Abstract
Oral submucous fibrosis (OSF) induced by chewing of the areca nut has been considered to be a precancerous lesion with a high probability of developing oral squamous cell carcinoma. Tanshinone (TSN) is the main component extracted from Salvia miltiorrhiza, a traditional Chinese medicine, which was found to have diverse pharmacological effects, such as anti-inflammatory and antitumor. In the current study, we aimed to identify the inhibitory effects and the underlying mechanism of TSN on OSF progress. We found that treatment with TSN inhibited the arecoline-mediated proliferation of primary human oral mucosal fibroblasts and reversed the promotive effects of arecoline on the EMT process. By RNA deep sequencing, we screened two possible targets for TSN: LSD1 and p53. We confirmed that p53 is much lower in OSF than in normal mucous tissues. In addition, p53 and its downstream molecules were decreased by arecoline treatment in oral mucosal fibroblasts, which was reversed by treatment with TSN in a dose-dependent manner. Our results also revealed that arecoline stimulation resulted in hypermethylation of the promoter of TP53 and subsequent downregulation of p53 levels, which was reversed by TSN. Furthermore, we identified that LSD1 could epigenetically activate TP53 by recruiting H3K27me1 and H3K4m2 to its promoter. Our findings provide new insights into the mechanism by which TSN influences arecoline-induced OSF and rationale for the development of clinical intervention strategies for OSF and even oral squamous cell carcinoma.
Key words: Oral submucous fibrosis (OSF), Areca nut, Oral squamous cell carcinoma, Tanshinone (TSN), Epithelial–mesenchymal transition (EMT), p53, Methylation
INTRODUCTION
The habit of chewing the areca nut (betel nut) is prevalent in South Asian populations and has also been observed in Europe and North America. Oral submucous fibrosis (OSF) is characterized by chronic inflammation and epithelial atrophy, and this has been associated with chewing the areca nut1. OSF causes the transformation of normal mucosal cells to a precancerous lesion with a high probability of developing oral squamous cell carcinoma2. Exposure of the oral cavity to areca nut induces several physical and molecular alterations, leading to pathological conditions such as OSF and oral cancer3. For example, chronic arecoline exposure (found in the areca nut) of human oral epithelial cells resulted in increased cluster of differentiation 44 (CD44) positivity and epithelial–mesenchymal transition (EMT) traits4, a crucial event in the formation of oral squamous cell carcinoma and OSF. Previous studies demonstrated that arecoline could also induce the expression of vimentin in human buccal mucosal fibroblasts5,6. Our previous study also showed that arecoline exposure resulted in a decrease in epithelial (E)-cadherin expression and upregulation of the expression of neural (N)-cadherin and vimentin in immortalized nontumorigenic human epidermal HaCaT cells7, suggesting that the EMT process is directly involved in the pathogenesis of OSF.
Salvia miltiorrhiza, or red sage, is a traditional Chinese medicine. It is used to promote blood circulation and regulate menstruation. Tanshinone (TSN) is the main component extracted from Salvia miltiorrhiza. TSN was found to have diverse pharmacological effects such as antioxidant, anti-inflammatory, and antitumor8. For example, TSN IIA can inhibit the growth of human gastric cancer cells through increasing the protein expression of phosphorylated mitogen-activated protein kinase (p-p38) and phosphorylated C-Jun N-terminal kinase (p-JNK) and activating tumor protein p53 (p53), followed by increasing the protein expression of cyclin-dependent kinase inhibitor 1A (p21)9. High concentrations of TSN IIA cause complete phosphorylation and degradation of RNAse polymerase II (RNAPII) followed by p53 activation and apoptosis in hepatic cancer cells10. These studies suggest that TSN may induce cancer cell death through activation of the p53 pathway. It is therefore possible that TSN inhibits the proliferation of human oral mucosal fibroblasts (hOMF) and the EMT process by activating the p53 pathway; however, there has been no evidence to date supporting this hypothesis.
In the current study, we aimed to identify the inhibitory effects of TSN on OSF progress with the aim of delineating a possible mechanism of action for the compound.
MATERIALS AND METHODS
Tissue Samples
A total of 10 oral submucosal fibrosis tissues and 10 normal oral mucous samples were obtained from The First Affiliated Hospital (Zhengzhou, P.R. China) according to the Legislation and Ethical Boards of Zhengzhou University. Informed consents were signed by all subjects. All samples were collected and identified by histopathological evaluation and stored in liquid nitrogen until use.
Cell Culture
Primary hOMFs were obtained from the normal gingiva around sound premolars using the previously described method11. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 12% fetal bovine serum (FBS; Life Technologies) in a humidified atmosphere of 5% CO2 air at 37°C. Cells between the fifth and eighth passages were used for the experiments.
The human oral cancer cell line SCC-9 and human embryonic kidney 293 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Life Technologies) supplemented with 10% FBS at 37°C in a humidified 5% CO2 incubator.
Cell Treatment
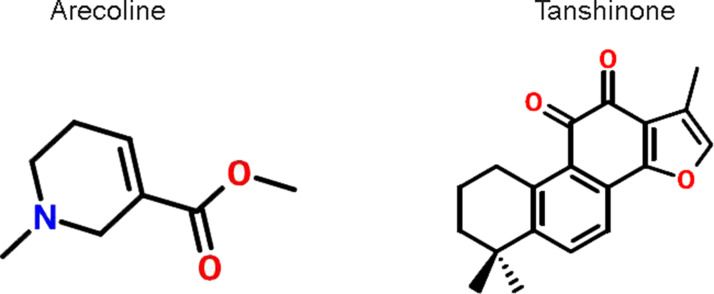
The lentivirus containing short hairpin RNA (shRNA)-p53 plasmid, shRNA-lysine-specific demethylase 1A (LSD1) plasmid, or LSD1-expressing plasmid were designed and purchased from GeneChem (Shanghai, P.R. China). Following 48 h of transfection, according to the manufacturer’s instructions (Lipofectamine 3000; Thermo Fisher Scientific, Waltham, MA, USA), the transfected cells were used for further analysis. Arecoline (Are; chemical formula: C8H13NO2; molecular weight: 155.2 kDa) and TSN IIA (TSN; chemical formula: C19H18O3; molecular weight: 294.3 kDa) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Their chemical structures are shown in Figure 1. The cells were treated with a range of concentrations of arecoline (20, 40, 80, and 160 μg/ml) or TSN (5, 10, 20, and 50 μM) for 48 h and then used for further analysis.
Figure 1.
The chemical structures of arecoline (Are) and Tanshinone (TSN).
Immunofluorescence Assays
Cells cultured on coverslips were fixed with 4% formaldehyde for 30 min and then permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 20 min. After that, cells were blocked with 5% nonfat dry milk in tris-buffered saline (TBS) and Tween-20 (TBST) for 60 min and incubated with vimentin or keratin antibody (1:100; Abcam, Cambridge, UK) at 4°C overnight. The cells were incubated with Alexa Fluor® 555 goat anti-rabbit IgG (1:200; Sigma-Aldrich) at 37°C for 1 h. The coverslips were stained with 4′,6-diamidino-2-phenylindole (DAPI; 1:1,000; SC-3598; Santa Cruz, Dallas, TX, USA) for 2 min and mounted on slides using antifade mounting medium. Immunofluorescence images were acquired on a Nikon ECLIPSE 80i microscope (Nikon, Tokyo, Japan).
MTT Assay
Cell proliferation was assessed every 24 h by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. After the indicated treatment, cells were seeded onto 96-well plates at 3 × 103 cells/well. Briefly, cells were exposed to MTT (Sigma-Aldrich) at a final concentration of 5 mg/ml and incubated for an additional 4 h at 37°C. The formazan generated in each well was dissolved in 150 ml of dimethyl sulfoxide (DMSO; Sigma-Aldrich). Absorbance of each well at 490 nm was then read using a microplate reader.
Western Blot
Radioimmunoprecipitation assay (RIPA) lysis buffer was used to extract protein from tissues and indicated cells. BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA) was used to measure the protein concentration. A total of 60 μg of proteins was separated on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto 0.22-μm nitrocellulose membranes. The membranes were blocked for 2 h with 5% nonfat dry milk diluted with TBS and incubated with primary antibodies [mouse monoclonal anti-p53 (1:8,000), mouse monoclonal anti-LSD1 (1:1,000), rabbit polyclonal anti-N-cadherin (1:3,000), rabbit polyclonal anti-vimentin (1:2,000), rabbit polyclonal anti-p21 (1:3,000), and rabbit polyclonal anti-p53 upregulated modulator of apoptosis (puma; 1:2,000) from Abcam; rabbit polyclonal anti-E-cadherin (1:1,000), rabbit polyclonal anti-B-cell CLL/lymphoma 2-associated X protein (BAX; 1:2,000), and mouse monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:2,000) from Santa Cruz] overnight at 4°C. The membranes were washed with TBST and then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, 1:2,000; goat anti-mouse, 1:2,000; Santa Cruz) for 1 h at 37°C. Enhanced chemiluminescence reagent (Millipore, Billerica, MA, USA) was used to detect the signal on the membrane. The data were analyzed via densitometry using Image-Pro Plus software version 6.0 (Bio-Rad, Cambridge, MA, USA) and normalized to the expression of the internal control (GAPDH).
mRNA Microarray Analysis
mRNA microarrays (HOA 7.1) were used for differentially expressed gene analysis. Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Shenzhen, P.R. China) according to the manufacturer’s instructions. The test sample and the reference were labeled with cyanine 5 (Cy5) and cohybridized to the mRNA arrays. The hybridized arrays were scanned by Agilent 0.1 XDR (Phalanx Biotech, Hsinchu City, Taiwan). Further analysis was done using the Rosetta Resolver 7.2. Expression profile clustering and visualization were performed with unsupervised hierarchical clustering analysis (Ernest Orlando Lawrence Berkeley National Laboratory, Berkeley, CA, USA).
Measurement of the TP53 Promoter CpG Island Methylation Status by Methylation-Specific Polymerase Chain Reaction (PCR; MSP)
Genomic DNA was extracted using the Qiagen FFPE DNA Kit (Qiagen, Valencia, CA, USA). Genomic DNA (1 μg per sample) was modified with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo, Orange County, CA, USA) according to the manufacturer’s instructions. MSP was performed on the bisulfate-treated DNA. The primers used were unmethylated TP53, 5’-GGTTTTAGGATTAGAGGATAGATTGA-3’ (forward) and 5′-CCTTCCTACAAAA AAAACAAACAAA-3′ (reverse); and methylated TP53, 5′-GGGTTTTAGGATTAGAG GATAGATC-3′ (forward) and 5′-CTTCCTACAAAAAAAACAAACGAA-3′ (reverse). The annealing temperature was 58°C for both methylated and unmethylated PCR, with 35 cycles used for each.
Luciferase Reporter Assay
The p53 promoter region (2,000 bp) was synthesized and inserted into a pGL3-basic vector (Promega, Madison, WI, USA). The successful constructs were verified by DNA sequencing. The Dual-Luciferase Assay Kit was used to assess luciferase activities, following the manufacturer’s protocol. The cells were plated into 96-well clusters and then cotransfected with 100 ng of pGL3-basic vector or pGL3-p53, together with LSD1 or the negative control. Forty-eight hours after transfection, luciferase activity was detected using a Dual-Luciferase Reporter Assay system (Promega) and normalized to Renilla luciferase activity.
RNA Immunoprecipitation (RIP)
RIP assay was used to determine whether TP53 interacts with or binds to the RNA-binding protein LSD1 in the human hypopharyngeal carcinoma cells. The EZMagna RIP Kit (Millipore) was used to conduct the RIP experiment, following the manufacturer’s protocol. The cells were lysed using complete RIP lysis buffer; then the extract was incubated with magnetic beads conjugated with LSD1 antibodies or control IgG (Millipore) for 8 h at 4°C. Next, the beads were washed with washing buffer and incubated with proteinase K at 55°C for 30 min to remove the proteins. Finally, purified RNA was reverse transcribed into cDNA and subjected to qualitative real-time PCR (qPCR) analysis to determine the presence of TP53 using specific primers.
Chromatin Immunoprecipitation (ChIP) Assay
The EZ-Magna ChIP Kit (Millipore) was used to conduct ChIP assays in accordance with the manufacturer’s protocol. The cells were fixed with 4% paraformaldehyde and incubated with glycine for 10 min to generate DNA–protein cross-links. The cells were then lysed with Cell Lysis Buffer and Nuclear Lysis Buffer and sonicated to generate chromatin fragments. Next, the lysates were immunoprecipitated with Magnetic Protein A Beads conjugated with methylated lysine 27 histone A3 (H3K27me1; Millipore), dimethylated lysine 4 histone A3 H3K4me2-specific antibodies (Millipore), or IgG as a control. Finally, the precipitated DNA was analyzed by qualitative real-time reverse transcriptase PCR (qRT-PCR).
Tumor Xenograft in Nude Mice
Animal experiments were approved by the Ethical Committee for Animal Research of Zhengzhou University (Protocol No. 2013-020). All nude mice (4–5 weeks old, male) were purchased from the Central Animal Facility of Zhengzhou University. To assess tumor growth, 200 ml of SCC-9 cells (1 × 106) was subcutaneously injected into the left side to the rear of each mouse (five mice per group). One day after injection, the mice were intragastrically administrated with either arecoline (100 mg/kg) or TSN (150 mg/kg), or cotreated with both arecoline (100 mg/kg) and TSN (150 mg/kg), once a day for 7 weeks. Tumor size was measured regularly and calculated using the formula 0.52 × L × W 2, where L and W are the long and short diameters of the tumor, respectively.
Immunohistochemical (IHC) Staining
The paraffin-embedded tumor sections of SCC-9 xenografts were serially cut at 4 μm. Slides were deparaffinized in dimethylbenzene and dehydrated in an alcohol gradient. Slides were immersed in 3% H2O2 to inactivate endogenous peroxidase, and then the antigens were retrieved by boiling in citric acid buffer (pH 6.0) in a microwave oven at high power for 15 min. After cooling, the slices were blocked with normal goat serum and incubated with primary antibody rabbit polyclonal anti-Ki-67 antibody (1:100 dilution; Cat No. GTX16667; Genetex, Hsinchu, Taiwan) overnight at 4°C. The sections were then washed with PBS and incubated with anti-rabbit secondary antibody (Zsbio, Beijing, P.R. China) for 2 h at 37°C. The sections were then washed with PBS and stained using a diaminobenzidine (DAB) detection kit (Zsbio). Finally, the sections were counterstained with hematoxylin. The images were obtained using a microscope (DMB5-2231P1; Motic)
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA), and the data are presented as the mean ± standard deviation. An unpaired two-tailed Student’s t-test or one-way analysis of variance (ANOVA) with Bonferroni adjusted t-test or Dunnett’s multiple comparison test performed post hoc was used to analyze the data depending on the conditions. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
Treatment With TSN Inhibits Arecoline-Mediated Increase in Cell Viability in Oral Mucosal Fibroblasts
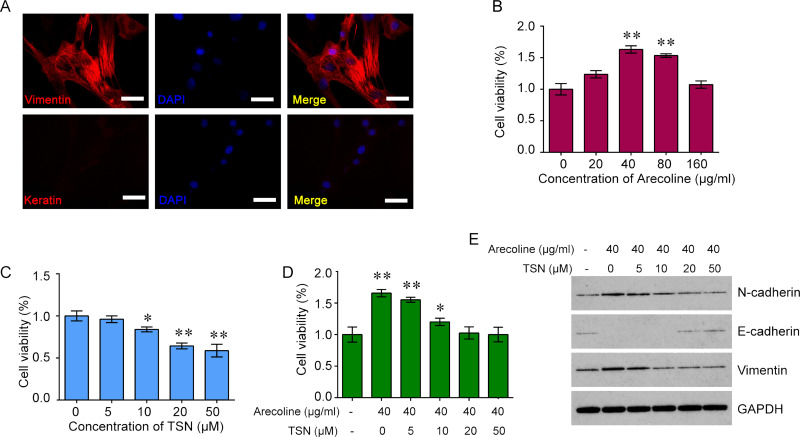
To investigate the role of TSN on arecoline-induced oral submucosal fibrosis, we first isolated the hOMFs from the normal gingiva around sound premolars. Immunofluorescence assay was performed to confirm the isolated cells. The cells were positive for vimentin but negative for keratin, verifying that the cells were oral mucosal fibroblasts (Fig. 2A). To determine the effects of arecoline and TSN on the cell viability of oral mucosal fibroblasts, we treated the cells with a range of concentrations of arecoline (20, 40, 80, and 160 μg/ml) or TSN (5, 10, 20, and 50 μM) and found that treatment with arecoline significantly increased the cell viability of oral mucosal fibroblasts at 40 and 80 μg/ml, whereas treatment with TSN reduced the cell viability of oral mucosal fibroblasts in a dose-dependent manner (Fig. 2B and C). In addition, we cotreated with arecoline (40 μg/ml) and TSN (5, 10, 20, and 50 μM) for 48 h in oral mucosal fibroblasts and found that treatment with TSN inhibited the arecoline-mediated increase in cell viability of oral mucosal fibroblasts (Fig. 2D). Furthermore, we analyzed the key molecules of the EMT pathway, including E-cadherin, N-cadherin, and vimentin. The results showed that treatment with arecoline decreased the expression of E-cadherin but increased the expression of N-cadherin and vimentin in oral mucosal fibroblasts, whereas cotreatment with TSN reversed the effects of arecoline on the expression of E-cadherin, N-cadherin, and vimentin in a dose-dependent manner (Fig. 2E).
Figure 2.
The effects of Are and TSN on cell proliferation in oral mucosal fibroblasts (OMFs). (A) The expression of two markers of fibroblasts, vimentin and keratin, in primary culture human oral mucosal fibroblasts (hOMFs). Representative immunofluorescence (IF) staining for vimentin (red) in fibroblasts (top). Keratin is negatively expressed in fibroblasts (bottom). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Original magnification: 400×; scale bar: 50 μm. (B) Treatment with Are increased the cell viability of OMFs in a dose-dependent manner. (C) Treatment with TSN reduced the cell viability of OMFs in a dose-dependent manner. (D) Treatment with TSN inhibited the Are-mediated increase in cell viability of OMFs. (E) Treatment with Are decreased the expression of epithelial (E)-cadherin but increased the expression of neural (N)-cadherin and vimentin in OMFs, whereas cotreatment with TSN reversed the effects of Are on the expression of E-cadherin, N-cadherin, and vimentin in a dose-dependent manner. One-way ANOVA and Dunnett’s multiple comparison test. Mean ± SEM. n = 5. *p < 0.05, **p < 0.01 compared to control (no drug).
TSN Rescues Arecoline-Mediated Downregulation of TP53
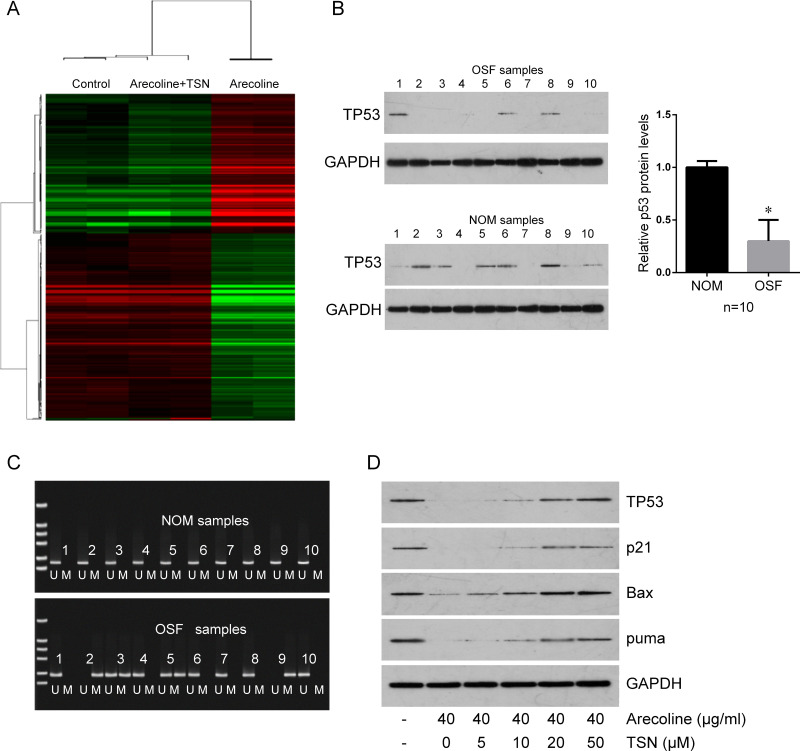
To further investigate the mechanism by which TSN inhibits arecoline-induced OSF, the primary oral mucosal fibroblasts were treated with arecoline alone or cotreated with arecoline and TSN. The untreated cells were used as a control. Total RNA was extracted and used for mRNA expression profile microarray screening. The microarray revealed 1,606 differentially expressed mRNAs between arecoline-treated cells and control cells (Fig. 3A). The top 10 altered genes are listed in Table 1. Of note, TP53 gene expression was significantly decreased in arecoline-treated cells compared with control cells, but rescued in cells cotreated with arecoline and TSN; LSD1 was significantly upregulated by arecoline, which was rescued by TSN (Table 1). We further confirmed that the expression of the wild type of p53 was significantly decreased in 10 cases of OSF tissues compared with 10 normal oral mucous samples (Fig. 3B). The methylation levels of the promoter of TP53 in OSF samples and normal oral mucous samples were determined by MSP. The methylation levels of the promoter of TP53 were higher in OSF tissues than in normal oral mucous tissues (40% vs. 0%) (Fig. 3C), indicating that the low levels of wild-type p53 in OSF tissues may be induced by hypermethylation of the promoter. Arecoline treatment also inhibited p53 downstream molecules (p21, Bax, and puma), which were rescued by TSN treatment (Fig. 3D).
Figure 3.
TSN rescues Are-mediated downregulation of tumor protein 53 (TP53) mRNA and p53 protein. (A) mRNA expression profile microarray screening. Heatmap of the genes differentially expressed in the control, cells treated with Are alone, and cells cotreated with Are plus TSN. Red represents upregulated mRNAs and green for downregulated mRNAs. (B) Western blot was used to determine the expression of p53 in 10 oral submucous fibrosis (OSF) samples (top) and 10 normal oral mucous (NOM) samples (bottom). Quantification is shown on the right. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for normalizing the expression of p53. (C) The methylation levels of the promoter of TP53 in the same 10 OSF samples and 10 NOM samples in (B) was determined by methylation-specific polymerase chain reaction (MSP). U, unmethylated; M, methylated. (D) Treatment with TSN reversed Are-mediated downregulation of p53, cyclin-dependent kinase inhibitor 1A (p21), B-cell CLL/lymphoma 2-associated X protein (Bax), and p53 upregulated modulator of apoptosis (puma) in primary culture OMFs in a dose-dependent manner. GAPDH was used as a protein-loading control. *p < 0.05 compared to NOM.
Table 1.
The Top 10 Significantly Altered (Upregulated and Downregulated) Genes in Cells Treated With Tanshinone (TSN) or Arecoline (Are)
| Relative Expression | |||
|---|---|---|---|
| Control | Are | Are + TSN | |
| Upregulated genes | |||
| CYTL1 | 1 | 3.7 | 1.77 |
| PDK4 | 1 | 4.5 | 0.74 |
| FAM49B | 1 | 2.1 | 0.58 |
| LUZP2 | 1 | 3.2 | 0.95 |
| LSD1 | 1 | 11.6 | 1.23 |
| TBX1 | 1 | 2.7 | 1.72 |
| APOOL | 1 | 2.5 | 0.81 |
| CHST13 | 1 | 9.3 | 0.93 |
| TCAM1P | 1 | 2.0 | 0.85 |
| ABCB1 | 1 | 2.4 | 0.99 |
| Downregulated genes | |||
| WFDC1 | 1 | 0.22 | 1.37 |
| ANO2 | 1 | 0.25 | 1.65 |
| MAGEA6 | 1 | 0.31 | 0.94 |
| TVP23A | 1 | 0.42 | 0.95 |
| GSTP1 | 1 | 0.37 | 0.73 |
| HAMP | 1 | 0.16 | 1.09 |
| KLHL13 | 1 | 0.27 | 1.49 |
| TP53 | 1 | 0.04 | 1.31 |
| GSTP1 | 1 | 0.18 | 1.10 |
| CGA | 1 | 0.23 | 1.19 |
TSN Reverses Arecoline-Mediated Hypermethylation of TP53 Promoter by Inhibiting LSD1
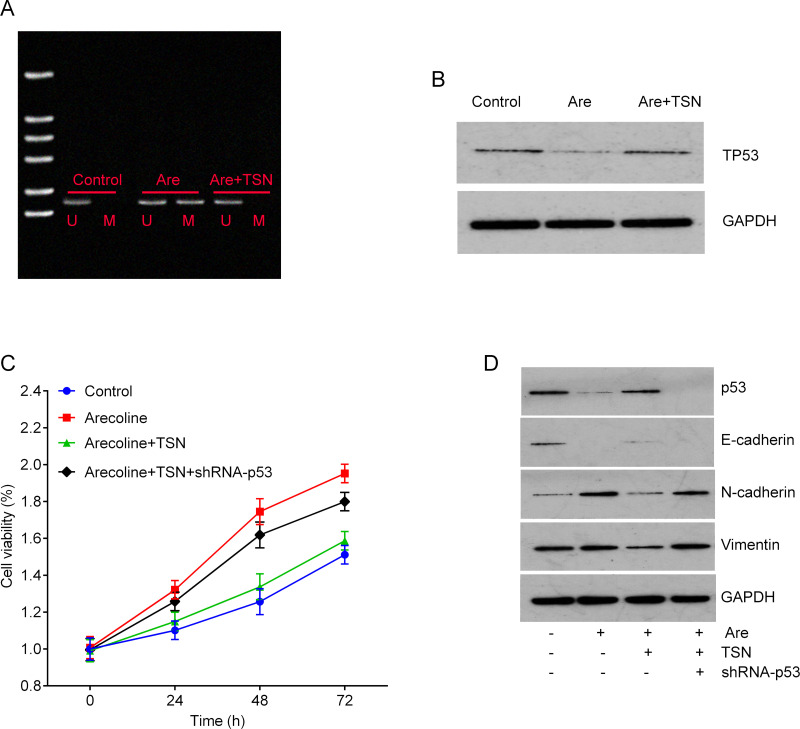
We further investigated whether arecoline and TSN treatment can affect the hypermethylation of the p53 promoter. Our results showed that the promoter of TP53 was hypermethylated by arecoline treatment, which was reversed by TSN treatment (Fig. 4A). In addition, treatment with TSN reversed arecoline-mediated downregulation of p53 in primary culture oral mucosal fibroblasts (Fig. 4B). Notably, TSN treatment abolished arecoline-induced fibroblast proliferation, and this effect was attenuated by knockdown of TP53 by shRNA (Fig. 4C). Predictably, knockdown of TP53 abolished the effects of TSN on the expression of E-cadherin, N-cadherin, and vimentin, suggesting that TSN regulates the EMT process through the p53 pathway (Fig. 4D).
Figure 4.
TSN suppresses cell viability of primary culture OMFs through increasing the expression of TP53. (A) The methylation levels of the promoter of TP53 in primary culture OMFs were determined by MSP. (B) Treatment with TSN reversed the Are-mediated downregulation of p53 in primary culture OMFs. GAPDH was used as a protein loading control. (C) MTT assay was used to measure cell viability after the indicated treatment. Knockdown of TP53 by short hairpin RNA (shRNA) abolished the inhibitory effects of TSN on the cell viability of OMFs induced by Are. (D) Knockdown of TP53 abolished the effects of TSN on the expression of E-cadherin, N-cadherin, and vimentin evaluated by Western blot.
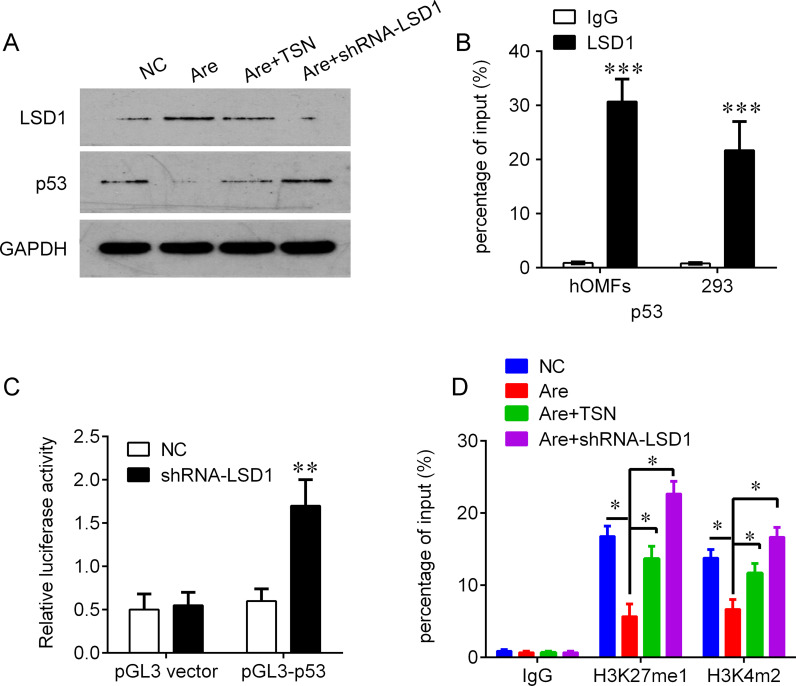
Because of the negative correlation between LSD1 and p53, we further investigated whether LSD1, a histone transmethylase, could regulate the expression of p53. We found that knockdown of LSD1 could significantly increase the protein levels of p53 under the arecoline challenge (Fig. 5A). We further investigated the mechanism by which LSD1 regulates p53. ChIP assays were performed to evaluate whether LSD1 could bind to the TP53 promoter. The results showed that LSD1 could bind to the promoter regions of TP53 (Fig. 5B). In addition, the promoter region of TP53 was inserted into a PGL3 luciferase reporter vector, and Dual-Luciferase Reporter analysis showed that knockdown of LSD1 could activate luciferase (Fig. 5C). Moreover, we found that knockdown of LSD1 could increase the binding of H3K27me1 and H3K4m2 on the TP53 promoter, which was similar to the TSN treatment (Fig. 5D). These results indicate that TSN may epigenetically activate p53 in OSF by inhibiting LSD1.
Figure 5.
Knockdown of lysine-specific demthylase 1 (LSD1) by shRNA epigenetically activates p53 by recruiting methylated lysine 27 histone 3 (H3K27me1) and dimethylated lysine 4 histone 3 (H3K4me2). (A) Knockdown of LSD1 abolished the effects of Are on the expression of p53 evaluated by Western blot. (B) Chromatin immunoprecipitation-qualitative real-time polymerase chain reaction (ChIP-qPCR) analysis of LSD1 occupancy in the TP53 promoter regions in the OMFs and human embryonic kidney 293 cells. Immunoglobulin G (IgG) was used as a negative control. (C) Luciferase reporter analysis of luciferase activity in the OMFs cotransfected with pGL3-TP53 and shRNA-LSD1 lentivirus or an empty lentivirus (pGL3 vector). (D) ChIP-qPCR analysis of H3K27me1 and H3K4me2 occupancy in the TP53 promoters in the OMFs after the indicated treatment. IgG was used as a negative control. *p < 0.05, **p < 0.01, ***p < 0.001 compared with IgG in (B), negative control (NC) in (C), and Are treatment in (D).
TSN Treatment Suppresses Tumor Growth of Oral Carcinoma In Vivo
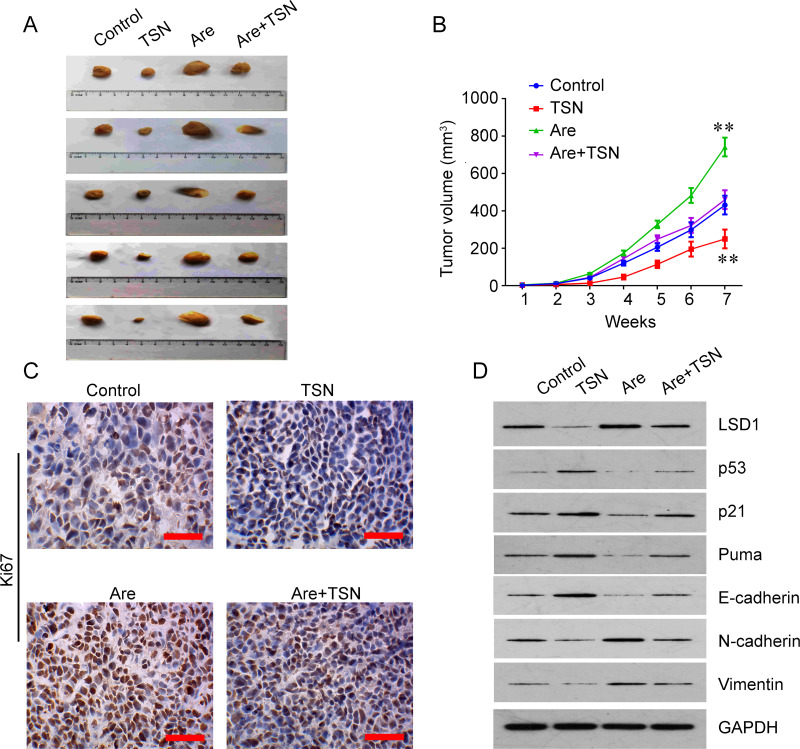
We next investigated whether TSN can suppress the tumor growth of oral carcinoma in vivo. A xenografted tumor assay was used to determine the suppressor role of TSN in vivo. TSN treatment decreased tumor growth, but arecoline treatment increased the tumor growth in nude mice, and TSN treatment had an antagonistic effect with arecoline on tumor growth in vivo (Fig. 6A and B). IHC staining for Ki-67 in tumor sections of SCC-9 xenografts showed that expression of the proliferative marker Ki-67 was decreased by TSN and increased by arecoline (Fig. 6C). Treatment with arecoline decreased the expression of LSD1 and E-cadherin but increased the expression of N-cadherin and vimentin, while TSN treatment had an opposite role. Cotreatment with TSN reversed the effects of arecoline on the expression of LSD1, p53, p21, puma, E-cadherin, N-cadherin, and vimentin (Fig. 6D).
Figure 6.
TSN treatment suppresses tumor growth of oral carcinoma in vivo. (A) Xenografted tumors collected on day 49 after subcutaneous implantation of SCC-9 cells. TSN treatment decreased tumor growth, but Are treatment increased the tumor growth in nude mice; TSN treatment had an antagonistic effect with Are on tumor growth in vivo. (B) Tumor growth curves (n = 5). Mean ± SEM. (C) Representative images of immunohistochemistry (IHC) staining for Ki-67 in tumor sections of SCC-9 xenografts. The expression of Ki-67 was decreased by TSN and increased by Are. Original magnification: 400×; scale bar: 50 μm. (D) Western blot was used to determine the expression of LSD1, p53, p21, puma, E-cadherin, N-cadherin, and vimentin in tumor tissues of SCC-9 xenografts. **p < 0.01 compared to control.
DISCUSSION
In this study, we found that treatment with TSN inhibited arecoline-mediated proliferation and EMT in oral mucosal fibroblasts. It is considered that OSF is a precancerous lesion. The association of areca nut chewing, OSF, and oral squamous cell carcinoma is quite profound, especially in Taiwan and South China, where up to 80% of oral squamous cell carcinoma is associated with the habit12. A range of case-control studies have demonstrated that there is a definite dose-dependent relation between the areca nut and causation of the disease, and it is well known that the onset of the disease is directly proportional to the concentration, incidence, and duration of chewing the nut13,14. Among the alkaloids present in areca nuts, arecoline is the main agent responsible for fibroblast proliferation. The localized mucosal inflammation caused by arecoline results in the recruitment of activated T cells that leads to the release of cytokines and transforming growth factor-β (TGF-β)15. In epithelial cells, TGF-β is able to induce EMT. Inflammation and wounding stress in epithelial cells can promote the EMT process, leading to fibroblast production and fibrogenesis16,17. Emerging evidence implies that EMT remains a very likely candidate in the processes of tumorigenesis and carcinoma metastasis18.
TSNs are potent cytotoxic agents that significantly inhibit the growth of various types of cancer cells including colon, breast, liver, and prostate and the blood cells in leukemia by inducing cell cycle arrest and apoptosis19–22. These proapoptosis effects are potentially involved in the activation of the p53 pathway, such as upregulation of proapoptosis proteins including p53, Bax, p21, puma, etc23. In addition, TSNs are able to inhibit the invasion and metastasis of cancer cells through the alteration of matrix metalloproteinases24. TSNs also have a synergistic anticancer effect with chemotherapeutic drugs such as cisplatin, doxorubicin, 5-flurouracil, and arsenic trioxide25. Notably, TSN IIA exerted a strong radiosensitizing effect on oral squamous carcinoma cells compared with the simple drug or single radiation treatment by enhancing reactive oxygen species (ROS) generation and autophagy26. Moreover, increasing evidence supports the important role of inflammatory factors for tumorigenesis and therapeutic responses. For example, TSNs at submicromolar and low micromolar levels inhibited interleukin-12 (IL-12) production in lipopolysaccharide (LPS)-activated mouse macrophages in a concentration-dependent manner27.
To explore the underlying mechanisms of TSN in oral submucosal fibrosis induced by arecoline, we exploited the integrated strategy consisting of RNA deep sequencing to investigate targeted genes. We found that the p53 pathway is the major cellular target of TSN in OSF. p53, one of the most frequently mutated and deleted tumor suppressors in a multitude of tumor types, has been frequently reported to be downregulated or deleted in oral squamous cell carcinoma, suggesting other mechanisms in OSF, the precancerous lesion likely responsible for the downregulation of p53. Consistently, we found that the expression of p53 was much lower in OSF than in normal mucous tissues. In addition, we found that arecoline treatment results in a significant decrease in p53 and its downstream molecules including p21, Bax, and puma. Treatment with TSN reversed the arecoline-mediated decrease in the expression of p53, p21, Bax, and puma in primary culture oral mucosal fibroblasts in a dose-dependent manner. Furthermore, our results demonstrate that arecoline stimulation resulted in hypermethylation of the promoter of TP53 and subsequent downregulation of p53 levels. Importantly, TSN treatment can epigenetically demethylate the promoter of TP53 and upregulate the p53 levels by inhibiting LSD1 expression.
One of the early genetic studies found a statistically significant increase in the size of C-band heteromorphism patterns on chromosome 1 in OSF and oral squamous cell carcinoma patients when compared with healthy subjects. OSF lesions abnormally express and stain positive for p53 with a specific clustered mutation in exon 5 of the p53 gene. Expression levels of p53 are progressively lower in oral squamous cell carcinoma than in submucosal fibrosis lesions28,29. In line with these findings, our RNA-deep sequencing results showed lower levels of TP53 in arecoline-treated cells than in control cells and in cells cotreated with arecoline and TSN.
It was demonstrated that TSN IIA could interact with the DNA minor groove, and in turn activate p53 to induce apoptosis, which was independent of ataxia–telangiectasia mutated (ATM) activation, suggesting that TSN-induced p53 activation in these cells was not through the ATM-mediated DNA damage response pathway10,30. Moreover, in vivo administration of TSN IIA (40 mg/kg of TSN IIA, every other day for 10 days) induced apoptosis in hepatic cancer cell H22 xenograft tumors10,30. Wang et al. found that dihydrotanshinone provoked mitochondrial dysfunction at an early stage by decreasing mitochondrial membrane potential and mitochondrial ROS levels followed by a time-dependent increase in intracellular ROS generation, which was reversed by pretreatment with free radical scavengers, suggesting that dihydrotanshinone induces apoptosis in colon cancer cells through a p53-independent pathway31. Lu et al. showed that TSN I-mediated Aurora A inhibition results in wild-type p53 upregulation, but not mutant p53, which is required for cell apoptosis in colorectal cancer cells23. Here our results demonstrated a novel mechanism that TSN epigenetically activates the p53 pathway by inhibiting LSD1. LSD1, the histone lysine-specific demethylase, acts as an epigenetic regulator and is overexpressed in various types of tumors32,33. LSD1 has been shown to demethylate histone H3 on lysine 4 (H3K4) and lysine 9 (H3K9). LSD1 is essential for mammalian development and is likely involved in many biological processes. LSD1 contains several defined domains and is associated with a number of protein complexes34. LSD1 can interact with p53 to repress p53-mediated transcriptional activation and to inhibit the role of p53 in promoting apoptosis by removing both monomethylation (K370me1) and dimethylation (K370me2) at K370. LSD1 represses p53 function through inhibition of the interaction of p53 with p53-binding protein 1 (53BP1)35. Interestingly, cryptotanshinone could increase the monomethylation and dimethylation of histone H3 lysine 9 (H3K9) in the promoter region of the androgen receptor (AR) by disrupting the interaction between AR and LSD1 to inhibit the transcriptional activity of AR and suppress the expression of several AR target genes at the mRNA and protein levels36. Here we showed that arecoline could induce LSD1 and result in p53 downregulation. This effect was reversed by increasing the binding of H3K27me1 and H3K4m2 on the TP53 promoter with TSN treatment. These results indicate that TSN may epigenetically activate TP53 in OSF by inhibiting LSD1.
In conclusion, we describe a novel mechanism whereby TSN reactivates p53 signaling silenced by arecoline via the functional inhibition of LSD1-mediated demethylation of H3K27me1 and H3K4me2. Our data suggest that TSN can be developed as a potential therapeutic agent for OSF and even oral squamous cell carcinoma.
ACKNOWLEDGMENT
The study was supported by a grant from the National Natural Science Foundation of China (Grant No. 81400492).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Wollina U, Verma SB, Ali FM, Patil K. Oral submucous fibrosis: An update. Clin Cosmet Investig Dermatol. 2015;8:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li M, Gao F, Zhou ZS, Zhang HM, Zhang R, Wu YF, Bai MH, Li JJ, Lin SR, Peng JY. Arecoline inhibits epithelial cell viability by upregulating the apoptosis pathway: Implication for oral submucous fibrosis. Oncol Rep. 2014;31:2422–8. [DOI] [PubMed] [Google Scholar]
- 3. Lee YH, Yang LC, Hu FW, Peng CY, Yu CH, Yu CC. Elevation of Twist expression by arecoline contributes to the pathogenesis of oral submucous fibrosis. J Formos Med Assoc. 2016;115:311–7. [DOI] [PubMed] [Google Scholar]
- 4. Wang TY, Peng CY, Lee SS, Chou MY, Yu CC, Chang YC. Acquisition cancer stemness, mesenchymal transdifferentiation, and chemoresistance properties by chronic exposure of oral epithelial cells to arecoline. Oncotarget 2016;7:84072–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang YC, Tsai CH, Tai KW, Yang SH, Chou MY, Lii CK. Elevated vimentin expression in buccal mucosal fibroblasts by arecoline in vitro as a possible pathogenesis for oral submucous fibrosis. Oral Oncol. 2002;38:425–30. [DOI] [PubMed] [Google Scholar]
- 6. Chang YC, Tsai CH, Lai YL, Yu CC, Chi WY, Li JJ, Chang WW. Arecoline-induced myofibroblast transdifferentiation from human buccal mucosal fibroblasts is mediated by ZEB1. J Cell Mol Med. 2014;18:698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng L, Jian X, Guo F, Li N, Jiang C, Yin P, Min AJ, Huang L. miR-203 inhibits arecoline-induced epithelial-mesenchymal transition by regulating secreted frizzled-related protein 4 and transmembrane-4 L six family member 1 in oral submucous fibrosis. Oncol Rep. 2015;33:2753–60. [DOI] [PubMed] [Google Scholar]
- 8. Chen Z, Xu H. Anti-inflammatory and immunomodulatory mechanism of Tanshinone IIA for atherosclerosis. Evid Based Complement Alternat Med. 2014;2014:267976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su CC. Tanshinone IIA inhibits gastric carcinoma AGS cells through increasing p-p38, p-JNK and p53 but reducing p-ERK, CDC2 and cyclin B1 expression. Anticancer Res. 2014;34:7097–110. [PubMed] [Google Scholar]
- 10. Zhang Z, Gao J, Wang Y, Song T, Zhang J, Wu G, Zhang T, Du G. Tanshinone IIA triggers p53 responses and apoptosis by RNA polymerase II upon DNA minor groove binding. Biochem Pharmacol. 2009;78:1316–22. [DOI] [PubMed] [Google Scholar]
- 11. Janke V, von Neuhoff N, Schlegelberger B, Leyhausen G, Geurtsen W. TEGDMA causes apoptosis in primary human gingival fibroblasts. J Dent Res. 2003;82:814–8. [DOI] [PubMed] [Google Scholar]
- 12. Chen HM, Hsieh RP, Yang H, Kuo YS, Kuo MY, Chiang CP. HLA typing in Taiwanese patients with oral submucous fibrosis. J Oral Pathol Med. 2004;33:191–9. [DOI] [PubMed] [Google Scholar]
- 13. Tilakaratne WM, Klinikowski MF, Saku T, Peters TJ, Warnakulasuriya S. Oral submucous fibrosis: Review on aetiology and pathogenesis. Oral Oncol. 2006;42:561–8. [DOI] [PubMed] [Google Scholar]
- 14. Jacob BJ, Straif K, Thomas G, Ramadas K, Mathew B, Zhang ZF, Sankaranarayanan R, Hashibe M. Betel quid without tobacco as a risk factor for oral precancers. Oral Oncol. 2004;40:697–704. [DOI] [PubMed] [Google Scholar]
- 15. Warnakulasuriya S, Kerr AR. Oral submucous fibrosis: A review of the current management and possible directions for novel therapies. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122:232–41. [DOI] [PubMed] [Google Scholar]
- 16. Yanjia H, Xinchun J. The role of epithelial-mesenchymal transition in oral squamous cell carcinoma and oral submucous fibrosis. Clin Chim Acta 2007;383:51–6. [DOI] [PubMed] [Google Scholar]
- 17. Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan J, Liu M, Yang L, Tu G, Zhu Q, Chen M, Cheng H, Luo H, Fu W, Li Z, Yang G. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: A new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and beta1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015;17:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jing X, Xu Y, Cheng W, Guo S, Zou Y, He L. Tanshinone I induces apoptosis and pro-survival autophagy in gastric cancers. Cancer Chemother Pharmacol. 2016;77:1171–81. [DOI] [PubMed] [Google Scholar]
- 20. Bai Y, Zhang L, Fang X, Yang Y. Tanshinone IIA enhances chemosensitivity of colon cancer cells by suppressing nuclear factor-kappaB. Exp Ther Med. 2016;11:1085–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim EO, Kang SE, Im CR, Lee JH, Ahn KS, Yang WM, Um JY, Lee SG, Yun M. Tanshinone IIA induces TRAIL sensitization of human lung cancer cells through selective ER stress induction. Int J Oncol. 2016;48:2205–12. [DOI] [PubMed] [Google Scholar]
- 22. Su CC. Tanshinone IIA decreases the migratory ability of AGS cells by decreasing the protein expression of matrix metalloproteinases, nuclear factor kappaB-p65 and cyclooxygenase-2. Mol Med Rep. 2016;13:1263–8. [DOI] [PubMed] [Google Scholar]
- 23. Lu M, Wang C, Wang J. Tanshinone I induces human colorectal cancer cell apoptosis: The potential roles of Aurora A-p53 and survivin-mediated signaling pathways. Int J Oncol. 2016;49:603–10. [DOI] [PubMed] [Google Scholar]
- 24. Akaberi M, Mehri S, Iranshahi M. Multiple pro-apoptotic targets of abietane diterpenoids from Salvia species. Fitoterapia 2015;100:118–32. [DOI] [PubMed] [Google Scholar]
- 25. Chang CC, Kuan CP, Lin JY, Lai JS, Ho TF. Tanshinone IIA facilitates TRAIL sensitization by up-regulating DR5 through the ROS-JNK-CHOP signaling axis in human ovarian carcinoma cell lines. Chem Res Toxicol. 2015;28:1574–83. [DOI] [PubMed] [Google Scholar]
- 26. Ding L, Wang S, Qu X, Wang J. Tanshinone IIA sensitizes oral squamous cell carcinoma to radiation due to an enhanced autophagy. Environ Toxicol Pharmacol. 2016;46:264–9. [DOI] [PubMed] [Google Scholar]
- 27. Kang BY, Chung SW, Kim SH, Ryu SY, Kim TS. Inhibition of interleukin-12 and interferon-gamma production in immune cells by tanshinones from Salvia miltiorrhiza. Immunopharmacology 2000;49:355–61. [DOI] [PubMed] [Google Scholar]
- 28. Mithani SK, Mydlarz WK, Grumbine FL, Smith IM, Califano JA. Molecular genetics of premalignant oral lesions. Oral Dis. 2007;13:126–33. [DOI] [PubMed] [Google Scholar]
- 29. Shen H, Liu L, Yang Y, Xun W, Wei K, Zeng G. Betulinic acid inhibits cell proliferation in human oral squamous cell carcinoma via modulating ROS-regulated p53 signaling. Oncol Res. 2017;25(7):1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Z, Zhang J, Jin L, Song T, Wu G, Gao J. Tanshinone IIA interacts with DNA by minor groove-binding. Biol Pharm Bull. 2008;31:2342–5. [DOI] [PubMed] [Google Scholar]
- 31. Wang L, Yeung JH, Hu T, Lee WY, Lu L, Zhang L, Shen J, Chan RL, Wu WK, Cho CH. Dihydrotanshinone induces p53-independent but ROS-dependent apoptosis in colon cancer cells. Life Sci. 2013;93:344–51. [DOI] [PubMed] [Google Scholar]
- 32. Yang CY, Lin CK, Tsao CH, Hsieh CC, Lin GJ, Ma KH, Shieh YS, Sytwu HK, Chen YW. Melatonin exerts anti-oral cancer effect via suppressing LSD1 in patient-derived tumor xenograft models. Oncotarget 2017;8(20):33756–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meng X, Li J, Zheng M, Zuo L, Sun C, Zhu Y, Fang L, Liu L, Zhou X. Stable H3 peptide was delivered by gold nanorods to inhibit LSD1 activation and induce human mesenchymal stem cells differentiation. Oncotarget 2017;8:23110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicholson TB, Chen T. LSD1 demethylates histone and non-histone proteins. Epigenetics 2009;4:129–32. [DOI] [PubMed] [Google Scholar]
- 35. Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature 2007;449:105–8. [DOI] [PubMed] [Google Scholar]
- 36. Wu CY, Hsieh CY, Huang KE, Chang C, Kang HY. Cryptotanshinone down-regulates androgen receptor signaling by modulating lysine-specific demethylase 1 function. Int J Cancer 2012;131:1423–34. [DOI] [PubMed] [Google Scholar]