To the Editor,
In this study we assessed viral factors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shedding, trying to prove that a RT-PCR crossing threshold (Ct) value superior to 33 on nasopharyngeal swab (NPS) samples could be a major criterion to enlighten infection prevention and control measures such as duration of eviction, contact tracing, or discharge from hospital wards. The SARS-CoV-2 replication cycle includes direct translation of genomic RNA into several viral proteins, synthesis of negative- and positive-stranded replicative intermediate RNAs (RIs), viral assembly, and release of mature virions [1].
To provide proof of active replication, we conducted real-time RT-PCR assays to detect specifically the presence of the viral E (envelope) subgenomic RNA and E negative-strand RNA in clinical samples. We also attempted to isolate virus from samples to associate the presence of RIs with the detection of viable virus. Data were obtained from 61 immunocompetent healthcare workers (HCWs) diagnosed with SARS-CoV-2 infection by RT-PCR on nasopharyngeal samples (12 from asymptomatic subjects and 49 from subjects with mild/moderate clinical disease). Detailed descriptions of the characteristics of the HCWs, clinical context, and sample processing methods are presented in the Supplementary Material.
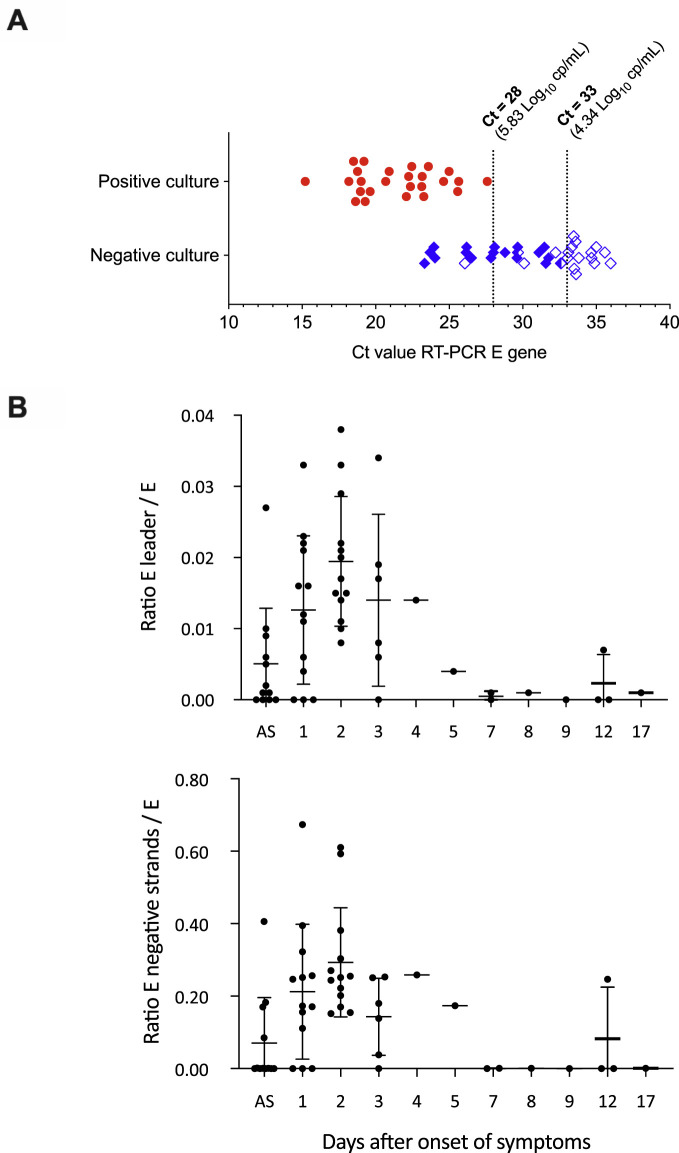
Overall, the median age was 28 (interquartile range (IQR) 23–36), and 38 were females. SARS-CoV-2 viral loads ranged from 9.64 to 3.57 log10 copies/mL (cp/mL) (Supplementary Material Table S1). Virus isolation was successful for 41.0% of clinical samples. Our data showed that the likelihood of recovering infectious virus correlates with high viral loads and the presence of RIs (Fig. 1 A). Strikingly, no isolate was recovered when viral load was below 5.83 log10 cp/mL (i.e. Ct >28), which is similar to cut-offs reported previously [[2], [3], [4]]. Moreover, no RIs were detectable in samples when the viral load was below 4.34 log10 cp/mL (i.e. Ct >33). Interestingly, the ratios of mean normalized RIs per genome indicate a high level of viral replication during the first 5 days after the onset of symptoms followed by a significant decline thereafter (Fig. 1B), as previously reported [4]. Among asymptomatic HCWs, high viral loads (>5 log10 cp/mL) were associated with detection of significant signals of RIs and virus isolation.
Fig. 1.
(A) Virus isolation success in relation to Ct value of the RT-PCR targeting the E viral gene. Red circles and blue diamonds represent positive and negative cell culture assays, respectively. Empty and half-filled symbols showed clinical samples with no E subgenomic or E negative-strand RNA signal detected, respectively. Crossing threshold (Ct) values of 28 and 33 correspond to 5.83 and 4.34 log10 copies/mL (cp/mL), respectively. (B) Subgenomic viral and negative-strand RNAs in relation to viralore E genomic RNA. The ratios are depicted according to the number of days after the onset of symptoms. Dots represent mean values of RT-PCR data obtained from at least two independent experiments on samples from individual healthcare workers (HCWs). Plots show median values with interquartile ranges. Data concerning date of symptom onset were confidently available for 54 HCWs (88.5%); HCWs with no precise data were excluded from the analysis. E subgenomic (‘E leader’) and E negative-strand RNAs were never detected beyond 7 days after the onset of symptoms, except at 12 days after the onset of symptoms for HCW N°35 who experienced persistent clinical signs at the time of sampling. Moreover, no isolate was recovered from the clinical sample of this patient (viral load 5.83 log10 copies/mL). AS, asymptomatic HCWs.
Although recent studies have expressed reservations [5], our findings confirm subgenomic viral RNAs as indicators of active replication in clinical samples [4]. Moreover, we demonstrated that detection of negative strands—never used hitherto—perfectly correlates with subgenomic viral RNA detection. With the accumulating evidence available thus far, our findings strengthen the possibility of using—in association with a symptom-based strategy—the RT-PCR Ct value cut-off of 33 as a value above which individuals would no longer be contagious.
Author contributions
SB initiated the study and coordinated all work carried out. SM did the targeted and strand-specific molecular assays, and cell culture isolation trials. ML provided clinical information about HCWs. SB, VC and A-GM drafted the initial manuscript with inputs. All authors contributed to the final submitted version. All authors have read and agreed to the final version of the manuscript.
Transparency declaration
The authors declare no potential conflicts of interest relevant to this article. This work was supported by Sorbonne Université, Paris, France.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.01.017.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. May 22:ciaa638; Epub ahead of print. PMID: 32442256; PMCID: PMC7314198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 5.van Kampen J., van de Vijver D., Fraaij P., Haagmans B.L., Lamers M.M., Okba N. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 doi: 10.1101/2020.06.08.20125310. 06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.