Abstract
Abstract
Bats as flying mammals are potent vectors and natural reservoir hosts for many infectious viruses, bacteria, and fungi, also detected in their excreta such as guano. Accelerated deforestation, urbanization, and anthropization hastily lead to overpopulation of the bats in urban areas allowing easy interaction with other animals, expansion, and emergence of new zoonotic disease outbreaks potentially harmful to humans. Therefore, getting new insights in the microbiome of bat guano from different places represents an imperative for the future. Furthermore, the use of novel high-throughput sequencing technologies allows better insight in guano microbiome and potentially indicated that some species could be typical guano-dwelling members. Bats are well known as a natural reservoir of many zoonotic viruses such as Ebola, Nipah, Marburg, lyssaviruses, rabies, henipaviruses, and many coronaviruses which caused a high number of outbreaks including ongoing COVID-19 pandemic. Additionally, many bacterial and fungal pathogens were identified as common guano residents. Thus, the presence of multi-drug-resistant bacteria as environmental reservoirs of extended spectrum β-lactamases and carbapenemase-producing strains has been confirmed. Bat guano is the most suitable substrate for fungal reproduction and dissemination, including pathogenic yeasts and keratinophilic and dimorphic human pathogenic fungi known as notorious causative agents of severe endemic mycoses like histoplasmosis and fatal cryptococcosis, especially deadly in immunocompromised individuals. This review provides an overview of bat guano microbiota diversity and the significance of autochthonous and pathogenic taxa for humans and the environment, highlighting better understanding in preventing emerging diseases.
Key points
Bat guano as reservoir and source for spreading of autochthonous and pathogenic microbiota
Bat guano vs. novel zoonotic disease outbreaks
Destruction of bat natural habitats urgently demands increased human awareness
Keywords: Bat guano, Microbiota, Zoonoses, Pathogens, Disease outbreaks
Introduction
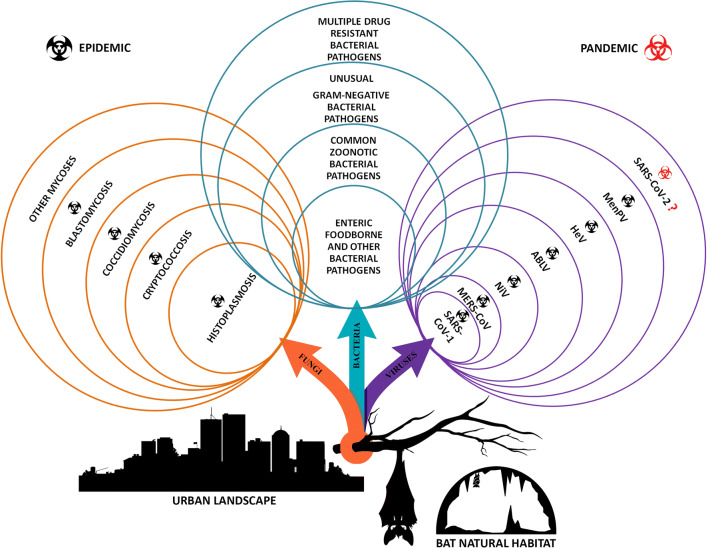
Bats (Chiroptera) are a diverse group of mammals with more than 1400 different species with abilities to inhabit various ecological niches and environments (Winter et al. 2016; Gorbunova et al. 2020). Although bats are geographically widespread, the ecology of their microbial communities and the role involved in bat health and behavior are scarce, especially on the microbiota residing in bat’s excreta accumulated in the form of guano, which could be potentially involved in pathogen transmission to humans (Dietrich et al. 2018; Dietrich and Markotter 2019). Bats revealed versatile feeding behavior; they have a flying capacity and inherent characteristics which makes them as highly potent vectors and natural reservoir hosts for many pathogens. Furthermore, it is quite clear that humans and bats are cross-visitors of each other’s habitats! The main assumption is that these flying mammals visit human settlements more often than humans visit their natural habitats. Based on this hypothesis, the question arising is “Could the frequent visitations of these animals in urban areas affect human health?” Accelerated urbanization and the destruction of natural habitats inevitably lead to increased risk of zoonotic diseases. It is known that zoonotic pathogens, especially new or still unexplored, can easily spread to humans through any contact point with animals or their excreta, as well as through markets selling the meat or by-products of wild animals (WHO 2020). Also, deforestation and encroachment of natural habitats leads to reduced feeding bat habitats forcing them to find alternative sites in peri-urban landscapes and with simultaneous less awareness in building communities’ resilience to future outbreaks can increase human exposure to zoonotic disease reservoirs allowing pathogens transfer to other species (United Nations Environment Programme and International Livestock Research Institute 2020). Additionally, climate changes as a force of nature can rapidly influence the geographic distribution and abundance of bats species and increase the incidence of the vector-borne and zoonotic diseases (Wells and Clark 2019). Inter-species transmission of the pathogens is a consequence of the contact between bats and other animals including humans, which potentially could result in disease outbreaks. Primarily, bats are contemplated as the reservoir of many viruses and diseases outbreaks caused by SARS coronavirus (SARS-CoV) (Tsang et al. 2003), Marburg virus (MARV) (Calisher et al. 2006), MERS coronavirus (MERS-CoV) (Kupferschmidt 2013), Nipah virus (NiV) (Luis et al. 2013), Ebola virus (EBOV) (WHO Ebola Response Team 2014), Australian bat lyssavirus (ABLV), Hendra (HeV) and Menangle (MenPV) viruses, and current COVID-19 pandemic, for which is not yet proved that bats transmit SARS-CoV-2 to humans (United Nations Environment Programme and International Livestock Research Institute 2020). Contrary to it, bat guano has been recognized as a potential source of bacterial and fungal pathogens. Furthermore, it is well known that, in many regions of the world, bat guano is preciously used in agriculture, frequently as patented and presented on the market, as organic biofertilizer in order to positively affect overall soil fertility (Grantina-Ievina and Ievinsh 2015; De Leon et al. 2018). However, in areas with a high use of guano as biofertilizer, agricultural workers could be at increased risk by contact with guano-harbored foodborne pathogens, which also could easily contaminate food products or infect livestock. Due to easy spread of pathogens into the environment with the interaction with other animals, intake of raw food, or water contamination and possible infection of humans, knowledge on the bats guano microbiota is highly important (Wolkers-Rooijackers et al. 2019). The presence of pathogenic enteric bacteria and other bacterial pathogens common in human and animal diseases such as species of Pasteurella, Salmonella, Shigella, Escherichia, Klebsiella, Proteus, Yersinia, Hafnia, Serratia, Staphylococcus, and Campylobacter genera among many others in different cave- and building–dwelling bat guanos (Sridhar et al. 2006; Mühldorfer 2013; Banskar et al. 2016; Wolkers-Rooijackers et al. 2019; Dimkić et al. 2020; Gerbáčová et al. 2020) has already been confirmed. Furthermore, some other bacterial species from genera Bartonella, Borrelia, Leptospira, Pseudomonas, Enterobacter, Acinetobacter, Bacillus, Arthrobacter, and Micrococcus, related to bat guano, can indicate novel species-specific bat-dwelling bacteria with medical importance to humans (Lei and Olival 2014; Veikkolainen et al. 2014; Banskar et al. 2016; Dimkić et al. 2020). Additionally, guano has also been recognized as a potential source of various dimorphic fungal pathogens, including endemic Histoplasma capsulatum, Coccidioides spp., Blastomyces dermatitidis, Cryptococcus neoformans, and opportunistic pathogens from genera Candida, Meyerozima, Trychophyton, Trychosporon, Microsporum, Sporotrix, Chrysosporium, Geomyces, Aspergillus, Penicillium, and Fusarium (Sugita et al. 2005; Ulloa et al. 2006; Ogórek et al. 2016; Cordero et al. 2016; Dimkić et al. 2020; Rodrigues et al. 2020). It should be pointed out that bats themselves are reservoirs for many infectious agents, including viruses, bacteria, and fungi, which could also be detected in guano (Fig. 1). Hence, the sampling of guano is a noninvasive and more convenient approach, in contrast to direct examination of bats, which can also be carried out even when bats hibernate (Ogórek et al. 2016).
Fig. 1.
Bat guano microbiome—a silent life-threatening pathogenic bacteria and epidemic/pandemic of fungal and viral diseases
This paper provides a new systematic review of the literature within the context of various quantitative and qualitative bat guano microbiota diversity researches, emphasizing the presence and significance of autochthonous phylogenetically different lineages and potential pathogenic taxa for humans and the environment. This could be very important in terms of biodiversity preservation and understanding how we can provide a protection of humans from emerging infectious diseases.
Bat (guano) virome
Bats are well known as a natural reservoir of many zoonotic viruses, including lyssaviruses, causative agents of rabies, henipaviruses, severe acute respiratory syndrome coronavirus, and Ebola virus (Li et al. 2010a). Their ability to host different viruses is enabled by their wide geographical distribution, migratory and feeding habits, and in many cases, high population density (Wibbelt et al. 2009). In several recent studies based on metagenomic analyses and other methods, more than 80 species of viruses were isolated or detected. Besides already mentioned, these species include adenoviruses, adeno-associated viruses, herpesviruses, astroviruses, and polyomaviruses (Chu et al. 2008; Misra et al. 2009; Li et al. 2010b). The study of the presence of viruses in guano samples from several insectivorous species of bats revealed a great diversity of identified virus species (Li et al. 2010a). These groups include animal viruses from the families Parvoviridae, Circoviridae, Adenoviridae, Poxviridae, Picornaviridae, Astroviridae, and Coronaviridae and bacteriophages from the families Siphoviridae and Microviridae. The same study also showed that bat guano virome contained plant and fungal viruses from Luteoviridae, Secoviridae, Tymoviridae, and Partitiviridae families, and the Sobemovirus genus. Within detected families, some species are particularly interesting; Cyclovirus, a new genus from the family Circoviridae contains viruses with covalently closed circular single strand DNA (ssDNA) genomes. These viruses were detected in cerebrospinal fluid samples from patients with unexplained paraplegia in Malawi (Smits et al. 2013). In the case of bat cyclovirus GF-4 identified in the samples of guano, a phylogenetic analysis of the Rep protein sequence placed this virus within the group of known cycloviruses as the distinctive species. In another study, metagenomic analysis of insectivorous bat guano in Hungary showed the presence of viruses from the order of Picornavirales. In tested samples, the aphid lethal paralysis virus (ALPV) and Big Sioux River virus (BSRV) were detected in Hungary for the first time (Zana et al. 2018). Additionally, among others, rabies viruses also are the part of the bats’ virome. Rabies is the zoonotic disease that is, if not treated, fatal for all mammals including humans. The causative agent of disease is the rabies virus (RABV), an RNA virus that belongs to the genus Lyssavirus within the family Rhabdoviridae. Bats are the main natural reservoirs of the rabies viruses; until now, 64 species of bats were reported positive for RABV worldwide, including insectivorous, frugivorous, and hematophagous species (Jiménez et al. 2017). Bat species that can carry rabies virus can be found in South and North America, as well as in Africa, in Western and Southeastern Europe. There have been only several recorded deaths of humans in Europe caused by the bite of rabies-infected bats (Lina and Hutson 2006). In most cases, humans can be infected if bitten by the domestic cats that are the main predators of bats. Although the rabies is a generally eradicated disease, in the United States, bats are responsible for 7 out of 10 rabies deaths, according to the U.S. Centers for Disease Control and Prevention.
Bat’s coronaviruses
Bat’s virome and relationships between bats and viruses, including more than 200 novel coronaviruses are very likely specific to the bat species (Calisher et al. 2006; Vijaykrishna et al. 2007). Since the new bat viruses, isolation has already proven to be difficult, current SARS-CoV-2 pandemic, still unexplored in transmission to humans, required extent detection in bats and sequencing of their genomes. Coronaviruses are the group of lipid-enveloped, single-strand RNA viruses (positive strand) with the genome size from 27 to 32 kb, among the largest of all RNA viruses’ genomes. Several recent studies indicated that bat coronaviruses represent the source of human coronaviruses such as SARS-CoV and MERS-CoV. Coronaviruses were isolated from the samples of bat guano of common flying fox (Pteropus medius) in Sri Lanka (Kudagammana et al. 2018). These viruses showed more than 97% of nucleotide sequence identity with coronaviruses detected in fruit bat Cynopterus sphinx and insectivorous bats Scotophilus heathii and Scotophilus kuhlii bats in Thailand. Although the lineage D of betacoronaviruses detected in this study represent the group of coronaviruses that was until now confined to bats, they might evolve to new strains potentially pathogenic to humans, since in some parts of South Asia, they are hunted and consumed as food. Besides that, bat guano is also collected and sold as fertilizer in some countries, including Thailand, Indonesia, Mexico, Cuba, and Jamaica. In samples of bat guano fertilizer in Thailand, a betacoronavirus from group C was isolated (Wacharapluesadee et al. 2013). This practice can represent the serious health risk to guano miners and, without the use of preventive measures, expose them to zoonotic pathogens. Coronaviruses were also detected in the bat guano samples and rectal swabs in Myanmar (Valitutto et al. 2020). In this study, three novel alphacoronaviruses, three novel betacoronaviruses, and one previously identified alphacoronavirus were detected for the first time in bats in Myanmar. Novel lineages of alphacoronaviruses were also detected in bat guano samples collected from caves on several locations in Kazakhstan (Mendenhall et al. 2019).
Bat (guano) bacteriome
In the last decade, composition of bacterial communities from bat guano has begun to intensively be characterized. In the focus of investigations were guano sampled from fresh fecal samples or guano piles in different places such as caves, buildings, and church towers. Also, surveys were based on bats’ gut as the primary source of guano bacteria. Significant bacterial diversity was detected in guano samples that varied in detected genera among analyzed bat species, sample types (fresh feces or guano piles), and sampling points of investigated caves or guano layers (Table 1).
Table 1.
Bacterial diversity in bat guano across the world
| Reference | Sample origin | Identification method | Distribution of most abundant taxa |
|---|---|---|---|
| Banskar et al. 2016 | Bat feces samples of Rousettus leschenaultii collected at the Robber’s Cave, Maharashtra, India | Cultivation method | Enterobacter, Enterococcus, Escherichia, Lactococcus, Staphylococcus, Serratia, Weissella, Citrobacter, Acinetobacter |
| High-throughput 16S rRNA sequencing | Proteobacteria, Tenericutes, Candidate division TM7, Firmicutes, Actinobacteria | ||
| Selvin et al. 2019 | Samples from Rhinolophus monoceros gut, The Arwah Cave, Meghalaya, India | Cultivation method | Staphylococcus, Bacillus, Hafnia, Brevibacillus, Pseudomonas, Serratia, Enterobacter |
| High-throughput 16S rRNA sequencing | Citrobacter, Lactococcus, Staphylococcus, Devosia, Acinetobacter, Arthrobacter, Streptomyces | ||
| Newman et al. 2018 | Bat guano sampled from different guano layers, lava tube cave, Sierra County, United States | Cultivation method | Microbacterium, Enterococcus, Lactococcus, Enterobacter, Bacillus, Pantoea, Staphylococcus, Weissella, Streptomyces, Deinococcus |
| High-throughput 16S rRNA sequencing | Enterococcus, Bacillus, Plesiomonas, Vespertiliibacter, Lactococcus, Paenibacillus, Clostridium, Mycoplasma, Pantoea, Weissella, Streptococcus | ||
| Gerbáčová et al. 2020 | Feces samples, building–dwelling insectivorous bat species (Myotis myotis, Rhinolophus hipposideros), twelve locations in central Slovakia | Cultivation method | Enterococcus, Lactococcus, Lactobacillus, Pseudomonas, Paenibacillus, Bacillus, Staphylococcus |
| 16S rRNA DGGE analysis | Rahnella, Micrococcus, Staphylococcus, Serratia | ||
| Dimkić et al. 2020 | Bat guano sampled from different layers in Ogorelička Pećina Cave, Serbia | Cultivation method | Bacillus, Enterococcus, Staphylococcus, Viridibacillus, Lysinibacillus, Paenibacillus, Pseudomonas, Escherichia, Serratia, Citrobacter |
| Borda et al. 2014 | Bat guano sampled from 7 caves in Romania | Cultivation method | Escherichia, Chryseomonas, Burkholderia |
| Vandžurová et al. 2013 | Guano samples from mixed Myotis myotis and M. blythii colony, the church tower in the Slovak Lupca village, Slovakia | Cultivation method | Staphylococcus nepalensis |
| Tomova et al. 2013 | Bat guano samples from the Gallery with prehistoric drawings in Magura Cave, Bulgaria | Cultivation method | Serratia, Pseudomonas, Enterobacter, Sphingobacterium |
| Wolkers-Rooijackers et al. 2019 | Bat guano samples of 10 insectivorous bats of different species sampled from limestone mines, province of Limburg, Netherlands | 16S rDNA cloning and sequencing | Carnobacterium, Serratia, Pseudomonas, Enterococcus, Yersinia |
| Afonso and Goydadin 2018 | Feces samples from 23 lesser horseshoe bat maternity roosts located in buildings (churches, barns), Franche-Comté region, France | msp2 (major surface protein gene) sequencing | Anaplasma phagocytophilum |
| Hornok et al. 2018 | Bat feces samples from 19 bat species collected at Hungary and Netherlands | PCR screening focused on vector-borne bacteria (rickettsia and hemotropic mycoplasmas) | Rickettsia, Neorickettsia, Mycoplasma |
| Vengust et al. 2018 | Feces samples of 12 different bat species, captured during autumn migration across central Europe at 8 different parts of Slovenia | High-throughput 16S rRNA sequencing | Pseudomonas, Staphylococcus, Carnobacterium, Acinetobacter, an unclassified Enterobacteriaceae |
| Veikkolainen et al. 2014 | Feces samples from Myotis daubentonii, Finland | High-throughput 16S rRNA sequencing | Leuconostoc, Enterobacter, Lactococcus, Chlamydia, Citrobacter, Aeromonas, Klebsiella |
| Li et al. 2018 | Fresh fecal pellets from bats with different diets randomly collected in Guangdong, Guangxi, and Yunnan, China | High-throughput 16S rRNA sequencing | Enterobacter, Fructobacillus, Ureaplasma, Klebsiella, Weissella, Plesiomonas, Enterococcus, Lactobacillus, Bacillus |
| Dietrich and Markotter 2019 | Bat feces sampled in a maternity colony of Rousettus aegyptiacus, Matlapitsi Cave, Limpopo province, South Africa | High-throughput 16S rRNA sequencing | Streptococcus, Actinobacillus, Nesterenkonia |
| De Leon et al. 2018 | Bat guano sampled from Cabalyorisa Cave, Philippines | High-throughput 16S rRNA sequencing | Unassigned Xanthomonadaceae, Mycobacterium, Bacillus, Luteibacter, Rhodococcus |
| De Mandal et al. 2015 | Bat guano sampled from three bat species in Pnahkyndeng cave, Meghalaya, Northeast India | High-throughput 16S rRNA sequencing | Chloroflexi, Crenarchaeota, Actinobacteria, Bacteroidetes, Proteobacteria, Planctomycetes |
In general, representatives from the phyla Proteobacteria and Firmicutes were detected as dominant groups in the bat guano or fresh feces samples collected from mist-netted bats. Culture-based analysis indicated that common genera in guano, regardless of the sample and sampling location, were Staphylococcus, Lactococcus, Enterobacter, and Bacillus (Banskar et al. 2016; Newman et al. 2018; Gerbáčová et al. 2020). These common genera are also detected as dominant in the gut of Rhinolophus monoceros (Selvin et al. 2019), with the exception of Lactococcus sp., which could be explained by culture method bias. Identified common taxa in guano from different bat species and sample types indicate that bat gut is the basis of the dominant bacterial genera. Other bacterial representatives are probably influenced by environmental factors. Besides valuable information provided by cultivation methods, traditional use of culturable media has limitations to comprehensively analyze whole microbial diversity (Abdelfattah et al. 2018). Thus, using high-throughput sequencing (HTS) as a tool to investigate bacterial communities of guano enabled detailed studies of these extreme microhabitats (Knight et al. 2018). In guano samples, originated from bats with different diets and different geographic regions (the USA, India, and China), by HTS genera Weissella, Lactococcus, Enterococcus, Bacillus, and Arthrobacter as common were detected (Banskar et al. 2016; Li et al. 2018; Newman et al. 2018). That indicated these species as potential typical members of guano.
From the aspect of clinical significance, bacteria originating from guano could be divided into four groups. Enteric foodborne and other pathogens includes genera Escherichia, Enterobacter, Yersinia, Hafnia, Serratia, Staphylococcus, Streptococcus, Pseudomonas, Rahnella, Micrococcus, Acinetobacter, and Arthrobacter. Among common zoonotic pathogens, following genera Bartonella, Borrelia, Leptospira, Campylobacter, Clostridium, and Bacillus were recognized. Group of unusual Gram-negative bacterial pathogens, in relation to atypical cell structure, includes genera Mycoplasma, Ureaplasma, Rickettsia, Anaplasma, and Chlamydia. Extended spectrum beta-lactamase (ESBL) and carbapenemase-producing Enterobacteriaceae (CPE) as a group of multiple drug-resistant bacterial pathogens are detected in several studies in guano which originated from different bat species. Many studies have, so far, identified pathogens as common guano residents (Veikkolainen et al. 2014; Banskar et al. 2016; Wolkers-Rooijackers et al. 2019; Gerbáčová et al. 2020). Therefore, guano piles are recognized as potential reservoirs of spreading zoonoses. Recently, two studies have characterized and compared the composition of bacterial communities in different layers of guano piles (Newman et al. 2018; Dimkić et al. 2020). Bacterial population shift was detected throughout the guano pile as nutritional composition and environmental factors have changed, from aerobic to anaerobic. Different distributions of bacterial taxa in layers were detected between studied caves, but common genera were identified as Bacillus and Enterococcus. Microbial diversity of bat guano is driven by different factors such as diet type and reproductive stage of the host (Phillips et al. 2012; Gaona et al. 2019). Also, caves as typical bat habitats are specific, extreme environments with low level of UV radiation, high gaseous ammonia levels, and could reach temperatures up to 43 °C (Newman et al. 2018). Different caves ensure the development of taxa specifically adapted for conditions in this environment. On the other hand, variations in the microbial community are conditioned by the geographic location, geochemical characteristics, and other environmental factors (Wiseschart and Pootanakit 2020).
Bacterial diversity of guano from building–dwelling bats was rarely studied (Vandžurová et al. 2012, 2013; Gerbáčová et al. 2020). Nevertheless, composition of their bacteriome is especially interesting considering the vicinity of human residence. Gerbáčová et al. (2020) reported presence of several possible human pathogens including Hafnia alvei, Staphylococcus nepalensis, Serratia fonticola, and S. liquefaciens in two building–dwelling bat species in Slovakia. Earlier, Staphylococcus nepalensis was detected by Vandžurová et al. (2013) from the summer colony bats from the church tower, also in Slovakia. Staphylococcus nepalensis is a potential human pathogen with presence of antibiotic-resistant strains (Nováková et al. 2006). Constant presence of S. nepalensis over a long period of time observed on bat populations in Slovakia indicates a persistence of potential pathogens in the bat population.
Bats as a natural reservoir of antibiotic multi-resistant bacteria
The existence of antibiotic multi-resistant bacteria in terrestrial mammals has been previously documented in multiple studies, providing an important insight into their potential role as reservoirs of resistant bacteria. Antibiotic-resistant bacteria have been detected in bat isolates across the globe, suggesting the possibility of them being one of the environmental reservoirs of resistant bacteria (Adesiyun et al. 2009; Allocati et al. 2016;).
McDougall et al. (2019) described the presence of bacteria resistant to several antibiotic families in gray-headed flying fox (Pteropus poliocephalus) populations across Australia. Isolated bacteria were found to carry genes responsible for resistance to aminoglycosides, trimethoprim, and beta-lactams. This study also provided evidence of resistome transfer between humans and bats, as well as from environment to bats. Studies in Nigeria also detected bacterial isolates from bat fecal droppings with resistance to multiple antibiotics (Oluduro 2012; Ajayi et al. 2020). Among Gram-positive isolates, most were resistant to amoxicillin, streptomycin, and erythromycin, while most of Gram-negative isolates were resistant to penicillins and cephalosporins.
Recent studies have detected presence of extended spectrum β-lactamases (ESBLs) in Enterobacteriaceae isolates from bat guano. Mbehang Nguema et al. (2020) reported for the first time the presence of multi-resistant ESBL-producing Enterobacteriaceae in fruit bats in Makokou. Their strains showed higher resistance to tetracycline, ciprofloxacin, and ofloxacin compared with the studies carried out in Nigeria (Oluduro 2012; Ajayi et al. 2020). Main enterobacterial species that showed resistance to ESBLs were Escherichia coli, followed by Klebsiella pneumoniae and E. cloacae. Genetic analysis of ESBL resistance genes by PCR and sequencing identified only two genes responsible for observed resistance phenotype: blaCTX-M-15 and blaSHV-11. Both genes are recognized as plasmid-mediated resistance genes (Cantón et al. 2013; Liakopoulos et al. 2016). In addition, blaCTX-M-15 beta-lactam resistance gene is the most widely distributed and the most prevalent ESBL gene in human strains worldwide (Ewers et al. 2012; Janatova et al. 2014). ESBL-producing E. coli isolates from bat guano, carrying the same resistance genes, were also detected in Peru, Poland, and Portugal (Benavides et al. 2018; Garcês et al. 2019; Nowakiewicz et al. 2020). During 2019, carbapenemase-producing Enterobacteriaceae (CPE) were detected in bat guano (Gharout-Sait et al. 2019). CPE isolates were identified as K. pneumoniae and were found to carry blaOXA-48 gene (CS34) and blaKPC-3 gene (CS63). K. pneumoniae isolate carrying blaKPC-3 gene was found to belong to the ST512 clonal group, subcluster of ST258 (Gharout-Sait et al. 2019). Strains belonging to this sequence types are recognized as a major contributor to the spread of carbapenemases and have been frequently associated with hospital outbreaks. In Gabon, phylogenetic analysis of bat guano isolates revealed that they were clustered with previously described human bacterial strains from Turkey and Tunisia, but without information on their relatedness to clonal groups (Mbehang Nguema et al. 2020). All these and other studies suggest that antibiotic resistance in bat microbiome may vary locally and could be influenced by contact with wastes of antibiotic treatments of animals and humans (Swift et al. 2019).
Bat (guano) mycobiota
Since many bat species are nocturnal animals that roost in caves, much more often than in tree cavities, caves could be considered natural habitats for Chiroptera (Kunz 1982). Although fungi in caves could colonize a variety of substrata, such as cave sediments, vermiculations on cave walls, carcasses, and droppings of troglobites or troglophiles (Nováková 2009), bat guano is according to Ogórek et al. (2016) the most suitable substrate for fungal growth and proliferation. Since caves are usually regarded as oligotrophic, without primary production, bat guano piles are an important source of organic matter (Ferreira 2019). Depending on Chiroptera diet, three types of bat guanos are described. The majority of troglophilous microbats (former Microchiroptera) from the northern temperate regions are insectivorous; hence, their guano is finely granulated and consists of small pieces of insect cuticle. Chitin-degrading saprobes could be regarded as the first colonizers of newly established guano piles in caves roosted with insectivorous bats. On the other hand, megabats (Pteropodoidea or Yinpterochiroptera) are predominantly frugivorous, and their geographical distribution encompasses tropical regions of the Old world as well as Australia. Guano of frugivorous bats contains small undigested seeds easily colonized by saprobes. Hematophagous or vampire bats (Desmodontinae) are native to Central and South America, and their guano is with a pasty consistency and reddish color or sometimes black with powdery consistency (Gnaspini 2012). All types of guano (from insectivorous, frugivorous, and hematophagous bats) are suitable substrata for fungal colonization. Investigation conducted in a sandstone cave Meu Rei located in the Catimbau National Park (Brazil) showed that fungal richness does not differ greatly between insectivorous, frugivorous, and hematophagous bats or between fresh and non-fresh guano, while regarding fungal propagules abundance guano types were in ascending order: frugivorous< haematophagous< insectivorous (Cunha et al. 2020). Once pioneer fungal community is established further heterotrophic succession goes on, followed by changes in community structure and physicochemical properties of guano (Ferreira 2019). Guano communities are very simple and consist of only a few trophic levels (mostly of two with guanophages and their predators); however, they can harbor a large number of fungal propagules (Gnaspini 2012). Previous studies suggest that fungal density in bat guano piles could be high. Ogórek et al. (2016) reported very high fungal density in guano samples in Harmanecká cave in Slovakia (3498.3 CFU/g), while even higher fungal density in guano samples from Ogorelička Pećina Cave (Serbia) of 6384.61 CFU/g were presented by Dimkić et al. (2020). Apart from high fungal density, the mycobiota documented in guano samples can also be moderate to high (Vaughan 2012). Previous studies suggest the presence of a broad spectrum of fungal species documented in guano (Nováková 2009). Although the majority of guano-dwelling species are saprobes, guano can also be a reservoir of pathogenic fungi harmful to animals and humans. Opportunistic pathogens, causative agents of severe or mild mycoses, and toxigenic molds can be documented in bat guano worldwide. Larcher et al. (2003) documented the high fungal diversity in bat guano samples from caves in the region of Pays de la Loire (Western France). Their findings included pathogenic yeasts (Candida glabrata and Meyerozyma guilliermondii) and eight species of pathogenic keratinophilic fungi (Trichophyton terrestre, Scopulariopsis brevicaulis, Aphanoascus fulvescens, Myceliophthora thermophila, Chrysosporium lobatum, Ch. merdarium, Ch. pseudomerdarium, and Ch. pruinosum). Sugita et al. (2005) reported the presence of nine species of genus Trichosporon, causative agent of white piedra and opportunistic trichosporonosis, from guano of bat-inhabited limestone and volcanic caves in Japan. Other opportunistic pathogens presented in this work were Candida lusitaniae and Debaryomyces hansenii. Ulloa et al. (2006) reported the presence of pathogens Aphanoascus fulvescens, Candida catenulata, C. ciferrii, M. guilliermondii, and Sporothrix sp. in tested guano samples from different caves in State of Guerrero in Mexico. The presence of Aspergillus fumigatus, known causative agent of invasive pulmonary mycosis in immunosuppressed individuals, was documented in guano samples from Domica Cave and Čertova Cave in Slovakia (Nováková 2009). Pathogenic yeast M. guilliermondii was isolated from guano samples collected in Ogorelička Pećina Cave in Serbia (Dimkić et al. 2020). According to Ogórek et al. (2016) bat guano is a “hidden treasure” of fungi that may be hazardous for mammal health, such as Candida, Geomyces, Microsporum, Trichosporon, and Trichophyton species.
The most important attention regarding guano-dwelling fungi is paid to dimorphic human fungal pathogens, which are the causative agents of severe diseases within specific geographic areas of known endemicity. It is already known that endemic mycoses manifested as focal and systemic diseases in a limited geographical area and remain a major public health problem in several countries. However, over the past few decades new epidemiological reports have indicated that endemic fungi in certain areas are thought to be “non-endemic” (Ashraf et al. 2020). Interestingly, these pathogens could be the cause of various infections outside these areas. Endemic mycoses diagnosed outside of their established geographic ranges could be the main causes of morbidity and mortality in immunocompromised individuals, especially AIDS patients and patients with other immunosuppressive medical conditions (Wheat 1995; Ashraf et al. 2020). In this regard, due to the significant fact that there are many asymptomatic cases, the occurrence and development of these mycoses in such patients can be an important benchmark for the detection of these pathogens beyond endemic areas. The most prominent guano-associated dimorphic fungus is Histoplasma capsulatum, causative agent of systemic histoplasmosis also known as cave disease (Rodrigues et al. 2020). In addition to histoplasmosis, endemic mycoses are also caused by other dimorphic fungi such as Coccidioides immitis, Co. posadasii, Blastomyces dermatitidis, Paracoccidioides brasiliensis, P. lutzii, and Talaromyces marneffei (formerly known as Penicillium marneffei) (Sil and Andrianopoulos 2014). The above-mentioned species are more or less related to the guano and bat habitats, but perhaps the largest number of reports and scientific papers in recent two decades is related to Coccidioides species (Barker et al. 2007; Brown et al. 2013; Engelthaler et al. 2016; Kollath et al. 2019; Van Dyke et al. 2019; Oltean et al. 2020) and B. dermatitidis (Saccente and Woods 2010; Smith and Gauthier 2015; Slomka and Doub 2020).
Coccidioides immitis and Co. posadasii are causative agents of potentially fatal coccidiomycosis, known as valley fever, endemic for southwestern USA, northern Mexico, Central America, and some regions of South America (del Rocío Reyes-Montes et al. 2016). The first report regarding the presence of Co. immitis in bat guano samples, originated from roosting places deep within deserted mine tunnels in southern Arizona (Krutzsch and Watson 1978). Later, Cordeiro et al. (2012) proved that several chiropteran species from Brazil (Carollia perspicillata, Glossophaga soricina, and Desmodus rotundus) could be infected with Co. posadasii and these authors also hypothesized that the ability of bats daily to travel long distances in their search for food and the social behavior could introduce this fungus in previously non-disease-endemic areas. The predominant route of infection with Coccidioides spp. is inhalation. The majority of cases are asymptomatic, but in some patients, a complicated form of pneumonia can also develop (Saubolle et al. 2007). In the last decades, the incidence of Coccidioides infections is increasing in endemic areas, with >90% in Arizona and California between 2001 and 2006 (Hector et al. 2011). The result of this increase has a significant impact on public health around the world.
Blastomyces dermatitidis is known as endemic species in the Mississippi and Ohio River Valleys, the Midwestern states, and the area of New York and Canada (Klein et al. 1986a, 1987; Cano et al. 2003; Dworkin et al. 2005; Sil and Andrianopoulos 2014). In the endemic region, this species is not uniformly distributed. It is known that it inhabits an ecological niche that is characterized by forested, sandy, and moist soils with an acidic pH; decaying vegetation or organic material; and rotting wood areas rich in organic debris, located near water sources (e.g., rivers and lakes) (Smith and Gauthier 2015). Similar to H. capsulatum and Cryptococcus species, this fungus may also persist in bird guano (Klein et al. 1986b; McBride et al. 2017). The first report on isolation of B. dermatitidis from liver of species Rhinopoma hardwickei (the “lesser rat-tailed bat”) found within the basement of Safdar-Jang Tomb, a historical monument in New Delhi, was reported by Randhawa et al. (1985). Previous means that Chiroptera could be the vectors of blastomycosis, a systemic disease caused by this pathogen, which is also called Gilchrist’s disease (Pappas and Dismukes 2002) or North American blastomycosis. The clinical manifestations of this mycosis are broad, ranging from asymptomatic infection to acute respiratory distress syndrome and lethal outcome (McBride et al. 2017).
In contrast to previously discussed fungi, there are only few reports regarding the presence of Cryptococcus neoformans, causative agent of potentially fatal cryptococcosis, in bats and bat guano. Cryptococcus neoformans var. neoformans was documented in an old abandoned house inhabited by bats in Rio de Janeiro, Brazil (Lazera et al. 1993). Furthermore, Montagna et al. (2003) reported the presence of Cr. neoformans in the upper layers of the bat guano in three caves in the Apulia region in Italy. Although cryptococcosis is regarded as a deadlier disease than histoplasmosis, Cr. neoformans does not spread as easily, and outbreaks have never been documented. Nevertheless, caves and bat guano generate sheltered conditions that prevent desiccation and protect Cr. neoformans from environmental stresses, i.e., UV radiation and extreme temperatures and wind (Montagna et al. 2003).
Histoplasma capsulatum
Histoplasma capsulatum is often regarded as “the main culprit from the Kingdom Fungi” that could be concealed in bat guano. There are two varieties which were initially distinguished on the basis of symptoms they cause. Histoplasma capsulatum var. capsulatum is found globally (mainly North, South, and Central America, Southeast Asia, and Africa) and is associated with pulmonary and systemic (i.e., classical) histoplasmosis, while H. capsulatum var. duboisii is predominantly found in Western and Central Africa and causes skin and bone lesions (Cockshott and Lucas 1964; Sil and Andrianopoulos 2014).
It is well known that spelunkers should be aware of the risk bat-inhabited caves pose for developing histoplasmosis caused by H. capsulatum, especially if they are immunocompromised in any way (Lyon et al. 2004). Pulmonary histoplasmosis is a common disease of residents of the Ohio and Mississippi river valleys, where the fungus prevails and is the cause of the most common endemic fungal infection in the USA (Paul and Kuhlenschmidt 2018). However, new epidemiological reports indicate that regions other than the disease-endemic areas (midwestern and southeastern USA and Central and South America) are also affected, which indicates that H. capsulatum is wider spread than originally acknowledged (Deepe Jr 2018). Furthermore, in response to deforestation and urbanization, bats populations are adapting and setting up in the anthropized rural and urbanized environments, closer to human dwellings, leading to shift in the pattern of disease outbreaks to cover urban areas as well, with the number of infected individuals exceeding that found in rural areas (Jung and Threlfall 2018; Deepe Jr 2018). Though most of these infections are clinically silent and resolve without any consequence in immunocompetent individuals, H. capsulatum has been known to be behind such unusual cases as puzzling sickness of the pastor that cleaned copious amounts of bat guano from belfry of the church (Wynbrandt and Crouser 2007), outbreak of an unknown severe febrile illness of workers in the Dominican Republic tasked with cleaning accumulated bat guano from access tunnels to a hydroelectric dam inhabited by large bat colonies (Armstrong et al. 2018) or an outbreak affecting American tourists in a hotel in Acapulco (Guerrero, Mexico) where hotel’s ornamental potted plants, fertilized with guano compost, were determined as the infectious source (Taylor et al. 2005). In the period from 1938 to 2013, a total of 105 outbreaks involving 2.850 cases were reported in 26 states and the territory of Puerto Rico (Benedict and Mody 2016). Two outbreaks of extreme proportions have occurred in Indianapolis in the twentieth century, first associated with construction of a tennis stadium in the downtown area and other construction of a new natatorium on the IUPUI campus, with approximately 200,000 infected individuals in total (Deepe Jr 2018). Since H. capsulatum is known to exist within environmental pockets, i.e., “hotspots” (Armstrong et al. 2018), disruption of these soil reservoirs and release of large quantities of microconidia into the air can be possible cause of these outbreaks. Monitoring and data on histoplasmosis occurrence in Europe, as a non-endemic disease area, is very scarce. Disease has been reported among different risk groups in Germany, Italy, United Kingdom, France, Belgium, Sweden, Switzerland, Austria, Bulgaria, and Turkey (Ashbee et al. 2008) with only one case report of rare pulmonary histoplasmosis complicated with bronchocentric granulomatosis in Greece (Botsa et al. 2017). Histoplasma capsulatum infections are quite heterogeneous and may have significant consequences for immunological response and disease progression. Co-infections are also another important aspect to consider, since some of them could have a lethal outcome, such as reported human co-infections with C. neoformans (Cordero et al. 2016) and Blastomyces and Coccidioides spp. (Jehangir et al. 2015). In light of current pandemic, it is important to emphasize that two cases of histoplasmosis co-infection on immunocompromised AIDS patients with COVID-19 have been reported recently (Messina et al. 2020; Basso et al. 2020).
Open questions for the future
The current pandemic demands thinking about human impact on the natural system that increases the contact between humans and animals. Disturbing natural bat habitats, often to expand agricultural land, unleash pathogens that were sealed in the wild. Bearing in mind that bats and bat guano are the natural reservoir of pathogenic microorganisms, causative agents of serious diseases with epidemic and pandemic potential, it is quite clear that mankind is almost always responsible for the spillover of new diseases. Expanding the knowledge of guano microbiome is nowadays of utmost importance considering the presence of antibiotic multi-resistant bacteria, as well as pathogenic fungi with potential of outbreaks and “silent” viruses queuing for epidemics or eventually pandemics. Advances in microbiome-enabling technologies will help in better understanding of the community composition, function, and activity of both cultivable and non-cultivable microorganisms in the bat (guano) microbiome globally and could give an answer on the important question: “How can a microbiome be disclosed to increase human awareness and decreased all potential new disease outbreaks in the future?”
Acknowledgments
Availability of data and material
Not applicable.
Code availability
Not applicable.
Author contribution
ID and MLG conceived and designed mini-review. ID, DF, TJ, JK, MS, MN, NU and MLG wrote the manuscript. ID, DF, TJ, JK, MS, MN, NU and MLG read and approved the manuscript.
Funding
The authors acknowledge the Ministry of Education, Science and Technological Development of the Republic of Serbia for the financial support of their research (Contract No. 451-03-68/2020-14).
Declarations
Ethics approval
Not applicable.
Consent to participate
All authors agreed to participate.
Consent for publication
All authors are informed and agreed to publish.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdelfattah A, Malacrinò A, Wisniewski M, Cacciola S, Schena L. Metabarcoding: a powerful tool to investigate microbial communities and shape future plant protection strategies. Biol Control. 2018;120:1–10. doi: 10.1016/j.biocontrol.2017.07.009. [DOI] [Google Scholar]
- Adesiyun AA, Stewart-Johnson A, Thompson NN. Isolation of enteric pathogens from bats in Trinidad. J Wildl Dis. 2009;45(4):952–961. doi: 10.7589/0090-3558-45.4.952. [DOI] [PubMed] [Google Scholar]
- Afonso E, Goydadin A. Molecular detection of Anaplasma phagocytophilum DNA in the lesser horseshoe bat (Rhinolophus hipposideros) guano. Epidemiol Infect. 2018;146:1253–1258. doi: 10.1017/s0950268818001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajayi AO, Owaboriaye M, Rufus J, Arowolo HM, Osunla CA. Enteric viruses and bacterial species from the faecal droppings of bats in selected bat colonies in Ondo State, Nigeria. J Viro Res Rep. 2020;1(1):3–10. doi: 10.47363/JVRR/2020-(1)106. [DOI] [Google Scholar]
- Allocati N, Petrucci AG, Di Giovanni P, Masulli M, Di Ilio C, De Laurenzi V. Bat-man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Dis. 2016;2:16048. doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong P, Beard J, Bonilla L, Arboleda N, Lindsley M, Chae S, Castillo D, Nuñez R, Chiller T, de Perio M, Pimentel R, Vallabhaneni S. Outbreak of severe histoplasmosis among tunnel workers—Dominican Republic, 2015. Clin Infect Dis. 2018;66:1550–1557. doi: 10.1093/cid/cix1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbee H, Evans E, Ashbee H, Evans E, Viviani M, Dupont B, Chryssanthou E, Surmont I, Tomsikova A, Vachkov P, Ener B, Zala J, Tintelnot K. Histoplasmosis in Europe: report on an epidemiological survey from the European Confederation of Medical Mycology Working Group. Med Mycol. 2008;46:57–65. doi: 10.1080/13693780701591481. [DOI] [PubMed] [Google Scholar]
- Ashraf N, Kubat RC, Poplin V, Adenis AA, Denning DW, Wright L, McCotter O, Schwartz IS, Jackson BR, Chiller T, Bahr NC. Re-drawing the maps for endemic mycoses. Mycopathologia. 2020;185:843–865. doi: 10.1007/s11046-020-00431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banskar S, Bhute S, Suryavanshi M, Punekar S, Shouche Y. Microbiome analysis reveals the abundance of bacterial pathogens in Rousettus leschenaultii guano. Sci Rep. 2016;6(1):1–13. doi: 10.1038/srep36948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker BM, Jewell KA, Kroken S, Orbach MJ. The population biology of Coccidioides: epidemiologic implications for disease outbreaks. Ann N Y Acad Sci. 2007;1111:147–163. doi: 10.1196/annals.1406.040. [DOI] [PubMed] [Google Scholar]
- Basso RP, Poester VR, Benelli JL, Stevens DA, Zogbi HE, Vasconcellos ICDS, Pasqualotto AC, Xavier MO. COVID-19-associated histoplasmosis in an AIDS patient. Mycopathologia. 2020;6:1–4. doi: 10.1007/s11046-020-00505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides J, Shiva C, Virhuez M, Tello C, Appelgren A, Vendrell J, Solassol J, Godreuil S, Streicker D. Extended-spectrum beta-lactamase-producing Escherichia coli in common vampire bats Desmodus rotundus and livestock in Peru. Zoonoses Public Health. 2018;65:454–458. doi: 10.1111/zph.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict K, Mody RK. Epidemiology of histoplasmosis outbreaks, United States, 1938-2013. Emerg Infect Dis. 2016;22(3):370–378. doi: 10.3201/eid2203.151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borda DR, Nåstase-Bucur RM, Spinu M, Uricariu R, Mulec J. Aerosolized microbes from organic rich materials: case study of bat guano from caves in Romania. J Caves Karst Stud. 2014;76(2):114–126. doi: 10.4311/2013mb0116. [DOI] [Google Scholar]
- Botsa E, Thanou I, Kabanarou S, Thanos L. Rare case of pulmonary histoplasmosis complicated with bronchocentric granulomatosis in a non-endemic area. Respir Med Case Rep. 2017;22:1–3. doi: 10.1016/j.rmcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Benedict K, Park BJ, Thompson GR. Coccidioidomycosis: epidemiology. Clin Epidemiol. 2013;5:185–197. doi: 10.2147/CLEP.S34434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C, Childs J, Field H, Holmes K, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/cmr.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano MV, Ponce-de-Leon GF, Tippen S, Lindsley MD, Warwick M, Hajjeh RA. Blastomycosis in Missouri: epidemiology and risk factors for endemic disease. Epidemiol Infect. 2003;131(2):907–914. doi: 10.1017/s0950268803008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2013;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DK, Poon LL, Guan Y, Peiris JS. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockshott WP, Lucas AO. Histoplasmosis duboisii. Q J Med. 1964;33:223–238. doi: 10.1093/oxfordjournals.qjmed.a067017. [DOI] [PubMed] [Google Scholar]
- Cordeiro RDA, de Castro e Silva KR, Brilhante RSN, Moura FBP, Duarte NFH, Marques FJDF, Cordeiro RDA, de Araújo B, Wagner R, Bandeira TDJPG, Rocha MFG, Sidrim JJC. Coccidioides posadasii infection in bats, Brazil. Emerg Infect Dis. 2012;18(4):668–670. doi: 10.3201/eid1804.111641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero R, Liedke S, de S. Araújo GR, Martinez LR, Nimrichter L, Frases S, Peralta JM, Casadevall A, Rodrigues ML, Nosanchuk JD, Guimaraes AJ. Enhanced virulence of Histoplasma capsulatum through transfer and surface incorporation of glycans from Cryptococcus neoformans during co-infection. Sci Rep. 2016;6:21765. doi: 10.1038/srep21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha AO, Bezerra JD, Oliveira TG, Barbier E, Bernard E, Machado AR, Souza-Motta CM. Living in the dark: bat caves as hotspots of fungal diversity. PLoS One. 2020;15(12):e0243494. doi: 10.1371/journal.pone.0243494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon MP, Montecillo AD, Pinili DS, Siringan MAT, Park DS. Bacterial diversity of bat guano from Cabalyorisa Cave, Mabini, Pangasinan, Philippines: a first report on the metagenome of Philippine bat guano. PLoS One. 2018;13(7):e0200095. doi: 10.1371/journal.pone.0200095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rocío Reyes-Montes M, Pérez-Huitrón MA, Ocaña-Monroy JL, Frías-De-León MG, Martínez-Herrera E, Arenas R, Duarte-Escalante E (2016) The habitat of Coccidioides spp. and the role of animals as reservoirs and disseminators in nature. BMC Infect Dis 16(1):1–8. 10.1186/s12879-016-1902-7 [DOI] [PMC free article] [PubMed]
- De Mandal S, Panda AK, Bisht SS, Kumar NS. First report of bacterial community from a bat guano using Illumina next-generation sequencing. Genom Data. 2015;4:99–101. doi: 10.1016/j.gdata.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepe GS., Jr Outbreaks of histoplasmosis: the spores set sail. PLoS Pathog. 2018;14(9):e1007213. doi: 10.1371/journal.ppat.1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M, Markotter W. Studying the microbiota of bats: accuracy of direct and indirect samplings. Ecol Evol. 2019;9(4):1730–1735. doi: 10.1002/ece3.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M, Kearney T, Seamark EC, Paweska JT, Markotter W. Synchronized shift of oral, faecal and urinary microbiotas in bats and natural infection dynamics during seasonal reproduction. R Soc Open Sci. 2018;5(5):180041. doi: 10.1098/rsos.180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimkić I, Stanković S, Kabić J, Stupar M, Nenadić M, Ljaljević-Grbić M, Žikić V, Vujisić P, Tešević V, Vesović N, Pantelić D, Savić-Šević S, Vukojević J, Ćurčić S. Bat guano-dwelling microbes and antimicrobial properties of the pygidial gland secretion of a troglophilic ground beetle against them. Appl Microbiol Biotechnol. 2020;104:4109–4126. doi: 10.1007/s00253-020-10498-y. [DOI] [PubMed] [Google Scholar]
- Dworkin MS, Duckro AN, Proia L, Semel JD, Huhn G. The epidemiology of blastomycosis in Illinois and factors associated with death. Clin Infect Dis. 2005;41(12):107–118. doi: 10.1086/498152. [DOI] [PubMed] [Google Scholar]
- Ebola Response Team WHO. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Roe CC, Hepp CM, Teixeira M, Driebe EM, Schupp JM, Gade L, Waddell V, Komatsu K, Arathoon E, Logemann H, Thompson GR, Chiller T, Barker B, Keim P, Litvintseva AP. Local population structure and patterns of western hemisphere dispersal for Coccidioides spp. the Fungal cause of Valley fever. mBio. 2016;7(2):550–566. doi: 10.1128/mBio.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C, Bethe A, Semmler T, Guenther S, Wieler L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- Ferreira RL. Guano communities. In: White WB, Culver DC, Pipan T, editors. Encyclopedia of caves. 3. Cambridge: Academic Press; 2019. pp. 474–484. [Google Scholar]
- Gaona O, Gómez-Acata ES, Cerqueda-García D, Neri-Barrios CX, Falcón LI. Fecal microbiota of different reproductive stages of the central population of the lesser-long nosed bat, Leptonycteris yerbabuenae. PLoS One. 2019;14(7):e0219982. doi: 10.1371/journal.pone.0219982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês A, Correia S, Amorim F, Pereira J, Igrejas G, Poeta P. First report on extended-spectrum beta-lactamase (ESBL) producing Escherichia coli from European free-tailed bats (Tadarida teniotis) in Portugal: A one-health approach of a hidden contamination problem. J Hazard Mater. 2019;370:219–224. doi: 10.1016/j.jhazmat.2017.12.053. [DOI] [PubMed] [Google Scholar]
- Gerbáčová K, Maliničová L, Kisková J, Maslišová V, Uhrin M, Pristaš P. The faecal microbiome of building-dwelling insectivorous bats (Myotis myotis and Rhinolophus hipposideros) also contains antibiotic-resistant bacterial representatives. Curr Microbiol. 2020;77(9):2333–2344. doi: 10.1007/s00284-020-02095-z. [DOI] [PubMed] [Google Scholar]
- Gharout-Sait A, Touati A, Ahmim M, Brasme L, Guillard T, Agsous A, de Champs C. Occurrence of carbapenemase-producing Klebsiella pneumoniae in bat guano. Microb Drug Resist. 2019;25:1057–1062. doi: 10.1089/mdr.2018.0471. [DOI] [PubMed] [Google Scholar]
- Gnaspini P. Guano communities. In: White W, Culver D, editors. Encyclopedia of caves. 2. Cambridge: Academic Press; 2012. pp. 357–364. [Google Scholar]
- Gorbunova V, Seluanov A, Kennedy BK. The world goes bats: living longer and tolerating viruses. Cell Metab. 2020;32(1):31–43. doi: 10.1016/j.cmet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantina-Ievina L, Ievinsh G (2015) Microbiological characteristics and effect on plants of the organic fertilizer from vermicompost and bat guano. In: Annual 21st International Scientific Conference: Research for rural development, vol 1, pp 95–101
- Hector RF, Rutherford GW, Tsang CA, Erhart LM, McCotter O, Anderson SM, Komatsu K, Tabnak F, Vugia DJ, Yang Y, Galgiani JN. The public health impact of coccidioidomycosis in Arizona and California. Int J Environ Res Public Health. 2011;8(4):1150–1173. doi: 10.3390/ijerph8041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornok S, Szőke K, Estók P, Krawczyk A, Haarsma AJ, Kováts D, Boldogh SA, Morandini P, Szekeres S, Takács N, Kontschán J, Meli ML, Fernández de Mera IG, de la Fuente J, Gyuranecz M, Sulyok KM, Weibel B, Gönczi E, de Bruin A, Sprong H, Hofmann-Lehmann R. Assessing bat droppings and predatory bird pellets for vector-borne bacteria: molecular evidence of bat-associated Neorickettsia sp. in Europe. Antonie Van Leeuwenhoek. 2018;111:1707–1717. doi: 10.1007/s10482-018-1043-7. [DOI] [PubMed] [Google Scholar]
- Janatova M, Albrechtova K, Petrzelkova KJ, Dolejska M, Papousek I, Masarikova M, Cizek A, Todd A, Shutt K, Kalousova B. Antimicrobial-resistant Enterobacteriaceae from humans and wildlife in Dzanga-Sangha Protected Area, Central African Republic. Vet Microbiol. 2014;171:422–431. doi: 10.1016/j.vetmic.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Jehangir W, Tadepalli GS, Sen S, Regevik N, Sen P. Coccidioidomycosis and blastomycosis: endemic mycotic co-infections in the HIV patient. J Clin Med Res. 2015;7(3):196–198. doi: 10.14740/jocmr2036w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez JFC, Lopéz RDP, Garcia NV. Bat reservoirs for rabies virus and epidemiology of rabies in Colombia: a review. CES Med Vet Zootec. 2017;12(2):134–150. doi: 10.21615/cesmvz.12.2.5. [DOI] [Google Scholar]
- Jung K, Threlfall CG. Trait-dependent tolerance of bats to urbanization: a global meta-analysis. Proc Biol Sci. 2018;285(1885):20181222. doi: 10.1098/rspb.2018.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BS, Vergeront JM, Davis JP. Epidemiologic aspects of blastomycosis, the enigmatic systemic mycosis. Semin Respir Infect. 1986;1(1):29–39. [PubMed] [Google Scholar]
- Klein BS, Vergeront JM, Weeks RJ, Kumar UN, Mathai G, Varkey B, Kaufman L, Bradsher RW, Stoebig JF, Davis JP. Isolation of Blastomyces dermatitidis in soil associated with a large outbreak of blastomycosis in Wisconsin. N Engl J Med. 1986;314(9):529–534. doi: 10.1056/NEJM198602273140901. [DOI] [PubMed] [Google Scholar]
- Klein BS, Vergeront JM, DiSalvo AF, Kaufman L, Davis JP. Two outbreaks of blastomycosis along rivers in Wisconsin. Isolation of Blastomyces dermatitidis from riverbank soil and evidence of its transmission along waterways. Am Rev Respir Dis. 1987;136:1333–1338. doi: 10.1164/ajrccm/136.6.1333. [DOI] [PubMed] [Google Scholar]
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- Kollath DR, Miller KJ, Barker B. The mysterious desert dwellers: Coccidioides immitis and Coccidioides posadasii, causative fungal agents of coccidioidomycosis. Virulence. 2019;10(1):222–233. doi: 10.1080/21505594.2019.1589363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzsch PH, Watson RH. Isolation of Coccidioides immitis from bat guano and preliminary findings on laboratory infectivity of bats with Coccidioides immitis. Life Sci. 1978;22(8):679–683. doi: 10.1016/0024-3205(78)90491-5. [DOI] [PubMed] [Google Scholar]
- Kudagammana HDWS, Thevanesam V, Chu DKW, Eriyagama NB, Peiris JSM, Noordeen F. Coronaviruses in guano from Pteropus medius bats in Peradeniya, Sri Lanka. Transbound Emerg Dis. 2018;65:1122–1124. doi: 10.1111/tbed.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz TH. Roosting ecology of bats. In: Kunz TH, editor. Ecology of bats. 1. Boston: Springer; 1982. pp. 1–55. [Google Scholar]
- Kupferschmidt K. Emerging infectious diseases. Link to MERS virus underscores bats’ puzzling threat. Science. 2013;341:948–949. doi: 10.1126/science.341.6149.948. [DOI] [PubMed] [Google Scholar]
- Larcher G, Bouchara JP, Pailley P, Montfort D, Béguin H, de Bièvre C, Chabasse D. Fungal biota associated with bats in Western France. J Mycol Med. 2003;13:29–34. [Google Scholar]
- Lazera MS, Wanke B, Nishikawa MM. Isolation of both varieties of Cryptococcus neoformans from saprophytic sources in the city of Rio de Janeiro, Brazil. J Med Vet Mycol. 1993;31(6):449–454. doi: 10.1080/02681219380000581. [DOI] [Google Scholar]
- Lei BR, Olival KJ. Contrasting patterns in mammal–bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLoS Negl Trop Dis. 2014;8(3):e2738. doi: 10.1371/journal.pntd.0002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84(14):6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, Yuan J, Wang LF, Shi Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li L, Jiang H, Yuan L, Zhang L, Ma JE, Zhang X, Cheng M, Chen J. Fecal bacteriome and mycobiome in bats with diverse diets in South China. Curr Microbiol. 2018;75(10):1352–1361. doi: 10.1007/s00284-018-1530-0. [DOI] [PubMed] [Google Scholar]
- Liakopoulos A, Mevius D, Ceccarelli D. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front Microbiol. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina PH, Hutson AM. Bat rabies in Europe: a review. Dev Biol (Basel) 2006;125:245–254. [PubMed] [Google Scholar]
- Luis AD, Hayman DT, O’Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, Mills JN, Timonin ME, Willis CK, Cunningham AA, Fooks AR. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. 2013;280(1756):20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GM, Bravo AV, Espino A, Lindsley MD, Gutierrez RE, Rodriguez I, Corella A, Carrillo F, McNeil MM, Warnock DW, Hajjeh RA. Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998–1999. Am J Trop Med Hyg. 2004;70(4):438–442. doi: 10.4269/ajtmh.2004.70.438. [DOI] [PubMed] [Google Scholar]
- Mbehang Nguema P, Onanga R, Ndong Atome G, Obague Mbeang J, Mabika Mabika A, Yaro M, Lounnas M, Dumont Y, Zohra Z, Godreuil S, Bretagnolle F. Characterization of ESBL-producing Enterobacteria from fruit bats in an unprotected area of Makokou, Gabon. Microorganisms. 2020;8:138. doi: 10.3390/microorganisms8010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JA, Gauthier GM, Klein BS. Clinical manifestations and treatment of blastomycosis. Clin Chest Med. 2017;38(3):435–449. doi: 10.1016/j.ccm.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall F, Boardman W, Gillings M, Power M. Bats as reservoirs of antibiotic resistance determinants: a survey of class 1 integrons in grey-headed flying foxes (Pteropus poliocephalus) Infect Genet Evol. 2019;70:107–113. doi: 10.1016/j.meegid.2019.02.022. [DOI] [PubMed] [Google Scholar]
- Mendenhall IH, Kerimbayev AA, Strochkov VM, Sultankulova KT, Kopeyev SK, Su YCF, Smith GJD, Orynbayev MB. Discovery and characterization of novel bat coronavirus lineages from Kazakhstan. Viruses. 2019;11(4):356. doi: 10.3390/v11040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina FA, Marin E, Caceres DH, Romero M, Depardo R, Priarone MM, Rey L, Vázquez M, Verweij PE, Chiller TM, Santiso G. Coronavirus disease 2019 (COVID-19) in a patient with disseminated histoplasmosis and HIV-a case report from Argentina and literature review. J Fungi (Basel) 2020;6(4):275. doi: 10.3390/jof6040275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V, Dumonceaux T, Dubois J, Willis C, Nadin-Davis S, Severini A, Wandeler A, Lindsay R, Artsob H. Detection of polyoma and corona viruses in bats of Canada. J Gen Virol. 2009;90:2015–2022. doi: 10.1099/vir.0.010694-0. [DOI] [PubMed] [Google Scholar]
- Montagna MT, Santacroce MP, Caggiano G, Tato D, Ajello L. Cavernicolous habitats harbouring Cryptococcus neoformans: results of a speleological survey in Apulia, Italy, 1999–2000. Med Mycol. 2003;41(5):451–455. doi: 10.1080/13693780310001602785. [DOI] [PubMed] [Google Scholar]
- Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Public Health. 2013;60(1):93–103. doi: 10.1111/j.1863-2378.2012.01536.x. [DOI] [PubMed] [Google Scholar]
- Newman MM, Kloepper LN, Duncan M, McInroy JA, Kloepper JW. Variation in bat guano bacterial community composition with depth. Front Microbiol. 2018;9:914. doi: 10.3389/fmicb.2018.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková A. Microscopic fungi isolated from the Domica Cave system (Slovak Karst National Park, Slovakia). A review. Int J Speleol. 2009;38:71–82. doi: 10.5038/1827-806X.38.1.8. [DOI] [Google Scholar]
- Nováková D, Pantůček R, Petráš P, Koukalová D, Sedláček I. Occurrence of Staphylococcus nepalensis strains in different sources including human clinical material. FEMS Microbiol Lett. 2006;263(2):163–168. doi: 10.1111/j.1574-6968.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- Nowakiewicz A, Zięba P, Gnat S, Trościańczyk A, Osińska M, Łagowski D, Kosior-Korzecka U, Puzio I (2020) Bats as a reservoir of resistant Escherichia coli: a methodical view. Can we fully estimate the scale of resistance in the reservoirs of free-living animals? Res Vet Sci 128:49–58. 10.1016/j.rvsc.2019.10.017 [DOI] [PubMed]
- Ogórek R, Dyląg M, Kozak B, Višnovska Z, Tančinová D, Lejman A. Fungi isolated and quantified from bat guano and air in Harmanecká and Driny Caves (Slovakia) J Cave Karst Stud. 2016;78(1):41–49. doi: 10.4311/2015MB0108. [DOI] [Google Scholar]
- Oltean HN, Springer M, Bowers JR, Barnes R, Reid G, Valentine M, Engelthaler DM, Toda M, McCotter OZ. Suspected locally acquired coccidioidomycosis in human. Emerg Infect Dis. 2020;26(3):606–609. doi: 10.3201/eid2603.191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluduro AO. Antibiotic-resistant commensal Escherichia coli in faecal droplets from bats and poultry in Nigeria. Vet Ital. 2012;48:297–308. [PubMed] [Google Scholar]
- Pappas PG, Dismukes WE. Blastomycosis: Gilchrist’s disease revisited. Curr Clin Top Infect Dis. 2002;22:61–77. [PubMed] [Google Scholar]
- Paul V, Kuhlenschmidt MS. Unusual case of cellulitis due to primary cutaneous histoplasmosis. BMJ Case Rep. 2018;11(1):e226621. doi: 10.1136/bcr-2018-226621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Delton Hanson J, Siles L, Ordóñez-Garza N, San Francisco M, Baker RJ. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol. 2012;21(11):2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- Randhawa HS, Chaturvedi VP, Kini S, Khan ZU. Blastomyces dermatitidis in bats: first report of its isolation from the liver of Rhinopoma hardwickei hardwickei gray. Sabouraud. 1985;23:69–76. doi: 10.1080/00362178585380111. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, Beale MA, Hagen F, Fisher MC, Della Terra PP, de Hoog S, Brilhante RSN, de Aguiar Cordeiro R, de Souza Collares Maia Castelo-Branco D, Rocha MFG, Sidrim JJC, de Camargo ZP (2020) The global epidemiology of emerging Histoplasma species in recent years. Stud Mycol 97:100095. 10.1016/j.simyco.2020.02.001 [DOI] [PMC free article] [PubMed]
- Saccente M, Woods GL. Clinical and laboratory update on blastomycosis. Clin Microbiol Rev. 2010;23(2):367–381. doi: 10.1128/CMR.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saubolle MA, McKellar PP, Sussland D. Epidemiologic, clinical, and diagnostic aspects of coccidioidomycosis. J Clin Microbiol. 2007;45(1):26–30. doi: 10.1128/JCM.02230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin J, Lanong S, Syiem D, De Mandal S, Kayang H, Kumar NS, Kiran GS. Culture-dependent and metagenomic analysis of lesser horseshoe bats’ gut microbiome revealing unique bacterial diversity and signatures of potential human pathogens. Microb Pathog. 2019;137:103675. doi: 10.1016/j.micpath.2019.103675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A, Andrianopoulos A. Thermally dimorphic human fungal pathogens--polyphyletic pathogens with a convergent pathogenicity trait. Cold Spring Harb Perspect Med. 2014;5(8):a019794. doi: 10.1101/cshperspect.a019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomka M, Doub J. A rare case of Blastomyces dermatitidis brain abscess in an immunocompetent host. Med Mycol Case Rep. 2020;28:8–11. doi: 10.1016/j.mmcr.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Gauthier G. New developments in Blastomycosis. Semin Respir Crit Care Med. 2015;36:715–728. doi: 10.1055/s-0035-1562898. [DOI] [PubMed] [Google Scholar]
- Smits SL, Zijlstra E, van Hellemond JJ, Schapendonk C, Bodewes R, Schürch AC, Haagmans BL, Osterhaus AD. Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg Infect Dis. 2013;19(9):1511–1513. doi: 10.3201/eid1909.130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar KR, Ashwini KM, Seena S, Sreepada KS. Manure qualities of guano of insectivorous cave bat Hipposideros speoris. Trop Subtrop Agroecosystems. 2006;6(2):103–110. [Google Scholar]
- Sugita T, Kikuchi K, Makimura K, Urata K, Someya T, Kamei K, Niimi M, Uehara Y. Trichosporon species isolated from guano samples obtained from bat-inhabited caves in Japan. Appl Environ Microbiol. 2005;71(11):7626–7629. doi: 10.1128/AEM.71.11.7626-7629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift B, Bennett M, Waller K, Dodd C, Murray A, Gomes R, Humphreys B, Hobman J, Jones M, Whitlock S, Mitchell L, Lennon R, Arnold K. Anthropogenic environmental drivers of antimicrobial resistance in wildlife. Sci Total Environ. 2019;649:12–20. doi: 10.1016/j.scitotenv.2018.08.180. [DOI] [PubMed] [Google Scholar]
- Taylor ML, Ruíz-Palacios GM, Rocío Reyes-Montes M, Rodríguez-Arellanes G, Carreto-Binaghi LE, Duarte-Escalante E, Hernández-Ramírez A, Pérez A, Suárez-Alvarez RO, Roldán-Aragón YA, Romero-Martínez R, Sahaza-Cardona JH, Sifuentes-Osornio J, Soto-Ramírez LE, Peña-Sandoval GR. Identification of the infectious source of an unusual outbreak of histoplasmosis, in a hotel in Acapulco, state of Guerrero, Mexico. FEMS Immunol Med Microbiol. 2005;45(3):435–441. doi: 10.1016/j.femsim.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Tomova I, Lazarkevich I, Tomova A, Kambourova M, Vasileva-Tonkova E. Diversity and biosynthetic potential of culturable aerobic heterotrophic bacteria isolated from Magura Cave, Bulgaria. Int J Speleol. 2013;42(1):65–76. doi: 10.5038/1827-806X.42.1.8. [DOI] [Google Scholar]
- Tsang T, Lai-Yin T, Pak-Yin L, Lee M. Update: outbreak of severe acute respiratory syndrome-worldwide, 2003. MMWR Morb Mortal Wkly Rep. 2003;52(12):241–241. [PubMed] [Google Scholar]
- Ulloa M, Lappe P, Aguilar S, Park H, Pérez-Mejía A, Toriello C, Taylor ML. Contribution to the study of the mycobiota present in the natural habitats of Histoplasma capsulatum: an integrative study in Guerrero, Mexico. Rev Mex Biodiv. 2006;77(2):153–168. doi: 10.22201/ib.20078706e.2006.002.328. [DOI] [Google Scholar]
- United Nations Environment Programme and International Livestock Research Institute . Preventing the next pandemic: zoonotic diseases and how to break the chain of transmission. Nairobi: UNEP; 2020. [Google Scholar]
- Valdez H, Salata RA. Bat-associated histoplasmosis in returning travelers: case presentation and description of a cluster. J Travel Med. 1999;6(4):258–260. doi: 10.1111/j.1708-8305.1999.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Valitutto MT, Aung O, Tun KYN, Vodzak ME, Zimmerman D, Yu JH, Win YT, Maw MT, Thein WZ, Win HH, Dhanota J, Ontiveros V, Smith B, Tremeau-Brevard A, Goldstein T, Johnson CK, Murray S, Mazet J. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15(4):e0230802. doi: 10.1371/journal.pone.0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke MCC, Thompson GR, Galgiani JN, Barker BM. The rise of Coccidioides: forces against the dust devil unleashed. Front Immunol. 2019;11(10):2188. doi: 10.3389/fimmu.2019.02188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandžurová A, Piliš V, Bačkor P, Júdová J, Javorský P, Faix ŠPP, Pristaš P. Microflora of the bat guano. Folia Vet. 2012;56(2):68–69. [Google Scholar]
- Vandžurová A, Bačkor P, Javorský P, Pristaš P. Staphylococcus nepalensis in the guano of bats (Mammalia) Vet Microbiol. 2013;164(1-2):116–121. doi: 10.1016/j.vetmic.2013.01.043. [DOI] [PubMed] [Google Scholar]
- Vaughan MJS. Fungal community diversity and structure from cave mineral surfaces and bat guano in Kartchner caverns, Arizona. Arizona: Dissertation, University of Arizona; 2012. [Google Scholar]
- Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT. Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg Infect Dis. 2014;20(6):960. doi: 10.3201/eid2006.130956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengust M, Knapic T, Weese JS. The fecal bacterial microbiota of bats; Slovenia. PLoS One. 2018;13(5):e0196728. doi: 10.1371/journal.pone.0196728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JSM, Chen H, Guan Y. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007;81(8):4012–4020. doi: 10.1128/JVI.01135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Sintunawa C, Kaewpom T, Khongnomnan K, Olival KJ, Epstein JH, Rodpan A, Sangsri P, Intarut N, Chindamporn A, Suksawa K, Hemachudha T. Group C betacoronavirus in bat guano fertilizer, Thailand. Emerg Infect Dis. 2013;19(8):1349–1351. doi: 10.3201/eid1908.130119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells K, Clark NJ. Host specificity in variable environments. Trends Parasitol. 2019;35(6):452–465. doi: 10.1016/j.pt.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Wheat J. Endemic mycoses in AIDS: a clinical review. Clin Microbiol Rev. 1995;8(1):146–159. doi: 10.1128/CMR.8.1.146-159.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) Zoonoses. https://www.who.int/news-room/fact-sheets/detail/zoonoses.
- Wibbelt G, Speck S, Field H. Methods for assessing diseases in bats. In: Kunz TH, Parsons S, editors. Ecological and behavioral methods for the study of bats. Baltimore: Johns Hopkins University Press; 2009. pp. 775–794. [Google Scholar]
- Winter AS, Kimble JC, Young JM, Buecher DC, Valdez EW, Hathaway JJ, Porras-Alfaro A, Read KJ, Northup DE. External bacterial diversity on bats in the southwestern United States: changes in bacterial community structure above and below ground. PeerJ Prepr. 2016;4:e2493v1. doi: 10.7287/peerj.preprints.2493v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseschart A, Pootanakit K. Metagenomic-based approach to a comprehensive understanding of cave microbial diversity. In: Bhatt P, de Mandal S, editors. Recent advancements in microbial diversity. Cambridge: Academic Press; 2020. pp. 561–586. [Google Scholar]
- Wolkers-Rooijackers J, Rebmann K, Bosch T, Hazeleger W. Fecal bacterial communities in insectivorous bats from the Netherlands and their role as a possible vector for foodborne diseases. Acta Chiropt. 2019;20:475. doi: 10.3161/15081109acc2018.20.2.017. [DOI] [Google Scholar]
- Wynbrandt J, Crouser E. Transformation of pulmonary histoplasmosis to sarcoidosis: a case report. Respir Med. 2007;101:863–864. doi: 10.1016/j.rmed.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Zana B, Kemenesi G, Urbán P, Földes F, Görföl T, Estók P, Boldogh S, Kurucz K, Jakab F. Metagenomic analysis of bat guano samples revealed the presence of viruses potentially carried by insects, among others by Apis mellifera in Hungary. Acta Vet Hung. 2018;66:151–161. doi: 10.1556/004.2018.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.