Abstract
Solid-phase peptide synthesis (SPPS) is usually performed with optically pure building blocks to prepare peptides as single enantiomers. Herein we report that SPPS using racemic amino acids provides stereorandomized (sr) peptides, containing up to billions of different stereoisomers, as well-defined single HPLC peaks, single mass products with high yield, which can be used to investigate peptide bioactivity. To exemplify our method, we show that stereorandomization abolishes the membrane-disruptive effect of α-helical amphiphilic antimicrobial peptides but preserves their antibiofilm effect, implying different mechanisms involving folded versus disordered conformations. For antimicrobial peptide dendrimers by contrast, stereorandomization preserves antibacterial, membrane-disruptive, and antibiofilm effects but reduces hemolysis and cytotoxicity, thereby increasing their therapeutic index. Finally, we identify partially stereorandomized analogues of the last resort cyclic peptide antibiotic polymyxin B with preserved antibacterial activity but lacking membrane-disruptive and lipopolysaccharide-neutralizing activity, pointing to the existence of additional targets.
Short abstract
Stereorandomized peptides obtained using racemic amino acids may retain the activity of the parent all-l or all-d sequence, as shown for antimicrobial peptides, peptide dendrimers, and polymyxin B.
Introduction
The high efficiency of solid-phase peptide synthesis (SPPS)1,2 allows for interesting variations in synthesis planning, such as the split-and-mix protocol to prepare one-bead-one-compound libraries3−5 or the synthesis of peptide mixtures by using several different amino acids in the same coupling step.6−8 Thanks to the availability of both l- and d-amino acids as synthetic building blocks, one can also vary residue stereochemistry and explore any number of stereoisomers of a given peptide sequence9−11 or investigate mixtures of enantiomeric peptides synthesized individually as all l- or all d- sequences as reported for amyloid and hydrogel forming peptides.12−15 Here we asked the question whether SPPS using racemic rather than single enantiomer amino acids to form stereorandomized (sr) peptides might provide new insights into the mechanism of action of peptides. As a case study, we focused on antimicrobial peptides (AMPs),16−19 antimicrobial peptide dendrimers (AMPDs),20−28 and polymyxin B,29 which are all membrane-disruptive compounds active on Gram-negative bacteria including multidrug resistant strains.
Results and Discussion
SPPS Provides sr-Peptides, sr-Peptide Dendrimers, and sr-Cyclic Peptides as Homogeneous Products
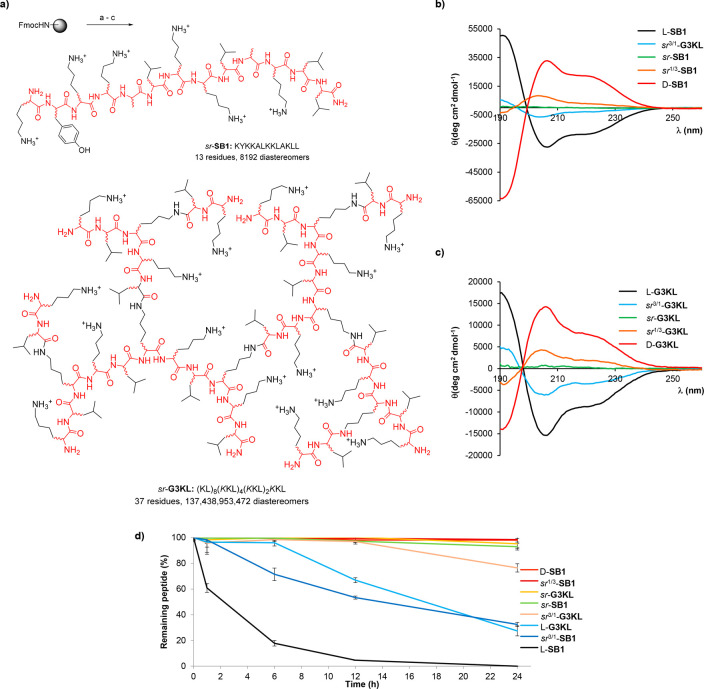
We first tested SPPS using racemic amino acids with AMP SB1, a short linear sequence of 13 residues,30 and with AMPD G3KL, which comprises 37 residues in a lysine-branched architecture accessible in 11 peptide coupling steps, to form sr-SB1 and sr-G3KL (Figure 1a).21 We also performed SPPS runs using 3:1 and 1:3 l/d ratios of each amino acid to form sr3/1-SB1, sr3/1-G3KL, sr1/3-SB1, and sr1/3-G3KL. To our delight, in each case, the crude as well as the HPLC-purified sr-product gave a single peak and a single mass indistinguishable from the homochiral parent compound when analyzed by LC/MS (Table 1, see also Supporting Information). Obtaining sr-peptides as homogeneous products is remarkable when considering that they contain a very large number of different stereoisomers but also reflects the fact that reversed-phase HPLC primarily separates compounds by hydrophobicity.
Figure 1.
Synthesis of stereorandomized peptides and peptide dendrimers. (a) Stereorandomized SPPS of AMP sr-SB1 and AMPD sr-G3KL. SPPS conditions: (a) 20% v/v piperidine in DMF, 2 min, 25 °C and 5 min, 50 °C; (b) FmocAAOH (5 equiv), Oxyma (7.5 equiv), DIC (10 equiv),10 min, 50 °C; (c) TFA/TIS/H2O (94:5:1), 4 h at room temperature. (b) CD spectra of l-, d- and sr-SB1 and (c) CD spectra of l-, d-, and sr-G3KL, at 200 μg/mL TFA salt of the compound in 6 mM aq. phosphate buffer pH 7.4 with the 20% of TFE added. (d) Serum stability assay. Conditions: 400 μM compound in aq. 0.1 M Tris buffer pH 7.5 containing 25% v/v human serum.
Table 1. Synthesis and Activity of Homochiral (l- or d-), Stereorandomized (sr-) and Racemic (-rac) Antimicrobial Peptides, Peptide Dendrimers, and Polymyxin B.
| Cpd.a | SPPS yield | MS analysis | analytical HPLC | P. aeruginosa | A.baumannii | E. coli | K. pneumoniae | EYPG vesicle leakage | EYPC vesicle leakage | hemolysis on hRBC, MHC | PAO1 biofilm MBIC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| mg (%)b | calc./obs.c | tR (min.)d | MIC (μg/mL)e | (%)f | (%)f | (μg/mL)g | (μg/mL)h | ||||
| Linear Peptides | |||||||||||
| SB1: KYKKALKKLAKLL (13 residues, 8192 diastereomers) | |||||||||||
| L-SB1 | 56 (30) | 1543.07/1543.08 | 1.53 | 2 | 2 | 0.5 | 32 | 60 | 52 | >2000 | 16 |
| D-SB1 | 74 (26) | 1543.07/1543.07 | 1.55 | 1 | 2 | 0.5 | 32 | 70 | 45 | >2000 | 8 |
| rac-SB1 | - | - | - | 2 | 2 | 1 | 16 | 91 | 51 | >2000 | 16 |
| sr-SB1 | 62 (22) | 1543.07/1543.08 | 1.53 | >64 | >64 | 32 | >64 | 12 | 1 | >2000 | 16 |
| sr3/1-SB1 | 48 (17) | 1543.07/1543.07 | 1.54 | >64 | >64 | 16 | >64 | 20 | 9 | >2000 | 16 |
| sr1/3-SB1 | 57(20) | 1543.07/1543.07 | 1.54 | >64 | >64 | 16 | >64 | 27 | 6 | >2000 | 16 |
| DJK5: VQWRAIRVRVIR (12 residues, 4096 diastereomers) | |||||||||||
| L-DJK5 | 22 (9) | 1549.98/1549.98 | 1.53 | 32 | 16 | 32 | >64 | 32 | 1 | >2000 | >32 |
| D-DJK5 | 19 (7) | 1549.98/1549.98 | 1.50 | 4–2 | 8 | 4–2 | 2 | 36 | 2 | 1000 | 8 |
| sr-DJK5 | 13 (8) | 1549.98/1549.98 | 1.52 | >64 | >64 | 16 | >64 | 8 | 1 | >2000 | 32 |
| Indolicidin: ILPWKWPWWPWRR (13 residues, 8192 diastereomers) | |||||||||||
| L-Indo | 28 (10) | 1905.05/1905.04 | 1.80 | >64 | 16 | 32 | 32 | 18 | 18 | 125 | >32 |
| D-Indo | 22 (8) | 1905.05/1905.05 | 1.82 | >64 | 16 | 16 | 32 | 35 | 18 | 125 | >32 |
| sr-Indo | 18 (7) | 1905.05/1905.05 | 1.87 | >64 | 8 | 16 | 16 | 18 | 37 | 125 | >32 |
| Peptide Dendrimers | |||||||||||
| G3KL: (KL)8(KKL)4(KKL)2KKL (37 residues, 137 438 953 472 possible diastereomers) | |||||||||||
| L-G3KL | 124 (23) | 4531.38/4531.43 | 1.47 | 2 | 4 | 1 | >64 | 42 | 6 | 1000 | 16 |
| D-G3KL | 142 (17) | 4531.38/4531.37 | 1.46 | 4 | 8 | 1 | 32 | 51 | 8 | 1000 | 16 |
| rac-G3KL | - | - | - | 4 | 8 | 2 | >64 | 77 | 1 | 1000 | 16 |
| sr-G3KL | 138 (24) | 4531.38/4531.43 | 1.53 | 8 | >64 | 8 | >64 | 38 | 1 | 1000 | 16 |
| sr3/1-G3KL | 94 (11) | 4531.38/4531.43 | 1.54 | 4 | >64 | 8 | >64 | 58 | 1 | 1000 | 16 |
| sr1/3-G3KL | 86 (10) | 4531.38/4531.44 | 1.52 | 8 | >64 | 4 | >64 | 66 | 1 | 1000 | 16 |
| TNS18: (OF)4(KBL)2KKLK(C10) (18 residues, 262 144 possible diastereomers) | |||||||||||
| L-TNS18 | 121 (42) | 2395.61/2395.61 | 1.64 | 2 | 2 | 4 | >64 | 64 | 1 | 1000 | 8 |
| D-TNS18 | 156 (14) | 2395.61/2395.60 | 1.63 | 4 | 8 | 4 | >64 | 65 | 5 | 500 | 8 |
| rac-TNS18 | - | - | - | 4 | 4 | 2 | >64 | 76 | 3 | 500 | 16 |
| sr-TNS18 | 8 (4) | 2395.61/2395.60 | 1.64 | 4 | 8 | 4 | >64 | 61 | 3 | 1000 | 16 |
| T25: (KL)8(KKL)4(KLL)2KKLL (38 residues, 274 877 906 944 possible diastereomers) | |||||||||||
| L-T25 | 26 (7) | 4614.44/4614.47 | 1.55 | 4 | 4 | 4 | 16 | 50 | 31 | 62.5 | 8 |
| D-T25 | 57 (7) | 4614.44/4614.45 | 1.59 | 4 | 2 | 8 | 16 | 45 | 28 | 125 | 8 |
| rac-T25 | - | - | - | 4 | 4 | 8 | 16 | 46 | 37 | 31.25 | 8 |
| sr-T25 | 3854 (23) | 4614.44/4614.44 | 1.59 | 2 | 8 | 4 | 32 | 40 | 1 | >2000 | 8 |
| sr-T25a: (KL)8(KKL)4(KLL)2KKLL (38 residues, G2 and G3 stereorandomized, 268 435 456 diastereomers) | |||||||||||
| 34 (6) | 4614.44/4614.45 | 1.61 | 4 | 4 | 4 | 32 | 58 | 8 | 62.5 | >16 | |
| sr-T25b: (KL)8(KKL)4(KLL)2KKLL (38 residues, G0 and G1 stereorandomized, 1024 diastereomers) | |||||||||||
| 26 (5) | 4614.44/4614.43 | 1.57 | 4 | 16 | 4 | >64 | 67 | 2 | >2000 | 16 | |
| Cyclic Peptides | |||||||||||
| polymyxin B ( 10 residues, 1024 possible diastereomers) | |||||||||||
| PMB | - | - | - | 0.25 | 0.25 | 0.25 | 0.25 | 53 | 2 | >2000 | 8 |
| PMB2 | 26 (3) | 1188.73/1188.74 | 1.58 | 0.25 | 0.25 | 0.25 | 0.5 | 39 | 1 | >2000 | 8 |
| sr-PMB2 | 148 (14) | 1188.73/1188.74 | 1.55 | 32 | 64 | 2 | >64 | 5 | 6 | >2000 | >16 |
| sr-PMB2a (10 residues, residues 1, 3, 4, 7, 9, and 10 racemic, 64 diastereomers) | |||||||||||
| 24 (5) | 1188.73/1188.74 | 1.56 | 4 | 4 | 1 | >64 | 1 | 1 | >2000 | 16 | |
| sr-PMB2b (10 residues, residues 2, 3, 4, 9, and 10 racemic, 32 diastereomers) | |||||||||||
| 18 (3) | 1188.73/1188.74 | 1.58 | 2 | 2 | 0.25 | 2 | 1 | 1 | >2000 | 16 | |
One-letter codes for amino acids, K = branching lysine l- and d- sequences are with only l- or only d-amino acids, sr-sequences have all positions individually racemized corresponding to the indicated number of diastereomers, and rac-sequences are a 1:1 mixture of all l- and all d-sequences, in sr-T25a and sr-T25b only the residues underlined are racemic, see Figures 1 and 4 for structural formulas of AMPDs and PMB.
Yields given for RP-HPLC purified product.
High-resolution electrospray ionization mass spectrometry (positive mode), the calculated monoisotopic mass, and the observed mass in the reconstructed spectrum are given.
Retention time in analytical RP-HPLC (A/D = 100/0 to 0/100 in 2.2 min, λ = 214 nm).
Minimum inhibitory concentration (MIC, μg/mL) was determined on P. aeruginosa PAO1, A. baumannii ATCC 19606, E. coli W3110 and K. pneumoniae NCTC418 in Müller-Hinton medium, after incubation for 16–20 h at 37 °C.
Lipid vesicles made of egg yolk phosphatidyl glycerol (EYPG) or egg yolk phosphatidyl choline (EYPC) were suspended in buffer (10 mM TRIS, 107 mM NaCl, pH 7.4). After 50 s, the indicated compound was added to reach the indicated concentration. After 300 s, 1.2% Triton X-100 was added for full fluorescein release. The percentage leakage observed with 10 μg/mL compound at 250 s is given. Full curves are given in Figures S8–S14.
Minimum hemolytic concentration (MHC) measured on human red blood cells in 10 mM phosphate, 150 mM NaCl, pH 7.4, 25 °C, 4.
Minimum biofilm inhibitory concentration (MBIC, μg/mL) was measured in 0.25% (w/v) nutrient broth no. 2, Oxoid on P. aeruginosa PAO1. All experiments were done in at least two independent duplicates, see the Supporting Information for details.
CD spectra of stereorandomized SB1 and G3KL in aqueous neutral buffer containing up to 20% v/v trifluoroethanol (TFE) as folding inducer31,32 showed an α-helical signal with intensity proportional to the enantiomeric purity of the building blocks, resulting in a flat signal at 1:1 d/l mixture of building blocks (Figure 1b,c, Figures S1 and S4). While the all l-enantiomers were susceptible to serum proteolysis (l-SB1: t1/2 ∼ 2 h, l-G3KL: t1/2 ∼ 15 h), incorporation of as little as 25% d-amino acids in the sr-samples significantly reduced (sr3/1-SB1) or entirely blocked (sr3/1-G3KL) serum proteolysis, reflecting the selectivity of proteases for l-peptides (Figure 1d).
Stereorandomized SPPS of AMPs DJK5 (12 residues)33 and indolicidin (indo, 13 residues),34−37 AMPDs TNS18 (18 residues, 10 steps)24 and T25 (38 residues, 12 steps),25 as well as polymyxin B2 (PMB2, 10 residues, cyclic peptide),38 similarly provided the corresponding sr-sequences as homogeneous products in excellent isolated yields, which showed similarly flat CD spectra (Figures S2, S3, S5, S6, and S7). We also obtained clean products when we stereorandomized only selected positions in the case of T25 and PMB2. Taken together, these different syntheses showed that partially or fully stereorandomized peptides are readily accessible in homogeneous form for linear, dendritic, and cyclic peptide sequences.
Stereorandomization Abolishes the Membrane-Disruptive and Antibacterial Activities of α-Helical AMPs SB1 and DJK5 but Does Not Affect Their Biofilm Inhibition or the Activity of the Disordered AMP Indolicidin
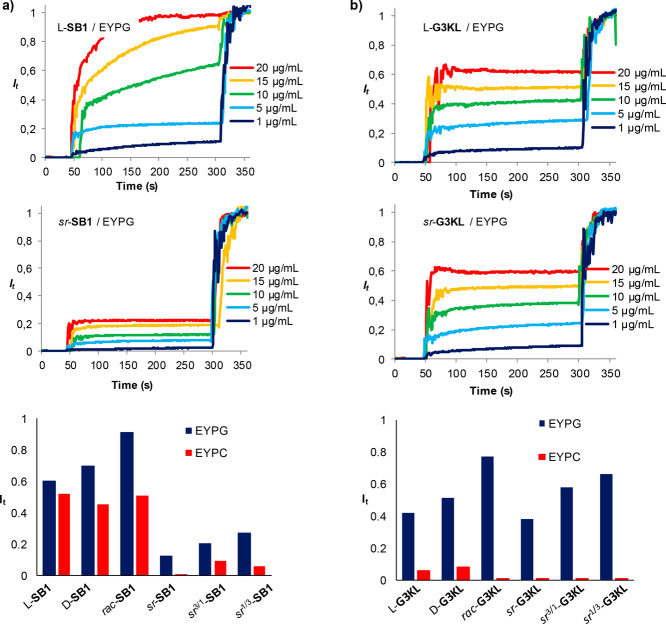
AMPs SB1 (as the all l- or all d-enantiomer) folds to an amphiphilic α-helix stable as 4-helix bundle within crystal structures as well as in a membrane environment.30 Stereorandomization of SB1 to form 1:3, 1:1, and 3:1 sr-SB1 entirely abolished its antimicrobial activity on Gram-negative strains (Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae) or strongly reduced it (Escherichia coli) (Table 1, columns 5–8). The membrane-disruptive activity of SB1 on synthetic vesicles containing either egg white phosphatidyl glycerol (EYPG, mimicking bacterial membranes) or egg white phosphatidyl choline (EYPC, mimicking eukaryotic membranes) was similarly strongly reduced upon stereorandomization (Table 1, columns 9–10, Figure 2a, Figure S8).39 By contrast, the racemate rac-SB1, the 1:1 mixture of all l- and all d-enantiomers, was as active as the individual enantiomers in the different assays. None of the samples showed any measurable hemolysis on human erythrocytes (Table 1, column 11). We observed similar effects with AMP DJK5,33 which also forms an amphiphilic α-helix (Figure S2) and essentially lost its antibacterial and EYPG vesicle leakage activity in its sr-version compared with the all l- or all d-enantiomers (Figure S9).
Figure 2.
Vesicle leakage assays with homochiral and stereorandomized SB1 (a) and G3KL (b). Lipid vesicles made of egg yolk phosphatidyl glycerol (EYPG) or egg yolk phosphatidyl choline (EYPC) were suspended in buffer (10 mM TRIS, 107 mM NaCl, pH 7.4). After 50 s, the indicated compound was added to reach the indicated concentration. After 300 s, 1.2% Triton X-100 was added for full fluorescein release. The bar plots report relative fluorescence signal intensity measured after 250 s at 10 μg/mL.
AMPs SB1 and DJK5 also exhibited a significant inhibitory activity against P. aeruginosa biofilms (Table 1, column 12). In contrast to the antibacterial effect, however, biofilm inhibition by SB1 was independent of stereochemical purity. For DJK5, antibiofilm effects required the presence of d-residues, an effect which has been shown to protect this peptide from proteolysis.33 By comparison, indolicidin (indo),34−37 for which CD spectroscopy indicates a disordered conformation (Figure S3), was more active as sr-indo than either l-indo or d-indo in terms of antibacterial and vesicle leakage activity (Figure S10). This data shows that indolicidin is membrane disruptive in a disordered conformation and does not require folding to be active. No antibiofilm activity was detected with any of the three indo peptides. Note that stereorandomization did not affect the hemolytic activity of any of the three linear AMPs discussed above, which was very weak for SB1 and DJK5 but quite significant for indo (Table 1, column 11).
Taken together, these experiments show that the antibacterial and membrane-disruptive activity of α-helical AMPs SB1 and DJK5 requires α-helical folding, while their antibiofilm activity does not and therefore must involve a disordered conformation. The AMP indo represents a useful control showing that membrane disruption is also possible for peptides with disordered conformation but in this case is unaffected by stereorandomization.
Stereorandomization of AMPDs G3KL and TNS18 Preserves Their Membrane-Disruptive, Antibacterial, and Antibiofilm Activities
AMPD G3KL exhibits an α-helical CD spectrum in membrane-like environments similar to linear AMPs.21 Modeling studies with l-G3KL indicate partial α-helical segments within the dendrimer, however without formation of a globally amphiphilic conformation.25 The same applies to TNS18, a smaller G2 analogue of G3KL optimized for activity by addition of a lipid tail at the core to enhance membrane interactions, and which also adopts an α-helical but not globally amphiphilic conformation in a membrane-like environment.24
Here we found that stereorandomization of AMPDs G3KL and TNS18 only very partially affected their antibacterial activity (Table 1, only G3KL lost activity against A. baumannii upon stereorandomization). Similarly, their membrane-disruptive activity as measured by vesicle leakage assays, their weak hemolytic activity, and their biofilm inhibition were all unaffected by stereorandomization (Figure 2b, Table 1, Figure S11 and S12). These data show that α-helical folding is not required for these activities and support our previous hypothesis, based on modeling studies, that membrane disruption by AMPDs is independent of helical folding and is triggered by a transition from a hydrophobically collapsed conformation in water to an open conformation exposing hydrophobic residues in contact with the bacterial membrane.25
Stereorandomization of AMDP T25 Increases Its Selectivity against Bacteria over Eukaryotic Cells
We next tested the effect of stereorandomization on AMPD T25, an analogue of G3KL with a more hydrophobic dendrimer core featuring one additional leucine residue in G0 (KL → KLL) and a lysine-leucine mutation in G1 (KL → LL). Compared with G3KL, the modified dendrimer core of T25 results in a broader activity spectrum against Gram-negative bacteria also including Klebsiella pneumoniae, against which G3KL is inactive, but also induces strong hemolysis (MHC = 62.5 μg/mL, Table 1) and toxicity against various human cell lines (Figure 3a, Figure S15).25
Figure 3.
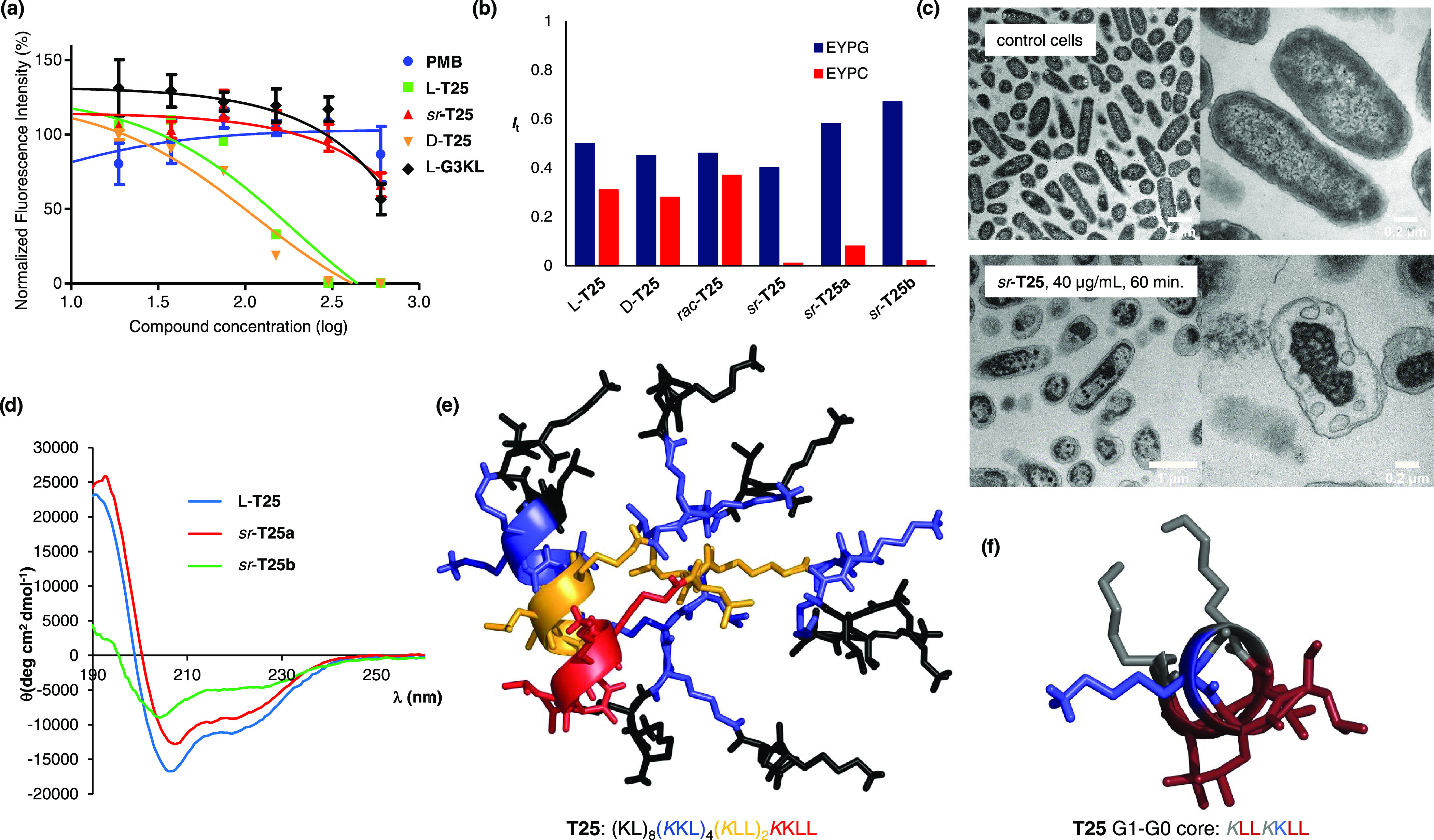
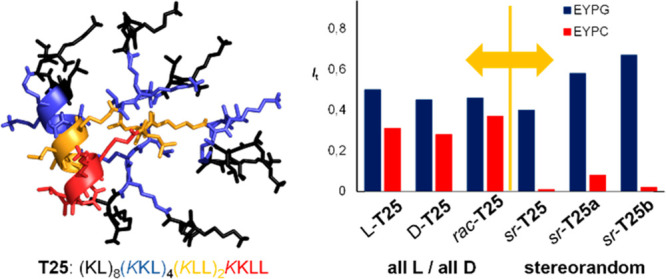
Full and partial stereorandomization of AMPD T25. (a) Cytotoxicity of G3KL, polymyxin B, and T25 analogues in HepG2 cells in DMEM containing 10% FBS. (b) Vesicle leakage assays. (c) TEM images of P. aeruginosa cells treated for 60 min at 10× MIC with sr-T25 (40 μg/mL). (d) CD spectra of T25 analogues (TFA salt 200 μg/mL) in aq. 6 mM phosphate pH 7.4 upon addition of 20% TFE. (e) Molecular dynamics simulation of dendrimer l-T25. Stick model of MD structures color-coded by dendrimer generation (red = G0, orange = G1, blue = G2, black = G3) highlighting the α-helical fold. (f) Cut-out view of (e) showing the G1-G0 α-helical core of l-T25 (brown = leucine, blue = lysine, gray = branching lysine).
Similar to the above AMPDs, stereorandomization to form sr-T25 preserved the vesicle leakage activity on anionic EYPG vesicles mimicking bacterial membranes (blue bars, Figure 3b, Figure S13). sr-T25 also displayed similar antibacterial activities as l- and d-T25 including a panel of MDR bacteria, except for K. pneumoniae NCTC418 and the P. aeruginosa clinical isolate ZEM9A (Table 1, Table S1). Transmission electron microscopy (TEM) images of P. aeruginosa cells exposed to sr-T25 showed extensive membrane damage, in line with its membrane-disruptive activity on EYPG vesicles (Figure 3c, Figure S16). However, stereorandomization of T25 entirely abolished its hemolysis and its vesicle leakage activity on neutral EYPC vesicles mimicking eukaryotic membrane (Table 1, Figure 3b, red bars). Furthermore, sr-T25 showed reduced cell toxicity compared to l- and d-T25 (Figure 3a and Figure S15).
Taken together, these data showed that stereorandomization can selectively affect membrane-disruptive properties leading to a higher selectivity for bacterial over eukaryotic membranes and a stronger activity against bacteria versus eukaryotic cells. This observation is partially related to the selective reduction of hemolysis while preserving antibacterial effects reported previously in AMP mixtures obtained by copolymerizing l-Lys and l-Phe upon switching to copolymerizing l-Lys and d-Phe,7,8 although these polymers are made from enantiomerically pure amino acids, while we are using racemic amino acids.
Partial Stereorandomization of T25 Shows That α-Helical Folding of the Dendrimer Core Triggers Hemolysis and Toxicity but Is Not Required for Antibacterial Effects
As for α-helical AMPs and other AMPDs, the CD spectrum of the all l- enantiomer l-T25 features a strong α-helical signal in the presence of TFE (Figure 3d, blue trace). Indeed, molecular dynamics (MD) simulation of AMPD l-T25 using GROMACS40 showed that an α-helix involving the α-peptide backbone spanning from the dendrimer core (G0) through the first generation (G1) and second generation (G2) branches should be stable and probably accounted for the α-helical CD signal (Figure 3e, Figure S18). Strikingly, the heptapeptide spanning through G0 and G1 formed an amphiphilic α-helix exposing a compact hydrophobic patch of four leucine residues on the dendrimer surface (Figure 3f). To test whether the α-helical conformation of the G1-G0 heptapeptide might be responsible for the hemolytic properties of l-T25, we prepared partially stereorandomized analogues featuring either a homochiral dendrimer core (G0 and G1) and stereorandomized outer branches (G2 and G3, sr-T25a), or vice versa (sr-T25b, Table 1).
Both sr-T25a and sr-T25b showed similar antibacterial, EYPG and EYPC vesicle leakage activities as l-T25 and sr-T25 (Table 1, Figure 3b). Furthermore, only AMPD sr-T25a with the homochiral core showed an α-helical CD spectrum similar to l-T25 (Figure 3d, red trace) and retained its strong hemolytic effect (Table 1). By contrast, analogue sr-T25b with a stereorandomized core and homochiral outer branches had a CD spectrum indicating a disordered conformation (Figure 3d, green trace), and completely suppressed EYPC vesicle leakage activity and hemolysis (Table 1, Figure 3b).
Taken together, these data showed that the α-helical core is responsible for the hemolytic activity and cell toxicity of l-T25, presumably mediated by formation of a hydrophobic patch of four leucine residues. The fact that sr-T25b largely preserves its antibacterial activity shows that the folded core is however not necessary in this case.
Full or Partial Stereorandomization of the Cyclic Peptide Polymyxin B2 Suppresses Membrane Disruption and LPS Binding but Partially Preserves Antibacterial Activity
To test whether stereorandomization might also affect the activity of cyclic peptides, we investigated the last resort antibiotic PMB, which is also a membrane-disruptive compound as the AMPs and AMPDs discussed above.41,42 In addition, PMB binds strongly to lipid A and thereby neutralizes the immune activating properties of the lipopolysaccharide (LPS) released upon lysis of bacterial cells.43 Structure–activity relationship studies have identified residues critical for the activity of PMB.29,44 However, in terms of stereochemistry, only the LPS binding activity of the cyclic portion of PMB and its enantiomer has been investigated.45
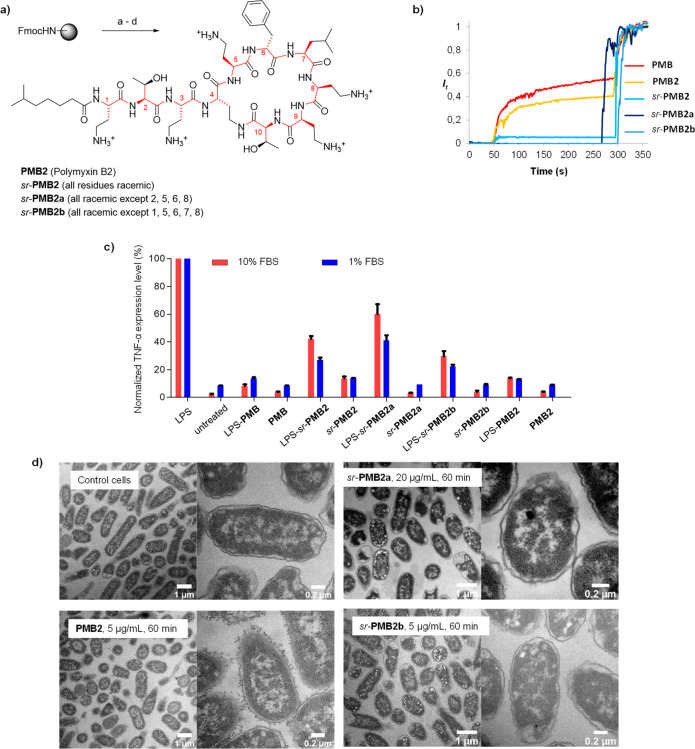
Commercial PMB is a mixture of PMB1, acylated with 6-methyl-octanoic acid at the N-terminus, and PMB2, acylated with 6-methyl-heptanoic acid at the N-terminus. Here we investigated PMB2, which we synthesized using the reported SPPS approach.38 We used the same method to obtain the fully stereorandomized (sr-PMB2) and two partially stereorandomized analogues fixing stereochemistry in positions 2, 5, 6, 8 (sr-PMB2a) or positions 1, 5, 6, 7, 8 (sr-PMB2b), which have been previously reported to be critical for activity (Figure 4a).29,45
Figure 4.
Stereorandomization of polymyxin B2. (a) Solid phase total synthesis of polymyxin B2. Conditions: a) linear chain synthesis by SPPS; b) on-resin cyclization: (i) Pd(Ph3)4 (0.25 equiv), PhSiH3 (25 equiv), in dry DCM, (ii) Fmoc deprotection with 20% v/v piperidine in DMF, (iii) Oxyma (7.5 equiv), DIC (10 equiv), 25 °C,12 h ; c) TFA cleavage with TFA/TIS/H2O (94:5:1), 4 h, 25 °C, then HPLC purification. (b) Membrane-disruptive properties of commercial polymyxin and synthetic polymyxin B analogues (10 mM TRIS, 107 mM NaCl, pH 7.4). After 50 s the indicated compound was added to reach the indicated concentration. After 300 s 1.2% Triton X-100 was added for full fluorescein release. (c) Determination of TNF-α expression level in Raw264.7 murine macrophages by ELISA immunoassay. Polymyxin derivatives were coincubated with or without 0.1 μg/mL of E. coli LPS containing 1 or 10% FBS for 4 h at 37 °C and 5% CO2. (d) TEM images of E. coli cells treated for 60 min at 20 × MIC with polymyxin B analogues. see the Supporting Information for details.
While our synthetic PMB2 had the similar activity as commercial PMB, sr-PMB2 essentially lost all of the antimicrobial and antibiofilm activity of the homochiral parent compound except against E. coli (MIC = 2 μg/mL, Table 1). Vesicle leakage activity on EYPG vesicles, reflecting membrane-disruptive activity, was also abolished in sr-PMB2 (Figure 4b, Figure S14). Furthermore, sr-PMB showed reduced LPS neutralizing activity, as assessed by LPS induced TNF-α release from mouse macrophages (Figure 4c).
The partially stereorandomized analogues sr-PMB2a and sr-PMB2b also lost their membrane disruptive and, in part, their LPS neutralizing activities (Figure 4b,c). However, both analogues retained significant antibacterial activity; in particular, sr-PMB2b had the same activity as PMB against E. coli (MIC = 0.25 μg/mL, Table 1). Strikingly, TEM images of E. coli cells exposed to the different PMB analogues at 20× MIC showed that only PMB2 triggered detectable membrane damage on the bacteria, in line with the vesicle leakage activity data (Figure 4d, Figure S17). Strikingly, sr-PMB2a and sr-PMB2b did not induce any visible damage despite of their activity against these bacteria.
The different membrane-disruptive and antibacterial effects observed with PMB2 and its full or partially stereorandomized analogues demonstrate that stereochemical purity is essential for membrane disruption and LPS neutralization by this antibiotic but also point to the existence of alternative targets at least in the case of E. coli.
Conclusions
In summary, we have shown, at the example of antimicrobial peptides, peptide dendrimers, and the cyclic peptide polymyxin B2, that SPPS with racemic amino acids readily delivers sr-analogues as well-defined homogeneous products. sr-Analogues exhibit distinct properties compared with the all l-, all d-, or rac- (1:1 mixture of all l- and all d-) compounds, in particular specific modulation of membrane disruption, hemolysis and cytotoxicity. Comparing sr- with all l- or all d-peptides generally offers a straightforward method to probe the role of stereochemical purity and folding in activity and potentially unravel diverse mechanisms of action involving different conformations of the same peptide. This new approach should be broadly applicable to better understand and improve the activity and properties of peptides for various types of activity.
Methods
Synthesis and characterization of peptide dendrimers and all assays, measurements, and modeling studies are described in the Supporting Information. No unexpected or unusually high safety hazards were encountered.
Acknowledgments
This work was supported financially by the Swiss National Science Foundation (Grant nos. 200020_178998 and 407240_167048)
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01135.
Details of synthesis and characterization of peptide dendrimers and all assays, measurements, and modeling studies (PDF)
Author Contributions
T.N.S. designed and carried out the study and wrote the paper. B.H.G. carried out TEM and cell culture experiments. T.K. and C.v.D. designed and supervised experiments with MDR bacteria. S.J designed and carried out molecular dynamics studies and wrote the paper. J.L.R. designed and supervised the study and wrote the paper. All authors read and commented the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Merrifield R. B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85 (14), 2149–2154. 10.1021/ja00897a025. [DOI] [Google Scholar]
- Amblard M.; Fehrentz J.-A.; Martinez J.; Subra G. Methods and Protocols of Modern Solid Phase Peptide Synthesis. Mol. Biotechnol. 2006, 33 (3), 239–254. 10.1385/MB:33:3:239. [DOI] [PubMed] [Google Scholar]
- Lam K. S.; Salmon S. E.; Hersh E. M.; Hruby V. J.; Kazmierski W. M.; Knapp R. J. A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature 1991, 354 (6348), 82–84. 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Komnatnyy V. V.; Nielsen T. E.; Qvortrup K. Bead-Based Screening in Chemical Biology and Drug Discovery. Chem. Commun. 2018, 54 (50), 6759–6771. 10.1039/C8CC02486C. [DOI] [PubMed] [Google Scholar]
- Schwaar T.; Lettow M.; Remmler D.; Börner H. G.; Weller M. G.. Efficient Screening of Combinatorial Peptide Libraries by Spatially Ordered Beads Immobilized on Conventional Glass Slides. High Throughput 2019, 8 ( (2), ). 10.3390/ht8020011. [DOI] [PMC free article] [PubMed]
- Houghten R. A.; Pinilla C.; Blondelle S. E.; Appel J. R.; Dooley C. T.; Cuervo J. H. Generation and Use of Synthetic Peptide Combinatorial Libraries for Basic Research and Drug Discovery. Nature 1991, 354 (6348), 84–86. 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Hayouka Z.; Chakraborty S.; Liu R.; Boersma M. D.; Weisblum B.; Gellman S. H. Interplay among Subunit Identity, Subunit Proportion, Chain Length, and Stereochemistry in the Activity Profile of Sequence-Random Peptide Mixtures. J. Am. Chem. Soc. 2013, 135 (32), 11748–11751. 10.1021/ja406231b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amso Z.; Hayouka Z. Antimicrobial Random Peptide Cocktails: A New Approach to Fight Pathogenic Bacteria. Chem. Commun. 2019, 55 (14), 2007–2014. 10.1039/C8CC09961H. [DOI] [PubMed] [Google Scholar]
- Durani S. Protein Design with L- and d-α-Amino Acid Structures as the Alphabet. Acc. Chem. Res. 2008, 41 (10), 1301–1308. 10.1021/ar700265t. [DOI] [PubMed] [Google Scholar]
- Albada H. B.; Prochnow P.; Bobersky S.; Bandow J. E.; Metzler-Nolte N. Highly Active Antibacterial Ferrocenoylated or Ruthenocenoylated Arg-Trp Peptides Can Be Discovered by an L-to-D Substitution Scan. Chem. Sci. 2014, 5 (11), 4453–4459. 10.1039/C4SC01822B. [DOI] [Google Scholar]
- Hazam P. K.; Jerath G.; Chaudhary N.; Ramakrishnan V. Peptido-Mimetic Approach in the Design of Syndiotactic Antimicrobial Peptides. Int. J. Pept. Res. Ther. 2018, 24 (2), 299–307. 10.1007/s10989-017-9615-3. [DOI] [Google Scholar]
- Nagy K. J.; Giano M. C.; Jin A.; Pochan D. J.; Schneider J. P. Enhanced Mechanical Rigidity of Hydrogels Formed from Enantiomeric Peptide Assemblies. J. Am. Chem. Soc. 2011, 133 (38), 14975–14977. 10.1021/ja206742m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanekamp R. J.; DiMaio J. T. M.; Bowerman C. J.; Nilsson B. L. Coassembly of Enantiomeric Amphipathic Peptides into Amyloid-Inspired Rippled β-Sheet Fibrils. J. Am. Chem. Soc. 2012, 134 (12), 5556–5559. 10.1021/ja301642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torbeev V.; Grogg M.; Ruiz J.; Boehringer R.; Schirer A.; Hellwig P.; Jeschke G.; Hilvert D. Chiral Recognition in Amyloid Fiber Growth. J. Pept. Sci. 2016, 22 (5), 290–304. 10.1002/psc.2861. [DOI] [PubMed] [Google Scholar]
- Dutta S.; Foley A. R.; Warner C. J. A.; Zhang X.; Rolandi M.; Abrams B.; Raskatov J. A. Suppression of Oligomer Formation and Formation of Non-Toxic Fibrils upon Addition of Mirror-Image Aβ42 to the Natural l-Enantiomer. Angew. Chem., Int. Ed. 2017, 56 (38), 11506–11510. 10.1002/anie.201706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T.; Haney E. F.; Vogel H. J. The Expanding Scope of Antimicrobial Peptide Structures and Their Modes of Action. Trends Biotechnol. 2011, 29 (9), 464–472. 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Mojsoska B.; Jenssen H. Peptides and Peptidomimetics for Antimicrobial Drug Design. Pharmaceuticals 2015, 8 (3), 366–415. 10.3390/ph8030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. D. T.; Sothiselvam S.; Lu T. K.; de la Fuente-Nunez C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547. 10.1016/j.jmb.2018.12.015. [DOI] [PubMed] [Google Scholar]
- Neundorf I. Antimicrobial and Cell-Penetrating Peptides: How to Understand Two Distinct Functions Despite Similar Physicochemical Properties. Adv. Exp. Med. Biol. 2019, 1117, 93–109. 10.1007/978-981-13-3588-4_7. [DOI] [PubMed] [Google Scholar]
- Stach M.; Maillard N.; Kadam R. U.; Kalbermatter D.; Meury M.; Page M. G. P.; Fotiadis D.; Darbre T.; Reymond J.-L. Membrane Disrupting Antimicrobial Peptide Dendrimers with Multiple Amino Termini. MedChemComm 2012, 3 (1), 86–89. 10.1039/C1MD00272D. [DOI] [Google Scholar]
- Stach M.; Siriwardena T. N.; Kohler T.; van Delden C.; Darbre T.; Reymond J. L. Combining Topology and Sequence Design for the Discovery of Potent Antimicrobial Peptide Dendrimers against Multidrug-Resistant Pseudomonas Aeruginosa. Angew. Chem., Int. Ed. 2014, 53 (47), 12827–12831. 10.1002/anie.201409270. [DOI] [PubMed] [Google Scholar]
- Pires J.; Siriwardena T. N.; Stach M.; Tinguely R.; Kasraian S.; Luzzaro F.; Leib S. L.; Darbre T.; Reymond J. L.; Endimiani A. In Vitro Activity of the Novel Antimicrobial Peptide Dendrimer G3KL against Multidrug-Resistant Acinetobacter Baumannii and Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2015, 59 (12), 7915–7918. 10.1128/AAC.01853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Sayed P.; Kaeppeli A.; Siriwardena T.; Darbre T.; Perron K.; Jafari P.; Reymond J. L.; Pioletti D. P.; Applegate L. A. Anti-Microbial Dendrimers against Multidrug-Resistant P. Aeruginosa Enhance the Angiogenic Effect of Biological Burn-Wound Bandages. Sci. Rep. 2016, 6, 1–10. 10.1038/srep22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena T. N.; Stach M.; He R.; Gan B.-H.; Javor S.; Heitz M.; Ma L.; Cai X.; Chen P.; Wei D.; Li H.; Ma J.; Köhler T.; van Delden C.; Darbre T.; Reymond J.-L. Lipidated Peptide Dendrimers Killing Multidrug-Resistant Bacteria. J. Am. Chem. Soc. 2018, 140 (1), 423–432. 10.1021/jacs.7b11037. [DOI] [PubMed] [Google Scholar]
- Siriwardena T. N.; Capecchi A.; Gan B. H.; Jin X.; He R.; Wei D.; Ma L.; Kohler T.; van Delden C.; Javor S.; Reymond J. L. Optimizing Antimicrobial Peptide Dendrimers in Chemical Space. Angew. Chem., Int. Ed. 2018, 57 (28), 8483–8487. 10.1002/anie.201802837. [DOI] [PubMed] [Google Scholar]
- Siriwardena T. N.; Lüscher A.; Köhler T.; van Delden C.; Javor S.; Reymond J.-L. Antimicrobial Peptide Dendrimer Chimera. Helv. Chim. Acta 2019, 102 (4), e1900034. 10.1002/hlca.201900034. [DOI] [Google Scholar]
- Gan B.-H.; Siriwardena T. N.; Javor S.; Darbre T.; Reymond J.-L. Fluorescence Imaging of Bacterial Killing by Antimicrobial Peptide Dendrimer G3KL. ACS Infect. Dis. 2019, 5 (12), 2164–2173. 10.1021/acsinfecdis.9b00299. [DOI] [PubMed] [Google Scholar]
- Jeddou F. B.; Falconnet L.; Luscher A.; Siriwardena T.; Reymond J.-L.; van Delden C.; Köhler T.. Adaptive and Mutational Responses to Peptide Dendrimer Antimicrobials in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2020, 64 ( (4), ), 10.1128/AAC.02040-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T.; Thompson P. E.; Nation R. L.; Li J. Structure–Activity Relationships of Polymyxin Antibiotics. J. Med. Chem. 2010, 53 (5), 1898–1916. 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeriswyl S.; Gan B.-H.; Siriwardena T. N.; Visini R.; Robadey M.; Javor S.; Stocker A.; Darbre T.; Reymond J.-L. X-Ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas Aeruginosa Lectin B Complexes. ACS Chem. Biol. 2019, 14 (4), 758–766. 10.1021/acschembio.9b00047. [DOI] [PubMed] [Google Scholar]
- Jasanoff A.; Fersht A. R. Quantitative Determination of Helical Propensities from Trifluoroethanol Titration Curves. Biochemistry 1994, 33 (8), 2129–2135. 10.1021/bi00174a020. [DOI] [PubMed] [Google Scholar]
- Arunkumar A. I.; Kumar T. K.; Yu C. Specificity of Helix-Induction by 2,2,2-Trifluoroethanol in Polypeptides. Int. J. Biol. Macromol. 1997, 21 (3), 223–230. 10.1016/S0141-8130(97)00064-0. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Nunez C.; Reffuveille F.; Mansour S. C.; Reckseidler-Zenteno S. L.; Hernandez D.; Brackman G.; Coenye T.; Hancock R. E. D-Enantiomeric Peptides That Eradicate Wild-Type and Multidrug-Resistant Biofilms and Protect against Lethal Pseudomonas Aeruginosa Infections. Chem. Biol. 2015, 22 (2), 196–205. 10.1016/j.chembiol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsted M. E.; Novotny M. J.; Morris W. L.; Tang Y. Q.; Smith W.; Cullor J. S. Indolicidin, a Novel Bactericidal Tridecapeptide Amide from Neutrophils. J. Biol. Chem. 1992, 267 (7), 4292–4295. 10.1016/S0021-9258(18)42830-X. [DOI] [PubMed] [Google Scholar]
- Falla T. J.; Karunaratne D. N.; Hancock R. E. W. Mode of Action of the Antimicrobial Peptide Indolicidin. J. Biol. Chem. 1996, 271 (32), 19298–19303. 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- Sitaram N.; Subbalakshmi C.; Nagaraj R. Indolicidin, a 13-Residue Basic Antimicrobial Peptide Rich in Tryptophan and Proline, Interacts with Ca2+-Calmodulin. Biochem. Biophys. Res. Commun. 2003, 309 (4), 879–884. 10.1016/j.bbrc.2003.08.095. [DOI] [PubMed] [Google Scholar]
- Hsu C.-H.; Chen C.; Jou M.-L.; Lee A. Y.-L.; Lin Y.-C.; Yu Y.-P.; Huang W.-T.; Wu S.-H. Structural and DNA-Binding Studies on the Bovine Antimicrobial Peptide, Indolicidin: Evidence for Multiple Conformations Involved in Binding to Membranes and DNA. Nucleic Acids Res. 2005, 33 (13), 4053–4064. 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.-L.; Cui A.-L.; Hu X.-X.; You X.-F.; Li Z.-R.; Zheng J.-S. A New Strategy for Total Solid-Phase Synthesis of Polymyxins. Tetrahedron Lett. 2015, 56 (33), 4796–4799. 10.1016/j.tetlet.2015.06.056. [DOI] [Google Scholar]
- Hennig A.; Gabriel G. J.; Tew G. N.; Matile S. Stimuli-Responsive Polyguanidino-Oxanorbornene Membrane Transporters as Multicomponent Sensors in Complex Matrices. J. Am. Chem. Soc. 2008, 130 (31), 10338–10344. 10.1021/ja802587j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Deris Z. Z.; Swarbrick J. D.; Roberts K. D.; Azad M. A. K.; Akter J.; Horne A. S.; Nation R. L.; Rogers K. L.; Thompson P. E.; Velkov T.; Li J. Probing the Penetration of Antimicrobial Polymyxin Lipopeptides into Gram-Negative Bacteria. Bioconjugate Chem. 2014, 25 (4), 750–760. 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund N. A.; Piggot T. J.; Jefferies D.; Sessions R. B.; Bond P. J.; Khalid S. Interaction of the Antimicrobial Peptide Polymyxin B1 with Both Membranes of E. Coli: A Molecular Dynamics Study. PLoS Comput. Biol. 2015, 11 (4), e1004180. 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harm S.; Gabor F.; Hartmann J. Low-Dose Polymyxin: An Option for Therapy of Gram-Negative Sepsis. Innate Immun. 2016, 22 (4), 274–283. 10.1177/1753425916639120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa K.; Sato Y.; Ohki K.; Okimura K.; Uchida Y.; Shindo M.; Sakura N. Contribution of Each Amino Acid Residue in Polymyxin B3 to Antimicrobial and Lipopolysaccharide Binding Activity. Chem. Pharm. Bull. 2009, 57 (3), 240–244. 10.1248/cpb.57.240. [DOI] [PubMed] [Google Scholar]
- Tsubery H.; Ofek I.; Cohen S.; Fridkin M. The Functional Association of Polymyxin B with Bacterial Lipopolysaccharide Is Stereospecific: Studies on Polymyxin B Nonapeptide. Biochemistry 2000, 39 (39), 11837–11844. 10.1021/bi000386q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.