Abstract
Recent studies revealed that emodin extracted from Chinese herbs exhibits an anticancer effect on different cancer types, including colon cancer. However, the mechanism is not well understood. In our study, we confirmed that emodin treatment inhibited cell viability and induced apoptosis in colon cancer cells. Further experiments found that emodin was also able to induce autophagy, which is indispensible for apoptosis induced by emodin. More interestingly, emodin treatment also results in mitochondrial dysfunction and ROS accumulation in colon cancer cells. Finally, we stressed that ROS accumulation is essential for autophagy and apoptosis induced by emodin. In conclusion, emodin induces apoptosis in colon cancer cells through induction of autophagy, during which ROS generation is of the essence. Our findings improve understanding of emodin’s effect on colon cancer suppression and provide a new theoretical basis for colon cancer therapy.
Key words: Emodin, Colon cancer, Apoptosis, Autophagy, Reactive oxygen species (ROS)
INTRODUCTION
Colorectal cancer is the third most commonly diagnosed cancer worldwide1 and one of the leading causes of cancer-related mortality2. Colorectal cancer remains an incurable disease due to lack of effective therapeutic treatments, especially for advanced colon cancer3. Therefore, detection and identification of a new therapeutic strategy are extremely urgent for colon cancer therapy.
Emodin, also referred to as 1,3,8-trihydroxy-6-methyl-anthraquinone, is extracted from Chinese herbs and has been traditionally used as an active constituent of many herbal laxatives4. Nevertheless, recent research highlighted that emodin also exhibits an anticancer effect on different cancer types. Emodin has been reported to inhibit cancer growth by inducing apoptosis in many cancer cell lines, such as lung cancer cells5, hepatocarcinoma cells6, and colon cancer cells7. However, no specific molecular mechanism has been established to explain how emodin affects apoptosis of colon cancer cells.
Autophagy was first defined as the process by which cytoplasmic materials were delivered to the lysosome for degradation and was considered to play an essential role in the control of cell metabolism8. Absence of autophagy in the liver tissues results in liver tumor formation9, indicating that autophagy is closely related to cancer suppression. Reactive oxygen species (ROS), composed of a range of oxygen-containing highly reactive species like hydrogen peroxide, free radicals, and oxygen anions10, contributes to mitochondrial dysfunction11 and cell injury12, indicating the potential role of ROS in cell survival. A recent study illustrated that ROS could promote the process of autophagy13; on the other hand, autophagy reduces ROS by removing damaged mitochondria and other materials contributing to ROS generation14. Thus, dysfunction of autophagy may result in abnormal mitochondrial function and oxidative stress. However, the exact mechanisms for the effect of ROS on autophagy are largely unknown.
Therefore, in colon cancer, the relationship among apoptosis, autophagy, and ROS is not well understood. Based on our study, we demonstrated that emodin induces apoptosis in colon cancer cells through an autophagy-dependent manner, during which ROS accumulation is indispensable.
MATERIALS AND METHODS
Cell Culture
Human colon cancer cell line HCT116 was maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, New York, NY, USA), and LOVO cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco). Medium was supplemented with 10% fetal bovine serum (FBS; Biological Industries, State of Israel) and 1% penicillin G/streptomycin sulfate. Cells were maintained at 37°C in a humidified incubator under 5% CO2. Other reagents used in this study, emodin, N-acetylcysteine (NAC; ROS scavenger), and chloroquine (CQ), were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Annexin V-FITC/PI Double Staining
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining of cells was performed by the FITC Annexin-V Apoptosis Detection Kit (KeyGEN Biotech, Nanjing, Jiangsu, P.R. China). Briefly, cells (5 × 105) were seeded onto six-well plates and incubated for 24 h at 37°C in culture medium and then washed with PBS twice, trypsinized, and resuspended in 500 μl of binding buffer. Cells were stained for 15 min with annexin V-FITC and PI and then analyzed by flow cytometry within 1 h.
MTT Assay
MTT assay was performed by Cell Growth Determination Kit MTT Based (Sigma-Aldrich). MTT solution was added in an amount equal to 10% of the culture volume and incubated for 3 to 4 h. The resulting MTT formazan crystals read as absorbance at a wavelength of 570 nm were spectrophotometrically measured.
Colony Formation Assay
For colony formation experiments, attached cells were trypsinized with 1 ml of trypsin (Sigma-Aldrich) and plated at low density (200 per well in a six-well plate). After 24 h, emodin was added at an indicated concentration for 48 h. After drug removal, cells were allowed to proliferate, and clones were allowed to grow in a humidified 5% CO2, 37°C environment for 15 days in growth medium. Cells were fixed with 70% ethanol and stained with 0.05% crystal violet (Sigma-Aldrich) for analysis.
Western Blot Analysis
Cells were lysed in EBC250 lysis buffer [50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 0.5% Nonidet P-40, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml leupeptin, 2 μg/ml aprotinin, 50 mM NaF, and 0.5 mM Na3VO4]. Proteins were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA), and hybridized to an appropriate primary antibody and horseradish peroxidase-conjugated secondary antibody for subsequent detection by enhanced chemiluminescence. Monoclonal antibodies specific for cleaved caspase 3 (380169), cleaved caspase 9 (614895), Bcl-2(220449), and GADPH (380627) were purchased from Zen BioScience (Chengdu, Sichuan, P.R. China). Monoclonal antibody specific for cleaved PARP (sc-8007) was purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-mouse IgG-HRP (Lot. 20150610) and anti-rabbit IgG-HRP (Lot. EE0814) secondary antibodies for Western blotting were obtained from Zen BioScience. Monoclonal antibodies specific for cytochrome (F0824A), Bax (Sc-6236), LC3-B (Sc-376404), Beclin-1 (Sc-11427), and p62 (sc-25523) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Immunofluorescence
For fluorescence microscopy, cells were seeded at a density of 7.5 × 104 per well in BD Biocoat collagen I-coated eight-well chamber slides and incubated at 37°C at 5% CO2 for 24 h before treatment. Following treatment, cells were fixed onto glass slides, permeabilized with 0.1% Triton X-100 in PBS, and stained by antibodies. Antibodies used were LC3-B (Sc-376404; Santa Cruz Biotechnology) and anti-FITC mouse mAb (201051-2B3; Zen BioScience). Cells were fixed onto glass slides. Images were acquired using an Olympus fluorescence microscope (BX43F) analyzed using photoshop CS2 software.
ROS Detection
Detection and measurement of intracellular ROS were assessed by Reactive Oxygen Species Assay Kit purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, Jiangsu, P.R. China). Intracellular ROS generation was assessed using dichlorofluorescin diacetate (DCFH-DA), a cell-permeant probe that is hydrolyzed by intracellular esterases to dichlorofluorescin (DCF), which is oxidized by ROS to form highly fluorescent DCF, and the fluorescence intensity is proportional to the amount of ROS produced in the cells. Briefly, cells were plated in a six-well plate and after 24 h were pretreated with or without emodin. Then DCFH-DA was added for 30 min and incubated at 37°C. Cells were then rinsed twice with PBS, and fluorescence intensity was monitored by flow cytometry.
Statistical Analysis
All values were reported as mean ± SD of three independent experiments. Statistical analysis was carried out using GraphPad Prism 6 software. Grayscale image analysis was performed by ImageJ software.
RESULTS
Emodin Induces Apoptosis in HCT116 and LOVO Cells
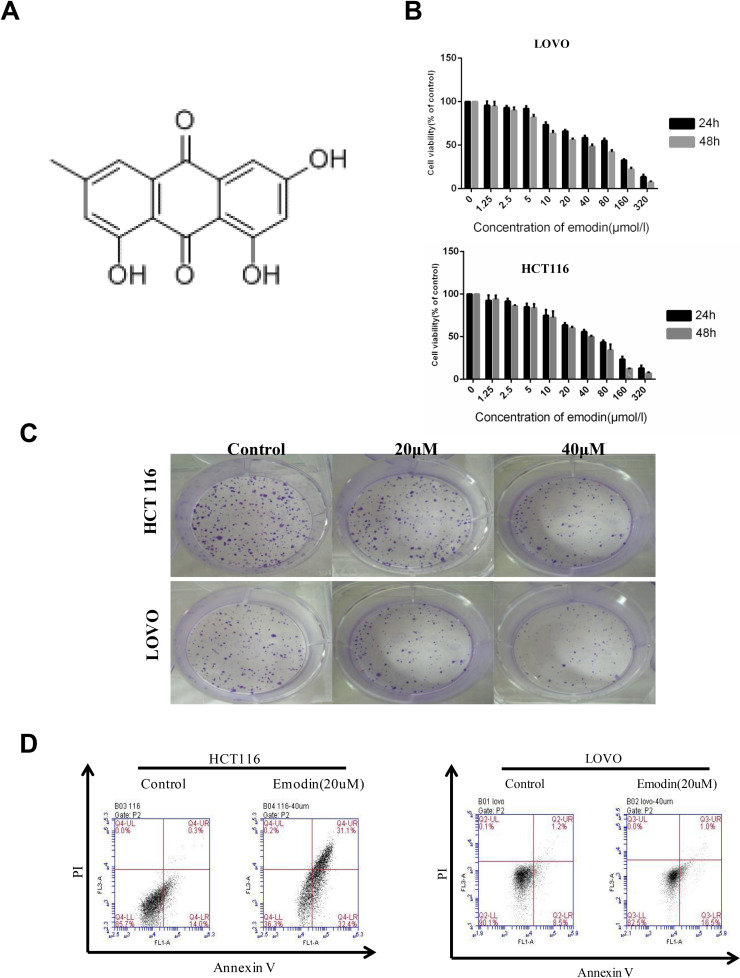
Colon cancer cell lines (HCT116 and LOVO) were treated with emodin (chemical structure shown in Fig. 1A) for indicated times under different concentrations. MTT assays were performed to determine its effect on cell proliferation (Fig. 1B). Emodin treatment significantly decreased cell variability in both HCT116 and LOVO cells in both a dose-dependent and time-dependent manner. Colony formation assays were performed in HCT116 and LOVO cells to provide further evidence of inhibitory function of emodin on cell proliferation (Fig. 1C). As shown, emodin treatment (20 and 40 μM) significantly inhibits colony formation of HCT116 and LOVO cells. Together, these data suggest that emodin can inhibit growth of colon cancer cells.
Figure 1.
Emodin inhibits growth of HCT116 and LOVO cells. HCT116 and LOVO cells were treated with emodin for the indicated concentrations and time. (A) Chemical structure of emodin. (B) MTT assays were performed to detect cell viability. (C) HCT116 and LOVO cells were subjected to colony formation assay after emodin treatment (20 and 40 μM, respectively) for 24 h. (D) HCT116 and LOVO cells after emodin treatment were subjected for annexin V/propidium iodide double staining.
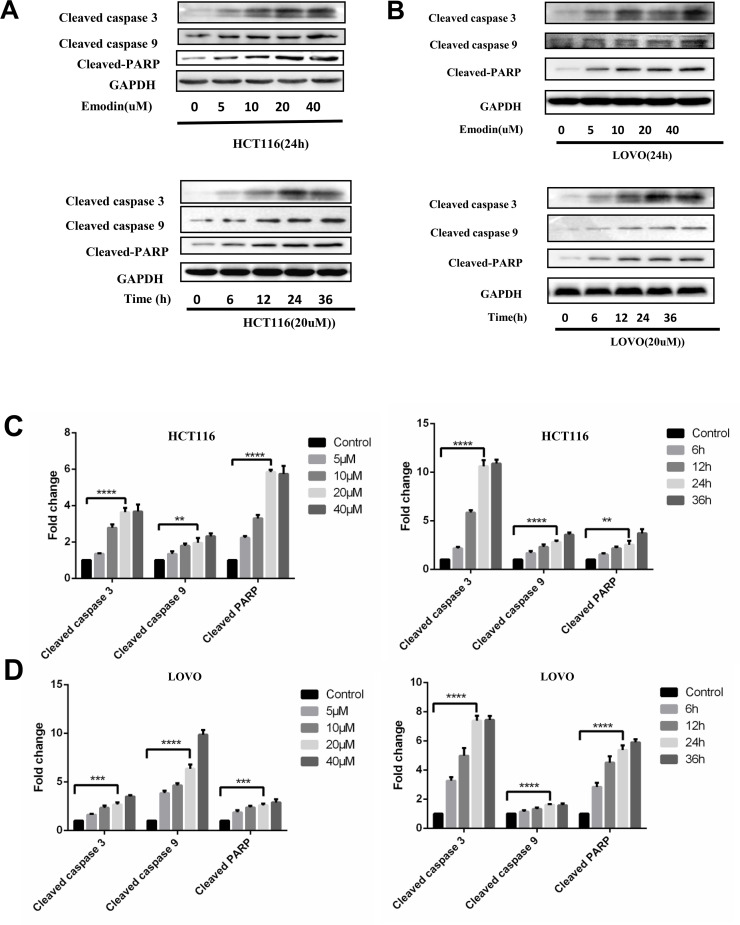
In order to illuminate the mechanism of emodin’s function on cell growth suppression, apoptosis of HCT116 and LOVO cell lines with emodin treatment (20 μM, 24 h) was determined by annexin V/PI double staining (Fig. 1D). Western blot analysis was also performed to detect caspase cleavage in HCT116 (Fig. 2A) and LOVO (Fig. 2B) cells. With emodin treatment, cleavage of caspase 3, caspase 9, and PARP was increased in both cells. For Western blotting experiments, results from three independent experiments were analyzed, and cleavages of three proteins were increased significantly compared to the control groups (Fig. 2C and D). These data demonstrated that emodin could induce apoptosis in both a dose-dependent and time-dependent manner.
Figure 2.
Emodin induces apoptosis in HCT116 and LOVO cells. HCT116 and LOVO cells were treated with emodin for the indicated concentration and time. (A) Cell lysates of HCT116 were collected for Western blotting as indicated. (B) Cell lysates of LOVO were collected for Western blotting as indicated. Grayscale analysis of Western blotting results for apoptosis was performed in HCT116 (C) and LOVO (D). Results from three independent experiments were quantitated and presented as means and SE. **p < 0.01; ***p < 0.001; ****p < 0.0001.
Emodin Induces Autophagy in HCT116 and LOVO Cells
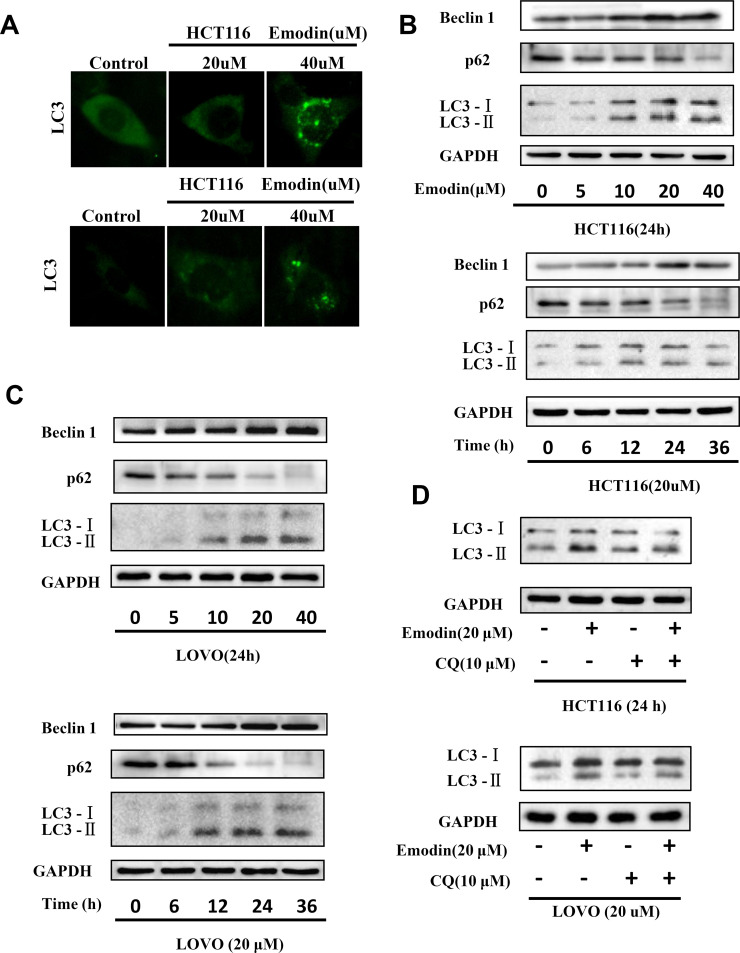
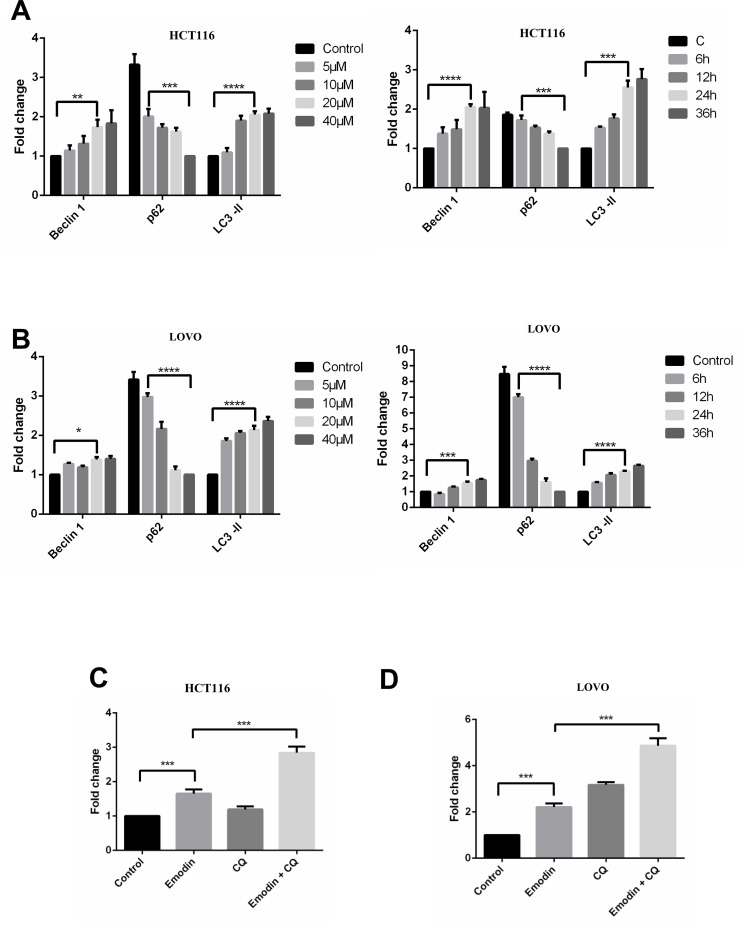
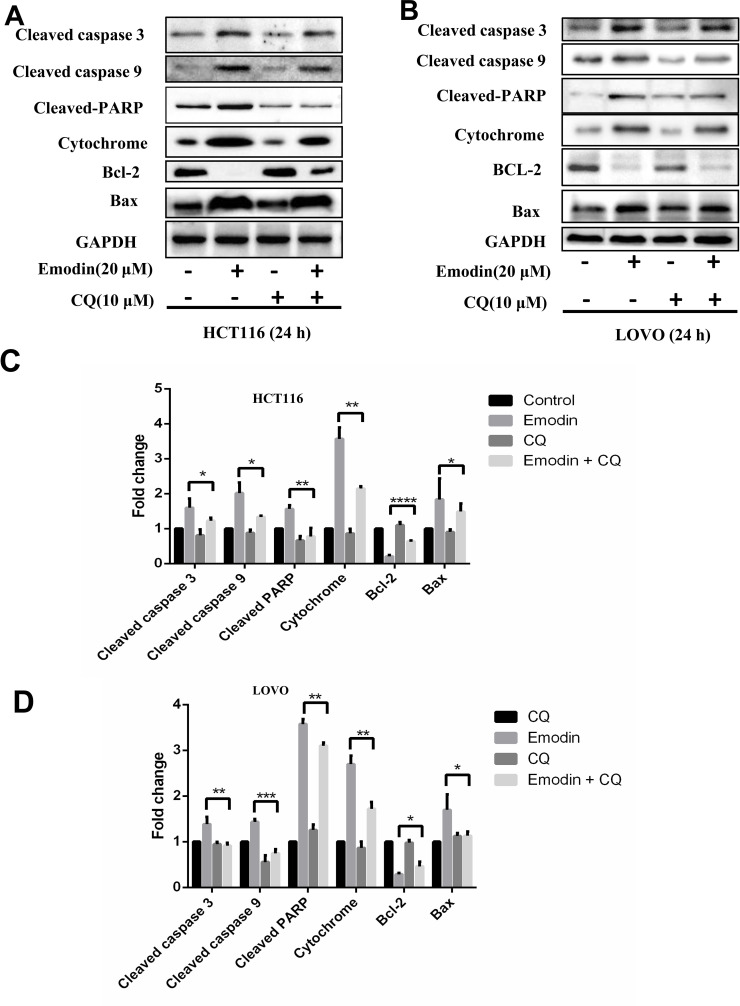
To investigate the mechanism by which emodin induces apoptosis, we then detected autophagy in HCT116 and LOVO cells, as a previous study highlighted the essential role of autophagy in the process of apoptosis9. Emodin treatment resulted in extensive GFP-LC3 localization (Fig. 3A), suggesting that emodin induces autophagy in both HCT116 and LOVO cells. Additionally, results from the Western blotting experiments showed that emodin treatment increases LC3-2 accumulation in HCT116 (Fig. 3B) and LOVO cells (Fig. 3C), respectively. Also, levels of p62 and Beclin-1 were changed corresponding to emodin treatment. Results from three independent experiments were analyzed to show the significance for each Western blotting experiment (Fig. 4A and B). Furthermore, an autophagy inhibitor, CQ, was used to further confirm that emodin did induce autophagy (Fig. 3D). Statistical data are shown in Figure 4C and D. Accumulation of LC3-2 induced by CQ suggests that emodin induces complete autophagy flux.
Figure 3.
Emodin induces autophagy in HCT116 and LOVO cells. HCT116 and LOVO cells were treated with emodin for the indicated concentration and time. (A) GFP-LC3 was visualized by fluorescence microscopy. (B) Cell lysates of HCT116 were collected for Western blotting as indicated. (C) Cell lysates of LOVO were collected for Western blotting as indicated. (D) HCT116 and LOVO treated with emodin were also treated with or without chloroquine (CQ), and cell lysates were subjected for Western blotting.
Figure 4.
Grayscale analysis of Western blotting results for autophagy induced by emodin. Cell lysates of HCT116 (A) and LOVO (B) cells were subjected for Western blotting, and grayscale analysis was performed as indicated. Cells treated with emodin were also treated with or without CQ. Cell lysates were subjected for Western blotting based on which grayscale analysis was assessed in HCT116 (C) and LOVO (D) cells. Results from three independent experiments were quantitated and presented as means and SE. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Emodin Induces Apoptosis Through an Autophagy Pathway
As reported, autophagy is closely related to apoptosis under some conditions15. Therefore, we speculated that emodin may induce apoptosis through an autophagy-dependent pathway. In order to prove this speculation, we detected apoptosis with emodin induced in the presence and absence of CQ treatment. CQ rescued emodin-induced apoptosis in HCT116 (Fig. 5A) and LOVO cells (Fig. 5B). Data based on three independent experiments are shown in Figure 5C and D. These data suggest that emodin induces apoptosis through an autophagy pathway.
Figure 5.
Emodin induces apoptosis through an autophagy pathway. HCT116 (A) and LOVO (B) cells treated with emodin (20 μM) were also treated with or without CQ (10 μM) for 24 h. Cell lysates were subjected to Western blotting. Grayscale analysis of Western blotting results was performed in HCT116 (C) and LOVO (D). Results from three independent experiments were quantitated and presented as means and SE. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
We also detect protein levels of cytochrome, Bcl-2, and Bax because mitochondrial dysfunction has been reported to influence apoptosis. Interestingly, emodin treatment did result in mitochondrial dysfunction, indicating the potential role of mitochondria dysfunction in apoptosis and autophagy induced by emodin.
ROS Mediates Emodin-Induced Autophagy and Apoptosis
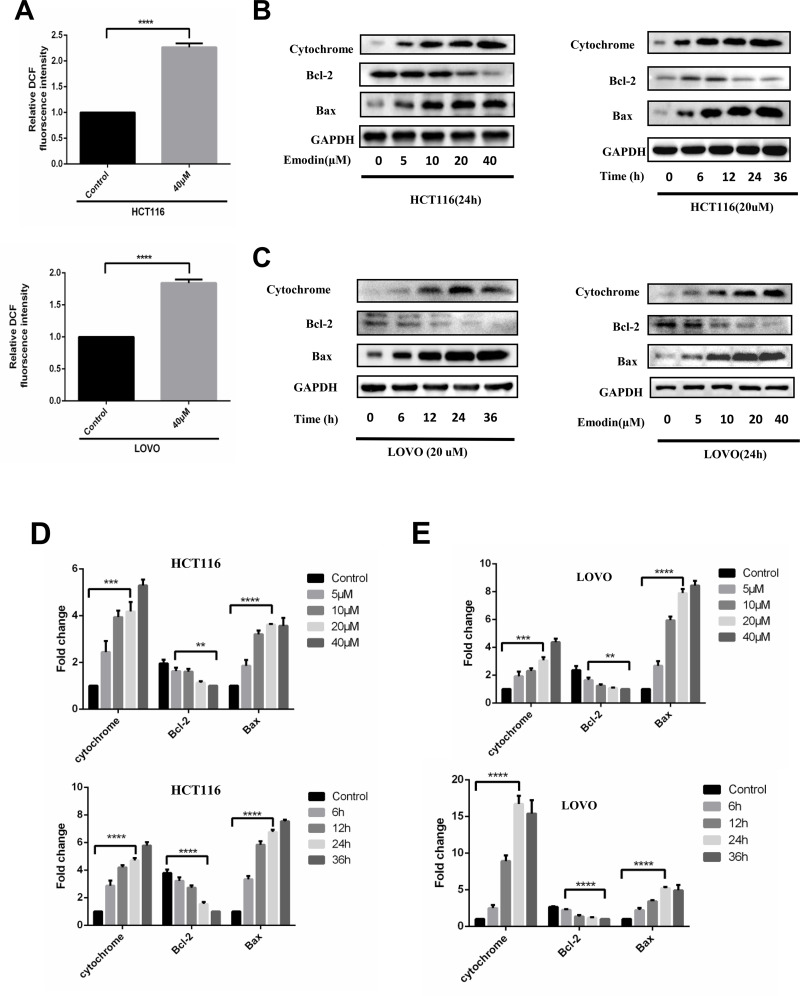
The data above showed that emodin induced cell death through an autophagy-dependent pathway, but the mechanism by which emodin induces autophagy is not yet clear. One thing to be noted, mitochondrial dysfunction is involved in such a process due to our data. As mitochondrial dysfunction may induce ROS accumulation, which is closely related to autophagy, we detected the ROS level after emodin treatment in both HCT116 and LOVO cells (Fig. 6A). ROS levels were increased with emodin treatment for 24 h, suggesting that emodin may induce autophagy and cell death though ROS accumulation. Further experiments were performed to detect levels of cytochrome, Bcl-2, and Bax in both HCT116 (Fig. 6B) and LOVO cells (Fig. 6C) under different conditions to stress mitochondrial dysfunction induced by emodin treatment. Results from three independent experiments were analyzed as shown in Figure 6D and E.
Figure 6.
Emodin induces reactive oxygen species (ROS) generation and mitochondrial dysfunction. HCT116 and LOVO cells were treated with emodin for the indicated time and concentration. (A) ROS accumulation was detected by flow cytometry. Fluorescence intensity from three independent experiments was quantitated and presented as means and SE. ***p < 0.001. (B) Cell lysates of HCT116 were collected for Western blotting as indicated. (C) Cell lysates of LOVO were collected for Western blotting as indicated. Grayscale analysis of Western blotting results was performed in HCT116 (D) and LOVO (E). Results from three independent experiments were quantitated and presented as means and SE. **p < 0.01; ***p < 0.001; ****p < 0.0001.
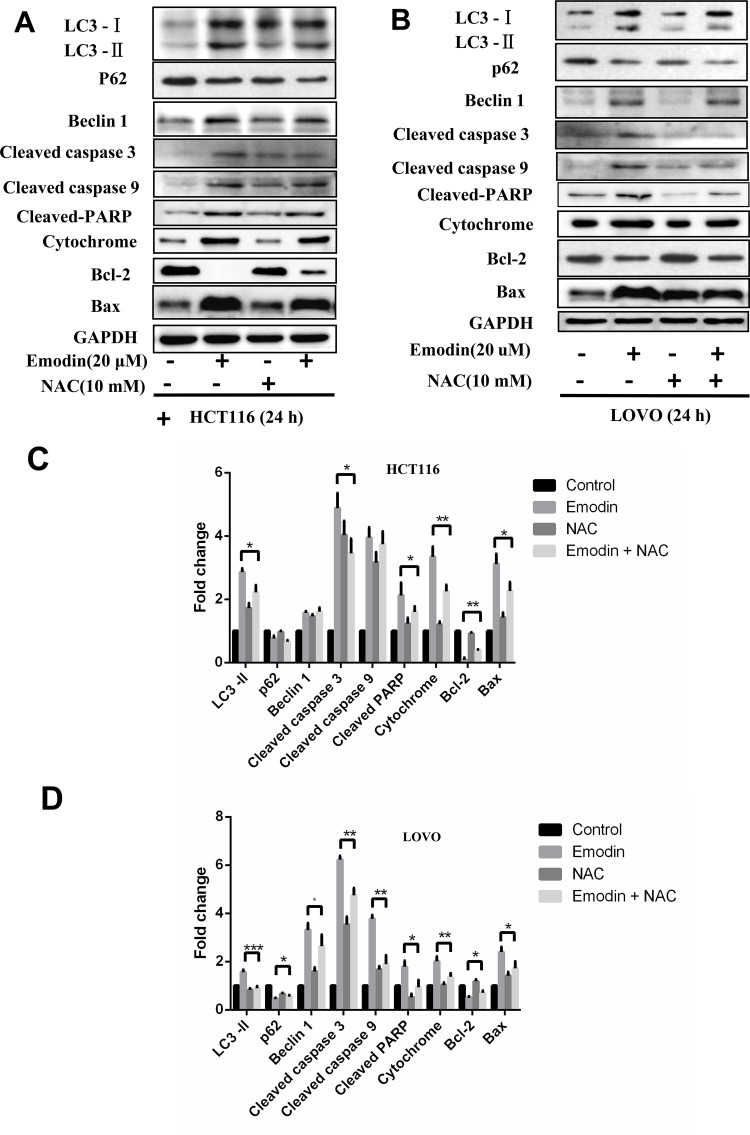
Furthermore, NAC, a ROS inhibitor, was used to confirm whether ROS accumulation was essential for emodin-induced apoptosis and autophagy. HCT116 and LOVO cells treated with emodin were also treated with or without NAC for 24 h, and cell lysates were collected for Western blot analysis. NAC rescued emodin-induced autophagy and apoptosis in both HCT116 (Fig. 7A) and LOVO cells (Fig. 7B), indicating that ROS mediated emodin-induced autophagy and apoptosis. Results from three independent experiments were analyzed to show the significance compared with control groups (Fig. 7C and D).
Figure 7.
ROS mediates emodin-induced autophagy and apoptosis. HCT116 (A) and LOVO (B) cells treated with emodin (20 μM) were also treated with or without NAC (10 μM) for 24 h. Cell lysates were subjected to Western blotting. Grayscale analysis of Western blotting results was performed in HCT116 (C) and LOVO (D) cells. Results from three independent experiments were quantitated and presented as means and SE. *p < 0.05; **p < 0.01; ***p < 0.001.
DISCUSSION
One of our results that emodin inhibits cell growth is in accordance with previous studies5–7, and we also confirmed that emodin induced apoptosis in colon cancer cells. Previous studies demonstrated that emodin induces apoptosis through different mechanisms. For example, in lung cancer, emodin induces apoptosis through ER stress and the TRIB3/NF-κB pathway5. Emodin also targets mitochondrial cyclophilin D to induce apoptosis of human hepatocarcinoma cells6. Specifically, emodin was reported to induce apoptosis and inhibit colon cancer proliferation through a FASN-involved pathway like AKT and Erk7. By detecting autophagy and ROS accumulation, we demonstrated that emodin could also induce colon cell apoptosis through an autophagy-dependent way during which ROS accumulation plays an essential role.
Autophagy affects cancer cell survival in various ways. Recent research has stressed the interconnection between autophagy and apoptosis. Bcl-2, Beclin-1, p62, caspase 3, and other related proteins are involved in the interaction between apoptosis and autophagy. For example, under nutrient-rich conditions, Bcl-2 binds to Beclin-1 and inhibits this autophagy inducer when autophagy is unnecessary16. However, when cells are subjected to starvation, this binding activity of Bcl-2 to Beclin-1 is weakened by JNK1-mediated Bcl-2 phosphorylation, and thus autophagy is initiated to ensure survival of cells17. Interestingly, our data showed that autophagy is essential for apoptosis, which inhibits cell viability. As reported, w09, a novel autophagy enhancer, could induce autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS–RAF1–MAP2K–MAPK1/3 pathway18. Thus, interaction between apoptosis and autophagy is quite complicated, and more effort should be taken to elucidate this interaction.
Based on our results, autophagy induces apoptosis via generation of ROS in colon cancer cells, providing new evidence to stress the essential role of the autophagy pathway in apoptosis. As reported, ROS generation led to apoptosis of cancer cells. A novel CDK1 inhibitor, BA-j, was able to selectively induce apoptosis in cancer cells by regulating ROS19. JS-K promotes apoptosis by inducing ROS production in human prostate cancer cells20. In our study, emodin promoted apoptosis of colon cancer cells also through a ROS-dependent way, further verifying the interaction between ROS accumulation and apoptosis. Altered mitochondrial morphology under metabolic excess results in ROS production and mitochondrial dysfunction, and conversely, ROS inside mitochondria causes mitochondrial dysfunction, which may affect mitochondrial morphology21, indicating a potential relation between ROS and mitochondrial dysfunction. According to the results of our experiments, emodin induced both ROS generation and mitochondrial dysfunction.
In conclusion, our data suggest a novel mechanism for apoptosis of colon cancer cells induced by emodin, adding to the accumulating evidence that suggests an anticancer effect of emodin. Our findings further suggest crosstalk among apoptosis, autophagy, and ROS generation, but more effort should be taken to stress the precise mechanism.
ACKNOWLEDGMENTS
This work was supported by the Key Fund Project of Sichuan Provincial Department of Education (No. 16ZA0291), the Discipline Building Project of Biology of Chengdu Medical College (CYXK2012011), and the Project of Research and Innovation Teams of Chengdu Medical College (CYTD15-02).
REFERENCES
- 1. Ferlay J SI, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Cunningham D AW, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet 2010;375(9719):1030–47. [DOI] [PubMed] [Google Scholar]
- 3. Zhang F, Zhang Y, Zhao W, Deng K, Wang Z, Yang C, Ma L, Openkova MS, Hou Y, Li K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget 2017;8(21):35460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monisha BA, Kumar N, Tiku AB. Emodin and its role in chronic diseases. Adv Exp Med Biol. 2016;928:47–73. [DOI] [PubMed] [Google Scholar]
- 5. Su J, Yan Y, Qu J, Xue X, Liu Z, Cai H. Emodin induces apoptosis of lung cancer cells through ER stress and the TRIB3/NF-κB pathway. Oncol Rep. 2017;37(3):1565–72. [DOI] [PubMed] [Google Scholar]
- 6. Zhang L, He D, Li K, Liu H, Wang B, Zheng L, Li J. Emodin targets mitochondrial cyclophilin D to induce apoptosis in HepG2 cells. Biomed Pharmacother. 2017;90:222–8. [DOI] [PubMed] [Google Scholar]
- 7. Lee KH, Lee MS, Cha EY, Sul JY, Lee JS, Kim JS, Park JB, Kim JY. Inhibitory effect of emodin on fatty acid synthase, colon cancer proliferation and apoptosis. Mol Med Rep. 2017;15(4):2163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Komatsu M, Waquir S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007;131(6):1149–63. [DOI] [PubMed] [Google Scholar]
- 10. Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S, Dhama K. Oxidative stress, prooxidants, and antioxidants: The interplay. Biomed Res Int. 2014;2014:761264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17(9):422–7. [DOI] [PubMed] [Google Scholar]
- 12. Fandy TE, Jiemjit A, Thakar M, Rhoden P, Suarez L, Gore SD. Decitabine induces delayed reactive oxygen species (ROS) accumulation in leukemia cells and induces the expression of ROS generating enzymes. Clin Cancer Res. 2014;20(5):1249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Navarro-Yepes J, Burns M, Anandhan A, Khalimonchuk O, del Razo LM, Quintanilla-Vega B, Pappa A, Panayiotidis MI, Franco R. Oxidative stress, redox signaling, and autophagy: Cell death versus survival. Antioxid Redox Signal. 2014;21(1):66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem J. 2012;441(2):523–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015;6(11):8474–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005;122(6):927–39. [DOI] [PubMed] [Google Scholar]
- 17. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008;30(6):678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang P, Zheng Z, Ling L, Yang X, Zhang N, Wang X, Hu M, Xia Y, Ma Y, Yang H, Wang Y, Liu H. w09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the EGFR-mediated RAS-RAF1-MAP2K-MAPK1/3 pathway. Autophagy 2017;17:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang S, Bao Y, Ju X, Li K, Shang H, Ha L, Qian Y, Zou L, Sun X, Li J, Wang Q, Fan Q. BA-j as a novel CDK1 inhibitor selectively induces apoptosis in cancer cells by regulating ROS. Sci Rep. 2015;5:13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu M, Chen L, Tan G, Ke L, Zhang S, Chen H, Liu J. JS-K promotes apoptosis by inducing ROS production in human prostate cancer cells. Oncol Lett. 2017;13(3):1137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galloway CA, Yoon Y. What comes first, misshape or dysfunction? The view from metabolic excess. J Gen Physiol. 2012;139(6):455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]