Abstract
Polymersomes are a class of synthetic vesicles composed of a polymer membrane surrounding an aqueous inner cavity. In addition to their overall size, the thickness and composition of polymersome membranes determine the range of potential applications in which they can be employed. While synthetic polymer chemists have made great strides in controlling polymersome membrane parameters, measurement of their permeability to various analytes including gases, ions, organic molecules, and macromolecules remains a significant challenge. In this Outlook, we compare the general methods that have been developed to quantify polymersome membrane permeability, focusing in particular on their capability to accurately measure analyte flux. In addition, we briefly highlight strategies to control membrane permeability. Based on these learnings, we propose a set of criteria for designing future methods of quantifying membrane permeability such that the passage of a variety of molecules into and out of their lumens can be better understood.
Short abstract
Herein, we discuss the recent advances in methods to quantify polymersome membrane permeability and propose five key principles to facilitate future developments of new, more quantitative strategies.
Introduction
Structural organization is an essential characteristic of nature. Semipermeable vesicular constructs—composed of a bilayer membrane surrounding an inner aqueous environment—perform a variety of functions by compartmentalizing molecules into disparate environments.1 Proteins and other (macro)molecules are distributed throughout the cell, released into or internalized from the extracellular space via the action of membrane-bound vesicles. The list of cellular compartments possessing membranes includes endosomes and exosomes, the endoplasmic reticulum, the cell nucleus, mitochondria, ribosomes, and the cell membrane itself.
There are many benefits to compartmentalization within living cells: (1) Sensitive entities (e.g., genetic material) are protected from degradation by external forces; (2) Processes are regulated via colocalization of interacting molecules; (3) High local concentrations within membrane-bound compartments increase reaction efficiencies; (4) Different chemical processes can occur simultaneously in separate environments without affecting each other; and (5) Highly reactive conditions (e.g., the low-pH environment found within endosomes) do not harm the rest of the cell (Figure 1).1,2 All of these benefits are contingent on the capability of membranes to regulate the flow of chemicals into and out of these disparate compartments.
Figure 1.
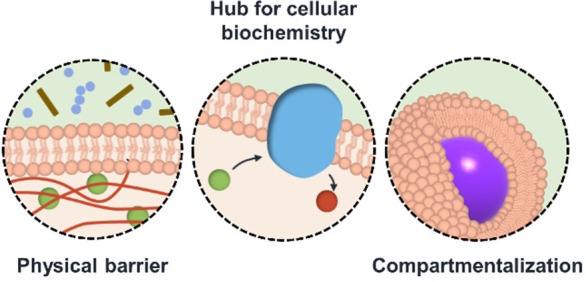
Selected functions of cellular membranes.
Nature is regarded as the expert in selective permeability, regulating with extreme precision the passage of a range of substrates from ions, gases, and small molecules to larger structures such as enzymes and proteins. This is achieved through the cohesive integration of several mechanisms of membrane transport. Small, moderately polar molecules, such as CO2, N2, O2, and ethanol, undergo passive diffusion through lipid membranes, driven by concentration or electrical gradients.3 However, this method is not sufficient for the transport of other molecules, such as charged ions or cellular metabolites, several of which are critical to key cellular processes.4 For these molecules, transfer across biological membranes is achieved via transporter-mediated entry.5−8 Selective cellular permeation of more complex molecules, such as polypeptides, neurotransmitters, and bacteria, occurs via more sophisticated mechanisms including peptide translocation domains, exocytosis, pinocytosis, and phagocytosis.3
Advances in controlled polymer synthesis have enabled chemists to create biomimetic vesicular structures comprising a wide range of amphiphilic block copolymers (known as polymersomes). When compared to traditional phospholipid vesicles, polymersomes benefit from improved stability against dilution as well as enhanced chemical diversity.9 These properties, in addition to their capacity to organize and confine molecules, make polymersomes excellent candidates for use as catalytic nanoreactors, drug delivery systems, cellular models, and nanosensors.1,10−12
While stability and functionality are key determinants of polymersome function, membrane permeability is perhaps the most important property that dictates their potential applications. For example, the membranes of catalytic nanoreactors must exhibit selective permeability to ensure catalyst retention while also allowing the permeation of substrate and product molecules across the membrane.9 In drug delivery systems, the membrane protects the therapeutic agent from decomposition (e.g., via binding to plasma proteins) while regulating its profile of release at a target location, so it is key in the efficacy of the therapeutic.13
Measuring Polymersome Permeability
The permeability of a membrane can be defined as the passive diffusion rate of analyte molecules across it. If two compartments, with concentration c1 and c2, are separated by a membrane, in the absence of other forces, Fick’s first law of diffusion relates the flux, J, and the concentration gradient, c, as
| 1 |
where D is the diffusion coefficient, and x is the position.14 Meyer and Overton later established a simple rule to predict lipid membrane permeabilities for passive diffusion using the partition coefficient, KP, of the membrane-permeating molecule from the aqueous phase into the organic phase, its diffusivity within the organic phase, DM, and the membrane thickness, d.15,16 Thus, rewriting Fick’s law of diffusion gives
| 2 |
where P is the permeability of the membrane, defined as
| 3 |
However, there are several analyte molecules that do not follow the Meyer–Overton rule, including H+, short chain carboxylic acids, and CO2.17 Due to the complexity associated with accurately determining membrane flux in polymer systems where dispersities in molecular weight, stereochemistry, and composition influence analyte permeation, this rule has yet to be shown to be directly transferable to polymeric membranes.
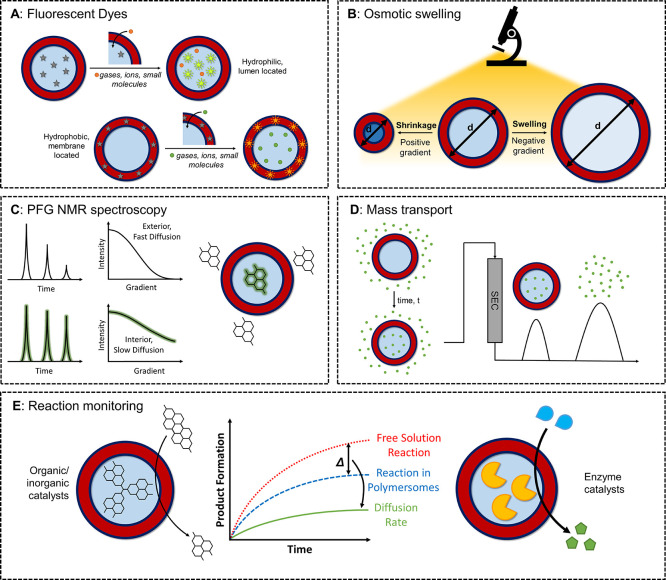
Several methodologies to investigate membrane permeability, in which an analyte molecule is tracked as it travels from one environment to another, have been explored. A schematic summary of different methods of assessing membrane permeability of polymersomes is shown in Figure 2. However, due to the difficulties in determining permeability coefficients—which are discussed in detail below—a limited number of reports containing quantitative permeability data have been published to date.18 As such, most studies report relative permeation rates for various probe molecules that cannot be directly compared between different systems.
Figure 2.
Methods for measuring polymersome membrane permeability.
Fluorescent Dyes
Many initial investigations into polymersome membrane permeability were based around the use of fluorescent dyes as a probe.19 These dyes can either be hydrophilic, and so encapsulated within the internal lumen or located in the exterior solution, or hydrophobic, thus situated within the polymeric membrane. There are two general approaches when using fluorescent dyes: encapsulation or release assays.
The first of these involves encapsulating the fluorescent dye within the lumen or membrane of the polymersome during formulation. Diffusion of an analyte across the membrane and its interaction with the probe results in a change in fluorescence output that can be detected using fluorescence spectroscopy. For this method, the assumption must be made that the fluorescent probe is retained in the desired location during polymersome preparation and purification. A wide range of probe molecules (e.g., H+, O2, R-SH, etc.) can be assessed with regards to their permeability by selecting an appropriately responsive dye. For example, the membrane permeability of H+ has been evaluated utilizing the hydrophobic dye boron-dipyrromethane (BODIPY),20 while Nile Red is effective for measuring the permeability of small molecules such as the quencher dichloroacetamide.21 Other analytes including Na+, Ca+, and adenosine triphosphate (ATP) have been investigated using this technique.22−24
For dye release assays, the analyte is first encapsulated within the polymersome, and a fluorescent probe molecule is introduced to the exterior solution. As the analyte molecule is released, a change in fluorescence output occurs that can be measured and quantified. For example, H+ diffusion across the membrane of poly(styrene)-b-poly(acrylic acid) (PS-b-PAA) polymersomes was quantified using a pH-sensitive fluorescent dye, 8-hydroxypyrene-1,3,6-trisulfonate (HPTS), in the solution outside of the polymersomes.25 In a similar study, the self-quenching dye fluorescein was encapsulated within poly(ethylene glycol)-b-poly(ε-caprolactone) (PEG-b-PCL) polymersomes above the quenching concentration.26 Upon permeation through the polymersome membrane into the exterior solution, the dye would fluoresce, as observed by spectrophotometry, allowing quantification of the polymersome membrane permeability. This method has also been applied to the release of other types of encapsulated cargo, such as the drug doxorubicin from PS-b-PAA polymersomes.27
A third approach has also been demonstrated that combines encapsulation and release, whereby fluorescent dyes of different sizes (e.g., fluorescein, PEGylated coumarin, or Rhodamine dyes, etc.) acted as both the permeating molecule and fluorescent probe simultaneously.28 However, unlike the above-mentioned reports, the rate of dye release was not assessed, and so quantification of the membrane permeability was not reported. Although the use of fluorescent dyes is endowed with simplicity, a broad scope of analyte molecules, and straightforward analysis, the method often suffers from low encapsulation percentages, high detection limits, significant dilution effects, and unwanted transfer of dye between the lumen, membrane, and exterior solution. In addition, the rate of permeation of the analyte is not measured directly in this method. Instead, the rate of fluorescence turn on/off is what is actually determined. When taken together, these limitations mean that an absolute value for permeability cannot be obtained, inhibiting an effective comparison of membrane permeability between different systems.
Osmotic Swelling
H2O transport into and out of artificial vesicular structures has been explored by observing the process of osmotic swelling and shrinking. Polymersomes loaded with salts are placed in solution such that a concentration gradient across the membrane is created.29 H2O molecules then diffuse into the lumen, causing the polymersomes to swell at a rate proportional to the concentration gradient across the polymersome membrane, which can be detected via microscopy. Concentration gradients can be established using glucose, sucrose, or NaCl.29,30 One challenge for this method is that there is difficulty in observing free-flowing objects such as polymersomes via microscopy, which possesses a restricted field of view and poor contrast. In addition, microscopic methods are currently restricted to micron-sized polymersomes.
Alternatively, Kumar et al. used stopped-flow light-scattering experiments to quantify the change in size of polymer triblock polymersomes upon introduction of an external NaCl solution.31
Although this method overcomes some of the limitations of microscopy, the technique is only relevant for cases when swelling occurs. As with the fluorescence method, the rate of diffusion of the analyte is not directly measured.
Pulsed-Field Gradient NMR Spectroscopy
More recently, researchers have used pulsed field gradient nuclear magnetic resonance (PFG-NMR) spectroscopy in probing polymersome membrane permeability. In this technique, the processional motion of the transverse magnetization, under the influence of gradient pulses, will depend on the spatial coordinates of the individual spins.32 Changes in the signal are correlated to a value of diffusivity.33 Through implementation of appropriate models, these diffusivity measurements can be used to calculate relative residence times of analyte molecules within the polymersome lumens, thus yielding an exchange rate across the membrane.
Mayer and co-workers initially explored the exchange of H2O molecules through different polymersome formulations.34 They concluded that the chemical composition of the polymeric membrane had a clear effect on the residence times of H2O within the polymersomes, with a positive correlation between polymer polarity and membrane permeability. Additionally, they found that membrane thickness was inversely proportional to membrane permeability, in agreement with results obtained from other methods.21 They later extended this work to include the permeation of poly(ethylene oxide), poly(vinyl alcohol), and glycerol as analytes across the membranes of polymersomes with varying hydrodynamic radii,35 showing the dependence of the size of the analyte molecules on permeability as well as the properties of the polymeric membrane itself. This technique has also been applied to polymersomes in ionic liquids with similar success, demonstrating expanded scope.36,37
Described as a universally applicable technique, PFG-NMR spectroscopy has been applied to measure permeability coefficients for a number of molecules across polymersome membranes, including imidazoles, glycerol, water, and polymers.34−38 The short time scale of NMR spectroscopic analysis reduces the lag time error associated with other permeability measurement techniques, as well as allowing for the calculation of diffusion coefficients for highly permeable membranes at equilibrium. It should be noted that two assumptions are typically made to simplify data analysis: (1) equal spin–spin and spin–lattice relaxations in both encapsulated and external domains; and (2) no influence of spin diffusion. However, unlike the other techniques addressed herein, PFG-NMR spectroscopy directly measures analyte diffusion rate. The key drawback in PFG-NMR spectroscopy with respect to the permeability measurement is the requirement for high-field instruments and specialized technical knowledge.
Influx Assays
Another method used to assess the membrane permeability of polymersomes is influx assays. This method places empty polymersomes into a solution of analyte and allows passive permeation to take place over a set time period, after which probe molecules in free solution are separated from polymersomes on the basis of size. The concentration of probe molecules in free solution is then measured, allowing the number of molecules that have permeated inside the polymersome to be quantified and provide an assessment of permeability.39
Several analytical instruments can be employed to measure solution concentration, allowing for a broad accessibility of this method to a range of researchers. Additionally, influx assays have been demonstrated to measure permeability for a range of molecules with different properties, including size and charge.18 However, such measurements are restricted to hydrophilic permeating molecules with negligible membrane retention. In addition, due to the changing concentration gradient over the course of the influx experiment, several assays with different time scales must be performed to provide an accurate picture of permeation rate. As with other methods, the time-averaged analyte flux is not directly measured.
Reaction Monitoring
Membrane permeability can also be quantified by physically separating one reagent in a bimolecular reaction within a polymersome lumen from a second reagent introduced into the surrounding solution. This relies on the second reagent permeating through the polymersome membrane and reacting with the encapsulated reagent, producing a measurable response (e.g., a change in solution absorbance). By choosing a reaction with fast kinetics in free solution, the overall rate of the reaction will be limited by the rate of diffusion of the second reagent. The kinetics of the polymersome reaction can then be compared to those in free solution, allowing for the calculation of the rate of diffusion, which is directly proportional to the membrane permeability.
A well-known chemical reaction was employed by Tomas and co-workers in determining the permeability of poly(ethylene oxide)-b-poly(butylene oxide) (PEO-b-PBO) polymersomes.40 The reaction between 3,3′,3″-phosphinidynetris(benzenesulfonic acid) (PH) and 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in free solution is rapid (k > 10 M–1 s–1), as measured by UV–vis spectroscopy. By encapsulating PH within the hydrophilic lumen of the polymersomes, they showed that PEO-b-PBO membranes were almost 1 order of magnitude more permeable to DTNB than phospholipids, contrary to results shown by other researchers when comparing polymeric and lipid membranes.41,42 This method benefits from the use of simple analytical equipment. Nevertheless, its scope is currently limited to assessing membrane permeability of a small number of analyte molecules, which gives minimal insight into application-specific permeability. Alongside this, it must be assumed that the analyte molecules instantaneously react with the encapsulant upon transfer into the internal lumen. As such, this assumption may generate a reduced permeability coefficient compared to the true value. In addition, this method is disadvantaged by the potential of encapsulant leakage and may cause inaccuracies in permeability measurements. Similar to the other methods, the rate of analyte permeation is not directly measured.
Enzyme Activity
As an extension to the above method, encapsulation of an enzyme within the polymersome lumen and subsequent analysis of kinetic activity has been employed as a method to measure permeability.1,43 The large molecular weight of enzymes compared to small molecule encapsulants improves their retention, especially over the time scales typical for a permeability experiment.44
The most widely adopted enzyme for permeability studies of polymersome membranes is horseradish peroxidase (HRP), likely due to its commercial availability in high purity and exceptional solution stability.45−48 A number of other biomacromolecules have been employed in such studies including superoxide dismutase (SOD),49Candida antarctica Lipase B (CALB),50 myoglobin,51 glucose oxidase (GOx),52,53 bovine serum albumin (BSA),54 laccase,55 and l-asparaginase (ASNS).56
Although many advantages have been demonstrated for the use of enzyme-catalyzed reactions to assess permeability, there are several limitations that remain. Most notable are the strict constraints with regards to environmental conditions imposed onto polymersome formulations when enzymes are involved. These include a narrow pH window, low temperature, and aqueous solvent—a problem when using certain self-assembly techniques, such as solvent exchange. This restriction also limits the method to the use of robust enzymes, inhibiting assessment of a wide scope of probe molecules.
Challenges in Quantifying Permeability
A key limitation is our inability to accurately compare permeability coefficients or permeation rates obtained via different methods across different systems. Up to this point, membrane permeability measurements have been used for relative comparisons between similar constructs. As described above, this limitation arises from (1) low analyte encapsulation percentages; (2) possible migration, or complete loss, of encapsulated analyte during purification alongside incomplete removal of unencapsulated material; (3) analyte molecule permeation on the time scale of the experiment; and (4) the occurrence of multiple processes ongoing during the experimental time scale. PFG-NMR spectroscopy appears to provide the most promise for achieving absolute permeability measurements; however, the development of simple and approachable alternative methods should also be pursued. We argue that the development of new methods would facilitate direct comparisons between particles prepared using different synthetic routes, self-assembly techniques, and/or with varied membrane chemistry.
Controlling Polymersome Permeability
Perhaps counterintuitively, control of membrane permeability is far simpler and more widely investigated than its measurement. For example, the chemical manipulation of monomers, introduction of stimuli-responsive groups, or cross-linking across membranes readily influence permeability.57 In addition, the incorporation of channel-forming transmembrane proteins or DNA nanopores to provide selective permeability has also been achieved.11
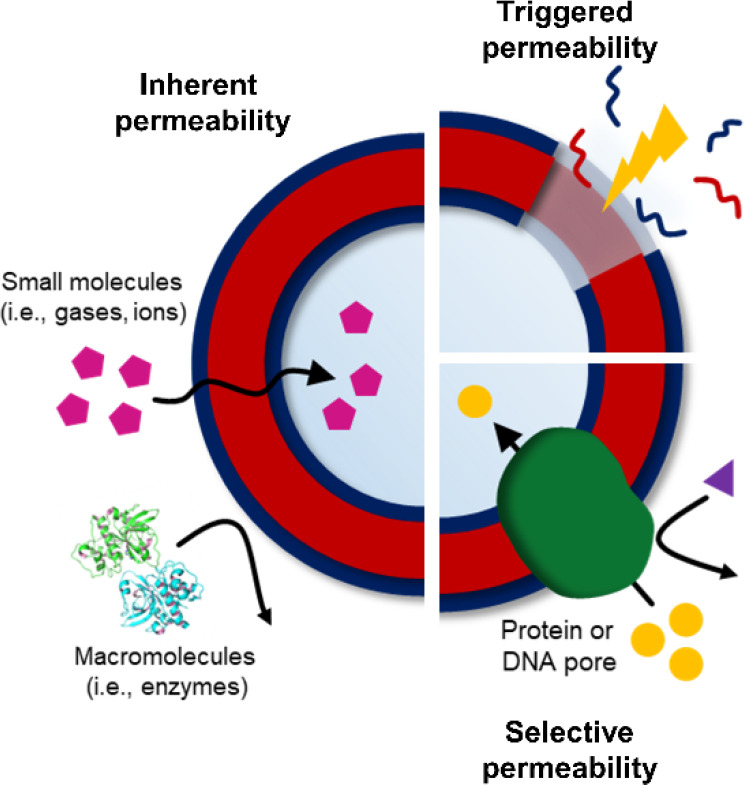
Control over the permeability of polymersome membranes can be categorized as follows: (1) the design of polymersomes with inherent permeability; (2) the introduction of chemistry for variable permeability; and (3) the formation of biohybrid polymersomes (Figure 3). Here, we summarize these individual concepts and explore the advantages and opportunities of each to give context to the importance of designed permeability.
Figure 3.
Manipulating polymersome permeability.
Polymersomes with Inherent Permeability
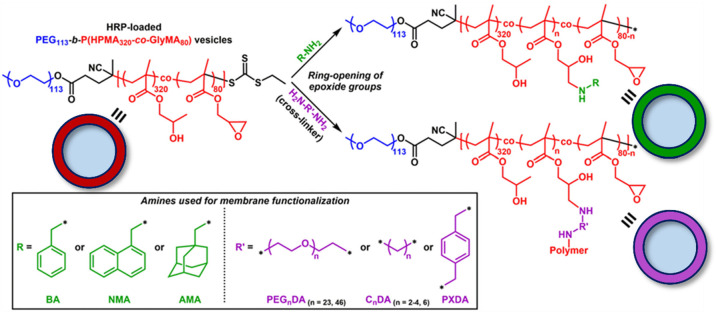
The most experimentally simple approach is to construct polymersomes with intrinsic permeability, meaning that small molecules can diffuse freely across as a consequence of membrane polymer chemistry, composition, or polymerization technique or through postpolymerization modulation. Membrane permeability can be controlled as a function of membrane thickness, dictated by polymer chain length or hydrophilic-to-lipophilic balance, which in turn influences diffusion in agreement with Fick’s first law.58 Increasing polymer molecular weight also favors chain entanglements, thereby restricting diffusion.29 Finally, the hydrophobicity and morphology of core-forming polymer blocks can influence diffusion, whereby glassy membranes slow rates of diffusion, and highly hydrophobic membranes are less hydrated and restrict passage of polar species.36,59 For example, poly(ethylene glycol)-b-poly(2-hydroxypropyl methacrylate) (PEG-b-PHPMA) polymersomes possess intrinsic permeability due to their highly hydrated membranes.48 Such polymersomes retain an encapsulated enzyme cargo but allow for size-selective transport of small molecule substrates.56 Permeability was tuned in this system through postpolymerization modification facilitated by the addition of a comonomer featuring pendant epoxy groups into the core-forming segment of the block copolymers.60 The resultant PEG-b-P(HPMA-co-GlyMA) polymersomes were subjected to systematic membrane modification using mono- or diamines (Figure 4). This strategy showed that modification of the membrane could result in an increase in thickness and a decrease in permeability as a function of cross-linking density and hydrophobicity.
Figure 4.
Schematic illustration of membrane modification to yield polymersomes with controllable permeability. Adapted with permission from ref (57). Copyright 2019 Royal Society of Chemistry.
Introducing Chemistry for Variable Permeability
Stimuli-responsive polymersomes, or “smart” polymersomes, have been extensively reported over the past decade.9,61,62 To date, polymersomes have been designed to be responsive to a number of different stimuli, including pH, temperature, light, redox species, and CO2. The advantage of this approach is that it not only allows for precise control over passage across the membrane as a result of a fixed size or chemistry effects as described above, but additionally gives tunable permeability only under certain external triggering conditions. These can be considered to fall within two categories: (1) the nonreversible disassembly of polymersomes in response to an applied stimulus; or (2) a reversible permeability switch in response to an environmental change. Perhaps the most widely explored stimulus to date is pH; for example, pH-responsive polymersomes were constructed through the self-assembly of block copolymers composed of PEG, poly(diethylaminoethyl methacrylate) (PDEAEMA) and poly(3,4-dimethyl maleic imido butyl methacrylate) (PDMIBMA), to provide both pH sensitivity and the ability to be photo-cross-linked.51,63 This design gave selective off/on permeation behavior, where at pH 8 the membrane was sufficiently hydrophobic to block passage of the enzyme substrate; however, at pH 6, the exchange of substrates and reaction products could freely occur. This cross-linking density and membrane permeability could be precisely tuned over a given range, where varying the irradiation time of polymersomes featuring photo-cross-linkable coumarin groups within the membrane resulted in different pore sizes and therefore size-selective transmembrane transport.64
An interesting application of stimuli-responsive polymersome membranes are so-called “breathing” polymersome nanoreactors (Figure 5).65,66 For example, polymersome membrane swelling (switching “on”) could be achieved as pH dropped in the presence of a “chemical fuel” due to a pH-responsive PDEAEMA core, allowing passage of substrates and reaction with encapsulated enzymes. This process subsequently increased the pH, causing shrinkage of the membranes into an “off” state, and therefore, this cycle was self-regulated and under complete control pending replenishment of the “chemical fuel”. This approach was also demonstrated in response to CO2, whereby polymersomes formed from diblock copolymers of PEG and a CO2-responsive building block, poly(N-amidino)dodecyl acrylamide (PAD), showed significant swelling and release of their encapsulated cargo in an enhanced CO2 environment. This behavior was reversible following introduction of O2; therefore, the permeability increase through modulation of CO2 treatment levels gave temporal control over cargo release.65 Triblock copolymer polymersomes of poly(N-vinylcaprolactam)-b-poly(dimethylsiloxane)-b-poly(N-vinylcaprolactam) (PVCL–PDMS–PVCL) showed temperature-mediated control over membrane permeability across a biologically relevant range.67 Sustained delivery of hydrophobic drugs was achieved above the lower critical solution temperature (LCST) of the PVCL blocks driven by a reversible shrinkage of the polymersomes, which caused a decrease in membrane hydration and constriction of the polymersome membrane, essentially forcing the release of the encapsulated cargo. Finally, light-responsive polymersomes were prepared from self-assembled amphiphilic block copolymers of PEG and a hydrophobic membrane featuring hexyl methacrylate (HMA) and donor–acceptor Stenhouse adducts (DASAs)68 Membrane permeability could be increased in the presence of visible light but returned to an impermeable state when the stimulus was removed.
Figure 5.
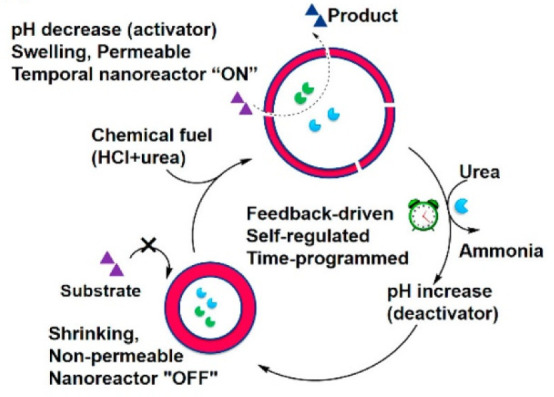
Schematic overview of feedback-induced temporal control of polymersome nanoreactors. Adapted with permission from ref (63). Copyright 2018 American Chemical Society.
Formation of Biohybrid Polymersomes
The final strategy is the biomimetic introduction of ion channels, transporters, pore-forming peptides, and membrane proteins into polymersomes, which combines the benefits of pore selectivity along with the robust and adaptable properties of polymersomes. One challenge is that polymer membranes are much thicker and generally more rigid compared to liposomes, resulting in a significant size mismatch between membrane proteins and the polymersome membranes.69,70 Modeling performed in this area suggests that inclusion of proteins perturbs the bilayer structure as a result of this thickness mismatch, but that polymer molecular weight, composition, and conformational freedom oppose this force, and thus, membrane compression can be achieved if polymer stretched-to-coil transformations are favorable.71 While the majority of work in this domain has centered on liposomes, polymer-based studies have steadily gained interest.11,72−75
An excellent example was the introduction of functional water channels into polymersome membranes by dialysis assembly in the presence of nonionic surfactants and membrane proteins, achieving a high incorporation of the water channel aquaporin-0 (AQP0) into the polymersome membranes.76 Similarly, the pore-forming outer membrane protein F (OmpF) was incorporated into PEG-b-PHPMA polymersomes.77 The self-assembly process was found to be tolerant to the presence of nonionic surfactants required for solubilization of the highly hydrophobic transmembrane protein, and additionally, the surfactant concentration could tune the size and membrane thickness of the final polymersomes.
Size-selective permeability can also be modulated through the introduction of membrane-spanning DNA nanopores, which are advantageous due to their structural robustness, modular construction, and easier fabrication compared with biologically derived proteins. Poly(2-(methacryloyloxy)ethyl phosphorylcholine-b-disisopropylamino) ethyl methacrylate (PMPC-b-PDPA) polymersomes were incubated with preformed DNA nanopores, and the number of pores could be tuned by feed ratio (Figure 6).78 The modified assemblies could encapsulate and retain an enzyme cargo, and the reaction kinetics of substrate conversion were enhanced following introduction of the nanopores, thus confirming an increase in construct permeability. In an attempt to create highly selective membrane permeability, ion channels were inserted into the membranes of polymersomes formed from PMOXA-b-PDMS-b-PMOXA triblock copolymers.23 Following incorporation of the biopore gramicidin (gA), the controlled passage of protons and monovalent cations such as Na+ and K+ was achieved. It was found that functionality of the ion channel could be retained up to a membrane thickness approximately 4× that of the pore itself, indicating good tolerance of thickness mismatch when using low-glass-transition (Tg) polymer constituents.
Figure 6.
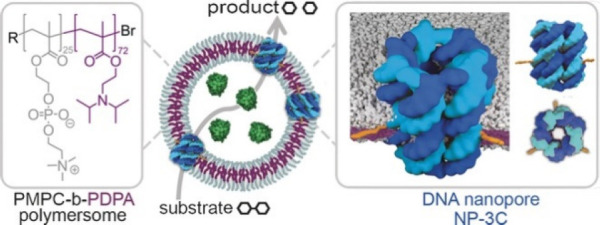
Functional hybrid polymersomes containing membrane-spanning DNA nanopores display size-selective permeability, permitting the transport of substrates and products through the DNA nanopores but retaining bioactive encapsulated enzymes. Adapted with permission from ref (75). Copyright John Wiley & Sons 2016.
Conclusions and Outlook
In the interest of developing more comprehensive methods, we have suggested five key principles as requisites to ensure the validity of polymersome membrane permeability measurements:
-
1)
rigorous removal of excess/unencapsulated probe molecules;
-
2)
appropriate control experiments—which may include kinetics of reactions in free solution or reactions of “blank” polymersomes without encapsulated cargo;
-
3)
reduced system complexity—methods to assess membrane permeability should be developed with specific applications in mind while limiting the number of processes ongoing in the experiment e.g., reaction kinetics, energy transfer, absorption/emission efficiencies, etc., and in addition, the generalizability of a method should be considered;
-
4)
emphasis on in situ measurements; and
-
5)
appropriate consideration for the time scale of the measurement relative to rates of diffusion and monitoring reactions.
We envision that these principles will facilitate the development of new methods to measure the membrane permeability of polymersomes and allow for quantitative and conclusive construction of structure–property relationships between polymersome chemistry, structure, and other descriptors and permeability, thus expediting advancement in the area of artificial, biomimetic vesicular structures.
Acknowledgments
This work was supported by the ERC (Grant 615142), EPSRC, and the University of Birmingham.
Author Contributions
† A.J.M. and A.K.P. contributed equally.
The authors declare no competing financial interest.
References
- Belluati A.; Craciun I.; Meyer C. E.; Rigo S.; Palivan C. G. Enzymatic reactions in polymeric compartments: nanotechnology meets nature. Curr. Opin. Biotechnol. 2019, 60, 53–62. 10.1016/j.copbio.2018.12.011. [DOI] [PubMed] [Google Scholar]
- Yewdall N. A.; Mason A. F.; van Hest J. C. M. The hallmarks of living systems: towards creating artificial cells. Interface Focus 2018, 8, 20180023. 10.1098/rsfs.2018.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N. J.; Hinner M. J. Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. Methods Mol. Biol. 2015, 1266, 29–53. 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl H. R.; Wheeler J. S.; Murray H. E. Sodium and Potassium in Health and Disease. Met. Ions Life Sci. 2013, 13, 29–47. 10.1007/978-94-007-7500-8_2. [DOI] [PubMed] [Google Scholar]
- Cooper G. M.Transport of Small Molecules. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, 2000. [Google Scholar]
- Baldwin S. A. Mammalian passive glucose transporters: members of an ubiquitous family of active and passive transport proteins. Biochim. Biophys. Acta, Rev. Biomembr. 1993, 1154 (1), 17–49. 10.1016/0304-4157(93)90015-G. [DOI] [PubMed] [Google Scholar]
- Alberts B.; Johnson A.; Lewis J.; Raff M.; Roberts K.; Walters P.. Carrier Proteins and Active Membrane Transport. In Mol. Biol. Cell, 4th ed.; Garland Science, 2002. [Google Scholar]
- Lodish H.; Berk A.; Zipursky S. L.; Matsudaira P.; Baltimore D.; Darnell J. E.. Active Transport by ATP-Powered Pumps. In Mol. Cell. Biol., 4th ed.; W. H. Freeman: New York, 2000. [Google Scholar]
- Che H.; van Hest J. C. M. Adaptive Polymersome Nanoreactors. ChemNanoMat 2019, 5 (9), 1092–1109. 10.1002/cnma.201900245. [DOI] [Google Scholar]
- Bordat A.; Boissenot T.; Nicolas J.; Tsapis N. Thermoresponsive polymer nanocarriers for biomedical applications. Adv. Drug Delivery Rev. 2019, 138, 167–192. 10.1016/j.addr.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Garni M.; Wehr R.; Avsar S. Y.; John C.; Palivan C.; Meier W. Polymer membranes as templates for bio-applications ranging from artificial cells to active surfaces. Eur. Polym. J. 2019, 112, 346–364. 10.1016/j.eurpolymj.2018.12.047. [DOI] [Google Scholar]
- Idrissi M. E.; Meyer C. E.; Zartner L.; Meier W. Nanosensors based on polymer vesicles and planar membranes: a short review. J. Nanobiotechnol. 2018, 16, 63–76. 10.1186/s12951-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoori S.; Leroux J.-C. Twenty-five years of polymersomes: lost in translation?. Mater. Horiz. 2020, 7, 1297–1309. 10.1039/C9MH01669D. [DOI] [Google Scholar]
- Fick A. Ueber Diffusion. Ann. Phys. 1855, 170, 59–86. 10.1002/andp.18551700105. [DOI] [Google Scholar]
- Meyer H. Naunyn-Schmiedeberg's Arch. Pharmacol. 1899, 42, 109–118. 10.1007/BF01834479. [DOI] [Google Scholar]
- Overton E.Studien über die Narkose; Fischer; Jena, 1901. [Google Scholar]
- Missner A.; Pohl P. 110 Years of the Meyer–Overton Rule: Predicting Membrane Permeability of Gases and Other Small Compounds. ChemPhysChem 2009, 10, 1405–1414. 10.1002/cphc.200900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poschenrieder S. T.; Klermund L.; Langer B.; Castiglione K. Determination of Permeability Coefficients of Polymersomal Membranes for Hydrophilic Molecules. Langmuir 2017, 33, 6011–6020. 10.1021/acs.langmuir.6b04598. [DOI] [PubMed] [Google Scholar]
- Garcia A. M. Determination of ion permeability by fluorescence quenching. Methods Enzymol. 1992, 207, 501–510. 10.1016/0076-6879(92)07035-M. [DOI] [PubMed] [Google Scholar]
- Quan L.; Ding H.; Pan C.; Wei Y.; Xie Z. Revealing membrane permeability of polymersomes through fluorescence enhancement. Colloids Surf., B 2018, 161, 156–161. 10.1016/j.colsurfb.2017.10.058. [DOI] [PubMed] [Google Scholar]
- Bai Z.; Zhao B.; Lodge T. P. Bilayer Membrane Permeability of Ionic Liquid-Filled Block Copolymer Vesicles in Aqueous Solution. J. Phys. Chem. B 2012, 116, 8282–8289. 10.1021/jp3033098. [DOI] [PubMed] [Google Scholar]
- Yildiz U. H.; Hoog H. P. M. D.; Fu Z.; Tomczak N.; Parikh A. N.; Nallani M.; Liedberg B. Third-Party ATP Sensing in Polymersomes A Label-Free Assay of Enzyme Reactions in Vesicular Compartments. Small 2014, 10, 442–447. 10.1002/smll.201300060. [DOI] [PubMed] [Google Scholar]
- Lomora M.; Garni M.; Itel F.; Tanner P.; Spulber M.; Palivan C. G. Polymersomes with engineered ion selective permeability as stimuli-responsive nanocompartments with preserved architecture. Biomaterials 2015, 53, 406–414. 10.1016/j.biomaterials.2015.02.080. [DOI] [PubMed] [Google Scholar]
- Lomora M.; Dinu I. A.; Itel F.; Rigo S.; Spulber M.; Palivan C. G. Does Membrane Thickness Affect the Transport of Selective Ions Mediated by Ionophores in Synthetic Membranes. Macromol. Rapid Commun. 2015, 36, 1929–1934. 10.1002/marc.201500289. [DOI] [PubMed] [Google Scholar]
- Wu J.; Eisenberg A. Proton Diffusion across Membranes of Vesicles of Poly(styrene-b-acrylic Acid) Diblock Copolymers. J. Am. Chem. Soc. 2006, 128, 2880–2884. 10.1021/ja056064x. [DOI] [PubMed] [Google Scholar]
- Scarpa E.; Bailey J. L.; Janeczek A. A.; Stumpf P. S.; Johnston A. H.; Oreffo R. O. C.; Woo Y. L.; Cheong Y. C.; Evans N. D.; Newman T. A. Quantification of intracellular payload release from polymersome nanoparticles. Sci. Rep. 2016, 6, 29460–19472. 10.1038/srep29460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choucair A.; Soo P. L.; Eisenberg A. Active Loading and Tunable Release of Doxorubicin from Block Copolymer Vesicles. Langmuir 2005, 21, 9308–9313. 10.1021/la050710o. [DOI] [PubMed] [Google Scholar]
- Kim J.; Kim K. T. Polymersome-Based Modular Nanoreactors with Size-Selective Transmembrane Permeability. ACS Appl. Mater. Interfaces 2020, 12, 23502–23513. 10.1021/acsami.0c05637. [DOI] [PubMed] [Google Scholar]
- Discher B. M.; Won Y. Y.; Ege D. S.; Lee J. C. M.; Bates F. S.; Discher D. E.; Hammer D. A. Polymersomes: Tough vesicles made from diblock copolymers. Science 1999, 284 (5417), 1143–1146. 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- Carlsen A.; Glaser N.; Meins J.-F. L.; Lecommandoux S. Block Copolymer Vesicle Permeability Measured by Osmotic Swelling and Shrinking. Langmuir 2011, 27, 4884–4890. 10.1021/la105045m. [DOI] [PubMed] [Google Scholar]
- Kumar M.; Grzelakowski M.; Zilles J.; Clark M.; Meier W. Highly permeable polymeric membranes based on the incorporation of the functional water channel protein Aquaporin Z. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (52), 20719–20724. 10.1073/pnas.0708762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärger J. NMR self-diffusion studies in heterogeneous systems. Adv. Colloid Interface Sci. 1985, 23, 129–148. 10.1016/0001-8686(85)80018-X. [DOI] [Google Scholar]
- Stilbs P. Fourier transform pulsed-gradient spin-echo studies of molecular diffusion. Prog. Nucl. Magn. Reson. Spectrosc. 1987, 19 (1), 1–45. 10.1016/0079-6565(87)80007-9. [DOI] [Google Scholar]
- Leson A.; Filiz V.; Forster S.; Mayer C. Water permeation through block-copolymer vesicle membranes. Chem. Phys. Lett. 2007, 444, 268–272. 10.1016/j.cplett.2007.07.023. [DOI] [Google Scholar]
- Leson A.; Hauschild S.; Rank A.; Neub A.; Schubert R.; Förster S.; Mayer C. Molecular Exchange through Membranes of Poly(2-vinylpyridine-block-ethylene oxide) Vesicles. Small 2007, 3, 1074–1083. 10.1002/smll.200600540. [DOI] [PubMed] [Google Scholar]
- So S.; Lodge T. P. Rate of Molecular Exchange through the Membranes of Ionic Liquid Filled Polymersomes Dispersed in Water. J. Phys. Chem. C 2014, 118, 21140–21147. 10.1021/jp505118h. [DOI] [Google Scholar]
- So S.; Yao L. J.; Lodge T. P. Permeability of Rubbery and Glassy Membranes of Ionic Liquid Filled Polymersome Nanoreactors in Water. J. Phys. Chem. B 2015, 119, 15054–15062. 10.1021/acs.jpcb.5b08425. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Hoffmann H.; Leson A.; Mayer C. Molecular Exchange through the Vesicle Membrane of Siloxane Surfactant in Water/Glycerol Mixed Solvents. J. Phys. Chem. B 2007, 111 (22), 6161–6166. 10.1021/jp0711848. [DOI] [PubMed] [Google Scholar]
- Gumz H.; Boye S.; Iyisan B.; Krönert V.; Formanek P.; Voit B.; Lederer A.; Appelhans D. Toward Functional Synthetic Cells In-Depth Study of Nanoparticle and Enzyme Diffusion through a Cross-Linked Polymersome Membrane. Adv. Sci. 2019, 6 (7), 1801299–1801312. 10.1002/advs.201801299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia G.; Ryan A. J.; Tomas S. Polymeric vesicle permeability: A facile chemical assay. Langmuir 2006, 22, 4910–4913. 10.1021/la060354p. [DOI] [PubMed] [Google Scholar]
- Rideau E.; Dimova R.; Landfester K. Liposomes and polymersomes: a comparative review towards cell mimicking. Chem. Soc. Rev. 2018, 47, 8572–8610. 10.1039/C8CS00162F. [DOI] [PubMed] [Google Scholar]
- Kim J.; Jeong S.; Korneev R.; Shin K.; Kim K. T. Cross-Linked Polymersomes with Reversible Deformability and Oxygen Transportability. Biomacromolecules 2019, 20, 2430–2439. 10.1021/acs.biomac.9b00485. [DOI] [PubMed] [Google Scholar]
- Che H.; van Hest J. C. M. v. Adaptive Polymersome Nanoreactors. ChemNanoMat 2019, 5, 1092–1109. 10.1002/cnma.201900245. [DOI] [Google Scholar]
- Lee J. C.-M.; Bermudez H.; Discher B. M.; Sheehan M. A.; Won Y.-Y.; Bates F. S.; Discher D. E. Preparation, stability, and in vitro performance of vesicles made with diblock copolymers. Biotechnol. Bioeng. 2001, 73 (2), 135–145. 10.1002/bit.1045. [DOI] [PubMed] [Google Scholar]
- Hu S.; Lu Q.; Xu Y.. Biosensors based on direct electron transfer of protein. In Electrochemical Sensors, Biosensors and their Biomedical Applications; Zhang X., Ju H., Wang J., Eds.; Academic Press: 2008; pp 531–581. [Google Scholar]
- Spulber M.; Najer A.; Winkelbach K.; Glaied O.; Waser M.; Pieles U.; Meier W.; Bruns N. Photoreaction of a Hydroxyalkyphenone with the Membrane of Polymersomes: A Versatile Method To Generate Semipermeable Nanoreactors. J. Am. Chem. Soc. 2013, 135, 9204–9212. 10.1021/ja404175x. [DOI] [PubMed] [Google Scholar]
- Einfalt T.; Witzigmann D.; Edlinger C.; Sieber S.; Goers R.; Najer A.; Spulber M.; Onaca-Fischer O.; Huwyler J.; Palivan C. G. Biomimetic artificial organelles with in vitro and in vivo activity triggered by reduction in microenvironment. Nat. Commun. 2018, 9, 1127. 10.1038/s41467-018-03560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman L. D.; Varlas S.; Arno M. C.; Fayter A.; Gibson M. I.; O’Reilly R. K. Permeable Protein-Loaded Polymersome Cascade Nanoreactors by Polymerization-Induced Self-Assembly. ACS Macro Lett. 2017, 6 (11), 1263–1267. 10.1021/acsmacrolett.7b00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaca O.; Hughes D. W.; Balasubramanian V.; Grzelakowski M.; Meier W.; Palivan C. G. SOD Antioxidant Nanoreactors: Influence of Block Copolymer Composition on the Nanoreactor Efficiency. Macromol. Biosci. 2010, 10 (5), 531–538. 10.1002/mabi.200900379. [DOI] [PubMed] [Google Scholar]
- Kim K. T.; Cornelissen J. J. L. M.; Nolte R. J. M.; van Hest J. C. M. A Polymersome Nanoreactor with Controllable Permeability Induced by Stimuli-Responsive Block Copolymers. Adv. Mater. 2009, 21, 2787–2791. 10.1002/adma.200900300. [DOI] [Google Scholar]
- Gaitzsch J.; Appelhans D.; Wang L.; Battaglia G.; Voit B. Synthetic Bio-nanoreactor: Mechanical and Chemical Control of Polymersome Membrane Permeability. Angew. Chem., Int. Ed. 2012, 51 (18), 4448–4451. 10.1002/anie.201108814. [DOI] [PubMed] [Google Scholar]
- Li J.; Dirisala A.; Ge Z.; Wang Y.; Yin W.; Ke W.; Toh K.; Xie J.; Matsumoto Y.; Anraku Y.; Osada K.; Kataoka K. Therapeutic Vesicular Nanoreactors with Tumor-Specific Activation and Self-Destruction for Synergistic Tumor Ablation. Angew. Chem. 2017, 129, 14213–14218. 10.1002/ange.201706964. [DOI] [PubMed] [Google Scholar]
- Li J.; Li Y.; Wang Y.; Ke W.; Chen W.; Wang W.; Ge Z. Polymer Prodrug-Based Nanoreactors Activated by Tumor Acidity for Orchestrated Oxidation/Chemotherapy. Nano Lett. 2017, 17, 6983–6990. 10.1021/acs.nanolett.7b03531. [DOI] [PubMed] [Google Scholar]
- Nishimura T.; Sasaki Y.; Akiyoshi K. Biotransporting Self-Assembled Nanofactories Using Polymer Vesicles with Molecular Permeability for Enzyme Prodrug Cancer Therapy. Adv. Mater. 2017, 29 (36), 1702406–1702413. 10.1002/adma.201702406. [DOI] [PubMed] [Google Scholar]
- Spulber M.; Baumann P.; Saxer S. S.; Pieles U.; Meier W.; Bruns N. Poly(N-vinylpyrrolidone)-Poly(dimethylsiloxane)-Based Polymersome Nanoreactors for Laccase-Catalyzed Biotransformations. Biomacromolecules 2014, 15, 1469–1475. 10.1021/bm500081j. [DOI] [PubMed] [Google Scholar]
- Blackman L. D.; Varlas S.; Arno M. C.; Houston Z. H.; Fletcher N. L.; Thurecht K. J.; Hasan M.; Gibson M. I.; O’Reilly R. K. Confinement of Therapeutic Enzymes in Selectively Permeable Polymer Vesicles by Polymerization-Induced Self-Assembly (PISA) Reduces Antibody Binding and Proteolytic Susceptibility. ACS Cent. Sci. 2018, 4 (6), 718–723. 10.1021/acscentsci.8b00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y. S.; van der Vlies A. J.; Gantz J.; Thacher T. N.; Antonijevic S.; Cavadini S.; Demurtas D.; Stergiopulos N.; Hubbell J. A. Micelles for Delivery of Nitric Oxide. J. Am. Chem. Soc. 2009, 131 (40), 14413–14418. 10.1021/ja905123t. [DOI] [PubMed] [Google Scholar]
- Battaglia G.; Ryan A. J. Bilayers and interdigitation in block copolymer vesicles. J. Am. Chem. Soc. 2005, 127 (24), 8757–8764. 10.1021/ja050742y. [DOI] [PubMed] [Google Scholar]
- Discher D. E.; Eisenberg A. Polymer vesicles. Science 2002, 297 (5583), 967–973. 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- Varlas S.; Foster J. C.; Georgiou P. G.; Keogh R.; Husband J. T.; Williams D. S.; O’Reilly R. K. Tuning the membrane permeability of polymersome nanoreactors developed by aqueous emulsion polymerization-induced self-assembly. Nanoscale 2019, 11 (26), 12643–12654. 10.1039/C9NR02507C. [DOI] [PubMed] [Google Scholar]
- Iqbal S.; Blenner M.; Alexander-Bryant A.; Larsen J. Polymersomes for Therapeutic Delivery of Protein and Nucleic Acid Macromolecules: From Design to Therapeutic Applications. Biomacromolecules 2020, 21 (4), 1327–1350. 10.1021/acs.biomac.9b01754. [DOI] [PubMed] [Google Scholar]
- Iyisan B.; Landfester K. Modular Approach for the Design of Smart Polymeric Nanocapsules. Macromol. Rapid Commun. 2019, 40 (1), 1800577–1800577. 10.1002/marc.201800577. [DOI] [PubMed] [Google Scholar]
- Gräfe D.; Gaitzsch J.; Appelhans D.; Voit B. Cross-linked polymersomes as nanoreactors for controlled and stabilized single and cascade enzymatic reactions. Nanoscale 2014, 6 (18), 10752–10761. 10.1039/C4NR02155J. [DOI] [PubMed] [Google Scholar]
- Zhang W. J.; Hong C. Y.; Pan C. Y. Artificially Smart Vesicles with Superior Structural Stability: Fabrication, Characterizations, and Transmembrane Traffic. ACS Appl. Mater. Interfaces 2017, 9 (17), 15086–15095. 10.1021/acsami.7b02966. [DOI] [PubMed] [Google Scholar]
- Yan Q.; Wang J.; Yin Y.; Yuan J. Breathing polymersomes: CO2-tuning membrane permeability for size-selective release, separation, and reaction. Angew. Chem., Int. Ed. 2013, 52 (19), 5070–5073. 10.1002/anie.201300397. [DOI] [PubMed] [Google Scholar]
- Che H.; Cao S.; van Hest J. C. M. Feedback-Induced temporal control of ″breathing″ polymersomes to create self-adaptive nanoreactors. J. Am. Chem. Soc. 2018, 140 (16), 5356–5359. 10.1021/jacs.8b02387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Kozlovskaya V.; Medipelli S.; Xue B.; Ahmad F.; Saeed M.; Cropek D.; Kharlampieva E. Temperature-Sensitive Polymersomes for Controlled Delivery of Anticancer Drugs. Chem. Mater. 2015, 27 (23), 7945–7956. 10.1021/acs.chemmater.5b03048. [DOI] [Google Scholar]
- Rifaie-Graham O.; Ulrich S.; Galensowske N. F. B.; Balog S.; Chami M.; Rentsch D.; Hemmer J. R.; Read de Alaniz J.; Boesel L. F.; Bruns N. Wavelength-Selective Light-Responsive DASA-Functionalized Polymersome Nanoreactors. J. Am. Chem. Soc. 2018, 140 (25), 8027–8036. 10.1021/jacs.8b04511. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Fu W.; Palivan C. G.; Meier W. Natural channel protein inserts and functions in a completely artificial, solid-supported bilayer membrane. Sci. Rep. 2013, 3 (1), 1–7. 10.1038/srep02196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itel F.; Najer A.; Palivan C. G.; Meier W. Dynamics of Membrane Proteins within Synthetic Polymer Membranes with Large Hydrophobic Mismatch. Nano Lett. 2015, 15 (6), 3871–3878. 10.1021/acs.nanolett.5b00699. [DOI] [PubMed] [Google Scholar]
- Pata V.; Dan N. The effect of chain length on protein solubilization in polymer-based vesicles (polymersomes). Biophys. J. 2003, 85 (4), 2111–2118. 10.1016/S0006-3495(03)74639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardin C.; Thoeni S.; Widmer J.; Winterhalter M.; Meier W. Nanoreactors based on (polymerized) ABA-triblock copolymer vesicles. Chem. Commun. 2000, 0 (15), 1433–1434. 10.1039/b004280n. [DOI] [Google Scholar]
- Yorulmaz Avsar S.; Kyropoulou M.; Di Leone S.; Schoenenberger C.-A.; Meier W. P.; Palivan C. G. Biomolecules turn self-assembling amphiphilic block co-polymer platforms into biomimetic interfaces. Front. Chem. 2019, 6 (JAN), 645–645. 10.3389/fchem.2018.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C. E.; Abram S. L.; Craciun I.; Palivan C. G. Biomolecule-polymer hybrid compartments: Combining the best of both worlds. Phys. Chem. Chem. Phys. 2020, 22 (20), 11197–11218. 10.1039/D0CP00693A. [DOI] [PubMed] [Google Scholar]
- Chimisso V.; Maffeis V.; Hürlimann D.; Palivan C. G.; Meier W. Self-Assembled Polymeric Membranes and Nanoassemblies on Surfaces: Preparation, Characterization, and Current Applications. Macromol. Biosci. 2020, 20 (1), 1900257–1900257. 10.1002/mabi.201900257. [DOI] [PubMed] [Google Scholar]
- Kumar M.; Habel J. E. O.; Shen Y. X.; Meier W. P.; Walz T. High-density reconstitution of functional water channels into vesicular and planar block copolymer membranes. J. Am. Chem. Soc. 2012, 134 (45), 18631–18637. 10.1021/ja304721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlas S.; Blackman L. D.; Findlay H. E.; Reading E.; Booth P. J.; Gibson M. I.; O’Reilly R. K. Photoinitiated Polymerization-Induced Self-Assembly in the Presence of Surfactants Enables Membrane Protein Incorporation into Vesicles. Macromolecules 2018, 51 (16), 6190–6201. 10.1021/acs.macromol.8b00994. [DOI] [Google Scholar]
- Messager L.; Burns J. R.; Kim J.; Cecchin D.; Hindley J.; Pyne A. L. B.; Gaitzsch J.; Battaglia G.; Howorka S. Biomimetic Hybrid Nanocontainers with Selective Permeability. Angew. Chem., Int. Ed. 2016, 55 (37), 11106–11109. 10.1002/anie.201604677. [DOI] [PMC free article] [PubMed] [Google Scholar]