function of miR-365 in CCA is still unclear. In our study, we found that the expression of miR-365 was significantly downregulated in CCA tissues compared with adjacent normal tissues. Low expression of miR-365 in CCA patients indicated poor prognosis. Furthermore, we found that overexpression of miR-365 markedly inhibited the proliferation but induced cell apoptosis. Mechanistically, we found that miR-365 targeted E2F transcription factor 2 (E2F2), an essential transcription factor in many biological processes, in CCA cells. By inhibiting the expression of E2F2, miR-365 suppressed tumor growth in CCA.
MATERIALS AND METHODS
Patient and Tissue Samples
Forty-nine paired CCA tissues were collected from the Xinjiang Uygur Autonomous Region People’s Hospital (Urumqi, P.R. China). Tissue samples were snap frozen in liquid nitrogen immediately after surgical resection and stored at −80°C; tissue samples were collected after obtaining written informed consent from all patients. The study was approved by the Institutional Review Board of the Xinjiang Uygur Autonomous Region People’s Hospital. No patients who had received radiotherapy and/or immunotherapy before or after surgical treatment were included in this study. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Xinjiang Uygur Autonomous Region People’s Hospital. All patients enrolled in the study gave written informed consent.
Cell Lines and Culture
CCA cell lines including noncancerous cholangiocyte cell line (H69) and CCA cell lines (SG231, CCALP1, HuCCAT1, and TFK1) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, P.R. China). All cells were cultured in RPMI-1640 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and grown in humidified 5% CO2 at 37°C. miR-365 mimic and relative controls were obtained from GenePharma (Shanghai, P.R. China). The transfection was conducted using Lipofectamine 2000 (Invitrogen).
Quantitative Real-Time PCR
Total RNA was extracted from CCA tissues and noncancerous cholangiocyte cells using the TRIzol reagent (Invitrogen Corp.) according to the manufacturer’s instructions. For miRNA analysis, quantitative real-time PCR (RT-qPCR) was performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and the corresponding primers. For mRNA analysis, RT-qPCR was performed using the TaqMan High-Capacity cDNA Reverse Transcription Kit, TaqMan Fast PCR Master Mix (Applied Biosystems) and the corresponding primers. RT-qPCRs were performed in triplicate on a RealPlex4 real-time PCR detection system.
Cell Proliferation Assay
Cells were seeded at 5,000 cells per well in 96-well plates at 24 h after transfection. Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) at 24, 48, and 72 h after the cells were seeded. Absorbance was determined at 450 nm using a microplate spectrophotometer (Thermo Labsystems, Vantaa, Finland).
Cell Cycle Analysis
Cells were harvested at 48 h after transfection. The cells were washed with PBS and fixed in ethanol at −20°C. The cells were then washed with PBS, rehydrated, and resuspended in propidium iodide (PI)–RNase A solution (Sigma-Aldrich; Merck KGaA) at 37°C for 30 min. The stained cells (1 × 105) were then analyzed for DNA content with a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Western Blot
Cells were collected and lysed using RIPA protein extraction reagent (Beyotime, Beijing, P.R. China) supplemented with a protease inhibitor cocktail (Roche, Belmont, CA, USA). Cell protein lysates were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto 0.22-μm NC membranes (Sigma-Aldrich). The membranes were incubated with specific primary antibodies. Membranes were washed three times with TBST, and then secondary antibodies (anti-rabbit) at dilutions of 1:5,000 were added. The protein bands were visualized by enhanced chemiluminescence (ECL) following the manufacturer’s instruction (Santa Cruz Biotechnology, Santa Cruz, CA, USA). GAPDH (Proteintech, Rosemont, IL, USA) served as the loading control.
Cell Apoptosis Analysis
Cells were harvested, washed with ice-cold PBS, and stained with Annexin-V-fluorescein isothiocyanate (FITC) apoptosis detection kits (KeyGEN Biotech, Nanjing, P.R. China). Cell apoptosis was analyzed in a flow cytometer (BD Biosciences).
Luciferase Activity Assay
The wild-type 3′-UTR of E2F2 mRNA was amplified from a human cDNA library. Mutations of the miR-365 binding site were introduced by site-directed mutagenesis. The PCR fragment was cloned into psiCHECK-2 vector (Promega, Madison, WI, USA) downstream of the firefly luciferase coding region. psiCHECK-2-control was used as internal control.
In Vivo Tumorigenicity
BALB/C athymic nude mice were housed under specific pathogen-free conditions. CCALP1 cells (2 × 106) transfected with miR-365 mimics or controls were injected into hind limbs of mice to generate xenograft tumors. Each group contained seven mice. Tumor size was measured and tumor volume was determined at indicative time points.
Statistical Analysis
Data were expressed as the mean ± standard deviation (SD) from at least three independent experiments. Differences were analyzed using the Student’s t-test for two group comparisons and one-way ANOVA for multiple group comparisons. A value of p < 0.05 or p < 0.01 was considered statistically significant.
RESULTS
miR-365 Was Downregulated in Human Cholangiocarcinoma Tissues
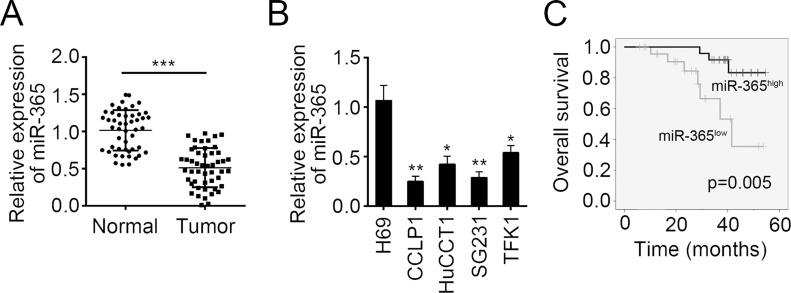
To explore the function of miR-365, we determined its expression pattern in CCA tissues. By RT-PCR, we found that miR-365 was significantly decreased in CCA tissues compared to adjacent normal tissues (Fig. 1A). Furthermore, we examined the expression of miR-365 in CCA cell lines. We found that miR-365 was markedly downregulated in all selected CCA cell lines compared to H69 cells (Fig. 1B). To further investigate the clinicopathological significance of miR-365 expression in patients with CCA, 49 patients were divided into two subgroups based on the median value: low miR-365 group (25 cases) and high miR-365 group (24 cases). Then we performed Kaplan–Meier survival analysis and found that lower expression of miR-365 in CCA patients indicated poorer prognosis (Fig. 1C).
Figure 1.
MicroRNA-365 (miR-365) was downregulated in human cholangiocarcinoma tissues. (A) Quantitative real-time PCR (RT-qPCR) analysis indicated that miR-365 was downregulated in cholangiocarcinoma (CCA) tissues (n = 49) compared to adjacent normal tissues (n = 49). (B) RT-qPCR analysis for miR-365 expression in a human cholangiocyte cell line (H69) and four human CCA cell lines (CCALP1, HuCCAT1, SG231, and TFK1). (C) CCA patients with higher expression of miR-365 showed better prognosis as shown by Kaplan–Meier survival analysis. Data are presented as means ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
miR-365 Overexpression Suppressed CCA Cell Proliferation but Induced Cell Apoptosis
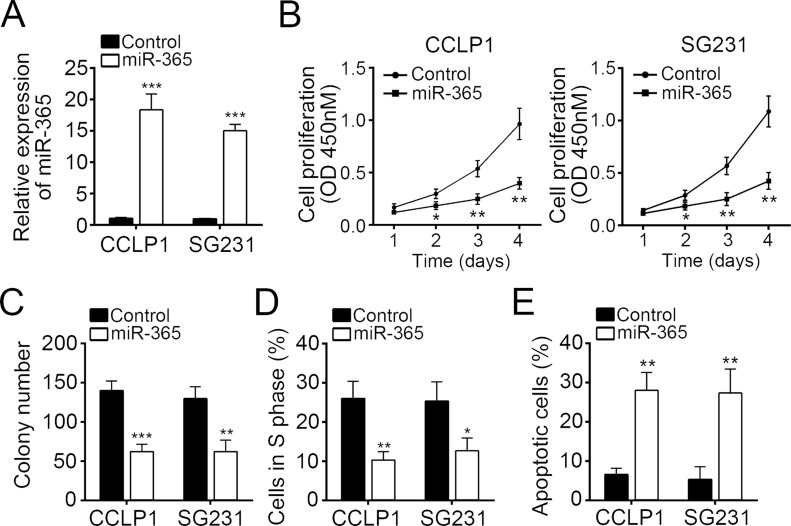
Then we overexpressed miR-365 in CCALP1 and SG231 cells by transducing a miR-365 mimic (Fig. 2A). Through CCK-8 proliferation assay, we found that overexpression of miR-365 significantly inhibited the cellular proliferation in CCALP1 and SG231 cells (Fig. 2B). Furthermore, overexpression of miR-365 markedly reduced the colony formation by CCALP1 and SG231 cells (Fig. 2C). To further explore the mechanism that miR-365 inhibited cell proliferation, we examined the effect of miR-365 on the cell cycle. By FACS analysis, we found that overexpression of miR-365 significantly decreased the cells in the S phase (Fig. 2D). We next explored cell apoptosis. By staining with annexin V/PI, we showed that overexpression of miR-365 remarkably promoted the rate of apoptotic CCALP1 and SG231 cells (Fig. 2E). In other words, our data demonstrated that overexpression of miR-365 significantly inhibited cell proliferation but promoted apoptosis in CCA.
Figure 2.
miR-365 overexpression suppressed CCA cell proliferation but induced cell apoptosis. (A) CCALP1 and SG231 cells were transfected with miR-365 mimic to overexpress miR-365. Cell proliferation was determined in CCALP1 and SG231 cells transfected with miR-365 mimic or controls by (B) cell counting kit-8 (CCK-8) assay and (C) colony formation assay. (D) Percentages of cells in the S phase were determined by FACS. (E) Cell apoptosis was measured in CCALP1 and SG231 cells transfected with miR-365 mimic or controls by staining with annexin V/propidium iodide (PI). Data are presented as means ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001.
miR-365 Inhibited Tumor Growth In Vivo
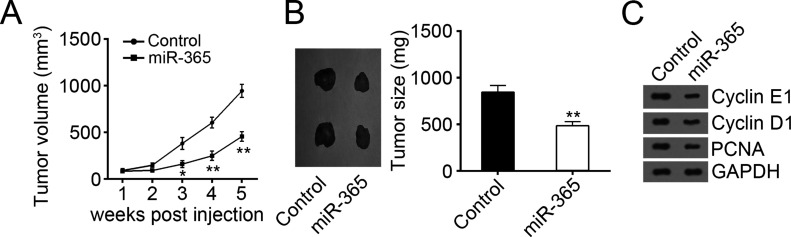
To further confirm the effect of miR-365 on cell proliferation, we conducted xenograft experiments. We injected CCALP1 cells transfected with miR-365 mimic into nude recipients. Every week, we measured tumor volumes and found that overexpression of miR-365 significantly inhibited tumor growth (Fig. 3A). After 5 weeks, we sacrificed these mice and checked tumor size. We found that overexpression of miR-365 remarkably suppressed the tumor weights (Fig. 3B). Then we analyzed cell cycle-related proteins (cyclin D1, cyclin E1, and PCNA) in the formed tumor tissues by Western blot. We found that overexpression of miR-365 inhibited the expression of these proteins (Fig. 3C).
Figure 3.
miR-365 inhibited tumor growth in vivo. (A) Tumor growth curves were established by measuring tumor volume every week for 5 weeks after injection. (B) Tumor weights isolated from nude mice in each treatment group were measured on week 5 after injection. (C) Protein levels of cyclin D1, cyclin E1, and PCNA in xenograft tumor tissues were determined by Western blot. GAPDH served as a loading control. Data are presented as means ± SD. *p < 0.05 and **p < 0.01.
E2F2 Was a Target of miR-365
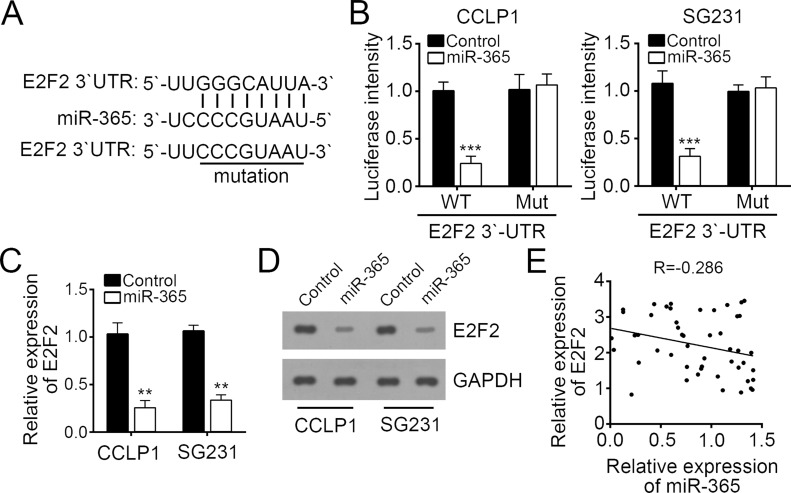
To explore the molecular mechanism, we predicted the target mRNAs of miR-365 by computational algorithms including TargetScan and miRanda. We found that E2F2 was the most potential target gene of miR-365. There was a potential interactive site in the 3′-UTR of E2F2 mRNA (Fig. 4A). To further validate the interaction between miR-365 and E2F2 mRNA, we performed dual-luciferase reporter assays. We found that overexpression of miR-365 significantly reduced the luciferase intensity in CCALP1 and SG231 cells transfected with E2F2-wt (Fig. 4B). Mutation of this binding site in E2F2 mRNA totally abrogated this inhibitory effect on luciferase intensity (Fig. 4B). Furthermore, we overexpressed miR-365 in CCALP1 and SG231 cells and found that overexpression of miR-365 significantly decreased the mRNA and protein levels of E2F2 (Fig. 4C and D). Finally, by Spearman’s rank test, we found that there was an inverse correlation between the expressions of miR-365 and E2F2 in CCA tissues (Fig. 4E).
Figure 4.
E2F2 was a target of miR-365. (A) The miR-365 binding site in the 3′-untranslated region (3′-UTR) region of E2F2 mRNA was predicted by bioinformatics analysis. (B) Luciferase reporter assays were performed using CCALP1 and SG231 cells cotransfected with the miR-365 mimic or control and E2F2-wt or E2F2-mut 3′-UTR reporter plasmid. Analysis of (C) E2F2 mRNA and (D) protein levels in CCALP1 and SG231 cells transfected with miR-365 mimic or controls by RT-qPCR. (E) Spearman’s correlation analysis was used to determine the correlations between the levels of miR-365 and E2F2 in human CCA tissues (n = 49). Data are presented as means ± SD. **p < 0.01 and ***p < 0.001.
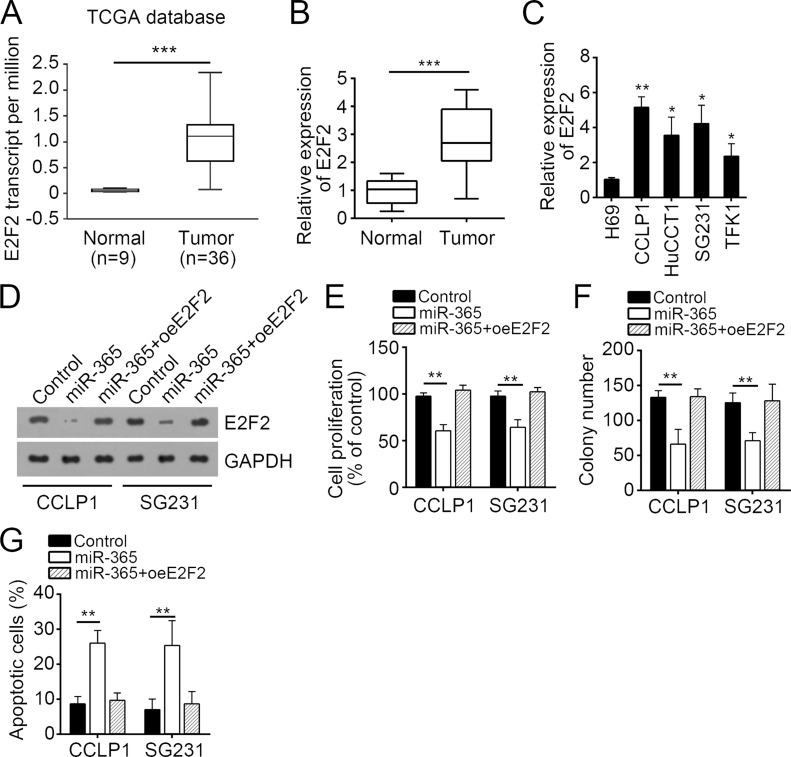
Restoration of E2F2 Rescued CCA Cell Proliferation and Reduced Apoptosis
The function of E2F2 has not been defined in CCA. To further determine the function of E2F2, we analyzed its expression in CCA tissues. According to the TCGA database, we found that E2F2 was significantly upregulated in CCA tissues compared to normal tissues (Fig. 5A). Furthermore, by RT-qPCR, we found that E2F2 was dramatically upregulated in 49 CCA tissues compared to paired normal tissues (Fig. 5B). The expression of E2F2 displayed a similar trend in CCA cell lines (Fig. 5C). To determine the role of E2F2 in CCA cells, we restored the protein levels in miR-365-overexpressing CCALP1 and SG231 cells (Fig. 5D). Then we performed CCK-8 assays. We found that overexpression of miR-365 significantly inhibited cell proliferation and colony formation, while restoration of E2F2 rescued CCALP1 and SG231 cell proliferation and colony formation (Fig. 5E and F). Furthermore, restoration of E2F2 also reversed the enhanced apoptosis of CCALP1 and SG231 cells mediated by overexpression of miR-365 (Fig. 5G). Taken together, our study demonstrated that miR-365 inhibited CCA cell proliferation and promoted apoptosis by targeting E2F2.
Figure 5.
Restoration of E2F2 rescued CCA cell proliferation and reduced apoptosis. (A) E2F2 was upregulated in CCA tissues compared to normal tissues according to the TCGA database. (B) RT-qPCR analysis showed that E2F2 was overexpressed in CCA tissues (n = 49). (C) E2F2 was significantly upregulated in CCA cell lines compared to normal cells. (D) The protein of E2F2 was restored in miR-365-overexpressing CCALP1 and SG231 cells. (E) CCK-8 and (F) colony formation assays were used to determine the effect of E2F2 overexpression on CCA cell proliferation. (G) Apoptotic cells were checked by staining with annexinV/PI, followed by FACS analysis. Data are presented as means ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
DISCUSSION
Several miRNAs have been identified as abnormally expressed in CCA tissues. For example, miRNA-34a was epigenetically silenced by EZH2-mediated DNA methylation in human CCA15. Suppression of miR-16 promotes tumor growth and metastasis through reversibly regulating yes-associated protein 1 (YAP1) in human CCA16. miRNA-191 is upregulated and acts as a tumor promoter by modulating the Ten-eleven translocation-1-tumor protein 53 (TET1-p53) pathway in intrahepatic CCA17. miRNA-101 is downregulated and inhibits the migration and invasion of intrahepatic CCA cells18. Expression of miRNAs is strongly associated with CCA growth, metastasis, and progression. In the present study, we showed that miR-365 is downregulated and its target gene E2F2 is upregulated in CCA tissues and cell lines. Overexpression of miR-365 inhibited cell proliferation and induced cell cycle arrest but promoted apoptosis by suppressing E2F2 expression, which suggested that miR-365 served as a tumor suppressor in CCA and might be a therapeutic target.
The expression of miR-365 has been shown to be closely related with various human tumors. For example, miR-365 is downregulated and induces hepatocellular carcinoma cell apoptosis via targeting Bcl-212. Serum miR-365 could be used as a potential diagnostic biomarker for breast cancer19. Serum miR-365 may also be a noninvasive prognostic marker for non-small cell lung cancer20. Bai et al. reported that miR-365 inhibits growth, invasion, and metastasis of malignant melanoma by targeting neuropilin 1 (NRP1) expression14. Besides, Zhou and colleagues reported that miR-365 promotes cutaneous squamous cell carcinoma (CSCC) by targeting nuclear factor I/B (NFIB)21. However, the role of miR-365 remains largely unknown. In our study, we found that miR-365 was also downregulated in CCA samples and cell lines. In addition, the expression of miR-365 was correlated with the prognosis of CCA patients. Consistent with previous conclusions, our data suggested miR-365 as a tumor suppressor in CCA.
E2F2 is a member of the E2F family of transcription factors, which play a crucial role in the control of the cell cycle and the action of tumor-suppressor proteins22,23. E2F proteins contain some conserved domains including a DNA-binding domain; a dimerization domain, which determines interaction with the differentiation-regulated transcription factor proteins (DP); a transactivation domain enriched in acidic amino acids; and a tumor suppressor protein association domain, which is embedded within the transactivation domain24. Emerging studies indicate that E2F2 serves as a promoter in many cancers. For instance, a recent report showed that E2F2 is highly expressed in lung cancer and correlates with the presence of vimentin and β-catenin25. E2F2 could promote tumor growth and epithelial-to-mesenchymal transition (EMT) in the lung. Tao et al. reported that E2F2 promotes cell proliferation in osteosarcoma26. Xie and colleagues indicated that E2F2 induces minichromosome maintenance complex component 4 (MCM4), cyclin E2 (CCNE2), and Wolf–Hirschhorn syndrome candidate 1 (WHSC1; nuclear receptor-binding SET domain protein 2; NSD2) upregulation in ovarian cancer and predicts poor overall survival27. Moreover, E2F2 is also reported to be involved in the development of gastric cancer28, hepatocellular carcinoma29, glioma30, and prostate cancer31. However, the role of E2F2 in CCA needs to be determined. In the present study, we found that E2F2 was the target gene of miR-365. Overexpression of miR-365 significantly inhibited the expression of E2F2 in CCA cells. Furthermore, we found that E2F2 was upregulated in CCA tissues and cell lines. Overexpression of E2F2 could markedly promote CCA cell proliferation and reduce cellular apoptosis. The downstream target genes of E2F2 in CCA cells still need further investigation.
In conclusion, our study, for the first time, identified miR-365 as a tumor suppressor in CCA. In addition, we also determined the molecular mechanism by which miR-365 inhibited tumor cell proliferation and induced apoptosis by directly targeting E2F2. Our study suggested that the miR-365/E2F2 axis might be a potential therapeutic target for CCA treatment.
ACKNOWLEDGMENT
We thank all patients involved in this study.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Shaib YH, Davila JA, McGlynn K, El-Serag HB. Rising incidence of intrahepatic cholangiocarcinoma in the United States: A true increase? J Hepatol. 2004;40(3):472–7. [DOI] [PubMed] [Google Scholar]
- 2. Ebata T, Ercolani G, Alvaro D, Ribero D, Di Tommaso L, Valle JW. Current status on cholangiocarcinoma and gallbladder cancer. Liver Cancer 2016;6(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fairweather M, Balachandran VP, D’Angelica MI. Surgical management of biliary tract cancers. Chin Clin Oncol. 2016;5(5):63. [DOI] [PubMed] [Google Scholar]
- 4. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383(9935):2168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Puik JR, Meijer LL, Le Large TY, Prado MM, Frampton AE, Kazemier G, Giovannetti E. miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics 2017;18(14):1343–58. [DOI] [PubMed] [Google Scholar]
- 6. Yu S, Xie H, Zhang J, Wang D, Song Y, Zhang S, Zheng S, Wang J. MicroRNA663 suppresses the proliferation and invasion of colorectal cancer cells by directly targeting FSCN1. Mol Med Rep. 2017;16(6):9707–14. [DOI] [PubMed] [Google Scholar]
- 7. Wang T, Cai Z, Hong G, Zheng G, Huang Y, Zhang S, Dai J. MicroRNA21 increases cell viability and suppresses cellular apoptosis in nonsmall cell lung cancer by regulating the PI3K/Akt signaling pathway. Mol Med Rep. 2017;16(5):6506–11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Yin J, Zhuang G, Zhu Y, Hu X, Zhao H, Zhang R, Guo H, Fan X, Cao Y. MiR-615-3p inhibits the osteogenic differentiation of human lumbar ligamentum flavum cells via suppression of osteogenic regulators GDF5 and FOXO1. Cell Biol Int. 2017;41(7):779–86. [DOI] [PubMed] [Google Scholar]
- 9. Yang R, Chen YJ, Tang C, Li HB, Wang B, Yan Q, Hu JB, Zou SQ. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b. BMC Cancer 2014;14:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Q, Tang HH, Yin SS, Dong C. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 2013;29(5):2046–52. [DOI] [PubMed] [Google Scholar]
- 11. Liu N, Jiang F, He TL, Zhang JK, Zhao J, Wang C, Jiang GX, Cao LP, Kang PC, Zhong XY, Lin TY, Cui YF. The roles of microRNA-122 overexpression in inhibiting proliferation and invasion and stimulating apoptosis of human cholangiocarcinoma cells. Sci Rep. 2015;5:16566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li MF, Yang Y, Huang Y, Gan XF, Zeng W, Liu YP, Guan H. miR-365 induces hepatocellular carcinoma cell apoptosis through targeting Bcl-2. Exp Ther Med. 2017;13(5):2279–85. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Wang YL, Xu CL, Wang Y, Zhang XM. MicroRNA-365 inhibits ovarian cancer progression by targeting Wnt5a. Am J Cancer Res. 2017;7(5):1096–106. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Bai JJ, Zhang ZL, Li X, Liu HF. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Cancer Biomark. 2015;15(5):599–608. [DOI] [PubMed] [Google Scholar]
- 15. Kwon H, Song K, Han C, Zhang JQ, Lu L, Chen WN, Wu T. Epigenetic silencing of miRNA-34a in human cholangiocarcinoma via EZH2 and DNA methylation impact on regulation of notch pathway. Am J Pathol. 2017;187(10):2288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han S, Wang D, Tang GH, Yang XX, Jiao CY, Yang RJ, Zhang YD, Huo LQ, Shao ZC, Lu ZF, Zhang JW, Li XC. Suppression of miR-16 promotes tumor growth and metastasis through reversely regulating YAP1 in human cholangiocarcinoma. Oncotarget 2017;8(34):56635–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, Nie J, Ding XT, Li B, Zhou GW, Zhang ZY. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology 2017;66(1):136–51. [DOI] [PubMed] [Google Scholar]
- 18. Deng G, Teng YL, Huang FZ, Nie WP, Zhu L, Huang W, Xu HB. MicroRNA-101 inhibits the migration and invasion of intrahepatic cholangiocarcinoma cells via direct suppression of vascular endothelial growth factor-C. Mol Med Rep. 2015;12(5):7079–85. [DOI] [PubMed] [Google Scholar]
- 19. Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92(2):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu YY, Zhang GX, Li HY, Han LH, Fu AG, Zhang NL, Zheng YG. Serum microRNA-365 in combination with its target gene TTF-1 as a non-invasive prognostic marker for non-small cell lung cancer. Biomed Pharmacother. 2015;75:185–90. [DOI] [PubMed] [Google Scholar]
- 21. Zhou MJ, Zhou L, Zheng L, Guo LH, Wang YH, Liu HX, Ou CS, Ding ZH. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting Nuclear Factor I/B (NFIB). PLoS One 2014;9(6):e100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Magae J, Wu CL, Illenye S, Harlow E, Heintz NH. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–26. [DOI] [PubMed] [Google Scholar]
- 23. Pierce AM, Schneider-Broussard R, Philhower JL, Johnson DG. Differential activities of E2F family members: Unique functions in regulating transcription. Mol Carcinog. 1998;22(3):190–8. [DOI] [PubMed] [Google Scholar]
- 24. Lee C, Chang JH, Lee HS, Cho YJ. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 2002;16(24):3199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feliciano A, Garcia-Mayea Y, Jubierre L, Mir C, Hummel M, Castellvi J, Hernandez-Losa J, Paciucci R, Sansano I, Sun YL, Cajal RY, Kondon H, Soriano A, Segura M, Lyakhovich A, LLeonart ME. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8:e3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tao TY, Shen QR, Luo JM, Xu Y, Liang WQ. MicroRNA-125a regulates cell proliferation via directly targeting E2F2 in osteosarcoma. Cell Physiol Biochem. 2017;43(2):768–74. [DOI] [PubMed] [Google Scholar]
- 27. Xie L, Li T, Yang LH. E2F2 induces MCM4, CCNE2 and WHSC1 upregulation in ovarian cancer and predicts poor overall survival. Eur Rev Med Pharmacol Sci. 2017;21(9):2150–6. [PubMed] [Google Scholar]
- 28. Wang HD, Zhang XT, Liu YX, Ni ZH, Lin Y, Duan ZP, Shi Y, Wang GQ, Li F. Downregulated miR-31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2. Oncotarget 2016;7(24):36577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang J, Yang J, Lei Y, Gao H, Wei T, Luo L, Zhang F, Chen H, Zeng Q, Guo L. An ANCCA/PRO2000-miR-520a-E2F2 regulatory loop as a driving force for the development of hepatocellular carcinoma. Oncogenesis 2016;5:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song H, Zhang Y, Liu N, Zhang DD, Wan C, Zhao S, Kong Y, Yuan LD. Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2. J Physiol Biochem. 2016;72(4):733–44. [DOI] [PubMed] [Google Scholar]
- 31. Dong QC, Meng P, Wang T, Qin WW, Qin WJ, Wang FL, Yuan JL, Chen ZN, Yang AG, Wang H. MicroRNA Let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One 2010;5(4):e10147. [DOI] [PMC free article] [PubMed] [Google Scholar]