Abstract
Background
EGFR is a major therapeutic target for colorectal cancer (CRC). Currently, extended RAS/RAF testing identifies only non-responders to EGFR inhibitors (EGFRi). We aimed to develop a mutation signature that further refines drug-sensitive subpopulations to improve EGFRi outcomes.
Methods
A pre-specified, 203-gene expression signature score measuring cetuximab sensitivity (CTX-S) was validated with two independent clinical trial datasets of cetuximab-treated CRC patients (n=44 and n=80) as well as an in vitro dataset of 147 cell lines. The CTX-S score was then used to decipher mutated genes that predict EGFRi sensitivity. The predictive value of the identified mutation signature was further validated by additional independent datasets.
Results
Here we report the discovery of a 2-gene (APC+TP53) mutation signature that was useful in identifying EGFRi-sensitive CRC subpopulations. Mutant APC+TP53 tumors were more predominant in left- vs right-sided CRCs (52% vs. 21%, p=0.0004), in MSS vs MSI cases (47% vs. 2%, p<0.0001), and in the consensus molecular subtype 2 vs. others (75% vs. 37%, p<0.0001). Moreover, mutant APC+TP53 tumors had favorable outcomes in two cetuximab-treated PDX datasets (p=0.0277, n=52; p=0.0008, n=98).
Conclusion
Our findings suggest that the APC and TP53 combination mutation may account for the laterality of EGFRi sensitivity and provide a rationale for refining treated populations. The results also suggest addition of APC+TP53 sequencing to extended RAS/RAF testing that may directly increase the response rates of EGFRi therapy in selected patients.
Impact
These findings, if further validate through clinical trials, could also expand the utility of EGFRi therapies that are currently underutilized.
Keywords: colorectal cancer, APC, TP53, mutations, cetuximab
INTRODUCTION
Two well-characterized EGFR inhibitors (EGFRi) (cetuximab, panitumumab) are FDA approved as first and second line targeted-therapies for metastatic colorectal cancer (CRC)(1–8). Despite approval, utilization has been modest, primarily because of drug restriction to the wild-type RAS subpopulation. Early CRC clinical trial studies involving cetuximab/panitumumab, either as monotherapies or as combination therapies, reported that a statistically significant drug response was generally observed in WT KRAS patients---but not in MUT KRAS patients(5, 8). On the other hand---despite selection---about half of patients with a WT KRAS still fail to respond to EGFRi treatments(9, 10), suggesting that additional genes, beyond KRAS, may negatively contribute to EGFRi response. Recently, mutations in NRAS and BRAF were reported to account for EGFRi therapy resistance in some WT KRAS CRCs(1–3). More recently, left-sided CRCs have been reported to be more favorably associated with response to cetuximab/panitumumab than right-sided tumors, as indicated by increased response rate (RR), better progression free survival (PFS) and/or overall survival (OS) (6, 11–13). A molecular basis of the laterality of anti-EGFR sensitivity, however, is still poorly understood.
We recently developed a new, robust molecular classification of CRC to help dissect this heterogeneous disease into 5 molecular subpopulations in order to improve treatment strategies(14, 15). This classification complements the recently reported consensus molecular subtypes (CMS) of colorectal cancer that were coalesced from six independent (gene expression) CRC classification systems(16). We performed an integrated analysis targeted gene sequencing for 1321 cancer-related genes, global gene expression, and MSI analyses across a large cohort of human CRC (n = 468). Among a number of mutated genes identified, striking pairwise, statistically significant, correlations were observed between APC, TP53, KRAS and BRAF that ultimately suggested a prognostic role for APC(15). Based on these results, we hypothesized there might also be a predictive role for APC and other associated genes.
Given the paucity of available clinical trial tissue samples with EGFRi exposure, we elected to use a cetuximab sensitivity (CTX-S) gene expression score as a surrogate for cetuximab response data in our CRC cohort, TCGA and other published data. This approach allowed us to develop a 2-gene “mutation signature” that is strongly-correlated with the CTX-S score and can be rapidly translated to the clinic.
MATERIALS AND METHODS
Datasets of Patient Samples, Cell Lines, and PDX Models
We previously analyzed 468 stages I-IV colorectal tumors, with global gene expression data from the surgical specimen, MSI status, and targeted gene sequencing of 1321 cancer-related genes(14, 15). A cohort of the 468 colorectal adenocarcinoma patients (including 367 primary lesions from stage 1–4 patients and 101 metastatic lesions) was accrued between October 2006 and September 2010, and written informed consent was obtained from participating patients as part of the Total Cancer Care® (TCC) project (17). The study was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule) and under the approval of the University of South Florida institutional review board (17). Primary and metastatic samples were both included based on our previous work demonstrating a high degree of mutation overlap between matched primary/metastatic samples(18). Here we further used this large, well-curated clinico-genomics/expression database of CRC patient samples to carry out mutation ranking analysis by the CTX-S score and other statistical analyses. We identified seven additional independent datasets, from Merck and public resources including Gene Expression Omnibus (GEO) and NCI Genomic Data Commons (GDC), for various validation and correlation analyses. These included WT KRAS CRC samples (n=44) selected from the control arm (cetuximab + irinotecan) of a Merck prospective clinical trial (MK0646)(19), a BMS trial cetuximab-treated CRC patient samples (n=80, Khambata-Ford et al.(4)), in vitro cetuximab-treated CRC cell lines (n=147, Medico et al.(20)), TCGA CRC patient samples (n=624 including 221 DNA-sequenced samples from TCGA(21)), and an additional set of Stages I-IV CRC patients samples (n=566, Marisa et al.(22)), as well as cetuximab-treated CRC PDX models (n=52, Julien et al.(23) and n=98, Bertotti et al.(24)). A summary of all eight datasets is given in Table 1, and detailed data description is given in Supplementary Methods and Tables S1–8.
Table 1.
List of eight colorectal cancer datasets used
| Datasets | Samples | N of samples | Stage | Cetuximab response | Prognosis | Gene Expression | MSI status | DNA sequencing | Driver Mutations | Primary tumor location |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. MK0646-PN004 CRCs(19) | patient tumors | 44 | IV | PFS, OS OR | Affymetrix | targeted-sequencing KRAS | KRAS | |||
| 2. Khambata-Ford et al. (2007) J Clin Onco (4) | patient tumors | 80 | IV | PFS, CR/PR, SD, PD | Affymetrix | targeted-sequencing EGFR, KRAS, BRAF | KRAS | |||
| 3. Medico et al. (2015) Nat Commun (20) | cell lines | 147 | In vitro growth inhibition | Affymetrix | MSI/MSS | targeted-sequencing KRAS, NRAS, BRAF | KRAS, NRAS, BRAF | |||
| 4. Moffitt CRCs---Schell et al. (2016) Nat Commun (15); Schell et al. (2016) Clin Cancer Res (14) | patient tumors | 468 | I, II, III, IV | OS | Affymetrix | MSI/MSS | targeted-sequencing (1321 genes) | APC, TP53, KRAS, NRAS, BRAF | Left/Right | |
| 5. TCGA CRCs--- TCGA Nature (2012) (21); NCI GDC | patient tumors | 624 | I, II, III, IV | RNAseq | MSI/MSS | whole exome sequencing on 221 tumors | APC, TP53, KRAS, NRAS, BRAF | Left/Right | ||
| 6. Marisa et al. (2013) PLoS Med (22) | patient tumors | 566 | I, II, III, IV | RFS | Affymetrix | targeted-sequencing KRAS, BRAF, TP53 | TP53, KRAS, BRAF | |||
| 7. Julien et al. (2012) Clin Cancer Res (23) | PDX models | 52 | I, II, III, IV | PR (+++), SD (++), PD (+, −) | MSI/MSS | targeted-sequencing 13 genes | APC, TP53, KRAS, BRAF | |||
| 8. Bettotti et al. (2015) Nature (24) | PDX models | 98 | I, II, III, IV | PR, SD, PD | whole exome sequencing | APC, TP53, KRAS, NRAS, BRAF |
Note: PFS---progression free survival; OS--- overall survival; OR--- objective response; CR---complete response; PR---partial response; SD---stable disease; PD---progressed disease; MSI (i.e. MSI-high)---microsatellite instable tumors; MSS---microsatellite stable tumors. Also see Supplementary Tables S1–S8 for detailed data description of individual datasets, respectively.
Expression Signatures (see Supplementary Table S9 for gene lists)
1. The CTX-S score: A pre-specified gene expression signature score that measures cetuximab sensitivity was initially constructed based on gene expression values from >800 cancer associated genes, each assessed in a set of 44 WT KRAS colon tumor samples from patients treated with cetuximab monotherapy. 203 genes with p-value <0.05 by PFS Cox Regression analysis were identified that included 94 UP and 109 DOWN expressed genes based on their association with response or resistance to cetuximab, respectively. Cetuximab sensitivity score (CTX-S) was defined as the average expression of the genes in the UP arm minus the average expression of genes in the DOWN arm. CTX-S was pre-specified for subsequent analysis of the validation sets. The signature score derivation and validation is summarized in Supplementary Fig. S1 and overall methodology used is similar to as we reported in previous studies(14, 25, 26). 2. The 18-gene RAS pathway score. This score was developed to measure the MEK functional output in association with RAS pathway activation(27). 3. The 64-gene Wnt pathway score. We previously adopted a set of 64 “consensus” β-catenin (upregulated) genes from a recent study of Herbst et al.(28) to assess differential activation of Wnt pathway in APC subgroups of Moffitt CRCs(15). The 64-gene Wnt pathway scores were calculated from the arithmetic mean expression of the 64 genes. 4. The 24-gene APC mutation-specific Wnt pathway score. We further selected 24 genes (out of the 64 genes) whose expression was significantly higher in APC-mutated tumors than those with wild-type APC (p<0.05 for two-tailed Welch t test) (see Supplementary Fig. S2A). The arithmetic mean expression of selected 24 genes is designated as the APC mutation-specific Wnt pathway score, which was validated by TCGA CRCs (n=221) (Supplementary Fig. S2B).
Statistical Methods
The statistical approaches used include (1) Survival Analysis, Correlation Analysis, and the t Test; (2) Mutation Ranking Analysis of Moffitt 468 CRCs; (3) Cochran-Mantel-Haenszel (CMH) Test, Barnard Test, and distribution analysis; (4) CMS classification. See Supplementary Methods for detailed description.
The statistical tests used in the article were given unadjusted p values for multiple testing, with an α = 0.05 chosen as the significance level, except for the mutation ranking analyses which use the Benjamini and Hochberg false discovery rate method(29). In addition, for the Welch t test in comparison among 7 or 5 MSI/MSS subgroups, those unadjusted p values remaining significant after adjustments for multi-comparisons by Holm-Bonferroni method(30) were highlighted by a maroon color. All tests were two-sided unless noted otherwise.
RESULTS
Development and validation of the CTX-S score
The CTX-S signature score was validated using two independent test sets of cetuximab-treated patients and a test set of cetuximab-treated CRC cell lines, as summarized in Supplementary Fig. S1. A detailed description of the results is given below:
Validation Set 1 (in vivo)
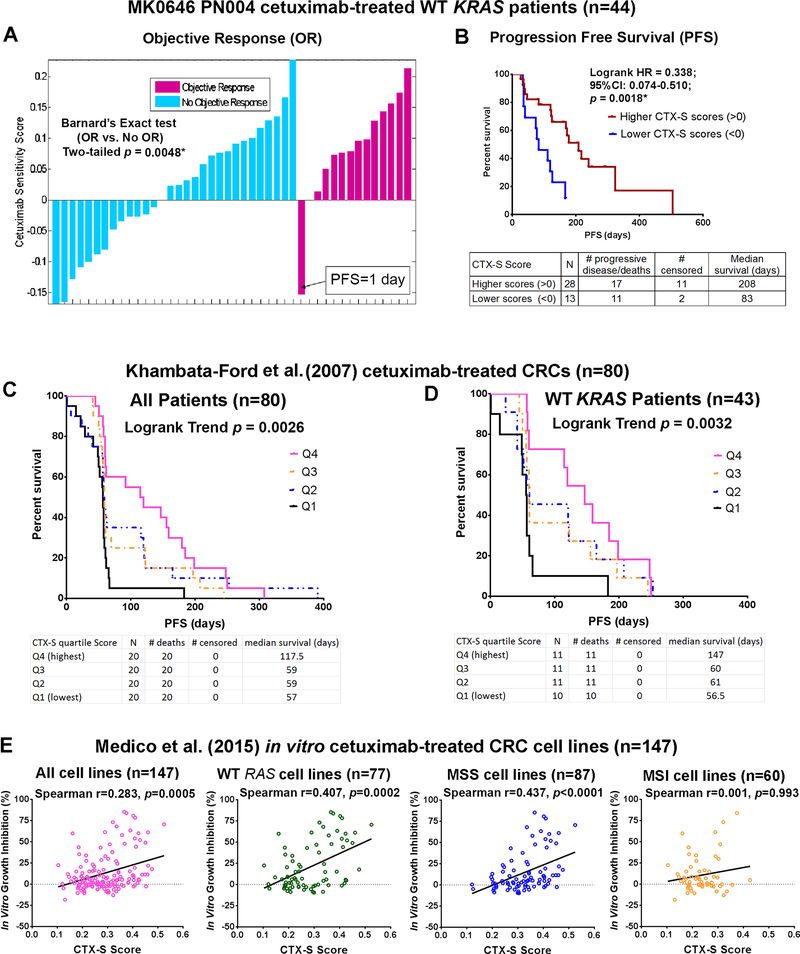
The CTX-S score was first validated using 44 CRC control arm samples and data from a Merck prospective clinical trial (MK0646), a randomized phase II/III study of dalotuzumab (IGF-1R inhibitor) in combination with cetuximab and irinotecan in chemo-refractory, KRAS wild-type, metastatic CRC(19). In the control arm “C” of MK0646, high IGF-1 expression was shown to be significantly associated with lower response rates to cetuximab + irinotecan and IGF-1 was considered to be a promising biomarker for differential response to anti-IGF1R therapies as well as anti-EGFR therapies(19). Here, we used the MK0646 control arm samples to further test whether the CTX-S signature score could predict positive outcomes to cetuximab + irinotecan therapy. Barnard’s exact test revealed that the CTX-S score was significantly associated with objective responses (OR) (vs. no objective response, p=0.0048; see Fig. 1A and Supplementary Table S10A). Moreover, Kaplan-Meier (KM) survival analysis showed that the CTX-S score was significantly associated with longer progression-free survival (PFS) (208 vs. 83 days, HR =0.34, 95% CI: 0.07–0.51, p=0.0018) (Fig. 1B) and longer overall survival (OS) (503 vs. 287 days, HR=0.30, 95% CI: 0.05–0.58, p=0.0052) (Supplementary Fig. S3).
Fig. 1. Validation analysis of the cetuximab sensitivity (CTX-S) signature score in two independent sets of CRC patient samples derived from clinical trials and one set of in vitro cetuximab-treated CRC cell lines.
A. A waterfall plot of objective response (OR) vs. adjusted CTX-S score in MK0646 PN004 wild-type (WT) KRAS CRCs (n=44) (see Supplementary Table S1 for detailed data description). The p value is for Barnard’s exact test (OR: No OR) (see Supplementary Table S10A). B. Kaplan Meier (KM) survival (PFS) analysis by higher (>0) vs. lower (<0) CTX-S scores was also performed the PN004 CRCs (n=41). Note: for A, B, of 44 CRCs, one sample with PFS of 1 day and two samples with CTX-S scores near 0.00 as shown in Fig. 1A were excluded from Barnard test and KM analysis. Also see Supplementary Fig. S3 for similar KM analysis on OS. C. KM survival (PFS) analysis by the CTX-S quartile scores was performed in Khambata-Ford et al. (2007) cetuximab-treated CRCs (n=80)(4). D. KM PFS analysis in Khambata-Ford WT KRAS patients (n=43). Also see Supplementary Table S10B,C for the Cochran-Mantel-Haenszel test showing the significant association of the CTX-S score with improved response (CR/PR and SD). E. Spearman correlation analysis of the CTX-S score with in vitro growth inhibition (%) by 10 μg/ml of cetuximab in Medico et al. (2015) cetuximab treated CRC cell lines (20). The analysis was performed in all cell lines (n=147) and WT RAS (KRAS/NRAS) cell lines (n=77) as well as MSS (n=87) and MSI cell lines (n=60), respectively (see detailed data description in Supplementary Table S3A). Note that the results for the other doses of cetuximab (1, 25, 50 and 100μg/ml) were similar, as 10 μg/ml had a 0.945 or higher (Pearson) correlation with these doses for in vitro growth inhibition (Supplementary Table S3B).
Validation Set 2 (in vivo)
The CTX-S score was subsequently validated using a second independent, well-characterized, dataset of 80 metastatic CRC patients prospectively treated with cetuximab monotherapy from a BMS clinical trial(4). This dataset was initially used to identify EREG and AREG as predictive markers whose high expression was significantly associated with longer PFS in cetuximab-treated patients(4), and later used in a variety of other gene expression classification and validation analyses associated with EGFRi treatment prediction(31–33). Notably, EREG is a member of the CTX-S score “UP genes”, whereas AREG is not included in the gene list of the CTX-S (see Supplementary Table S9). Five of six CR (complete response) + PR (partial response) samples and a majority of SD (stable disease) samples (13/19) had higher CTX-S scores (above the median) (see Supplementary Fig. S4), supporting the CTX-S score as a reasonable measure of cetuximab disease control response (DCR). This notion was supported by Cochran-Mantel-Haenszel (CMH) testing on cetuximab response vs. CTX-S quartile scores (q4 (highest), q3, q2 and q1 (lowest)) (Supplementary Table S10B,C). Moreover, KM analysis found that higher scores were significantly associated with better PFS in patients, regardless of KRAS mutation status (logrank trend test, p=0.0026, n=80) (Fig. 1C) as well as in patients with only WT KRAS (p=0.0320, n=43) (Fig. 1D).
Validation Set 3 (in vitro)
Recently, 147 CRC cell lines with heterogeneous genetic backgrounds were analyzed by Medico et al.(20) for KRAS, NRAS, and BRAF genotypes, MSI (MSI-high) status, as well as global gene expression and in vitro cetuximab sensitivity. Our further analysis of these cell lines showed that the CTX-S score was significantly correlated with in vitro cetuximab sensitivity (Fig. 1E), supporting the validity of the CTX-S score.
The CTX-S score was not prognostic
We have previously developed various gene expression signatures that were prognostic for CRC outcomes(14, 26). To assess prognostic potential, a KM analysis of the CTX-S score by quartiles was performed similarly as we did for the ΔPC1.EMT score in the same set of 468 CRCs(14). Results showed that unlike ΔPC1.EMT that predicted poor OS(14), the CTX-S score was not prognostic, as shown in either all patients (logrank trend test, p=0.969, n=468), or in WT RAS patients (p=0.273, n=264) (Supplementary Fig. S5A,B). This result was confirmed using a second large, independent, set of stage I-IV CRCs (n=557) reported by Marisa et al. (2013) for a classification analysis of CRC in association with prognosis (relapse-free survival, RFS)(22) (Supplementary Fig. S6A,B).
Correlation of the CTX-S score with EREG/AREG, RAS and Wnt signatures
Spearman correlation analysis showed that while the CTX-S score was positively correlated with EREG or AREG expression in all four patient tumor datasets (p<0.0001), it was negatively correlated with 18-gene RAS signature score in the three largest datasets (p<0.0001) (Table 2A). Notably, an 18-gene RAS pathway gene expression signature score was previously developed to measure RAS/RAF/MEK pathway activation (27). We recently adapted this signature from use in fresh frozen CRC samples to more clinically-available, FFPE tissues(34) as a means to identify cetuximab non-responders.
Table 2. A.
Spearman Correlations with the cetuximab sensitivity (CTX-S) score
| All CRC tumors/cell lines (regardless of RAS status) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spearman Corr. with CTX-S score | Moffitt CRCs (n=458)* | TCGA CRCs (n=624) | Marisa et al (2013) CRCs (n=566) | Khambata-Ford et al (2007) CRCs (n=80) | Medico et al (2015) CRC cell lines (n=147) | |||||
| Gene expression/Signature scores | r | p value | r | p value | r | p value | r | p value | r | p value |
| EREG | 0.562 | <0.0001 | 0.592 | <0.0001 | 0.655 | <0.0001 | 0.711 | <0.0001 | 0.266 | 0.001 |
| AREG | 0.463 | <0.0001 | 0.469 | <0.0001 | 0.542 | <0.0001 | 0.624 | <0.0001 | −0.173 | 0.036 |
| 18-gene RAS pathway score | −0.361 | <0.0001 | −0.209 | <0.0001 | −0.491 | <0.0001 | 0.123 | 0.277 | −0.106 | 0.201 |
| 64-gene Wnt pathway score | 0.069 | 0.140 | 0.023 | 0.573 | 0.036 | 0.655 | 0.276 | 0.013 | 0.311 | 0.0001 |
| 24-gene Wnt pathway score | 0.553 | <0.0001 | 0.457 | <0.0001 | 0.558 | <0.0001 | 0.666 | <0.0001 | 0.488 | <0.0001 |
Note
Of Moffitt 468 CRCs, 10 samples lacking appropriate gene probe values for some signature scores were excluded from correlation analysis.
The Wnt pathway is another major dysregulated signaling pathway in CRC, ~70% of which have one or two APC truncating mutations(15, 21, 35). We found a strong correlation with an APC mutation-dependent 24-gene Wnt pathway score across all five datasets tested. In addition, we found that the APC mutation-dependent score was significantly correlated with the in vitro cetuximab inhibitory effect in 147 cell lines (Supplementary Fig. S7).
These data led us to further investigate a potential association of the CTX-S score with APC mutations using Moffitt CRCs. We found that CTX-S scores were significantly higher (p<0.0001 for two-tailed Welch t test) in mutant APC (n=312, 67%) than wild-type APC tumors (n=156, 33%) (Supplementary Fig. S8A). Similar results were obtained when tumors were further divided into WT and MUT RAS (KRAS/NRAS) or MSI and MSS (Supplementary Fig. S8B,C). It was also true when the effect of MUT APC was examined in Moffitt Stage IV patients (n=110) (Supplementary Fig. S8D–F) and in TCGA CRCs (Supplementary Fig. S9A).
Identification of APC and TP53 as the highest-ranked CTX-S associated mutated genes
The finding of a potentially significant role of mutant APC in predicting cetuximab sensitivity prompted us examine additional genes. We applied an analytical approach fusing RNA-based gene expression signatures with DNA mutations to rank mutated genes in Moffitt CRCs (see Methods). The top 20 ranked mutated genes are shown in Table 2B, and a full rank list of mutated genes is given in Supplementary Table S11. While MSI status, BRAF (i.e. BRAF(V600E)) and TGFBR2 were the most negatively correlated with the CTX-S score, TP53 and APC were the highest ranked mutated genes that were strongly (positively) correlated with the scores (Table 2B, left). Here we treated the MSI status as “a mutated gene”. Notably, many negatively correlated genes, such as BRAF and TGFBR2, were strongly associated with MSI(15, 21, 35). After MSI tumors were removed, all the negatively correlated mutated genes had statistically non-significant, adjusted P values while TP53 and APC remained the only statistically significant positively correlated genes (Table 2B, right and Supplementary Table S12). This is supported by an analysis showing striking trends by multiple APC genotypes and TP53 mutations (Supplementary Table S13).
Table 2. B.
Ranking of cetuximab sensitivity score-associated mutated genes using Moffitt CRCs
| All Patients (n=468) | MSS Patients (n=407) | |||||||||||
| gene | p value | mutation | Asso Dir (+/−) | Adjusted p | gene | p value | mutation | Asso Dir (+/−) | Adjusted p | |||
| N | PCT | N | PCT | |||||||||
| TP53 | 1.89E-24 | 277 | 59.2% | + | 5.8E-22 | TP53 | 4.64E-22 | 261 | 64.1% | + | 4.09E-19 | |
| MSI_high* | 6.74E-24 | 61 | 13.0% | - | 2.06E-21 | APC | 8.55E-12 | 296 | 72.7% | + | 7.52E-09 | |
| TGFBR2 | 1.93E-23 | 62 | 13.2% | - | 5.9E-21 | BRAF | 0.000709 | 18 | 4.4% | - | 0.6227 | |
| APC | 7.96E-21 | 312 | 66.7% | + | 2.44E-18 | KRAS | 0.001858 | 177 | 43.5% | - | 0.998439 | |
| BRAF | 5.67E-17 | 53 | 11.3% | - | 1.74E-14 | SMAD3 | 0.00204 | 14 | 3.4% | - | 0.998439 | |
| CELSR1 | 1.56E-06 | 72 | 15.4% | - | 0.000478 | BARD1 | 0.002188 | 7 | 1.7% | - | 0.998439 | |
| HDLBP | 4.03E-06 | 23 | 4.9% | - | 0.001234 | CTNNB1 | 0.003312 | 13 | 3.2% | - | 0.998439 | |
| ITGB4 | 6.05E-06 | 45 | 9.6% | - | 0.001852 | BRCA1 | 0.005571 | 25 | 6.1% | - | 0.998439 | |
| PML | 6.3E-06 | 22 | 4.7% | - | 0.001926 | IDH1 | 0.006636 | 5 | 1.2% | - | 0.998439 | |
| HSPA2 | 1.23E-05 | 14 | 3.0% | - | 0.003749 | RAD18 | 0.006847 | 6 | 1.5% | - | 0.998439 | |
| MLL2 | 2.29E-05 | 84 | 17.9% | - | 0.006995 | SMAD2 | 0.006852 | 12 | 2.9% | - | 0.998439 | |
| MICAL1 | 2.36E-05 | 26 | 5.6% | - | 0.007217 | TLR7 | 0.011293 | 5 | 1.2% | - | 0.998439 | |
| PTPRS | 2.58E-05 | 47 | 10.0% | - | 0.007885 | HECW1 | 0.012886 | 24 | 5.9% | + | 0.998439 | |
| MAP3K9 | 2.72E-05 | 21 | 4.5% | - | 0.008312 | TRIB3 | 0.013672 | 8 | 2.0% | - | 0.998439 | |
| ITPR1 | 4.23E-05 | 34 | 7.3% | - | 0.012936 | NRP2 | 0.016995 | 15 | 3.7% | - | 0.998439 | |
| HDAC4 | 5.35E-05 | 20 | 4.3% | - | 0.016377 | TAF15 | 0.02442 | 5 | 1.2% | - | 0.998439 | |
| DOT1L | 5.87E-05 | 24 | 5.1% | - | 0.017956 | GRM1 | 0.02554 | 18 | 4.4% | - | 0.998439 | |
| RPS6KA2 | 5.95E-05 | 12 | 2.6% | - | 0.018212 | AFF4 | 0.026074 | 7 | 1.7% | - | 0.998439 | |
| CHD5 | 5.96E-05 | 23 | 4.9% | - | 0.018239 | FLT4 | 0.027436 | 18 | 4.4% | + | 0.998439 | |
| MLL4 | 7.03E-05 | 54 | 11.5% | - | 0.021519 | EGFR | 0.0279 | 11 | 2.7% | + | 0.998439 | |
Note: p value from normal scores test for comparing CTX-S scores of mutated and wild-type tumors for the given gene; N (PCT) − the number (percentage) of mutated tumors; APC − APC truncated mutation; BRAF − BRAF (V600E)
patients with MSI-high status. “Asso Dir” − the directionality of the association (+/ -); Adjusted p value was calculated using the Hochberg and Benjamini method(29). Here top 20 genes are listed according to their adjusted p values. See a full rank list of 1321 cancer-associated genes in Supplementary Tables S11 and S12.
APC+TP53 doubly-mutated (AP) tumors had the highest CTX-S scores
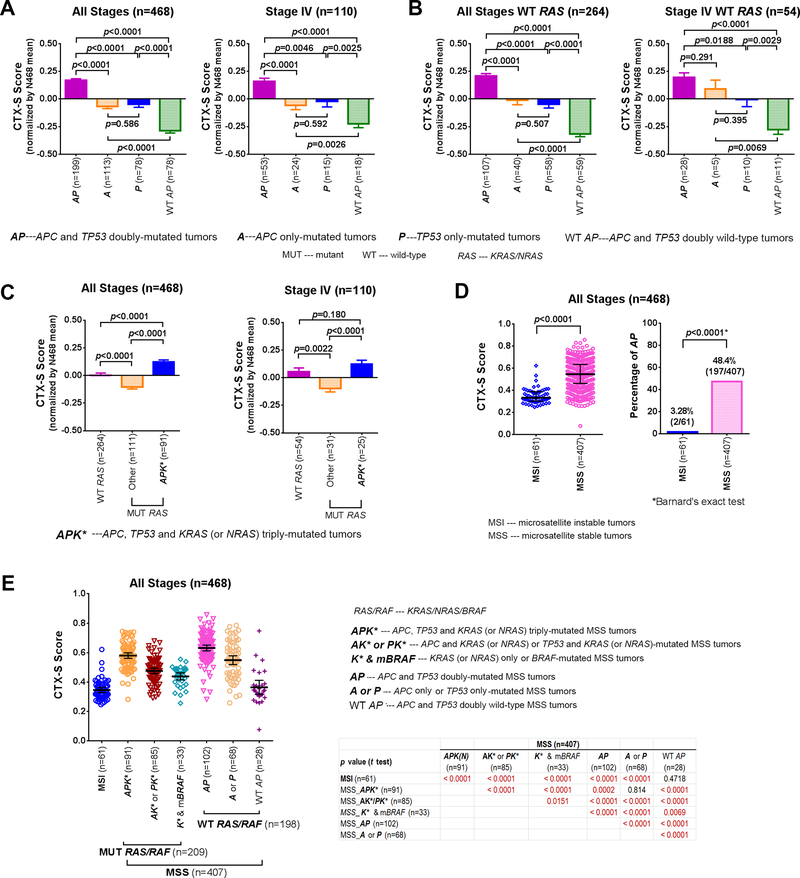
Since APC and TP53 were frequently co-mutated in CRC tumors(15, 21, 35), we examined whether mutant APC+TP53 together might cooperatively predict cetuximab sensitivity. For this purpose, Moffitt CRCs were divided into four subgroups: (1) MUT APC + MUT TP53 (AP); (2) MUT APC + WT TP53 (A); (3) WT APC + MUT TP53 (P); and (4) WT APC + WT TP53 (WT AP). Analysis indicates that doubly-mutated AP tumors had significantly higher CTX-S scores than all other three subgroups including the A and P mutant groups (p<0.0001 for two-tailed Welch t test) (Fig. 2A left panel). Notably, A and P mutant groups were not significantly different from each other, but had significantly higher scores than WT AP tumors (p<0.0001). The same pattern was observed in the 110 Stage IV tumors (Fig. 2A right panel). Similar results were obtained in Moffitt WT RAS tumors for all stages (n=264), and for Stage IV (n=54) patients (Fig. 2B). The significant association was validated using the TCGA CRC dataset (Supplementary Fig. S9B).
Fig. 2. The “MUT APC + MUT TP53” (AP) doubly-mutated tumors had significantly higher cetuximab sensitivity (CTX-S) scores than other tumors with “MUT APC only” (A), “MUT TP53 only” (P) or WT APC + WT TP53” (WT AP) in Moffitt CRCs.
A. Comparison for all stage patients (n=468) and Stage IV patients (n=110), respectively, regardless of RAS mutation status; B. Comparison for all stage WT RAS patients (n=266) and Stage IV WT RAS patients (n=55), respectively; C. Comparison of the CTX-S scores were also performed between WT RAS, APK* (MUT APC + MUT TP53 + MUT KRAS(or NRAS)) and other MUT RAS tumors without APK*. MUT – mutant; WT – wild-type; WT RAS – patients with wild-type KRAS/NRAS. The CTX-S scores were normalized by the mean of 468 CRCs, and bars represent Mean with standard errors (SEM). p values for two-tailed Welch t test are shown. D. The CTX-S scores were compared between MSI (i.e. MSI-high) and MSS tumors (left panel) in all patients (n=468). Bars represent Median with interquartile range. The comparison was also made for the percentage of “MUT APC + MUT TP53” (AP), with “*” representing two-tailed p values for Barnard’s exact test (right panel) (see Supplementary Table S14A). E. Comparison of CTX-S scores among 1 MSI and 6 MSS subgroups in Moffitt CRCs (n=468). Bars represent Median with interquartile range. Unadjusted p values for two-tailed Welch t test are shown and those remaining being significant after adjustments for multi-comparisons by Holm-Bonferroni method are highlighted by maroon color.
APK* triply-mutated tumors were associated with higher CTX-S scores than other RAS-mutated tumors
We found that the presence of MUT AP also had a striking positive effect on the CTX-S scores of KRAS/NRAS-mutated (K/N)-tumors in Moffitt CRCs. Note that because the great majority of RAS mutations in CRCs are KRAS mutations (~40%) and the frequency of NRAS mutations is much lower (~5%), for simplicity, we used K* to represent both KRAS and NRAS mutations (K/N). Although WT RAS tumors (n=264) had significantly higher CTX-S scores (p<0.0001 for two-tailed Welch t test) than MUT RAS tumors (n=111) in which the mutant APK* subpopulation was excluded, the APK* triply-mutated tumors (n=91) had even higher scores (p<0.0001) (Fig. 2C left panel).
When restricted to Moffitt Stage IV tumors (n=110), a striking difference remained for mutated K* tumors between the APK* and non-APK* subpopulations (Fig. 2C right panel). In the TCGA CRCs, the mutant APK* tumors and WT RAS tumors had no significant difference in CTX-S scores but both had significantly higher scores than other RAS-mutated tumors (Supplementary Fig. S9C). These data suggest that some APK* patients (heretofore not treated) may be sensitive to CTX treatment.
Comparison of the CTX-S scores among seven MSI and MSS subgroups
We next examined the association of APC+TP53 doubly-mutated (AP) tumors with the MSI/MSS status. In the 468 Moffitt CRCs, very strikingly, there were only two mutant AP tumors (1 APK and 1 APB) identified out of 61 MSI-H tumors (Fig. 3D). All 197 other AP tumors were MSS tumors. Similarly, the TCGA dataset had only one mutant AP tumor (APK) out of 28 MSI cases (Supplementary Fig. S9D). The statistical significances of these associations are given using Barnard’s exact test (Supplementary Table S14).
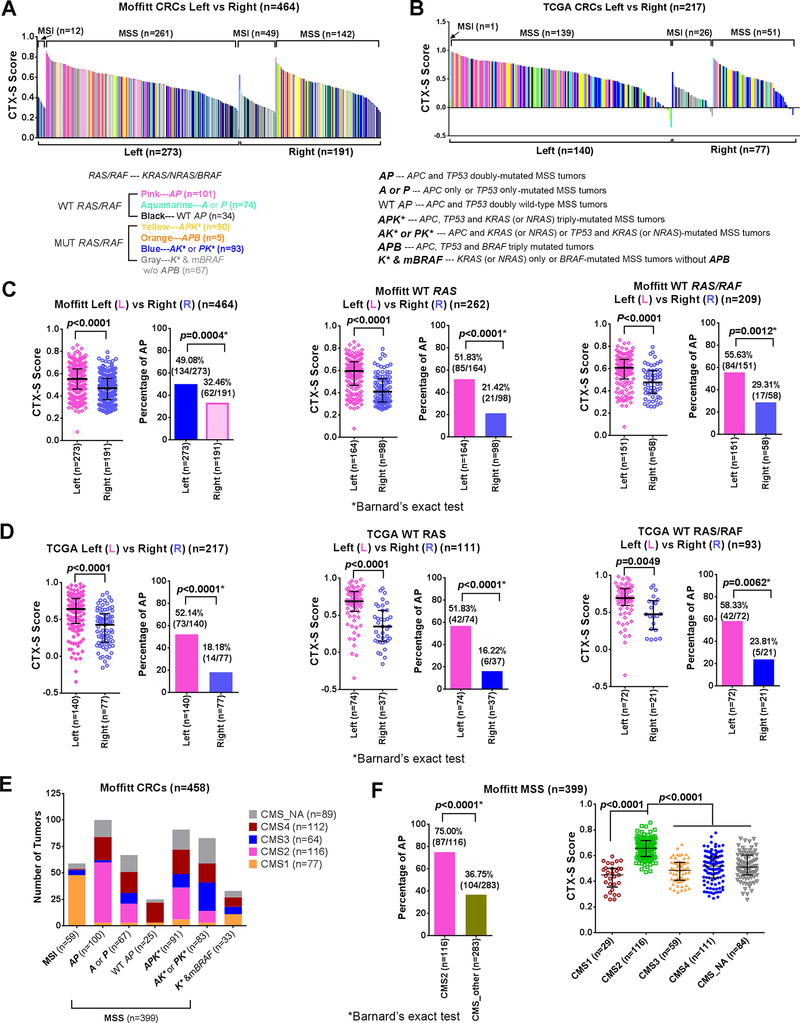
Fig. 3. Both the CTX-S score and the percentage of “MUT APC + MUT TP53” (AP) tumors were significantly higher in left-sided tumors in Moffitt and TCGA CRCs.
A diagram of the CTX-S scores (high to low) vs. 7 MUT/WT APC/TP53/RAS(KRAS/NRAS)/BRAF subgroups in MSI and MSS tumors was displayed for Moffitt (n=464) (A) or TCGA (n=217) (B) datasets. MUT – mutant; WT – wild-type. Note: Of 468 CRCs, 4 samples without tumor location information were excluded; Of 221 CRCs, 3 samples without tumor location information and 1 sample with MSI status information were excluded. Comparison of the CTX-S scores between “Left” and “Right” CRCs was made in all (available-data) patients, WT RAS patients, and WT RAS/RAF patients on Moffitt (C) and TCGA (D) CRCs, respectively. Note: WT RAS---patients with wild-type KRAS/NRAS; WT RAS/RAF---patients with wild-type KRAS/NRAS/BRAF; Bars represent Median with interquartile range. p values are for two-tailed Welch t test. The comparison was also made for the percentage of “MUT APC + MUT TP53” (AP) (Left vs. Right). * two-tailed p values for Barnard’s exact test (see Supplementary Table S15 and S16). The AP tumors were predominantly CMS2 subtype. E. The plot of the number of tumors of 1 MSI and 6 MSS subgroups (similarly as defined in Fig. 2E) vs. CMS1–4 and CMS_NA in Moffitt CRCs (n=458). Note: Of 468 CRCs, 10 samples without appropriate microarray data for the consensus molecular subtypes (CMS) classification analysis(16) were excluded as described previously(15). F. Left panel, the comparison of the percentage of “MUT APC + MUT TP53” (AP) (regardless of RAS mutation status) between CMS2 and other CMS subclasses in Moffitt MSS CRCs (n=399), with “*” representing a two-tailed p value of Barnard’s exact test. CMS_other includes CMS1, CMS3, CMS4 and CMS_NA (indeterminate). Right panel, comparison of the CTX-S scores among CMS1–4 and CMS_NA subclasses in MSS CRCs (n=399). Bars represent Median with interquartile range. p values are for two-tailed Welch t test.
Our data led us to postulate that AP mutations might cooperatively play a role in modulating cetuximab sensitivity in MSS tumors. To more specifically examine this, we divided the Moffitt CRCs into 1 MSI and 6 MSS subgroups. The groups can be statistically ordered with decreasing CTX-S scores as: AP > [APK*, A_P] > AK*_PK* > K*_MUT BRAF > [WT AP, MSI], where the groups inside the brackets are not significantly different from each other (Fig. 2E). Similar results were also obtained in 220 TCGA CRCs (Supplementary Fig. S10A) and in Moffitt Stage IV patients (n=110) (Supplementary Fig. S10B). Taken together, these data suggest the clinically relevant, provocative possibility that some APK* RAS-mutant MSS tumors might benefit from EGFRi therapies.
Mutant AP genotype association with left-sided, MSS tumors explains biology and left-sided CTX sensitivity
We assessed if the effect of mutant AP might be associated with the tumor sidedness. A plot of the CTX-S scores (high to low) of individual tumors with sidedness was produced to illustrate frequencies of the seven MSI and MSS subgroups (Left vs. Right) (see Fig. 3 (A) Moffitt and (B) TCGA, respectively). The CTX-S score was significantly higher in left-sided tumors than in right-sided tumors in all Moffitt patients (n=464, p<0.0001, two-tailed Welch t test), WT RAS only patients (n=262, p<0.0001), and WT RAS/RAF patients (n=209, p<0.0001) (Fig. 3C). Similar results were obtained for the TCGA dataset (Fig. 3D). The CMH test or Barnard’s exact test were performed on the frequencies of MSI and mutant AP by the sidedness (Left vs Right) in Moffitt and TCGA CRCs (Supplementary Tables S15 and S16). As expected, MSI was strongly associated with right-sided tumors (p<0.0001) in both datasets. Moreover, in close association with significantly differing CTX-S scores, the percentage of AP tumors was significantly higher in left-sided than right-sided cases in both datasets (Fig. 3C, D). We further performed the distribution analysis and CHM tests of the CTX-S score by quartiles for different A/P/K*/B subgroups (Left vs. Right) in the Moffitt464 patients (see Supplementary Table S17 A–E). The quartiles with higher scores (Q4 or Q3) are predicted to be more responsive to cetuximab. Briefly, this subset analysis shows while LEFT-sided AP tumors have 92% (77/84) of cases with cetuximab scores greater than the median, 82% (14/17) of RIGHT-sided AP tumors also exceed the median. Moreover, LEFT- and RIGHT-sided APK* tumors have cetuximab sensitivity scores greater than the median in 76% (37/49) and 73% (30/41) of cases, respectively. These data compare to all other tumors without AP mutations, where only 26% (72/276) of cases greater than the median, including only 41% (44/108) when restricted to KRAS/NRAS/BRAF wildtype tumors. Thus it appears that cetuximab sensitivity is driven by APC + TP53 mutations that may overcome sidedness and/or KRAS/NRAS mutation status to some extent. Consequently, some right and left sided tumors not currently considered for therapy may benefit. In addition, analysis using the two-tailed Welch t test showed no significant left vs. right CTX-S score difference in any particular subgroup in Moffitt dataset (Supplementary Fig. S11). These data suggest that the mutation frequencies, rather than the CTX-S scores, may be responsible for the sidedness effect.
Unlike the AP, single driver mutations were not consistently associated with the sidedness
Single driver mutations such as KRAS, NRAS and BRAF mutations have been used in predicting EGFRi responses(5, 6, 8). We performed Barnard’s test to examine if there is a potential association of the frequencies of four single drivers APC, TP53, KRAS/NRAS and BRAF with the sidedness in the Moffitt dataset (Supplementary Table S18) and the TCGA dataset (Supplementary Table S19). We observed a few, but scattering, significant associations that (i) MUT APC was significantly more left-sided in TCGA among all patients (n=217, p=0.0002), but not for MSS patients (n=190, p=0.23) and (ii) the same is also true for MUT TP53; (iii) MUT KRAS/NRAS was only significantly associated with right-sided tumors in Moffitt MSS patients; (iv) As expected, MUT BRAF (that was strongly associated with MSI tumors) was significantly associated with right-sided tumors in both Moffitt and TCGA all patients. However, when MSI cases were removed, the association of BRAF mutations became insignificant in MSS patients of both datasets, with very low counts of BRAF-mutated tumors.
APK* patients were more often distant metastatic whereas WT AP MSS tumors were associated with mucinous histotypes
We also examined the distribution of age, stages and histotypes as well as other clinic-pathological parameters in the MSI and 6 MSS subgroups of Moffitt CRCs (see Supplementary Table S20). MSI was significantly associated with Stage I-II (60%, p<0.01, individual chi-square (⌈2) contribution) and less associated with Stage IV (10%, p<0.05) and distant metastasis (11%, p<0.01). By contrast, the APK* patients were more often Stage III (45%, p<0.05) and distantly metastatic (49%, p<0.05). Interestingly, the WT AP subgroup (having low CTX-S scores) was significantly more associated with the mucinous histotype (32%, p<0.001), whereas the “cetuximab-sensitive” AP tumors appeared to be least mucinous (3%, p<0.05).
AP mutations were predominant in the CMS2 subtype linked to Wnt and MYC activity
The consensus molecular subtypes (CMS) were recently created from a comprehensive molecular analysis of thousands (n=4151) of human tumors to best define CRC(16). We performed CMS classification in Moffitt CRCs (see Supplementary Methods and Table S21). Results show that the majority of AP MSS tumors (57%) were the CMS2, whereas APK* tumors were also more associated with the CMS2 subtype than all other CMS classes, whereas most of the 59 MSI tumors were the CMS1 subtype (Fig. 3E). The strong association of the AP/APK* MSS tumors with the CMS2 was confirmed by Barnard’s exact test (Fig. 3F left panel). Furthermore, we found that the CTX-S scores of the CMS2 type were significantly higher (p<0.0001) than all other CMS classes (Fig. 3F right panel).
AP mutations associated with better outcomes in PDX models treated with cetuximab
To test our hypothesis, we identified two published cetuximab-treated CRC Patient-derived tumor xenograft (PDX) datasets (Julien et al., n=52 PDX models(23); Bertotti et al. n=98 PDX models(24)) which also had APC and TP53 mutation data (see Supplementary Tables S7 and S8). We performed the CMH trend test on cetuximab response by frequencies of A and/or P mutations in these two datasets. (i) For the Julien dataset, when the frequencies of AP mutations vs. A or P mutations vs. WT AP were compared between CR/PR/SD vs. PD tumors, the CMH trend P value was 0.0277 in 52 all PDX models or 0.0592 (marginally significant) in 25 WT RAS models (Table 3). (ii) For the larger Bertotti dataset, however, when compared between PR & SD vs. PD tumors, the CMH trend P value was 0.0008 in 98 PDX models that had WT KRAS, NRAS, BRAF and PIK3CA (Table 3). In addition, the CMH test was also performed when PR (n=23) and SD (n=50) cases were separated, with a p value of 0.0133 (Supplementary Table S22). Notably, the frequency of AP mutations had no difference between PR and SD cases (83% vs. 84%). These data again support a cooperative role of AP mutations in positively predicting cetuximab response (CR/PR and SD).
DISCUSSION
We have developed and validated a cetuximab sensitivity (CTX-S) signature score, using outcomes data from two independent, prospective clinical trials, as well as by an in vitro cetuximab-treated cell line dataset. The robustness of the CTX-S score is also supported by the findings that: (1) the score was not prognostic; (2) the score had a strong correlation with EREG and AREG, the predictive biomarkers of cetuximab response(4, 6, 36, 37); (3) the score was significantly associated with the CMS2 subtypes, which were recently reported to be associated with better cetuximab response(38, 39).
Due to the limited availability of clinical trial tissue samples with cetuximab exposure (especially mutant RAS patients), clinical outcomes, and deep molecular analysis beyond RAS/RAF testing, we used the CTX-S score as a “proxy” for clinical response. Using an integrated analytic approach we previously described(40), with the “lens” of gene expression, we identified a 2-gene mutation signature (APC+TP53)(AP) to predict cetuximab sensitivity, which was subsequently validated using two independent PDX datasets.
Identification of the 2-gene mutation signature (AP) has also provided new molecular insight into the recent observation that patients with MSI (often right-sided) almost uniformly lack AP mutations, and thus are resistant to EGFRi treatment, whereas patients with left-sided tumors more commonly harbor AP mutations, and are thus more responsive(6, 12, 13, 41, 42). Notably, it is the combination of AP mutations, rather than single driver mutations (APC, TP53, KRAS/NRAS, or BRAF), that was consistently and significantly associated with the sidedness. Currently, only left-sided CRCs with a WT KRAS/NRAS/BRAF status are considered eligible for EGFRi therapy(6). However, our further analysis suggests that mutations---rather than sidedness---may ultimately determine sensitivity to cetuximab. This is supported by the distribution analysis of the CTX-S score by quartiles for Left vs. Right A/P/K*/B subgroups in the Moffitt464 patients (see Supplementary Table S17 A–E). In either-sided tumors, AP and APK* were the most “sensitive” subgroups, whereas WT AP (that also had WT RAS/RAF and often had mucinous histotypes) was one of the highly “resistant” subgroups.
These data have two important clinical implications that may facilitate achieving the goal of precision cancer care. First, they could re-define the current clinical strategy guided by extended RAS/RAF testing and sidedness by excluding currently eligible, left-sided WT AP tumors, but including currently ineligible, right-sided MUT AP tumors. Second, the unexpected finding of a strong association of APK* with predicted CTX sensitivity suggests a potential therapeutic opportunity ---requiring further clinical validation--- to a subpopulation of previously excluded RAS patients with a poor prognosis who harbor APK* mutations(15). For example, a recent analysis of gene expression markers on CALGB 80203 (Alliance) trial data reported that high expression of CD73 was associated with longer PFS from cetuximab both in wild-type KRAS patients (chemo + cetuximab: HR (hazard ratio)=0.91 versus chemo only: HR=1.57, Pinteraction=0.026) as well as in mutant KRAS patients (chemo + cetuximab: HR=0.80 versus chemo only: HR=1.29, Pinteraction=0.025)(43), suggesting an intriguing and provocative hypothesis that some fraction of mutant KRAS patients may actually benefit from EGFRi therapies. Moreover, a number of clinical trial analyses have indicated that a substantial percentage (10–60%) of mutant KRAS patients treated with cetuximab/panitumumab have achieved stable disease (SD)(4, 36, 37, 44–51). Furthermore, analysis of the FIRE-3 (AIO KRK-0306) study showed that for the FOLFIRI + cetuximab treatment, compared to the wild-type KRAS exon 2 patients (n=297) who had 13 (4.4%) CR, 171 (57.6%) PR and 52 (17.5%) SD), RAS mutant patients (n=97) achieved 1 (1.0%) CR, 36 (37.1%) PR and significantly higher SD (31, 32.0%)(51). Finally, preclinical studies have indicated that EGFRi resistance can be reversed in some mutant KRAS CRC cell lines(52), and some KRAS-mutated CRC tumors may be decoupled from RAS pathway activation(25).
Our data suggest a hypothesis by which mutations in APC+TP53 might enable Wnt and p53 pathway “crosstalk” to transactivate the EGFR pathway, essentially addicting tumors to EGF ligands or enhance antibody-dependent cellular cytotoxicity (ADCC). Activation of the EGFR-PI3K-AKT signaling pathway has been clearly demonstrated in the APCMin/+ mouse by a mechanism involving upregulation of PGE2(53, 54). Similar to WNT, the p53 pathway has crosstalk with the EGFR pathway. Specifically, mutant TP53 has been shown to induce ERG1 transcription that is driven by p-ERK(55, 56). We found that the APC+TP53 double-mutated tumors were predominantly the CMS2 subtype that was associated with WNT and MYC activation and frequent mutations in either APC or TP53(16).
In conclusion, we have identified a 2-gene signature in identifying cetuximab-sensitive subpopulations. The signature may be useful in refining the appropriate subpopulation of patients for EGFRi treatment. Moreover, our findings provide a rationale for further prospective clinical studies that add sequencing of APC+TP53 to extended RAS/RAF testing to improve and expand the clinical utility of EGFRi---even potentially to some previously excluded patients harboring mutant RAS.
Supplementary Material
Table 4.
Cochran-Mantel-Haenszel (CMH) trend test on cetuximab (CTX) response by frequencies of MUT APC (A) and/or MUT TP53 (P) in cetuximab-treated CRC PDX models
| Julien all PDX models (n=52) | Julien WT KRAS (n=25) | Bertotti PDX models (n=98)* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Response | Mutation | Mutation | Mutation | ||||||||||
| Frequency Expected Cell Chi-Square Col PCT | AP | A or P | WT AP | Total | AP | A or P | WT AP | Total | AP | A or P | WT AP | Total | |
| CR/PR & SD | 10 6·92 1·37 50·00 | 8 9·35 0·19 29·63 | 0 1·73 1·73 0·00 | 18 | 7 4·8 1·00 70·00 | 5 6·72 0·44 35·71 | 0 0·48 0·48 0·00 | 12 | 61 54·38 0·81 83·56 | 6 8·94 0·97 50·00 | 6 9·68 1·40 46·15 | 73 | |
| PD | 10 13·08 0·72 50·00 | 19 17·65 0·10 70·37 | 5 3·27 0·92 100·00 | 34 | 3 5·2 0·93 30·00 | 9 7·28 0·41 64·29 | 1 0·52 0·44 100·00 | 13 | 12 18·62 2·36 16·44 | 6 3·06 2·82 50·00 | 7 3·32 4·09 53·85 | 25 | |
| Total | 20 | 27 | 5 | 52 | 10 | 14 | 1 | 25 | 73 | 12 | 13 | 98 | |
| CMH (nonzero corr.) | DF | 1 | 1 | 1 | |||||||||
| value | 4.85 | 3·56 | 11.23 | ||||||||||
| P | 0.0277 | 0·0592 | 0.0008 | ||||||||||
Note: MUT – mutant; WT – wild-type; A – APC mutation; P – TP53 mutation; AP – APC mutation + TP53 mutation; WT AP – WT APC + WT TP53; CR – complete response; PR – partial response; SD – stable disease; PD – progressed disease. For illustration purposes, the antitumor activities “+++”, “++” and “+”/“-” in Julien et al. PDX models were re-expressed as “CR/PR”, “SD” and “PD”, respectively, according to the pharmacological annotation as described (23). See Supplementary Tables S7 and S8 for detailed data description of Julien et al. (23) and Bertotti et al.(24) CTX-CRC PDX models, respectively.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grant U01CA157960 (to TJY). We thank Sabine Tejpar (UZ Leuven) for her collaboration with Merck (West Point, PA) in the initial development of the cetuximab sensitivity (CTX-S) signature score.
This work was supported by National Institutes of Health grant U01CA157960 (to TJY).
Abbreviations
- CRC
colorectal cancer
- EGFRi
EGFR inhibitor
- CTX
cetuximab
- PFS
progression free survival
- OS
overall survival
- RFS
relapse free survival
- CR
complete response
- PR
partial response
- SD
stable disease
- PD
progressed disease
- ORR
objective response rate
- PDX
Patient-derived tumor xenografts
For APC, TP53, KRAS, NRAS and BRAF mutations, single bolded letters exclusively refer to specific individual gene mutations: A = APC mutation; P = TP53 mutation; K = KRAS mutation; N = NRAS mutation; K* = K or N; B = BRAF mutation (e.g. AP = APC mutation + TP53 mutation; APK* = APC mutation + TP53 mutation + KRAS/NRAS mutation). WT AP = WT APC + WT TP53. When HUGO gene names are identified, the identified gene may not be exclusive (e.g. MUT APC = any tumor harboring APC mutations that could include KRAS, BRAF, NRAS, TP53). WT = wild-type; MUT = mutant
Footnotes
The authors declare no competing financial interests.
REFERENCES
- 1.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The lancet oncology. 2010;11(8):753–62. [DOI] [PubMed] [Google Scholar]
- 2.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27(35):5924–30. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369(11):1023–34. [DOI] [PubMed] [Google Scholar]
- 4.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–7. [DOI] [PubMed] [Google Scholar]
- 5.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28(7):1254–61. [DOI] [PubMed] [Google Scholar]
- 6.Mahipal A, Grothey A. Role of Biologics in First-Line Treatment of Colorectal Cancer. J Oncol Pract. 2016;12(12):1219–28. [DOI] [PubMed] [Google Scholar]
- 7.Yang M, Yeatman TJ. Molecular stratification of colorectal cancer populations and its use in directing precision medicine. Expert Review of Precision Medicine and Drug Development. 2017;2(4):205–15. [Google Scholar]
- 8.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27(12):2091–6. [DOI] [PubMed] [Google Scholar]
- 9.Mosakhani N, Lahti L, Borze I, Karjalainen-Lindsberg ML, Sundstrom J, Ristamaki R, et al. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer genetics. 2012;205(11):545–51. [DOI] [PubMed] [Google Scholar]
- 10.Malapelle U, Carlomagno C, de Luca C, Bellevicine C, Troncone G. KRAS testing in metastatic colorectal carcinoma: challenges, controversies, breakthroughs and beyond. Journal of clinical pathology. 2014;67(1):1–9. [DOI] [PubMed] [Google Scholar]
- 11.von Einem JC, Heinemann V, von Weikersthal LF, Vehling-Kaiser U, Stauch M, Hass HG, et al. Left-sided primary tumors are associated with favorable prognosis in patients with KRAS codon 12/13 wild-type metastatic colorectal cancer treated with cetuximab plus chemotherapy: an analysis of the AIO KRK-0104 trial. Journal of cancer research and clinical oncology. 2014;140(9):1607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van Cutsem E, Beier F, et al. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA Oncol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. [DOI] [PubMed] [Google Scholar]
- 14.Schell MJ, Yang M, Missiaglia E, Delorenzi M, Soneson C, Yue B, et al. A Composite Gene Expression Signature Optimizes Prediction of Colorectal Cancer Metastasis and Outcome. Clin Cancer Res. 2016;22(3):734–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schell MJ, Yang M, Teer JK, Lo FY, Madan A, Coppola D, et al. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun. 2016;7:11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nature medicine. 2015;21(11):1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer journal. 2011;17(6):528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim R, Schell MJ, Teer JK, Greenawalt DM, Yang M, Yeatman TJ. Co-evolution of somatic variation in primary and metastatic colorectal cancer may expand biopsy indications in the molecular era. PloS one. 2015;10(5):e0126670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sclafani F, Kim TY, Cunningham D, Kim TW, Tabernero J, Schmoll HJ, et al. A Randomized Phase II/III Study of Dalotuzumab in Combination With Cetuximab and Irinotecan in Chemorefractory, KRAS Wild-Type, Metastatic Colorectal Cancer. Journal of the National Cancer Institute. 2015;107(12):djv258. [DOI] [PubMed] [Google Scholar]
- 20.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS medicine. 2013;10(5):e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(19):5314–28. [DOI] [PubMed] [Google Scholar]
- 24.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC medical genomics. 2010;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, et al. EMT is the dominant program in human colon cancer. BMC medical genomics. 2011;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 2010;70(6):2264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Goke B, et al. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC genomics. 2014;15:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. [DOI] [PubMed] [Google Scholar]
- 30.Holm S A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6(2):65–70. [Google Scholar]
- 31.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nature medicine. 2013;19(5):614–8. [DOI] [PubMed] [Google Scholar]
- 32.Guinney J, Ferte C, Dry J, McEwen R, Manceau G, Kao KJ, et al. Modeling RAS phenotype in colorectal cancer uncovers novel molecular traits of RAS dependency and improves prediction of response to targeted agents in patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(1):265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, et al. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62(4):540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omolo B, Yang M, Lo FY, Schell MJ, Austin S, Howard K, et al. Adaptation of a RAS pathway activation signature from FF to FFPE tissues in colorectal cancer. BMC medical genomics. 2016;9(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker JB, Dutta D, Watson D, Maddala T, Munneke BM, Shak S, et al. Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer. Br J Cancer. 2011;104(3):488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pentheroudakis G, Kotoula V, De Roock W, Kouvatseas G, Papakostas P, Makatsoris T, et al. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes. BMC Cancer. 2013;13:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenz H-J, Ou F-S, Venook AP, Hochster HS, Niedzwiecki D, Goldberg RM, et al. Impact of consensus molecular subtyping (CMS) on overall survival (OS) and progression free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance). Journal of Clinical Oncology. 2017;35(15_suppl):3511-. [Google Scholar]
- 39.Okita A, Takahashi S, Ouchi K, Inoue M, Watanabe M, Endo M, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis TB, Yang M, Schell MJ, Wang H, Ma L, Pledger WJ, et al. PTPRS Regulates Colorectal Cancer RAS Pathway Activity by Inactivating Erk and Preventing Its Nuclear Translocation. Sci Rep. 2018;8(1):9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modest DP, Stintzing S, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, et al. Exploring the effect of primary tumor sidedness on therapeutic efficacy across treatment lines in patients with metastatic colorectal cancer: analysis of FIRE-3 (AIOKRK0306). Oncotarget. 2017;8(62):105749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold D, Lueza B, Douillard JY, Peeters M, Lenz HJ, Venook A, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2017;28(8):1713–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cushman SM, Jiang C, Hatch AJ, Shterev I, Sibley AB, Niedzwiecki D, et al. Gene expression markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: results from CALGB 80203 (Alliance). Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(5):1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hecht JR, Mitchell E, Chidiac T, Scroggin C, Hagenstad C, Spigel D, et al. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol. 2009;27(5):672–80. [DOI] [PubMed] [Google Scholar]
- 45.Modest DP, Jung A, Moosmann N, Laubender RP, Giessen C, Schulz C, et al. The influence of KRAS and BRAF mutations on the efficacy of cetuximab-based first-line therapy of metastatic colorectal cancer: an analysis of the AIO KRK-0104-trial. International journal of cancer Journal international du cancer. 2012;131(4):980–6. [DOI] [PubMed] [Google Scholar]
- 46.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67(6):2643–8. [DOI] [PubMed] [Google Scholar]
- 47.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–20. [DOI] [PubMed] [Google Scholar]
- 48.Perkins G, Lievre A, Ramacci C, Meatchi T, de Reynies A, Emile JF, et al. Additional value of EGFR downstream signaling phosphoprotein expression to KRAS status for response to anti-EGFR antibodies in colorectal cancer. International journal of cancer Journal international du cancer. 2010;127(6):1321–31. [DOI] [PubMed] [Google Scholar]
- 49.Tabernero J, Van Cutsem E, Diaz-Rubio E, Cervantes A, Humblet Y, Andre T, et al. Phase II trial of cetuximab in combination with fluorouracil, leucovorin, and oxaliplatin in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2007;25(33):5225–32. [DOI] [PubMed] [Google Scholar]
- 50.Weickhardt AJ, Price TJ, Chong G, Gebski V, Pavlakis N, Johns TG, et al. Dual targeting of the epidermal growth factor receptor using the combination of cetuximab and erlotinib: preclinical evaluation and results of the phase II DUX study in chemotherapy-refractory, advanced colorectal cancer. J Clin Oncol. 2012;30(13):1505–12. [DOI] [PubMed] [Google Scholar]
- 51.Stintzing S, Miller-Phillips L, Modest DP, Fischer von Weikersthal L, Decker T, Kiani A, et al. Impact of BRAF and RAS mutations on first-line efficacy of FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab: analysis of the FIRE-3 (AIO KRK-0306) study. Eur J Cancer. 2017;79:50–60. [DOI] [PubMed] [Google Scholar]
- 52.Lee CG, McCarthy S, Gruidl M, Timme C, Yeatman TJ. MicroRNA-147 induces a mesenchymal-to-epithelial transition (MET) and reverses EGFR inhibitor resistance. PLoS One. 2014;9(1):e84597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moran AE, Hunt DH, Javid SH, Redston M, Carothers AM, Bertagnolli MM. Apc deficiency is associated with increased Egfr activity in the intestinal enterocytes and adenomas of C57BL/6J-Min/+ mice. The Journal of biological chemistry. 2004;279(41):43261–72. [DOI] [PubMed] [Google Scholar]
- 54.Pai R, Soreghan B, Szabo IL, Pavelka M, Baatar D, Tarnawski AS. Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nature medicine. 2002;8(3):289–93. [DOI] [PubMed] [Google Scholar]
- 55.Stoddart A, Fernald AA, Wang J, Davis EM, Karrison T, Anastasi J, et al. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood. 2014;123(7):1069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauer L, Gitenay D, Vo C, Baron VT. Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene. 2010;29(18):2628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.