Summary
Indoleamine 2,3-dioxygenases (IDOs) degrade l-tryptophan to kynurenines and drive the de novo synthesis of nicotinamide adenine dinucleotide. Unsurprisingly, various invertebrates, vertebrates, and even fungi produce IDO. In mammals, IDO1 also serves as a homeostatic regulator, modulating immune response to infection via local tryptophan deprivation, active catabolite production, and non-enzymatic cell signaling. Whether fungal Idos have pleiotropic functions that impact on host-fungal physiology is unclear. Here, we show that Aspergillus fumigatus possesses three ido genes that are expressed under conditions of hypoxia or tryptophan abundance. Loss of these genes results in increased fungal pathogenicity and inflammation in a mouse model of aspergillosis, driven by an alternative tryptophan degradation pathway to indole derivatives and the host aryl hydrocarbon receptor. Fungal tryptophan metabolic pathways thus cooperate with the host xenobiotic response to shape host-microbe interactions in local tissue microenvironments.
Keywords: aspergillosis, indoles, Aspergillus fumigatus, IDO, tryptophan, AhR, IL-33, NAD, inflammation
Graphical Abstract
Highlights
-
•
Aspergillus Idos contribute to de novo NAD+ biosynthesis and fungal growth
-
•
In conditions of tryptophan abundance, Aspergillus releases indole derivatives via Aro
-
•
Activation of lung AhR enhances harmful lung inflammation during fungal infection
-
•
Pharmacological induction of Ido in Aspergillus improves infection outcome
Mammalians use the kynurenine pathway for tryptophan degradation inducing tolerogenic immune responses. Zelante et al. report that similarly, the fungus Aspergillus fumigatus uses the kynurenine pathway. Pathogenic immune response occurs when the fungus catabolizes tryptophan via the indolepyruvate pathway, which targets host AhR. Administration of Ido inducers ameliorates infection outcome.
Introduction
Heme dioxygenase-encoding genes are widely distributed across species, from metazoans to bacteria and fungi (Yuasa and Ball, 2011; Ball et al., 2014; Choera et al., 2018). During evolution, these genes have diverged through speciation and genetic duplication events, resulting in the emergence of paralogous genes in vertebrates (Ball et al., 2014). One such enzyme, indoleamine 2,3-dioxygenase (IDO) (IDO1 and IDO2 in mammals and idoA, idoB, and idoC in fungi), degrades l-tryptophan (Trp) to generate a series of catabolites collectively known as kynurenines. In mammals, this Trp degradation pathway contributes to immune tolerance (Grohmann et al., 2003; Metz et al., 2014; Munn and Mellor, 2013) that is mediated by the amino acid, l-kynurenine. In fungi, Idos primarily supply nicotinamide adenine dinucleotide (NAD+) via the kynurenine pathway (Wang et al., 2016; Yuasa and Ball, 2013). Because both fungal Idos and mammalian IDO1 have a high affinity for Trp and high catalytic efficiency, these two enzymes might have similar metabolic roles (Yuasa and Ball, 2015).
The activation of various metabolic pathways in different eukaryotic clades can lead to de novo NAD+ biosynthesis. But Trp can be degraded to parallel catabolite products other than kynurenines. For example, Trp is degraded by aromatic aminotransferases (termed Aro8 and Aro9 in Saccharomyces cerevisiae) to indolepyruvate via the Ehrlich pathway (Choera et al., 2018; Jain et al., 2012). These aromatic aminotransferases in S. cerevisiae are also involved in the synthesis of the essential amino acids phenylalanine (Phe) and tyrosine (Tyr). Consequently, loss of aro8 and aro9 results in Phe and Tyr auxotrophy (Urrestarazu et al., 1998; Iraqui et al., 1998). In Candida spp., products of the Ehrlich pathway can influence both filamentation and pigment production, which both have a role in mediating virulence (Brunke et al., 2010). For example, loss of aro8 in C. glabrata results in reduced pigment production and an increased sensitivity to hydrogen peroxide, thus linking Trp catabolism to fungal pathogenicity (Brunke et al., 2010).
The opportunistic fungal pathogen Aspergillus fumigatus possesses three putative ido genes (idoA, idoB, and idoC) and two putative aro8 and aro9 genes (aroH and aroI) (Yuasa and Ball, 2013; Wang et al., 2016; Dindo et al., 2018). Enzymatic studies suggest that Idos of A. oryzae participate in Trp degradation (Yuasa and Ball, 2011). Meanwhile, A. fumigatus grown on Trp upregulate ido gene expression (Wang et al., 2016). Researchers recently purified a recombinant form of Aspergillus AroH and examined the biochemical features of the protein (Dindo et al., 2018). The data indicated that AroH displays structural properties and a pyridoxal-5′-phosphate (PLP) binding capacity reminiscent of the yeast ortholog Aro8. However, the relative contributions of individual Idos or Aros to Aspergillus metabolic pathways and adaptation to the host environment are unclear.
Xenobiotic receptors (XRs) serve as cellular sensors for both endogenous and exogenous stimuli by regulating the transcription of genes encoding drug-metabolizing enzymes and those involved in energy homeostasis, cell proliferation, and/or immune responses. XRs are activated via ligand-dependent (direct) and ligand-independent (indirect) mechanisms by an excess of structurally unrelated chemicals. The XR family includes the constitutive androstane receptor (CAR), pregnane X receptor (PXR), aryl hydrocarbon receptor (AhR), and peroxisome proliferator-activated receptors (PPARs). These receptors permit host-microbe interaction in the gut, by sensing and responding to metabolites released by symbiotic microbiota during an innate immune response or as a result of host drug metabolism (Ranhotra et al., 2016).
Conserved xenobiotic-sensing and endobiotic-sensing mechanisms also exist in the intestinal mucosa to detect metabolic products. These mechanisms are thought to provide an additional level of defense in the gastrointestinal tract of multicellular organisms (Venkatesh et al., 2014). Indeed, dietary tryptophan-derived bacterial metabolites (indole-3-propionic acid) can activate the PXR found in intestinal epithelial cells in a crypt-villus gradient to regulate intestinal homeostasis. This effect modulates the abundance of tumor necrosis factor alpha (TNF-α), which in turn modulates intestinal barrier function (i.e., permeability). By this model, restoring gut homeostasis by reconstituting the intestine with indole-metabolite-producing bacteria and/or PXR-activating bacterial metabolites could result in abrogating pro-inflammatory signals and barrier permeability in the context of intestinal inflammation. The role played by the AhR in regulating the immune response and during lung infectious diseases has been debated (Beamer and Shepherd, 2013). One study showed, however, that AhR activation leads to interleukin-33 (IL-33) expression and increased lung inflammation in a mouse model of allergic airway inflammation (Tajima et al., 2020).
Here, we aimed to characterize the function of A. fumigatus Idos and Aros by generating Aspergillus mutants with the respective gene deletions. We then analyzed the growth, metabolic, and pathogenic profiles of these A. fumigatus mutants in vitro and in vivo in aspergillosis. We found that Idos were responsible for l-kynurenine production and contributed to NAD+ biosynthesis. Loss of all three ido genes activated the Ehrlich pathway, involving AroH. This metabolic switch compromised fungal fitness (i.e., growth ability in the lung) and host fitness, in which the release of other Trp metabolites promoted AhR activation, IL-33 release, allergy, and subverted disease-tolerance mechanisms in the lungs of A. fumigatus-colonized mice.
Results
Structure, function, and regulation of A. fumigatus Idos
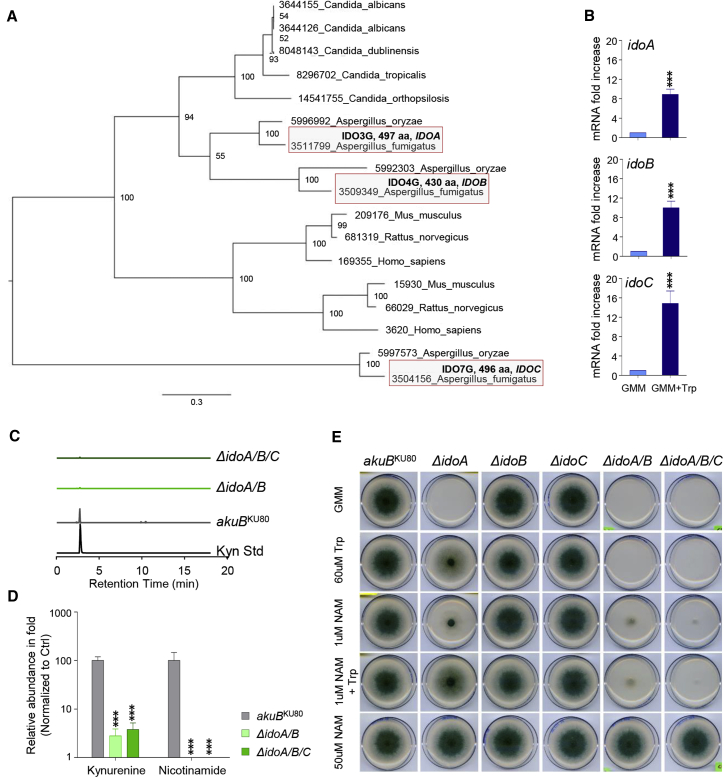
We first wanted to understand the sequence and homology of ido genes in A. fumigatus. BLAST analysis showed that A. fumigatus has three ido-related genes: idoA (Afu3g14250), idoB (Afu4g09830), and idoC (Afu7g02010) (Figure 1A). Phylogenetic analysis revealed that IdoA and IdoB were more closely related to each other than to IdoC (Figure 1A; Figure S1), indicating that they might represent paralogs that evolved from a common ancestral gene, as previously suggested for A. oryzae (Yuasa and Ball, 2013). IdoA and IdoB also showed the highest level of similarity with human IDO1 and IDO2, both of which showed considerable identity to IdoA (49%–49.5%) and IdoB (49%–49.9%) (Figure S2A). By contrast, IdoC showed a low level of homology with the other two proteins (37%–38%) and with human IDO1 and IDO2 (38.1%) (Figure S2A).
Figure 1.
Aspergillus Idos catabolize Trp for NAD+ and kynurenines biosynthesis
(A) Maximum likelihood phylogenetic analysis of IDOs.
(B) mRNA fold increase of Aspergillus idoA, idoB, and idoC in GMM and GMM + Trp at 24 h of culture.
(C and D) Peak levels and relative abundance of l-kynurenine (Kyn) and nicotinamide (D) in WT (Aspergillus akuBKU80) and mutant strains inoculated on solid GMM supplemented with NAM and Trp and cultured at 37°C for 84 h in triplicates.
(E) Growth of parental akuBKU80and mutant strains in GMM, GMM + Trp (60 μM), and NAD supplementation (NAM) to the growth medium.
(B and D) Data are represented as mean ± SD. Graphs are representative of data collected from three independent replicate experiments.
(B) Statistical significance (∗∗∗p < 0.0001) was determined against the untreated (GMM) (two-tailed Student’s t test unpaired parametric).
(D) Statistical significance (∗∗∗p < 0.0001) was determined against the Aspergillus akuBKU80 strain (two-way ANOVA and Bonferroni’s akuBKU80 versus ΔidoA/B and versus ΔidoA/B/C).
(C and E) Experiments were repeated three times.
See also Figures S1–S4.
Computational models of IdoA and IdoB indicated that 42% pocket residues in IdoA and IdoB, which mediate ligand binding, were identical to human IDO1 and IDO2, contributing to 60% pocket similarity overall (Figure S2B). Analyses of the binding-site residues showed that the differences were due to the presence of hydrophobic residues surrounding heme in human and fungal IDOs (Figures S2B–S2D). However, tridimensional inspection confirmed that these differences did not affect the overall hydrophilic/hydrophobic properties of the binding-site area (Figures S2B and S2E).
A striking difference that we identified between IdoA and human IDO1 was a reduction in the volume proximal to heme Fe2+ in IdoA (Figures S2B and S2F) due to the replacement of an alanine residue (Ala264) in IDO1 by an asparagine residue (Asn276; IdoA numbering scheme; in a loop close to the heme). This Asn residue likely affects the activation cycle of IDO by promoting the alternative pathway, where a resonance hybrid of IDO-Fe(III)/superoxide and IDO-Fe(II)/oxygen (the latter being the catalytically active form) has been described (Macchiarulo et al., 2009).
To functionally analyze the specific roles of the different A. fumigatus Idos, we deleted the respective ido genes (Figure S3) and assessed the biological activity of single, double, and triple mutants. All three ido mRNAs were upregulated in glucose minimal medium (GMM) enriched in Trp (GMM + Trp) (Figure 1B) or in Trp-rich media (Figure S4A). These genetic ablation studies showed that idoA and idoB mainly contribute to kynurenine production (Figure 1C), de novo NAD+ biosynthesis (Figure 1D), and fungal growth (Figure 1E). Accordingly, NAD supplementation (NAM) to the growth medium rescued the lethal phenotype (Figure 1E).
Next, we assessed fungal physiology on GMM + NAM media to determine the germination rate of the mutant strain (Figures S4B–S4D). Here, we observed no significant difference between GMM enriched in NAM (NAM) for the wild-type (WT) strain (Figure S4B) or between WT and Δido mutant strains on GMM + NAM media (Figure S4C; Videos 1, 2, 3, and 4). Thus, NAD auxotrophy as a result of loss of idoA and idoB can be rescued with NAM supplementation (Figure 1E; Videos 1, 2, 3, and 4).
akuBKU80 fungal strain was cultured in GMM for 22h (18 hours of imaging) at 37°C.
akuBKU80 fungal strain was cultured in GMM+NAM for 22h (18 hours of imaging) at 37°C.
ΔidoA/B/C fungal strain was cultured in GMM for 22h (18 hours of imaging) at 37°C
ΔidoA/B/C fungal strain was cultured in GMM+NAM for 22h (18 hours of imaging) at 37°C.
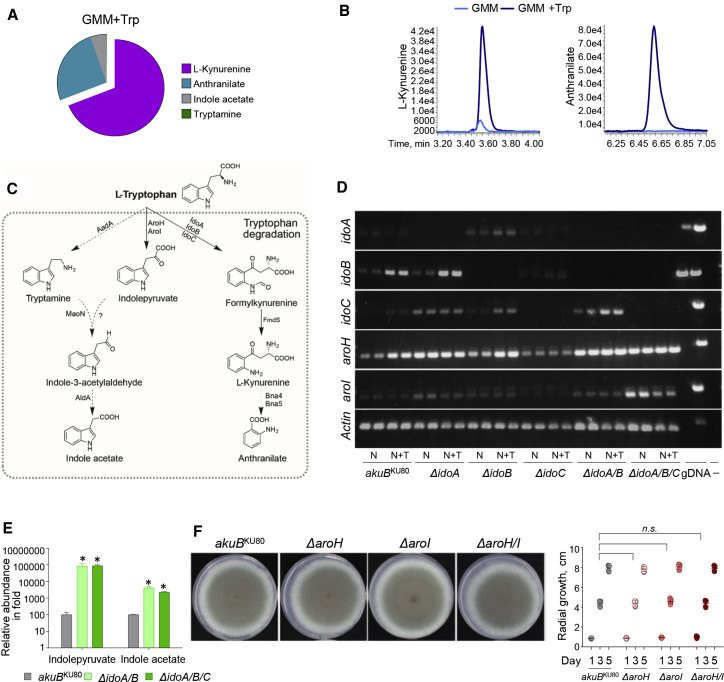
We also wanted to determine the type of metabolites released under Trp supplementation. Target metabolomic analyses on fungal supernatants showed that WT A. fumigatus growing in GMM + Trp released l-kynurenine and, consistent with the occurrence of kynureninase activity (Kynu, XP_751842.1) in the fungus, anthranilate (Figures 2A and 2B). In addition, we detected traces of indole derivatives (e.g., indole acetate, tryptamine) when the fungus was grown in GMM + Trp (Figure 2A). Indeed, the indolepyruvate pathway is parallel to the Ido/kynurenine pathway (Figure 2C).
Figure 2.
Aspergillus Trp alternative metabolism: the Aro pathway
(A and B) Pie diagram and peak levels of Trp-derived metabolites performed at 24 h of culture in GMM + Trp by targeted metabolomics of Aspergillus WT strain.
(C) Trp metabolic pathways, enzymes, and molecular structures of Trp-indole derivatives.
(D and E) RT-PCR of Aspergillus genes involved in the catabolic pathway (D) and relative abundance of indole-derivatives in WT (Aspergillus akuBKU80) and mutant strains inoculated on solid GMM supplement with NAM and Trp and cultured at 37°C for 84 h (E).
(F) Radial growth of Aspergillus akuBKU80 and A. fumigatus aro mutants. Strains were inoculated with 104 conidia onto the solidified GMM supplemented with 5 mM l-Trp. Colony diameters of each strain were measured after 1, 3, and 5 days of growth at 37°C.
(A, B, and D) Experiments were repeated three times. Statistical significance (∗∗p < 0.001) was determined against the Aspergillus akuBKU80 strain (unpaired t test: akuBKU80 versus ΔidoA/B and versus ΔidoA/B/C).
See also Figures S4 and S5.
Although the ido genes regulate the kynurenine pathway, the aro genes, aroH (Afu2g13630) and aroI (Afu5g02990), encode putative PLP-dependent aromatic aminotransferases that transaminate Trp to generate indolepyruvate, a precursor of the indol acetate pathway (Dindo et al., 2018; Choera et al., 2018). We thus assessed aroH and aroI expression in response to Trp in the A. fumigatus WT and ido mutants (Figure 2D). We observed higher aroH expression, especially in the presence of Trp in the WT control, ΔidoA, ΔidoA/B, and ΔidoA/B/C mutants. Although aroI expression was transient, it was particularly increased in the ΔidoA/B/C mutant (Figure 2D). These aroH and aroI expression patterns complement indolepyruvate and indole acetate metabolite production (Figure 2E; Figure S4D).
Overall, these findings suggest that the indolepyruvate pathway is activated under conditions in which Idos are inactive (Figures 2D and 2E) or in the presence of an excess of Trp (Figure 2D). As previously suggested, AroH likely serves as a backup for energy production (Jain et al., 2012). To further validate this hypothesis, we generated ΔaroH, ΔaroI, and ΔaroH/I mutant strains and observed similar in vitro colony growth rates and physiology with the WT strain (Figure 2F; Figure S5). These results point to a previously undescribed mechanism of fungal adaptation, involving Trp catabolism via Ido and Aro. IdoA and IdoB mainly contribute to Trp catabolism and l-kynurenine/NAD+ production. Loss of IdoA and IdoB drives Trp catabolism toward insdolepyruvates via Aros.
Deletion of the Ido pathway in Aspergillus induces AhR-dependent inflammation
In invasive pulmonary aspergillosis, A. fumigatus spores are inhaled into the lungs, where they germinate and undergo hyphal growth. The fungus ultimately obstructs and disrupts the blood vessels, leading to hypoxia and eventual tissue necrosis. The ability of this mold to adapt to hypoxia is regulated, in part, by the sterol regulatory element binding proteins (Vaknin et al., 2016). Loss of the genes encoding these proteins results in avirulence (Vaknin et al., 2016), as the ability to adapt to hypoxia is a key virulence attribute of A. fumigatus and other fungi. In host tissues, Ido activity reduces under hypoxic conditions (Elbers et al., 2016; Romani et al., 2008), but the impact of this reduced activity on fungal metabolism is unknown.
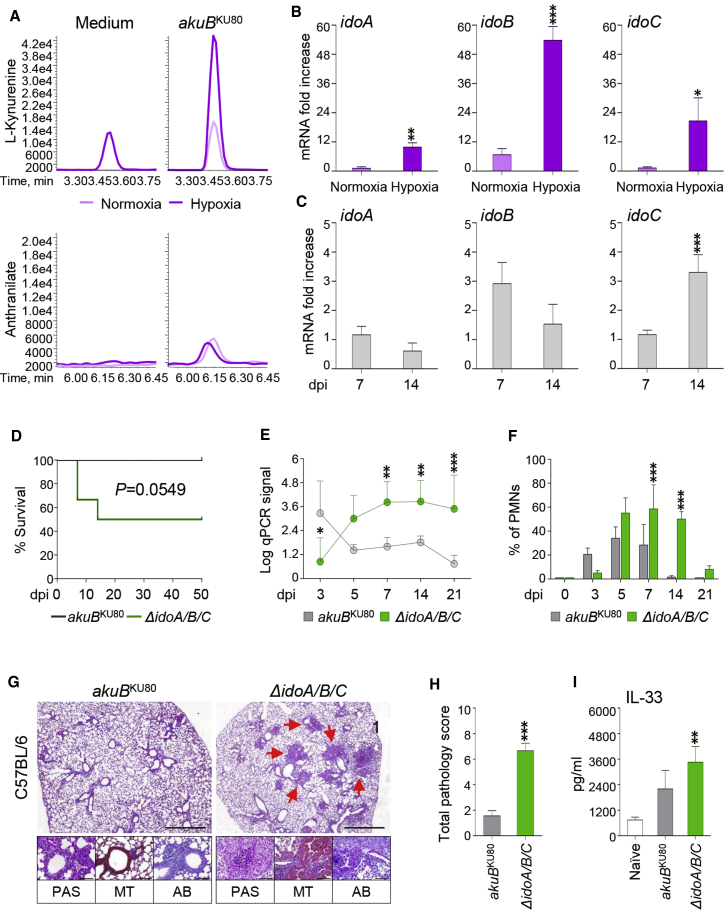
We performed targeted metabolomics to investigate whether A. fumigatus adapts to hypoxic microenvironments by modulating Trp metabolism. Using an oxygen-controlled fermenter system, we measured the dynamic changes of the Trp-derived metabolites from WT Aspergillus strains grown in RPMI 1640 maintained under hypoxia over a period of 24 h (Figure 3A). Surprisingly, we saw abundant l-kynurenine production (Figure 3A) and increased expression of the three ido genes (Figure 3B) under hypoxic conditions; however, we detected only trace amounts of Aro-derived metabolites (data not shown). These data suggest that the Ido-dependent pathway participates in fungal adaptation to hypoxia.
Figure 3.
Ido deficiency increases A. fumigatus pathogenicity in vivo
(A) Peak levels of Trp-derived metabolites performed at 24 h of culture in RPMI 1640 in normoxia or hypoxia by targeted metabolomics of Aspergillus WT strain.
(B) mRNA expression (fold increase) of idoA, idoB, and idoC in Aspergillus grown in normoxia or hypoxia after 24 h of culture.
(C–I) C57BL/6 mice (n = 9) were infected intranasally with 2 × 107 resting conidia of WT akuBKU80 or ΔidoA/B/C A. fumigatus mutant and sacrificed at different days postinfection (dpi).
(C) mRNA fold change of idoA, idoB, and idoC at 7 and 14 dpi in the lung of C57BL/6 mice. (D) Survival rates of C57BL/6 mice. (E) 18S rRNA expression measured by qPCR from C57BL/6 lungs.
(F) Bronchoalveolar lavage (BAL) cell count analysis of C57BL/6 showing the percentage of pulmonary neutrophils (PMNs).
(G) Histopathological analyses (periodic acid-Schiff [PAS], Alcian blue, and Masson’s Trichrome) in C57BL/6 lung tissue at 7 dpi. Scale bars, 10 μm and 100 μm.
(H) Total pathology score on lung histology at 7 dpi.
(I) Cytokine levels by ELISA.
(B, C, E, F, H, and I) Data are represented as means ± SD. Statistical significance for (B) (∗p < 0.01, ∗∗p < 0.001, ∗∗∗p < 0.0001) was determined against Aspergillus samples cultured in normoxia (Twotwo- tailed Student’s t Test test unpaired parametric). (C) Statistical significance, (∗∗p < 0.001) was determined against mice infected with the Aspergillus akuBKU80 strain at 7 dpi (Twotwo- tailed Student’s t Test test unpaired parametric). (D) Nonparametric test of infected mice with AkuBKU80 versus ΔidoA/B/C. (E and F) Statistical significance (∗p < 0.01, ∗∗p < 0.001) was determined against mice infected with the Aspergillus akuBKU80 strain (E) or naive mice (F) (two-way ANOVA and Bonferroni post hoc test).
(H–I) Statistical significance (∗∗p < 0.001, ∗∗∗p < 0.0001) was determined against mice infected with the Aspergillus akuBKU80 strain (two-tailed Student’s t test unpaired parametric). For (I), statistical significance (∗∗p < 0.001) was determined against mice uninfected (naive) (Two two-tailed Student’s t Test test unpaired parametric). Micrographs are representative of three replicate experiments.
See also Figures S6 and S7.
To assess the impact of the metabolic adaptation to hypoxia on fungal pathogenicity in vivo, we used a clinically relevant model of invasive Aspergillosis (Romani et al., 2008; Cenci et al., 2000; Mazaki et al., 2006). In brief, we infected immunocompetent C57BL6 mice intranasally with Aspergillus WT or ido mutants and evaluated parameters of infection, inflammation, and immune responses (Figures 3C–3I; Figures S6 and S7). The animal model we used is a typical standardized aspergillosis model of invasive infection. Because our study is focused on metabolic changes of Aspergillus in the mammalian host, the choice of inoculating mice intranasally is due to the need of reproducing a completely physiological entry of the pathogen in the lung. This model is also characterized by the repeated inoculation to avoid acute tissue damage as shown previously (Cenci et al., 2000; Desoubeaux and Cray, 2017). We found that all three fungal ido genes were transcriptionally expressed in infected C57BL6 mice (Figure 3C): idoA and idoB expression were upregulated early (7 days post infection [dpi]), whereas idoC was upregulated later in infection (14 dpi). As expected, the mice were relatively resistant to WT A. fumigatus but were susceptible to infection with the ΔidoA/B/C mutant, as shown by decreased survival (Figure 3D), increased fungal burden as revealed by the detected amount of Aspergillus-specific DNA (Figure 3E), inflammatory cell recruitment (Figure 3F), and pathology (Figures 3G and 3H) in the lungs. In addition, we detected increased release of the cytokine IL-33 (Figure 3I). IL-33 release underlines the capacity of the ΔidoA/B/C mutant strain to cause cell damage/injury. Consistently, IL-33 has been detected in bronchoalveolar lavage fluids collected after acute lung injury or after exposure to the fungal aeroallergen Alternaria alternata (Haenuki et al., 2012) and in the lungs in response to chitin or helminths (Haenuki et al., 2012). The contribution of IL-33 to asthma development occurs via a Th2-immune response. Thus, the increased secretion of the alarmin IL-33 in mice infected with the ΔidoA/B/C mutant might also indicate the activation of an allergic response to metabolites released via the indolepyruvate pathway. Indeed, we observed a high level of collagen deposition and goblet cells in lungs infected with the ΔidoA/B/C mutant (Figure 3G).
To understand the mechanisms underlying the higher level of virulence of the ΔidoA/B/C mutant strain, we measured the susceptibility to phagocytosis and killing by effector phagocytes. Here, we observed no differences between the ΔidoA/B/C mutant and the WT strain (Figures S6A and S6B). Interestingly, mice infected with each individual mutant survived infection and cleared the fungus to a similar level as mice infected with WT Aspergillus (Figures S6C–S6H). This finding argues some degree of redundancy among the three Idos and is consistent with their growth patterns (Figure 1E). More similarly to the ΔidoA/B/C mutant, the ΔidoA/B mutant induced higher polymorphonuclear neutrophil recruitment and lung pathology (Figures S6E–S6G). We obtained similar results in p47phox−/− mice that recapitulate chronic granulomatous disease (Figures S7A–S7C) and develop notable lung hypoxia resulting in a predisposition to Aspergillosis infection (Rees et al., 2017; Shepardson et al., 2014). Here, these mice were highly susceptible to the ΔidoA/B/C mutant strain compared with the WT Aspergillus strain.
Trp metabolites along the kynurenine pathway make a pivotal contribution to the host’s mucosal immune homeostasis in response to luminal microbiota (Santhanam et al., 2016). l-kynurenine, in particular, contributes to immune tolerance at the host-fungus interface (Romani et al., 2008). To assess whether the observed failure of the ΔidoA/B/C mutant to produce kynurenine in vitro could impact on fungal pathogenesis in vivo, we infected mice not competent for l-kynurenine production, namely, Ido1–/– mice, with the A. fumigatus WT or ΔidoA/B/C mutant strain (Figures S7D–S7F). After comparing the responses of Ido1–/– mice challenged with either the WT or ΔidoA/B/C mutant, we found that pathology (Figures S7D and S7E) and, therefore, fungal burden (Figure S7F) were increased in the Ido1–/– mice challenged with the ΔidoA/B/C mutant strain. Interestingly, lung pathology was more evident in Ido1–/– mice infected with ΔidoA/B/C compared with WT mice infected with ΔidoA/B/C (Figures 3G and 3H). These data suggest that both the host and the fungus produce immunologically active l-kynurenine with anti-inflammatory properties during infection, as already shown (Gargaro et al., 2016). In addition, because a higher availability in Trp has been shown in Ido1–/– mice (Huang et al., 2013), the ΔidoA/B/C mutant strain may be more active in Trp metabolism via the Aros pathway.
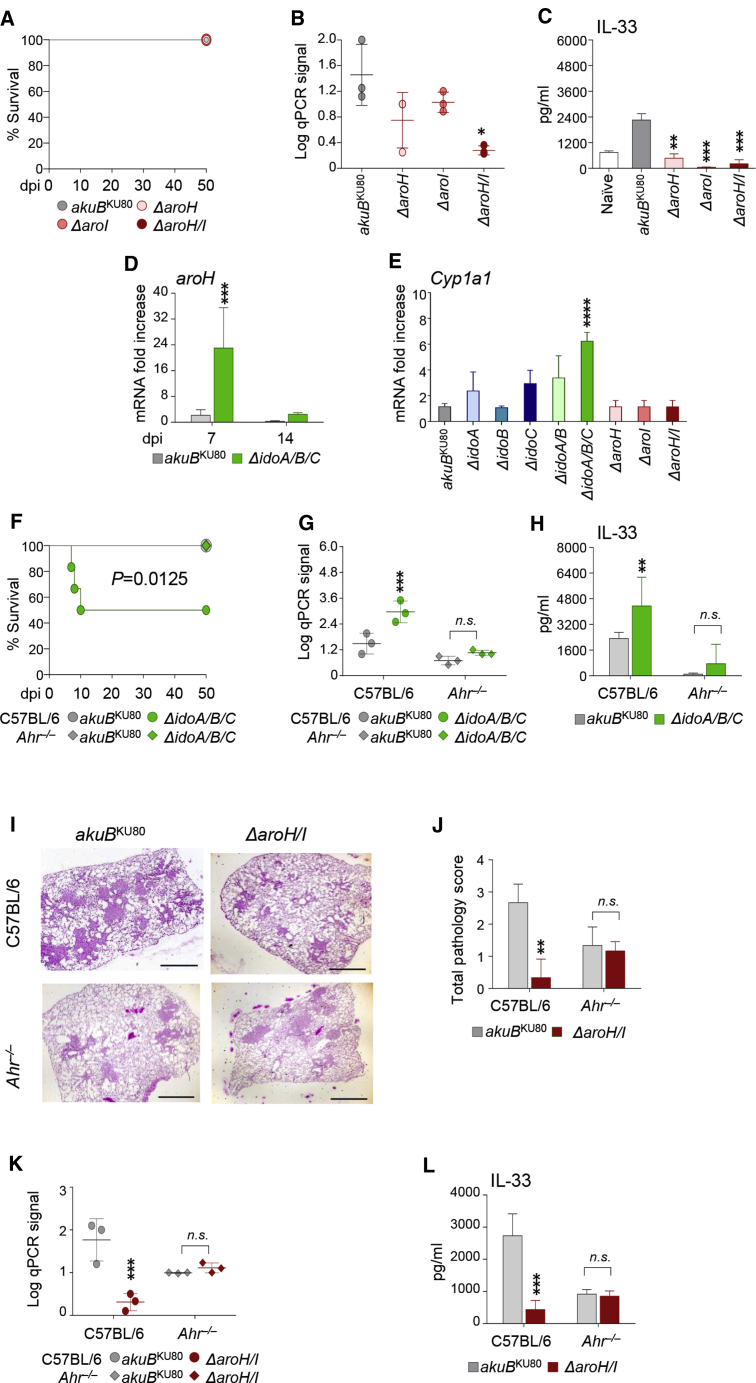
The ability of the ΔidoA/B/C mutant to produce metabolites driven by AroH/I (Figure 2E) led us to assess the physiological role of the aro genes in infection. In vivo, each single mutant for aro genes showed similar or less pathogenicity compared with the WT strain, in terms of survival (Figure 4A), colonization (Figure 4B), and IL-33 production in infected WT mice (Figure 4C). These data suggest that Aspergillus elicits a more infectious response in the mammalian host when the Ido pathway is abrogated and the AroH/I pathway remains active. To confirm this, we measured aroH expression in mice infected with the ΔidoA/B/C mutant, and we found that aroH mRNA did increase in the lung, particularly at 7 dpi, when the Ido pathway is fully abrogated in the fungus (Figure 4D). Because we found that the ΔidoA/B/C mutant can release indole derivatives via the indolepyruvate route (Figure 2E), and knowing also that indoles coming from the microbiota are effective AhR ligands (Agus et al., 2018), we also measured the main AhR downstream target gene Cyp1a1 at 7 dpi (Figure 4E). Of note, increased AhR-dependent Cyp1a1 transcription is a hallmark of AhR activation (Weems and Yost, 2010). We observed this upregulation of Cyp1a1 transcription in the lungs (Figure 4E) upon infection with the ΔidoA/B/C mutant strain, supporting a role for AhR in the biological effects of AroH-derived indoles (Chowdhury et al., 2009).
Figure 4.
The AroH/I pathway promotes lung inflammation via AhR
C57BL/6 and Ahr–/– mice (n = 9) were infected intranasally with 2 × 107 resting conidia of indicated A. fumigatus strains and sacrificed at 7 dpi.
(A) Survival rates.
(B) 18S rRNA expression measured by qPCR from C57BL/6 lungs.
(C) Cytokine levels by ELISA.
(D) mRNA fold change of aroH at 7 and 14 dpi in the lung of C57BL/6 mice.
(E) mRNA fold change of Cyp1a1 at 7 dpi in the lung of C57BL/6 mice.
(F) Survival rates of C57BL/6 and Ahr–/– mice.
(G) 18S rRNA expression measured by qPCR from C57BL/6 lungs.
(H) Cytokine levels by ELISA from C57BL/6 and Ahr–/– mouse lungs.
(I) Histopathological analyses (PAS) in C57BL/6 lung tissue at 7 dpi. Scale bars, 10 μm.
(J) Total pathology score on lung histology at 7 dpi.
(K) 18S rRNA expression measured by qPCR from C57BL/6 lungs at 7 dpi.
(L) Cytokine levels by ELISA at 7 dpi. Data are represented as means ± SD. Graphs are representative of data collected from three independent replicate experiments.
(B, C, and E) Statistical significance (∗p < 0.01, ∗∗p < 0.001, ∗∗∗p < 0.0001) was determined against mice infected with the Aspergillus akuBKU80 strain (one-way ANOVA and Bonferroni post hoc test). (D, G, H, J–L) Statistical significance (∗∗p < 0.001, ∗∗∗p < 0.0001) was determined against mice infected with the Aspergillus akuBKU80 strain (two-way ANOVA and Bonferroni post hoc test). (E) statistical significance (∗∗∗p < 0.0001, ∗∗p < 0.001) was determined against mice infected with the Aspergillus akuBKU80 strain (one-way ANOVA and Bonferroni post hoc test). (F) Nonparametric test AkuBKU80 versus ΔidoA/B/C.
To validate our hypothesis that fungal-mediated Cyp1a1 activation in the lung may mediate ΔidoA/B/C mutant in vivo pathogenicity, we infected WT and Ahr-deficient mice with the control and ΔidoA/B/C strains (Figures 4F–4H). We found that ido-deficient Aspergillus showed the same pattern as WT A. fumigatus in Ahr–/–mice (Figures 4F and 4G), thus suggesting that AhR ligands produced during infection may contribute to fungal virulence and to host lung pathology. Interestingly, Il33 mRNA was not induced in the lung when we infected Ahr–/–mice with ΔidoA/B/C but was induced when we infected WT mice with ΔidoA/B/C (Figure 4H). Il33 might, therefore, be a possible downstream AhR target in the lung, as recently suggested (Ishihara et al., 2019). We propose that activation of the alternative AroH pathway of Trp catabolism on the one hand favors fungal survival, but on the other hand might be responsible for the inflammatory response mounted by the host via AhR. Indeed, we demonstrated this possibility when we infected Ahr–/– mice with the ΔaroH/I strain (Figures 4I–4L). As expected, histopathological analysis of WT infected mice with the ΔaroH/I strain was reduced compared with infection with the Aspergillus WT strain. The lack of reduced histopathology in infected Ahr–/– mice (Figures 4I and 4J) supports an AhR-dependent mechanism underlying that the deletion of aro genes in Aspergillus reduces AhR activation and therefore lung histopathology. Moreover, the amount of ΔaroH/I Aspergillus DNA and IL-33 release in the lungs did not decrease in Ahr–/– mice (Figures 4K and 4L) compared with WT mice upon infection with the ΔaroH/I strain. These results demonstrate that when the Ido pathway is inactive in A. fumigatus, Trp degradation occurs via Aro, which may activate AhR as a host mechanism of detoxification in the lung resulting in IL-33 release.
Enhancing fungal Ido expression reduces virulence and lung inflammation
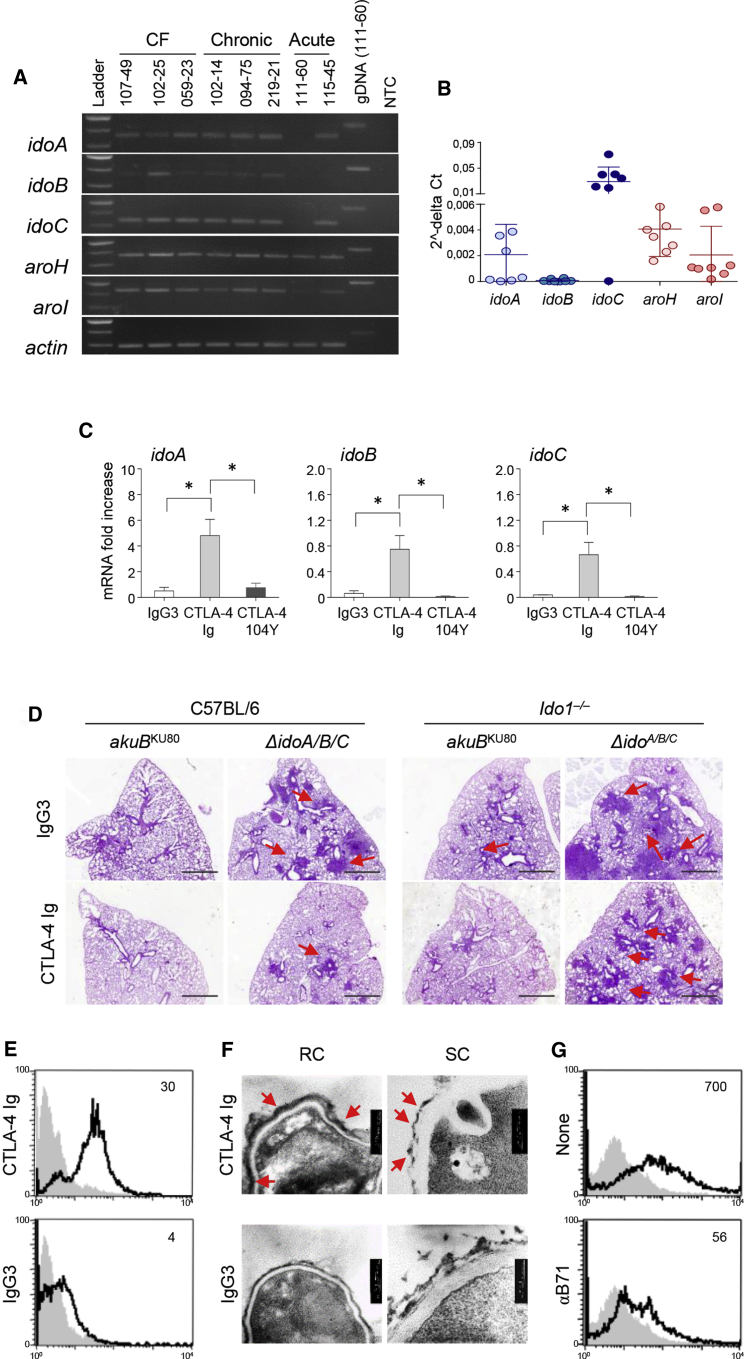
Our findings thus far support that fungal Ido activation affects A. fumigatus pathogenicity in vivo. Importantly, upon analyzing a panel of fungal isolates derived from different clinical contexts (e.g., cystic fibrosis [CF] patients [n = 3], patients with acute invasive aspergillosis [n = 2], and patients with chronic pulmonary aspergillosis [n = 3]), we consistently observed defective ido (particularly idoB) expression (Figures 5A and 5B). It thus seems that the negative correlation between fungal virulence and Ido expression could be of clinical relevance. Of note, the membrane protein B71 serves as an IDO activator in mammals (Bozec et al., 2014; Munn et al., 2004). Because of the similarity between mammalian IDOs and fungal Idos, and considering that we could detect a B71-like protein in fungal cells (Figure S8), we speculated that a mammalian IDO activator might also induce idos transcription in fungi.
Figure 5.
Targeting fungal Idos by CTLA-4 Ig reduces Aspergillus pathogenicity
(A and B) Clinical isolates were grown overnight in liquid GMM at 37°C rotating at 200 rpm. Clinical isolates were obtained from CF patients, patients with acute invasive aspergillosis, and patients with chronic pulmonary aspergillosis. RT-PCR of Aspergillus genes involved in the catabolic pathway (Figure 2C) and (B) relative abundance (2−ΔCT) of gene expression by qPCR analysis.
(C) idoA, idoB, and idoC Aspergillus mRNA expression in Aspergillus akuBKU80 strain exposed to CTLA4-Ig, isotype control IgG3, or mutated CTLA-4 Ig (CTLA-4 104Y) overnight at 37°C.
(D) Histopathological analyses (PAS) in infected C57BL/6 and Ido1–/– mice (n = 9) upon infection with the indicated Aspergillus strains in lung tissue at 7 dpi. Mice were treated intranasally with 50 μg/mouse of CTLA-4-Ig or IgG3 for 2 days, commencing on the second day of infection. Scale bars, 10 μm.
(E) FACS analysis of Aspergillus akuBKU80 resting conidia (RC) exposed to CTLA-4-Ig or IgG3 and reacted with secondary anti-IgG3-fluorescein isothiocyanate (FITC) antibody (continuous black line). The gray histogram denotes unstained cells.
(F) Representative transmission electron microscopy (TEM) images of Aspergillus akuBKU80 RC or swollen conidia (SC) treated with CTLA-4-Ig or IgG3 followed by secondary antibodies conjugated with gold particles.
(G) Fluorescence-activated cell sorting (FACS) analysis of Aspergillus akuBKU80 RC treated with CTLA-4-Ig in the presence of anti-B71 antibody. The gray histogram denotes unstained cells.
(B and C) Data are represented as mean ± SD. Plots are representative of data collected from three independent replicate experiments. (C) Statistical significance (∗p < 0.01) was determined with one-way ANOVA and Bonferroni post hoc test. (D–G) Micrographs and FACS overlay analysis are representative of three replicate experiments.
See also Figure S8.
To test our hypothesis, we exposed to Aspergillus in vitro a known inducer of IDO in mammalians. We used a known inducer of human IDO1 (Grohmann et al., 2002), Cytotoxic T Lymphocyte Antigen-4 (CTLA-4)-immunoglobulin (Ig), which is a fusion protein consisting of the extracellular domain of CTLA-4 and the Ig Fc portion that activates the B71 co-stimulatory molecule. We exposed WT A. fumigatus to CTLA-4-Ig in vitro and assessed the expression of the various fungal ido transcripts. CTLA-4-Ig treatment resulted in a significant induction of idoA/B/C gene expression (Figure 5C). As a control, a mutated CTLA-4-Ig protein affecting the B71-binding motif (CTLA-4 104Y) failed to induce gene expression (Figure 5C). Next, we infected WT and Ido1–/– mice with either Aspergillus WT or ΔidoA/B/C strains and treated them with CTLA-4-Ig or an isotype control intranasally (Figure 5D). CTLA-4-Ig treatment significantly reduced the lung histopathology in WT mice infected with ΔidoA/B/C and Ido1–/– mice infected with the WT strain. CTLA-4-Ig treatment, therefore, fails when both the host and fungal IDOs are deleted (Figure 5D); this result also implies that the fusion protein functions independently of IDO1 expression by the mammalian host. Together, these results suggested that fungal Idos enable the fungus to establish a close functional metabolic relationship with their mammalian host. This process avoids the undesired activation of the indolepyruvate pathway, which could damage the host through AhR binding/translocation in the host lung tissue (Figure 6).
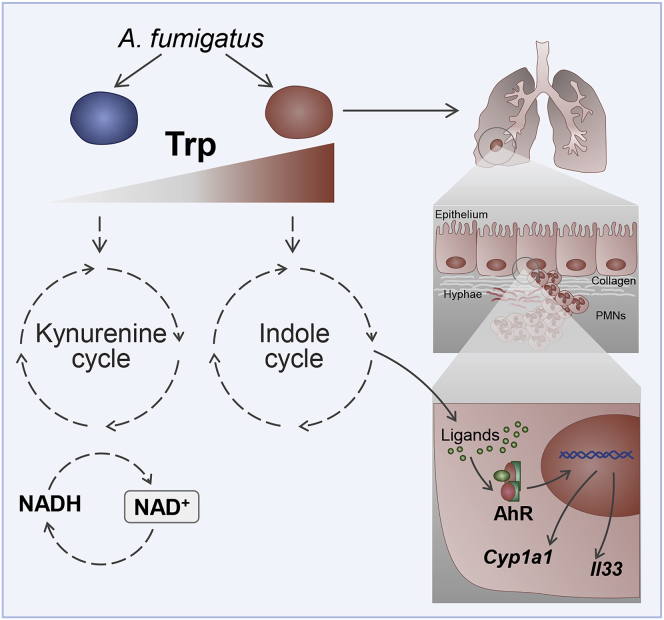
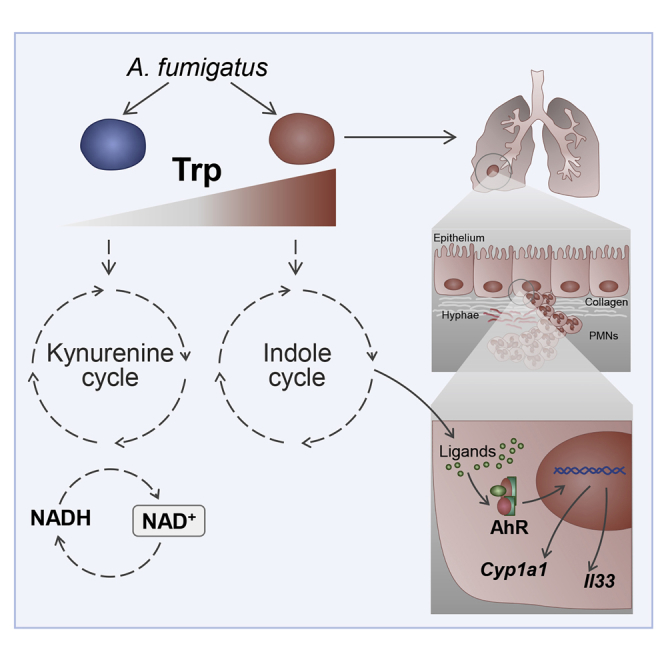
Figure 6.
Trp degradation models and host response to fungal metabolites
A. fumigatus shows two different routes of Trp catabolism (the kynurenine and the indole cycles). In conditions of reduced degradation of Trp via IDO abundance (e.g., host lung IDO deficiency) or genetic ido fungal deficiency, Aspergillus may activate mainly the indole cycle, releasing metabolites, which activate the AhR-xenobiotic response IL-33-mediated, which contributes to epithelial damage and lung inflammation.
Discussion
Originally identified in mammals, IDO-related proteins have been subsequently found in lower vertebrates, several invertebrates, fungi, and a number of bacterial species (Yuasa and Ball, 2015). The affinity and catalytic efficiency for Trp catabolism of IDOs varies greatly among vertebrates and invertebrates (Yuasa and Ball, 2015); however, the high affinity and catalytic efficiency for Trp catabolism exhibited by both fungal and mammalian IDOs suggests that these enzymes have similar biological roles (Ball et al., 2014). Here we found that, similar to mammalian IDO1 and besides fulfilling the need for NAD+ supply, fungal Idos markedly impact on host-fungal interactions. In physiological conditions and in the absence of any infection or tissue damage, the availability of NAD in mammalian host tissues/fluids is sufficient to allow the growth of pathogens (Kulkarni and Brookes, 2019). Indeed, NAM supplementation restored the growth of the mutants.
Of the three A. fumigatus idos, we saw that despite being differently induced by Trp (Wang et al., 2016), idoA and idoB were both induced by hypoxia, a condition frequently encountered by the fungus in the lungs (Grahl et al., 2012). Because mammalian IDO1 and kynurenine production are inhibited by hypoxia (Schmidt et al., 2013), our findings suggest a complementary role for the two sources (host and fungus) of kynurenine in maintaining lung immune homeostasis.
In addition to amino acid homeostasis (Krappmann et al., 2004), oxygen is crucial for A. fumigatus metabolism. However, under proper nutrient conditions, the fungus exploits fermentation and other mechanisms (Grahl et al., 2012) to adapt to low-oxygen environments, and this is critical for growth in the host and virulence (Grahl et al., 2011). Strain fitness in low oxygen correlates with virulence (Chung et al., 2014; Kowalski et al., 2016), and hypoxic fungal growth is associated with major transcriptional (Chung et al., 2014) and metabolic changes (Barker et al., 2012).
Our data support for a role of fungal Idos in establishing a metabolic balance with the mammalian host, revealing a mutualistic/symbiotic relationship between the fungus and its mammalian host in the lungs. Opposing AroH pathway activation via Idos might further contribute to the host-fungus interplay in the lung.
Taking our findings together, we propose a model explaining the impact of Aspergillus Idos on the immune and microbial tolerance in the lung (Figure 6). In short, the Ido/kynurenine pathway of Trp degradation promotes local control of inflammation and a status of eubiosis in the tissue, whereas the alternative AroH pathway may have antagonistic effects by promoting allergy, inflammation, dysbiosis, and epithelial damage (mediated by IL-33) (Figure 6).
As discussed, a negative correlation between fungal virulence and Ido expression has been shown in clinical isolates. Therapeutic induction of fungal Idos with currently available therapies, such as CTLA-4 Ig, thus confers clinical benefit. Our findings emphasize how knowledge of evolutionarily conserved metabolic pathways involving IDO and AhR between the host and fungi may translate into clinical advances in terms of fungal pathogenesis and antifungal therapy. They also underscore the importance of defining environmental parameters in vivo to better predict clinical outcomes of infection. In addition, our findings may open up avenues to identify novel antifungal drugs based on components of the Aro pathway.
We recently proposed that mathematical modeling and reverse-engineered gene-regulatory networks from experimental data could be used to determine the interconnectivity of multiple metabolic fungal pathways (Acerbi et al., 2020). Using continuous time Bayesian networks over a time-course gene expression dataset, we inferred the global regulatory network controlling Trp metabolism. The model revealed that high Trp levels activate the Trp catabolic routes independently, whereas low levels may interconnect the different metabolic pathways (Acerbi et al., 2020). Going forward, it is becoming evident that Aro pathway activation can disrupt lung tolerogenic responses. Studies that improve our understanding on how to switch on tolerogenic or non-tolerogenic Trp degradation routes in different host conditions are now warranted. We will then be in position to propose the modulation of the pathogenicity of the fungus during infection without necessarily destroying the mutualistic relationship between the fungus and the host but eventually acting on amino acid/nutrients tissue availability during infection and on activation of selected metabolic fungal profiles.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| PAS | BIO OPTICA | Cat. # 04-130802 |
| MASSON | BIO OPTICA | Cat. # 04-010802 |
| SABOURAUD AGAR | MILLIPORE | Cat. # 89579 |
| ALCIAN BLUE | N/A | N/A |
| Critical commercial assays | ||
| Itaq Universal SYBR® Green Supermix | BIORAD | Cat# 1725124 |
| PrimeScript RT Reagent Kit with gDNA Eraser | TAKARA | Cat. # RR047A and RR047B |
| Mouse IL-33 ELISA Ready-SET-Go | EBIOSCIENCE | Cat. # 501125201 |
| Experimental models: organisms/strains | ||
| C57BL/6ncrl | CHARLES RIVER, ITALIA | Cat# 6152460 |
| pyrG1, ΔakuB::pyrG, pyrG1, argB::pyrG, ΔAFUB_066940::argB | THIS STUDY, Zelante’s lab | TTC38.13 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔAFUB_034980::pyrG | THIS STUDY, Zelante’s lab | TTC37.1 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔargB::pyrG, ΔpyrG, ΔAFUB_088580::six, pyrG, argB | THIS STUDY, Zelante’s lab | TXL3.2 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔargB::pyrG, ΔpyrG, ΔAFUB_034980::pyrG, ΔAFUB_066940::argB | THIS STUDY, Zelante’s lab | TTC39.5 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔargB::pyrG, ΔpyrG, ΔAFUB_034980::pyrG, ΔAFUB_066940::argB, ΔAFUB_088580::six | THIS STUDY, Zelante’s lab | TTC42.1 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔargB::pyrG, ΔakuB::pyrG, ΔAFUB_051500::argB | THIS STUDY, Zelante’s lab | TTC 21.6 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔAFUB_029280::pyrG | THIS STUDY, Zelante’s lab | TTC 22.7 |
| pyrG1, ΔakuB::pyrG, pyrG1, ΔargB::pyrG, ΔpyrG, ΔAFUB_051500::argB, ΔAFUB_029280::pyrG | THIS STUDY, Zelante’s lab | TTC 23.1 |
| Software and algorithms | ||
| Graphpad Prism | SCR_002798 | RRID:SCR_002798 |
| Flowjo | SCR_008520 | RRID:SCR_008520 |
| AnalySIS FIVE | OLYMPUS | RRID:SCR_014430 |
Resource availability
Lead contact
Further information and requests for resources and reagents will be directed and fulfilled by the lead contact, Teresa Zelante (teresa.zelante@unipg.it).
Materials availability
Aspergillus fumigatus strains generated in this study are available under request to Teresa Zelante’s lab.
Data and code availability
The published article includes all datasets generated or analyzed during this study.
Experimental model and subject details
Fungal strains
Strains used or created in this study are listed in Table S1. The genetic background of the primary strain used in this study is A. fumigatus CEA17 (Osherov et al., 2001; da Silva Ferreira et al., 2006; Wiemann et al., 2017). All strains were maintained as glycerol stocks at −80°C, and activated on solid glucose minimal media (GMM) at 37°C (Shimizu and Keller, 2001; da Silva Ferreira et al., 2006). Growth media was supplemented with 1.26 g/L uridine and 0.56 g/L uracil for pyrG auxotrophs, 1 g/L L-arginine for argB auxotrophs, and 50uM Nicotinamide (NAM) in case of auxotrophy. CEA17 akuBKU80 was also used in hypoxic conditions by growing fungi in N2 with additional 5% mol/mol CO2 and 5% mol/mol O2. Supplemental Trp (Sigma Aldrich) resulted in a final concentration of 60 μM.
Mice
Female C57BL/6 8-10 week-old mice were purchased from Charles River. Homozygous Ido1–/–and Ahr–/– mice and mice deficient for p47 (p47phox–/–) on a C57BL/6 background were as described (Romani et al., 2008) were bred under specific pathogen-free conditions at the Animal Facility of Perugia University.
Ethics statement
The animal studies described in this manuscript were performed according to the Italian Approved Animal Welfare Authorization 360/2015-PR and Legislative Decree #26/2014, which provided ad hoc clearance by the Italian Ministry of Health over a 5-year period (2015–2020). Animals were assessed twice daily for physical conditions and behavior. Animals ranked as moribund were humanely euthanized by CO2 asphyxiation.
Infections
Mice were anesthetized before instillation (for three consecutive days) of a suspension of 2 × 107 resting conidia per 20 μL of saline intranasally of the Aspergillus CEA17 akuBKU80 WT or the different mutant strains. Mice were monitored for fungal growth and lung inflammation and histology. The extent of infection was measured by euthanizing mice at 3, 5, 7, 14, 21 days post infection, removing the lungs and culturing serial dilutions of the homogenized tissue on Sabouraud agar plates, in triplicate, to allow calculation of the mean number of colony-forming units per organ (c.f.u. per organ; mean ± s.d.). For qPCR measurements of the abundance of the 18S fungal rRNA coding gene, primary homogenates were prepared by adding 3.6 volumes of sterile saline per gram of tissue, as described (Bowman et al., 2001).
Method details
Computational analysis
The comparative model was built using Prime software (Schrödinger Release 2016-4), using the 2D0T crystal structure as a template, chain A (Sugimoto et al., 2006), the best available structure on commencing this study. The overall goodness of the models was checked by Ramachandran plots, via PROCHECK (Laskowski et al., 1996), whereby > 97% of the IdoA and IdoB residues fitted in the allowed region of the plot (Figure S2D). A further comparison with other human crystal structures made available while this study was underway was also performed to check for appropriateness of the original template in the light of such new pieces of information (Peng et al., 2016; Tojo et al., 2014). In terms of secondary structure and spatial arrangement, no critical issues were raised by the analysis of the new structures. The right protonation state at pH 7.4 was assigned using Protein Preparation wizard from Schrödinger. Maps of the binding sites were generated using SiteMap (SiteMap, version 3.0, Schrödinger).
Genetic manipulations for A. fumigatus ido and aro mutants by protoplasting method
Fungal DNA extraction, gel electrophoresis, restriction enzyme digestion, Southern blotting, hybridization and probe preparation were performed according to standard methods (Sambrook and Russell, 2001). For DNA isolation, A. fumigatus strains were grown for 24 h at 37°C in static liquid GMM, supplemented as needed for auxotrophs. DNA isolation was performed as described by Sambrook and Russell (2001). Gene deletion mutants in this study were constructed by targeted integration of the deletion cassette through transformation (Szewczyk et al., 2006; Lim et al., 2012). The deletion cassettes were constructed using a double-joint fusion PCR (DJ-PCR) approach (Szewczyk et al., 2006; Lim et al., 2012). A. fumigatus protoplast generation and transformation were carried out as previously described (Szewczyk et al., 2006; Lim et al., 2012). The primers used in this work are listed in Table S1 and the plasmids in Table S2. An A. fumigatus idoB (AFUB_034980) disruption consisted for the following: two 1 kb fragments flanking the ORF were amplified from CEA10 genomic DNA using the primer pair TC-1113/1114 and TC-1115/1116, respectively (Table S2). The selection marker, A. parasiticus pyrG, was PCR amplified from plasmid pJW24 (Calvo et al., 2004) using the primer pair TC-4/TC-5 (Table S2). The deletion construct was transformed into CEA17 (pyrG1) and TMN21 (argB1, pyrG1). Transformants were selected for pyrimidine prototrophy in media without any supplements and amended with 50uM NAM in case of auxotrophy. Transformants obtained were verified by PCR using primers pairs TC-1119/TC-1120, and Southern analysis using the flanking probes (Figure S1). The obtained mutants were named TTC36.x (ΔidoB, argB-), and TTC37.x (ΔidoB) (Figure S1). TTC36.15 (ΔidoB, argB-) and TTC37.1 (ΔidoB) were chosen for further experiments.
The A. fumigatus idoA (AFUB_066940) disruption cassette was constructed similarly utilizing a 2.7 kb selectable auxotrophic marker, A. fumigatus argB, cloned from plasmid pJMP4 (Palmer et al., 2013) with primers TC-20/TC-21 and the primer pairs TC-1131/1134 and TC-1132/1135 for flanking regions. A 3rd round PCR product was amplified with primer pairs TC-1133/1136. The deletion construct was transformed into TJG1.6 (argB1) and TTC36.15 (ΔidoB, argB-). Transformants obtained were verified by PCR using primers pairs TC-1137/TC-1138, and Southern analysis using the flanking probes (Figure S2). The obtained mutants were named TTC38.x (ΔidoA) (Figure S2), and TTC39.x (ΔΔidoAidoB) (Figure S4). TTC38.13 (ΔidoA) and TTC39.5 (ΔΔidoAidoB) were chosen for further experiments.
The A. fumigatus idoC (AFUB_088580) disruption cassette was constructed with the self-excising selection marker, six-B-rec-hygroR-six, which was purified from plasmid pKS29 (Calvo et al., 2004) (Table S2) by digestion with Fsp I. Two 1 kb fragments flanking the ORF were amplified using the primer pair XL-1/XL-2 and XL-3/XL-4, respectively (Table S2). The deletion construct was transformed into TMN2.1 (argB-, pyrG-) and TTC39.6 (ΔΔidoAidoB). Transformants were selected for hygromycin resistance in media with hygromycin at 200 μg/ml and amended with Nicotinamide in case of auxotrophy. Transformants were further screened by PCR (primer pair XL-1/XL-5 for ΔidoC mutants,) and Southern analysis using probe of Flanking regions (Figure S3). The obtained mutants were named TXL2.x (ΔidoC, hphR, argB-, pyrG-) and TTC41.x (ΔΔΔidoAidoBidoC, hphR). The self-excising selection marker was recycled by growing TXL2.2 and TTC41.1 on Xylose (2%) Minimal Media and transformants were further screened by PCR (primer pair XL-1/ XL-4) and Southern analysis using probe for Flanking regions (Figure S3). A construct containing argB-pyrG was introduced into the Ku locus, yielding transformant TXL3.x (ΔidoC, hphS) (Figure S3 and TTC42.x (ΔΔΔidoAidoBidoC, hphs) (Figure S5). TXL3.2 (ΔidoC) and TTC42.1 (ΔΔΔidoAidoBidoC) were chosen for further experiments.
To construct the A. fumigatus aroI (AFUB_051500) disruption cassette, the 2.7 kb selectable auxotrophic marker, A. fumigatus argB, cloned from plasmid pJMP4 was fused with 1 kb DNA fragment upstream and downstream of the aroH ORF were amplified from CEA10 genomic DNA using the primer pairs DAFUB_051500F1/DAFUB_051500R1 and DAFUB_051500F2/DAFUB_051500R2. The deletion construct was transformed into TJG1.6 (argB-) and TMN2.1 (pyrG-, argB-). Transformants obtained were verified by PCR using primers TC-1147 and TC-1148, and Southern analysis using the flanking probes (Figure S11). The obtained mutants were named TTC20.x (ΔaroI, pyrG-), and TTC21.x (ΔaroI) (Figure S11). TTC21.6 (ΔaroI) and TTC20.1 (ΔaroI, pyrG-) were chosen for further experiments.
An A. fumigatus aroH (AFUB_029280) disruption consisted for the following: A. parasiticus pyrG as a selection marker amplified from plasmid pJW24(Calvo et al., 2004) using the primer pair TC-4/TC-5 (Table S2), two 1 kb fragments flanking the ORF were amplified from CEA10 genomic DNA using the primer pairs TC-1104/TC-1105 and TC-1106/TC-1107, respectively (Table S2). The deletion construct was transformed into CEA17 (pyrG-) and TTC20.1 (ΔaroI, pyrG-). Transformants were further screened by PCR using primer paor TC-1110/TC-1111 and Southern analysis using probe for Flanking regions (Figure S11). The obtained mutants were named TTC22.x (ΔaroH) and TTC23.x (ΔΔaroHaroI) (Figure S11). TTC22.4 (ΔaroH) and TTC23.2 (ΔΔaroHaroI) were chosen for further experiments.
Aspergillus metabolomics on supernatants
A. fumigatus conidia (108/ml-1) were grown in GMM (1-L composition—20 g Glucose, 5 g (NH4)2SO4, 40 mg l-Arg HCl, 20 mg l-Leu, 2 μg Biotin, Trace Elements (as found in standard Yeast Nitrogen Base), or RPMI 1640 at 37°C for 24h. Supernatants were collected upon centrifugation at 3000 rpm for 15 min. All solvents and reagents were LC-MS grade and were supplied from Sigma-Aldrich (Milano, Italia). Sodiumdihydrogenphosphate-1-hydrate was from Merk (Darmstadt, Germany). Stock solutions of each isotopically-labeled internal standard were prepared in metanol (MeOH) and stored at –80°C. Individual stock solutions of stable isotope-labeled standards were prepared at a concentration of 1 mg/ml: l-kynurenine sulfate (ring-d4,3,3-d2, 97%) 95% (Cambridge Isotope Laboratories, CIL, USA); Trp (indole-d5, 98%, CIL); indole-3-acetic acid (indole-d5, 97%–98%, CIL); dl-3-hydroxykynurenine:HCl (13C2, 99%; 15N, 98%, CIL); indole-2,4,5,6,7-d5-3-acetic-2,2-d2 acid (99,2%, CDN Isotope, Point-Claire, Quebec, Canada); anthranilic-3,4,5,6-d4 acid (98%, CDN). A 200 μg/ml stock solution of each isotope-labeled internal standard was prepared in MeOH and stored at –80°C. Individual stock solutions of unlabeled analytes were also prepared in MeOH and stored at –80°C. Individual stock solutions of each standard were prepared at concentration of 1 mg/ml. All the compounds were from Sigma-Aldrich (Milan, Italy). Two separate working standard solutions, one containing all the isotope-labeled standards and one containing all the unlabeled standards, were prepared at 60 μg/ml in MeOH and stored at –80°C. The standard solution and the isotope-labeled standard solution were diluted at 2.5 μg/ml in water with 5 mM ammonium acetate (AmAc) and 0.2% formic acid (FoAc) (eluent A) and stored at –20°C. These solutions were used to prepare a calibration curve in the range of concentration of interest for quantitative measurements. Cell culture supernatants were thawed at room temperature. Sample preparation started within 40 min of thawing the samples. Twenty μl of the 2.5 μg/ml internal standard solution were added to 0.5 mL of each sample in cell culture medium. The sample was than diluted at 1 mL with phosphate buffer 0.2 M, pH 6.5 and loaded onto an OASIS HLB (1 ml, 30 mg, Waters) cartridge, wetted with 1 mL MeOH, conditioned with 1 mL phosphate buffer 0.2 M, pH 6.5 and then eluted with 2 mL of a MeOH:H2O:acetic acid (90:8:2, v:v:v) solution. The eluate was dried using a vacuum evaporator (Jouan, Thermo Electron) and the residue was dissolved in 100 μL of HPLC eluent A. A Dionex Ultimate 3000 HPLC system was used (Thermo Fisher Scientific) coupled to an API 3000 LC-MS/MS (AB Sciex, Toronto Canada) equipped with a Turbo Ion Spray source operating in positive ion mode. Analyst software (v.1.6.2) from AB Sciex was used for data acquisition and analysis. All MS parameters were optimized by direct infusion and source parameters (gas flows and temperatures) by flow injection. The ion source operated with ion spray voltage set at 5.5 kV, curtain gas at 8, ion source temperature at 500°C, ion source gas 7,000 ml/min; collision gas was nitrogen at 3.4 ∗ 10-5 bar pressure. Analytes were detected using scheduled multiple reaction monitoring (MRM) acquisition; two transitions were monitored for each molecule. All the acquisition parameters are listed in table S5. A Synergi 2.5 Hydro RP column (100 × 2 mm, 3 μm, 100 Å, Phenomenex, USA) was used; eluents were 5 mM AmAc in water (A) and acetonitrile (B), both containing 0.2% FoAc. The column temperature was kept at 35°C. Chromatographic separation of the analytes was performed using a linear gradient as reported in Tables S5 and S6 10 μl injection volume was used. The column effluent was delivered to the mass spectrometer with no split.
Primary metabolites extraction and analysis
Briefly, 104 A. fumigatus conidia from control and ido mutant strains were inoculated on solid GMM supplement with NAM and Trp and cultured at 37°C for 84 h in triplicates. Fungal tissue was collected and immediately frozen in liquid nitrogen. The fungal tissue was homogenized in 3 mL extraction solvent (2/2/1 (v/v/v) acetonitrile/methanol/water) cooled on dry ice. After centrifugation, 2 mL of supernatant was filtered using a 0.45 μm PTFE Mini-UniPrep filter vial (Agilent). The supernatant of these hyphal metabolites was used for metabolite analysis. For metabolite measurement, samples were dried under N2 and resuspended to 20ug/ml in LC-MS grade water (Sigma-Aldrich). Samples were analyzed using a HPLC-MS system consisting of Thermo Scientific Vanquish UHPLC coupled by electrospray ionization (ESI; negative mode) to a hybrid quadrupole-high-resolution mass spectrometer (Q Exactive orbitrap, Thermo Scientific) operated in full scan mode. Liquid chromatography separation was achieved using an ACQUITY UPLC® BEH C18 (2.1 × 100 mm column, 1.7 μm particle size, Waters). Solvent A was 97:3 water: methanol with 10 mM tributylamine and 10 mM acetic acid, pH 8.2; solvent B was methanol. The gradient was: 0 min, 5% B; 1.5 min, 5% B, 11.5 min, 95% B; 12.5 min, 95% B; 13 min, 5% B; 14.5 min, 5% B. Autosampler and column temperatures were 4°C and 25°C, respectively. Metabolite peaks were identified by their exact mass and matching retention time to those of pure standards (Figure S8) (Sigma-Aldrich).
RNA extraction and Semiquantitative RT-PCR analysis
Total RNA was extracted with Trizol reagent (Invitrogen) according to manufacturer’s protocol. Semiquantitative RT-PCR analysis was performed using 10 μg RNA, which was digested with DNase I (NEB catalog no. M0303L) to remove any contaminating genomic DNA. cDNA synthesis reactions were performed using the Bio-Rad iScript cDNA synthesis kit (catalog no. 170-8891) according to the manufacturer’s protocols. Fifty nanograms of cDNA was used per reaction to amplify specific fragments using gene-specific primers. The primers used are listed in Table S2 where Actin cDNA served as a loading control. Control and mutant strains were grown on solid inoculated on solid GMM supplement with NAM and GMM supplemented with NAM and Trp and cultured at 37°C for 84 h in triplicates.
Physiology experiments
Colony diameters of strains were measured after 3 days of growth at 37°C on solidified GMM and GMM supplemented with NAM (50uM) respectively. Strains were point-inoculated onto the media at 104 conidia total (in 5 μL). Germination was assessed as described in Fischer et. al in static liquid GMM and GMM supplemented with NAM (50uM) respectively as well. Briefly 1 × 105 spores/mL in GMM and GMM supplement with 50uM NAM were inoculated into each well of a Costar® 24-well dish (Corning, Corning, NY, USA). Microscopic images were captured using a Nikon Eclipse Ti inverted microscope equipped with an OKO-Lab microscopic enclosure to maintain the temperature at 37°C for A. fumigatus (OKO Lab, Burlingame, CA, USA). Germinated spores were observed using a Nikon Plan Fluor 20xPh1 DLL objective and phase-contrast images captured every 1–2 h using the Nikon NIS Elements AR software package (v. 4.13). A spore is noted to be germinating if an emerging germ tube was clearly present. One hundred spores were observed for each strain (n = 3) and growth condition. Values in figures represent the average percentage of spores germinated ± SEM. The Student t test was carried out to determine statistical significance using the GraphPad Prism software (La Jolla, CA, USA). Growth on various media was also assessed to display the phenotype of mutants. Strains were inoculated with 104 conidia onto the solidified GMM and GMM supplemented with 5mM L-Trp. Colony diameters of each strain were measure after 1day, 3days, and 5days of growth at 37°C.
Chemicals and reagents
All solvents and reagents were LC-MS grade and were supplied from Sigma-Aldrich (Milano, Italia). Sodiumdihydrogenphosphate-1-hydrate was from Merk (Darmstadt, Germany). Stock solutions of each isotopically-labeled internal standard were prepared in metanol (MeOH) and stored at –80°C. Individual stock solutions of stable isotope-labeled standards were prepared at a concentration of 1 mg/ml: l-kynurenine sulfate (ring-d4,3,3-d2, 97%) 95% (Cambridge Isotope Laboratories, CIL, USA); Trp (indole-d5, 98%, CIL); indole-3-acetic acid (indole-d5, 97%–98%, CIL); dl-3-hydroxykynurenine:HCl (13C2, 99%; 15N, 98%, CIL); indole-2,4,5,6,7-d5-3-acetic-2,2-d2 acid (99,2%, CDN Isotope, Point-Claire, Quebec, Canada); anthranilic-3,4,5,6-d4 acid (98%, CDN). A 200 μg/ml stock solution of each isotope-labeled internal standard was prepared in MeOH and stored at –80°C. Individual stock solutions of unlabeled analytes were also prepared in MeOH and stored at –80°C. Individual stock solutions of each standard were prepared at concentration of 1 mg/ml: indole-3-acetamide 98%; indole-3-carboxaldehyde 97%; indole-3-acetaldehyde-sodium bisulfite addition compound; anthranilic acid 99.5%; indol-3-acetonitrile 98%; 3-hydroxyanthranilic acid 97%; serotonin hydrochloride 98%; l-kynurenine; 3-hydroxy-dl-kynurenine. All these compounds were from Sigma-Aldrich (Milan, Italy). Two separate working standard solutions, one containing all the isotope-labeled standards and one containing all the unlabeled standards, were prepared at 60 μg/ml in MeOH and stored at –80°C. The standard solution and the isotope-labeled standard solution were diluted at 2.5 μg/ml in water with 5 mM ammonium acetate (AmAc) and 0.2% formic acid (FoAc) (eluent A) and stored at –20°C. These solutions were used to prepare a calibration curve in the range of concentration of interest for quantitative measurements.
Bronchoalveolar lavage (BAL)
BAL was performed on sacrificed animals by cannulating the trachea and washing the airways with PBS to collect the BAL fluid. Differential cell counts were generated on BAL smears stained with May-Grünwald Giemsa reagents (Sigma-Aldrich) observed using a BX51 microscope equipped with a high-resolution DP71 camera (Olympus).
Flow cytometry
Flow cytometry was performed on a Fortessa (Becton Dickinson) and data analyzed with FlowJo software (Tree Star). For cell-surface labeling, single-cell suspensions were incubated for 20 min at 4°C with cocktails of antibodies. Samples were washed and analyzed by flow cytometry.
In vivo treatments
Each mouse was slightly anesthetized, then given an intranasal administration for two days, commencing on the second day of infection, of 50 μg/mouse of CTLA-4–Ig or IgG3 (a generous gift from Louis Boon, Bioceros, Utrecht, the Netherlands).
Histology
For histology, lungs were removed and immediately fixed in 10% neutral buffered formalin (Bio-optica) for 24 h. Lungs were then dehydrated, embedded in paraffin, sectioned into 3–4 μm slices and stained with periodic acid-Schiff reagent (PAS), Alcian blue reagent or Masson reagent (Bio-optica). All images were visualized using a BX51 Olympus equipped with a high-resolution DP71 camera (Olympus) with a × 4, × 20 and × 40 objective with the analySIS image processing software (Olympus) or EVOS® FL Color Imaging System with a × 40 objective. The pathology scale considered both the severity and the distribution of morphologic changes within the lungs (Hubbs et al., 1997). The total pathology score is the sum of the severity and the distribution and potentially range from 0 to 10. The severity scores are: none = 0; minimal = 1; mild = 2; moderate = 3; marked = 4; severe = 5. The distribution scores are: none = 0; focal = 1; locally extensive = 2; multifocal = 3; multifocal and coalescent = 4; diffuse = 5 (Hubbs et al., 1997).
Cytokine detection
ELISAs were performed on lung homogenates or lung supernatants to measure cytokine production. IL-33 was assayed using commercially available antibody pairs and standards from Biolegend according to the manufacturer’s protocol.
qPCR
Real-time RT-PCR was performed using the 7500 Fast Real-Time PCR System (ThermoFisher) and FAST SYBR Green Master Mix (Applied Biosystems) following the manufacturer’s directions. Real-time PCR reactions for single genes were performed using 100ng of reverse transcribed RNA, FAST SYBR Green Master Mix and gene-specific primers (sequence reported in Table S3). Total murine RNA was extracted from purified cells or different organs by the TRIzol method (Invitrogen) according to the manufacturer’s protocol. All reactions were repeated at least three times independently and normalized with β-actin gene expression.
RT-PCR on clinical isolates
Eight clinical A. fumigatus strains (Nijmegen culture collection) isolated from CF patients (n = 3), and patients with acute invasive aspergillosis (n = 2) and chronic pulmonary aspergillosis (n = 3) were used. Clinical isolates were grown overnight in liquid glucose minimal media at 37°C rotating at 200 rpm. Clinical isolates were grown overnight in liquid glucose minimal media at 37°C rotating at 200 rpm. Fungal biomass was isolated via vacuum filtration and approximately 100 mg was placed in a screw top vial with 500 μl TRIzol (Invitrogen, UK) and 200 μl acid washed glass beads (Sigma Aldrich, UK). Fungal biomass was disrupted using a FastPrep-24TM5G (MPBiomedicals, USA) at 4.5 m/s for 30 s. Samples were left at room temperature for 5 min and subsequently centrifuged for 10 min at 13000 rpm at 4oC. The supernatant was transferred to a new Eppendorf and 0.4 volumes of chloroform was added. The samples were shaken by hand for 15 s and then left for 10 min at room temperature. Samples were subsequently centrifuged for 5 min at 13000 rpm at 4oC. The top colorless layer was transferred to a new Eppendorf and 0.5 volumes of isopropanol was added. RNA was left to precipitate at room temperature for 15 min. The sample was then centrifuged for 10 min at 13000 rpm at 4oC. The supernatant was removed and the pellet was air-dried for 15 min. The pellet was resuspended in DEPC-treated H2O and 500 μL LiCl buffer (4 M LiCl, 20 mM Tri HCl pH7.4, 10 mM EDTA) was added. The solution was vortexed and left to precipitate at −20 oC for 1 h. The precipitated solution was centrifuged for 30 min at 13000 rpm at 4oC. The pellet was washed twice using ice-cold 70% ethanol and centrifuged for 10 min at 1300 rpm at 4oC. Ethanol was removed by pipette and the pellet was air-dried for 15 min. RNA was resuspended in 100 μl DEPC-treated H2O. Total RNA was treated with DNase I RNase-free (New England Biolabs, UK) following the manufacturer’s instructions. cDNA was synthesized using the iScriptTM Reverse Transcription kit (Bio-Rad, UK). PCR reactions were performed using Phusion Flash High-Fidelity PCR Master Mix (ThermoFischer Scientific, UK) and cDNA as a template. PCR reactions were run on a 1% agarose gel with a 1 kb DNA ladder (Promega).
mCTLA4 fusion proteins
Expression plasmids and 293-F Transfection Mouse CTLA4 was expressed as Ig fusion protein. A wild-type version and a mutant version was generated. The mutant version contained a Y104A mutation in the CTLA4 amino acid sequence, giving a molecule not binding to its counterpart. As extracellular domain (amino acid 36 – 161; GenBank accession code NP_033973) of mouse CTLA4 (w/o and w/ the Y104A mutation) was linked to Hinge-CH2-CH3 domain (amino acid 94 – 329; GenBank accession code BAA11356) of mouse IgG3. Furthermore, a mouse Ig 19 amino acid signal peptide was included N-terminal. Therefore, cDNA encoding mCTLA4-Ig and mCTLA4-Y104A-Ig were ordered at Geneart (Germany). cDNA constructs were optimized for mammalian expression. After synthesis, each cDNA was cloned using HindIII and XhoI into an expression plasmid. Using an EndoFree Plasmid Maxi Kit (QIAGEN, Germany) plasmid DNA was generated of both constructs for the transfection.
Using the FreeStyle™ 293 Expression System (ThermoFisher Scientific; USA) mCTLA4-Ig and mCTLA4-Y104A-Ig were transient expressed according to manufacturer’s procedure. Briefly, plasmid DNA (μg) was mixed with 293fectin (μl) (ratio 1:2) and added to the 293-F cells (1 μg DNA/ 106 viable cells/ml). Transfected cells were cultured for 6 days under standard conditions in an humidified CO2 incubator (5% CO2; 36.5°C) on an orbital shaker (125 rpm). Using protein G affinity chromatography expressed Fc-fusion proteins were purified from the harvested supernatant. Produced materials were dialyzed against PBS pH 7.4, A280 nm measured for quantification, and aliquoted (stored at −20°C).
Killing assay
Thioglycolate-induced peritoneal polymorphonuclear (PMN) cells were obtained from naive C57BL/6 mice. PMN were plated at final concentration of 1 × 106 cells/well in 96-well plates, and fungicidal assays were performed incubating cultures at 37°C for 2h at 1:1 A. fumigatus:PMN ratio. The wells were washed with PBS-diluted TRITON X-100 (Sigma Aldrich) to lyse PMNs. Lysate suspensions were diluted and grown in Sabouraud agar overnight at 37°C in individual plates. The percentage of CFU inhibition (mean ± SE) was determined as: percentage of colony formation inhibition = 100 − (CFU experimental group/CFU control cultures) × 100. All assays were done with five wells per condition in more than three independent experiments.
Phagocytosis assay
PMN were challenged with 1 × 106 cells at 1:1 PMN: A. fumigatus ratio and were subsequently incubated at 4°C for 2 h. After culture, cells were put under centrifugation (7 min, 700 rpm), and cytospins were stained with May-Grünwald-Giemsa (Bio Optica). At least 200 PMNs per sample were counted under oil immersion microscopy (100 × ). All images were visualized using a BX51 Olympus equipped with a high-resolution DP71 camera (Olympus). One hundred randomly chosen cells were examined microscopically to calculate the number of PMNs with at least one associated conidium. Results are expressed as phagocytic index, where the average number of conidia phagocytized per 100 neutrophils, or as percentage of phagocytosis, the percentage of neutrophils containing at least 1 conidium. The experiments were repeated twice for each fungal species with two different primary cultures.
CTLA-4 binding and TEM
For flow cytometry experiments, Aspergillus akuBKU80 1 × 106 resting conidia were incubated first with CTLA-4 Ig or isotype control IgG3 (1 μg/ml) (Biolegend) or in combination with αB71 antibody (Biolegend) for 15 min at 4°C and second, with secondary FITC-labeled antibody (PharMingen). For transmission electron microscopy, Aspergillus akuBKU80 resting conidia or swollen (obtained in YPD at a density of 1 × 107 conidia/ml and incubated at 37°C in a rotary shaker, 300 rpm for 4 h) cells were washed with 0.2M sodium cacodylate buffer (pH 7.4). Cells were then fixed in cacodylate fixative buffer (0.1M sodium cacodylate, 2% paraformaldehyde, 3% gluteraldehyde) overnight at 4°C. The cells were then washed with 0.2M sodium cacodylate buffer and dehydrated on an alcohol series (30%, 50%, 70%, 80%, 90% and 100%) for 15 min each. Specimens were then embedded into acrylic resin. Ultrafine sections were obtained by cutting into the resin specimens with a glass blade on an ultramicrotome, and mounted on nickel grids. The grids were then washed with phosphate buffered saline, and then stained with CTLA-4 Ig or isotype control IgG3 (1 μg/ml) (Biolegend) for 15 min at 4°C followed by secondary antibodies that have been conjugated with either 15nm gold particles (Aurion). All antibody incubations were done in PBS containing 1% bovine serum albumin. After antibody staining, grids were post-fixed with cacodylate fixative buffer for 15 min, and then stained with 2% uranyl acetate. Micrographs were taken with an EM 208 transmission electron microscope (Phillips). For protein identification, Aspergillus akuBKU80 equal protein concentrations of resting conidia and swollen conidia were prepared with enzymatic Assay of Lyticase (Sigma Aldrich). Murine HTR B71+/+ and B72+/+cell lysate was obtained in Laemmli buffer. The lysate was separated in SDS-PAGE and transferred to a nitrocellulose membrane. Blot of cell lysates were incubated with monoclonal α-mouse CD80 antibody (PharMingen). Chemiluminescence detection was performed with LiteAblot Plus chemiluminescence substrate (EuroClone), using the ChemiDocTM XRS+ Imaging System (Bio-Rad), and quantification was obtained by densitometry image analysis using Image Lab 5.1 software (Bio-Rad). For immunoprecipitation assay, equimolar amounts of cell lysate or swollen conidia lysate were immunoprecipitated using 2 μg/ml anti-CD80 and secondary HRP-conjugated antibody. Proteins were detected with the ECL chemiluminescence detection system (Amersham Corp) followed by exposure to Bio-Rad ChemiDoc XRS+ Imaging system with Image Lab software. Control experiments included western blottings on immunoprecipitates with an irrelevant antibody. Protein identification was performed by MALDI-TOF. The gel bands of interest were chopped in small pieces, washed with water and shrinked with acetonitrile. Reduction and alkylation were performed in a solution of 10 mM DTT (56°C, 45 min) and iodoacetamide 55 mM (room temperature, 30 min, in the dark), respectively. Shrinking and rehydratating were carried out with acetonitrile and 100 mM ammonium bicarbonate in order to completely remove staining. Proteins were digested by adding a solution of 12 ng/μl of trypsin (type IX-S, from porcine pancreas, Sigma-Aldrich) in 10 mM ammonium bicarbonate (37°C, overnight). Digestions were stopped with 10% TFA and the supernatants were recovered for subsequent mass spectrometry analyses. A MALDI-TOF/TOF spectrometer Ultraflex III (Bruker Daltonics, Bremen, Germany) set up in reflectron positive mode was used to perform the analyses. Spectra were acquired in the mass range of 900-4000 m/z and processed using Flex Analysis software 3.0 from Bruker Daltonics. For the analyses of the proteolytic digests 1 μl of a 1:1 solution of the matrix (10 g/L α-cyano-4-hydroxycinnamic acid in 70/30 acetonitrile/0.1% TFA) and of the sample was spotted on a AncorChip (Bruker Daltonics, Bremen, Germany) target plate and allowed to dry. Peptide mass fingerprint data were obtained for each sample. Mascot searches were done against the Swiss-Prot database (released version) calculating 2 maximum miss cleavage for trypsin, carbamidomethylation of cysteine as fixed modification, oxidation of methionine as variable modifications.
Quantification and statistical analysis
Data are expressed as mean ± sd. Horizontal bars indicate the means. Statistical significance was calculated by one or two-way ANOVA (Bonferroni’s post hoc test) for multiple comparisons and by a two tailed Student’s t test for single comparison. Statistical analysis was performed with GraphPad Prism 6 software (GraphPad Software). All of the statistical details of experiments can be found in the figure legends, including the statistical tests used, exact value of n, which represents the number of animals.
Acknowledgments
We thank Dr. Cristina Massi-Benedetti for digital art and editorial assistance. This work was supported by Italian grant “Programma per Giovani Ricercatori - Rita Levi Montalcini 2013” (project number: PGR13XNIDJ to T.Z.); the Specific Targeted Research Project FunMeta (ERC-2011-AdG-293714 to L.R.); the Italian Fondazione Cassa di Risparmio di Perugia (project number 2018.0412.021 to T.Z.); AIRC Investigator grant 16851 (to P.P.); Telethon Research grant GGP17094 (to F.F.); and NIH 5R01AI065728-10 (to N.P.K.). A.W. and E.B. are supported by the MRC Centre for Medical Mycology (grant MR/N006364/1) at the University of Aberdeen. Z.S.’s research was supported by the Grant Agency of the Czech Republic (GAČR no. 17-24592Y) and by the Czech Ministry of Education, Youth and Sports (CETOCOEN PLUS CZ.02.1.01/0.0/0.0/15_003/0000469, LM2015051, and CZ.02.1.01/0.0/0.0/16_013/0001761).
Author contributions
Conceptualization, T.Z. and T.C.; investigation, F.F., A.B., and J.-P.L.; methodology, G. Paolicelli, G. Pieraccini, R.R., F.B., M. Pieroni, C.B., G.C., and C.G.; validation, A.D.L., C.V., V.O., X.L., A.W., E.B., P.E.V., M.B., M. Pariano, and M.C.; resources, M.G., A.Y.W., L.B., M.d.H., and Z.S.; project administration, T.Z., N.P.K., and P.P.; funding acquisition, L.R.
Declaration of interests
The authors declare no competing interests.
Published: January 26, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108673.
Supplemental information
References
- Acerbi E., Hortova-Kohoutkova M., Choera T., Keller N., Fric J., Stella F., Romani L., Zelante T. Modeling Approaches Reveal New Regulatory Networks in Aspergillus fumigatus Metabolism. J. Fungi (Basel) 2020;6:108. doi: 10.3390/jof6030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agus A., Planchais J., Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Ball H.J., Jusof F.F., Bakmiwewa S.M., Hunt N.H., Yuasa H.J. Tryptophan-catabolizing enzymes - party of three. Front. Immunol. 2014;5:485. doi: 10.3389/fimmu.2014.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker B.M., Kroll K., Vödisch M., Mazurie A., Kniemeyer O., Cramer R.A. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genomics. 2012;13:62. doi: 10.1186/1471-2164-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer C.A., Shepherd D.M. Role of the aryl hydrocarbon receptor (AhR) in lung inflammation. Semin. Immunopathol. 2013;35:693–704. doi: 10.1007/s00281-013-0391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.C., Abruzzo G.K., Anderson J.W., Flattery A.M., Gill C.J., Pikounis V.B., Schmatz D.M., Liberator P.A., Douglas C.M. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 2001;45:3474–3481. doi: 10.1128/AAC.45.12.3474-3481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozec A., Zaiss M.M., Kagwiria R., Voll R., Rauh M., Chen Z., Mueller-Schmucker S., Kroczek R.A., Heinzerling L., Moser M. T cell costimulation molecules CD80/86 inhibit osteoclast differentiation by inducing the IDO/tryptophan pathway. Sci. Transl. Med. 2014;6:235ra60. doi: 10.1126/scitranslmed.3007764. [DOI] [PubMed] [Google Scholar]
- Brunke S., Seider K., Almeida R.S., Heyken A., Fleck C.B., Brock M., Barz D., Rupp S., Hube B. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol. Microbiol. 2010;76:25–47. doi: 10.1111/j.1365-2958.2010.07052.x. [DOI] [PubMed] [Google Scholar]
- Calvo A.M., Bok J., Brooks W., Keller N.P. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci E., Mencacci A., Bacci A., Bistoni F., Kurup V.P., Romani L. T cell vaccination in mice with invasive pulmonary aspergillosis. J. Immunol. 2000;165:381–388. doi: 10.4049/jimmunol.165.1.381. [DOI] [PubMed] [Google Scholar]
- Choera T., Zelante T., Romani L., Keller N.P. A Multifaceted Role of Tryptophan Metabolism and Indoleamine 2,3-Dioxygenase Activity in Aspergillus fumigatus-Host Interactions. Front. Immunol. 2018;8:1996. doi: 10.3389/fimmu.2017.01996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G., Dostalek M., Hsu E.L., Nguyen L.P., Stec D.F., Bradfield C.A., Guengerich F.P. Structural identification of Diindole agonists of the aryl hydrocarbon receptor derived from degradation of indole-3-pyruvic acid. Chem. Res. Toxicol. 2009;22:1905–1912. doi: 10.1021/tx9000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D., Barker B.M., Carey C.C., Merriman B., Werner E.R., Lechner B.E., Dhingra S., Cheng C., Xu W., Blosser S.J. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 2014;10:e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Ferreira M.E., Kress M.R., Savoldi M., Goldman M.H., Härtl A., Heinekamp T., Brakhage A.A., Goldman G.H. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoubeaux G., Cray C. Rodent Models of Invasive Aspergillosis due to Aspergillus fumigatus: Still a Long Path toward Standardization. Front. Microbiol. 2017;8:841. doi: 10.3389/fmicb.2017.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindo M., Costanzi E., Pieroni M., Costantini C., Annunziato G., Bruno A., Keller N.P., Romani L., Zelante T., Cellini B. Biochemical Characterization of Aspergillus fumigatus AroH, a Putative Aromatic Amino Acid Aminotransferase. Front. Mol. Biosci. 2018;5:104. doi: 10.3389/fmolb.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers F., Woite C., Antoni V., Stein S., Funakoshi H., Nakamura T., Schares G., Däubener W., Eller S.K. Negative Impact of Hypoxia on Tryptophan 2,3-Dioxygenase Function. Mediators Inflamm. 2016;2016:1638916. doi: 10.1155/2016/1638916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargaro M., Pirro M., Romani R., Zelante T., Fallarino F. Aryl Hydrocarbon Receptor-Dependent Pathways in Immune Regulation. Am. J. Transplant. 2016;16:2270–2276. doi: 10.1111/ajt.13716. [DOI] [PubMed] [Google Scholar]
- Grahl N., Puttikamonkul S., Macdonald J.M., Gamcsik M.P., Ngo L.Y., Hohl T.M., Cramer R.A. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 2011;7:e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N., Shepardson K.M., Chung D., Cramer R.A. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot. Cell. 2012;11:560–570. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U., Orabona C., Fallarino F., Vacca C., Calcinaro F., Falorni A., Candeloro P., Belladonna M.L., Bianchi R., Fioretti M.C., Puccetti P. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat. Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Fallarino F., Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Haenuki Y., Matsushita K., Futatsugi-Yumikura S., Ishii K.J., Kawagoe T., Imoto Y., Fujieda S., Yasuda M., Hisa Y., Akira S. A critical role of IL-33 in experimental allergic rhinitis. J. Allergy Clin. Immunol. 2012;130:184. doi: 10.1016/j.jaci.2012.02.013. 94.e11. [DOI] [PubMed] [Google Scholar]
- Huang L., Li L., Klonowski K.D., Tompkins S.M., Tripp R.A., Mellor A.L. Induction and role of indoleamine 2,3 dioxygenase in mouse models of influenza a virus infection. PLoS ONE. 2013;8:e66546. doi: 10.1371/journal.pone.0066546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs A.F., Castranova V., Ma J.Y., Frazer D.G., Siegel P.D., Ducatman B.S., Grote A., Schwegler-Berry D., Robinson V.A., Van Dyke C. Acute lung injury induced by a commercial leather conditioner. Toxicol. Appl. Pharmacol. 1997;143:37–46. doi: 10.1006/taap.1996.8053. [DOI] [PubMed] [Google Scholar]
- Iraqui I., Vissers S., Cartiaux M., Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- Ishihara Y., Haarmann-Stemmann T., Kado N.Y., Vogel C.F.A. Interleukin 33 expression induced by aryl hydrocarbon receptor in macrophages. Toxicol. Sci. 2019;170:404–414. doi: 10.1093/toxsci/kfz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V.K., Divol B., Prior B.A., Bauer F.F. Effect of alternative NAD+-regenerating pathways on the formation of primary and secondary aroma compounds in a Saccharomyces cerevisiae glycerol-defective mutant. Appl. Microbiol. Biotechnol. 2012;93:131–141. doi: 10.1007/s00253-011-3431-z. [DOI] [PubMed] [Google Scholar]
- Kowalski C.H., Beattie S.R., Fuller K.K., McGurk E.A., Tang Y.W., Hohl T.M., Obar J.J., Cramer R.A., Jr. Heterogeneity among Isolates Reveals that Fitness in Low Oxygen Correlates with Aspergillus fumigatus Virulence. MBio. 2016;7:e01515-16. doi: 10.1128/mBio.01515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S., Bignell E.M., Reichard U., Rogers T., Haynes K., Braus G.H. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 2004;52:785–799. doi: 10.1111/j.1365-2958.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni C.A., Brookes P.S. Cellular Compartmentation and the Redox/Nonredox Functions of NAD. Antioxid. Redox Signal. 2019;31:623–642. doi: 10.1089/ars.2018.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lim F.Y., Sanchez J.F., Wang C.C., Keller N.P. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 2012;517:303–324. doi: 10.1016/B978-0-12-404634-4.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchiarulo A., Camaioni E., Nuti R., Pellicciari R. Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease. Amino Acids. 2009;37:219–229. doi: 10.1007/s00726-008-0137-3. [DOI] [PubMed] [Google Scholar]
- Mazaki Y., Hashimoto S., Tsujimura T., Morishige M., Hashimoto A., Aritake K., Yamada A., Nam J.M., Kiyonari H., Nakao K., Sabe H. Neutrophil direction sensing and superoxide production linked by the GTPase-activating protein GIT2. Nat. Immunol. 2006;7:724–731. doi: 10.1038/ni1349. [DOI] [PubMed] [Google Scholar]
- Metz R., Smith C., DuHadaway J.B., Chandler P., Baban B., Merlo L.M., Pigott E., Keough M.P., Rust S., Mellor A.L. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int. Immunol. 2014;26:357–367. doi: 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D.H., Mellor A.L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D.H., Sharma M.D., Mellor A.L. Ligation of B7-1/B7-2 by human CD4+ T cells triggers indoleamine 2,3-dioxygenase activity in dendritic cells. J. Immunol. 2004;172:4100–4110. doi: 10.4049/jimmunol.172.7.4100. [DOI] [PubMed] [Google Scholar]
- Osherov N., Kontoyiannis D.P., Romans A., May G.S. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemother. 2001;48:75–81. doi: 10.1093/jac/48.1.75. [DOI] [PubMed] [Google Scholar]
- Palmer J.M., Bok J.W., Lee S., Dagenais T.R.T., Andes D.R., Kontoyiannis D.P., Keller N.P. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus. PeerJ. 2013;1:e4. doi: 10.7717/peerj.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y.H., Ueng S.H., Tseng C.T., Hung M.S., Song J.S., Wu J.S., Liao F.Y., Fan Y.S., Wu M.H., Hsiao W.C. Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J. Med. Chem. 2016;59:282–293. doi: 10.1021/acs.jmedchem.5b01390. [DOI] [PubMed] [Google Scholar]
- Ranhotra H.S., Flannigan K.L., Brave M., Mukherjee S., Lukin D.J., Hirota S.A., Mani S. Xenobiotic Receptor-Mediated Regulation of Intestinal Barrier Function and Innate Immunity. Nucl. Receptor Res. 2016;3:101199. doi: 10.11131/2016/101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees C.A., Stefanuto P.H., Beattie S.R., Bultman K.M., Cramer R.A., Hill J.E. Sniffing out the hypoxia volatile metabolic signature of Aspergillus fumigatus. J. Breath Res. 2017;11:036003. doi: 10.1088/1752-7163/aa7b3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L., Fallarino F., De Luca A., Montagnoli C., D’Angelo C., Zelante T., Vacca C., Bistoni F., Fioretti M.C., Grohmann U. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–215. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Santhanam S., Alvarado D.M., Ciorba M.A. Therapeutic targeting of inflammation and tryptophan metabolism in colon and gastrointestinal cancer. Transl. Res. 2016;167:67–79. doi: 10.1016/j.trsl.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S.K., Ebel S., Keil E., Woite C., Ernst J.F., Benzin A.E., Rupp J., Däubener W. Regulation of IDO activity by oxygen supply: inhibitory effects on antimicrobial and immunoregulatory functions. PLoS ONE. 2013;8:e63301. doi: 10.1371/journal.pone.0063301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepardson K.M., Jhingran A., Caffrey A., Obar J.J., Suratt B.T., Berwin B.L., Hohl T.M., Cramer R.A. Myeloid derived hypoxia inducible factor 1-alpha is required for protection against pulmonary Aspergillus fumigatus infection. PLoS Pathog. 2014;10:e1004378. doi: 10.1371/journal.ppat.1004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Keller N.P. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Oda S., Otsuki T., Hino T., Yoshida T., Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc. Natl. Acad. Sci. USA. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C.E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S.A., Oakley B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tajima H., Tajiki-Nishino R., Watanabe Y., Kurata K., Fukuyama T. Activation of aryl hydrocarbon receptor by benzo[a]pyrene increases interleukin 33 expression and eosinophil infiltration in a mouse model of allergic airway inflammation. J. Appl. Toxicol. 2020;40:1545–1553. doi: 10.1002/jat.4017. [DOI] [PubMed] [Google Scholar]
- Tojo S., Kohno T., Tanaka T., Kamioka S., Ota Y., Ishii T., Kamimoto K., Asano S., Isobe Y. Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors. ACS Med. Chem. Lett. 2014;5:1119–1123. doi: 10.1021/ml500247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrestarazu A., Vissers S., Iraqui I., Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol. Gen. Genet. 1998;257:230–237. doi: 10.1007/s004380050643. [DOI] [PubMed] [Google Scholar]
- Vaknin Y., Hillmann F., Iannitti R., Ben Baruch N., Sandovsky-Losica H., Shadkchan Y., Romani L., Brakhage A., Kniemeyer O., Osherov N. Identification and Characterization of a Novel Aspergillus fumigatus Rhomboid Family Putative Protease, RbdA, Involved in Hypoxia Sensing and Virulence. Infect. Immun. 2016;84:1866–1878. doi: 10.1128/IAI.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.M., Choera T., Wiemann P., Pisithkul T., Amador-Noguez D., Keller N.P. TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus. Fungal Genet. Biol. 2016;89:102–113. doi: 10.1016/j.fgb.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems J.M., Yost G.S. 3-Methylindole metabolites induce lung CYP1A1 and CYP2F1 enzymes by AhR and non-AhR mechanisms, respectively. Chem. Res. Toxicol. 2010;23:696–704. doi: 10.1021/tx9004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Perevitsky A., Lim F.Y., Shadkchan Y., Knox B.P., Landero Figueora J.A., Choera T., Niu M., Steinberger A.J., Wüthrich M. Aspergillus fumigatus Copper Export Machinery and Reactive Oxygen Intermediate Defense Counter Host Copper-Mediated Oxidative Antimicrobial Offense. Cell Rep. 2017;19:2174–2176. doi: 10.1016/j.celrep.2017.05.075. [DOI] [PubMed] [Google Scholar]
- Yuasa H.J., Ball H.J. Molecular evolution and characterization of fungal indoleamine 2,3-dioxygenases. J. Mol. Evol. 2011;72:160–168. doi: 10.1007/s00239-010-9412-5. [DOI] [PubMed] [Google Scholar]
- Yuasa H.J., Ball H.J. Indoleamine 2,3-dioxygenases with very low catalytic activity are well conserved across kingdoms: IDOs of Basidiomycota. Fungal Genet. Biol. 2013;56:98–106. doi: 10.1016/j.fgb.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Yuasa H.J., Ball H.J. Efficient tryptophan-catabolizing activity is consistently conserved through evolution of TDO enzymes, but not IDO enzymes. J. Exp. Zoolog. B Mol. Dev. Evol. 2015;324:128–140. doi: 10.1002/jez.b.22608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
akuBKU80 fungal strain was cultured in GMM for 22h (18 hours of imaging) at 37°C.
akuBKU80 fungal strain was cultured in GMM+NAM for 22h (18 hours of imaging) at 37°C.
ΔidoA/B/C fungal strain was cultured in GMM for 22h (18 hours of imaging) at 37°C
ΔidoA/B/C fungal strain was cultured in GMM+NAM for 22h (18 hours of imaging) at 37°C.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.