While previous reports had suggested that the typhoid toxin (TT) could potentially use ArtB as an alternate binding subunit, this was thought to play a minor role in the evolution and biology of the toxin. In this study, we establish that both TT genes and artB are widespread among Salmonella enterica subsp. enterica, suggesting that TT likely plays a broader role in Salmonella virulence that extends beyond its proposed role in typhoid fever.
KEYWORDS: typhoid toxin, nontyphoidal Salmonella, cytolethal distending toxin, ArtB
ABSTRACT
Salmonella enterica encodes a wide array of virulence factors. One novel virulence factor, an A2B5 toxin known as the typhoid toxin (TT), was recently identified among a variety of S. enterica serovars. While past studies have shown that some serovars encode both the TT (active subunits CdtB and PltA and binding subunit PltB) and a second binding subunit (ArtB), these serovars were thought to be the exception. Here, we show that genes encoding the TT are detected in more than 100 serovars representing distinct phylogenetic lineages of S. enterica subsp. enterica, although clade B and section Typhi are significantly more likely to encode TT genes than serovars from other clades. Furthermore, we show that 81% of these TT-positive serovars also encode artB, suggesting that the cooccurrence of both toxin binding subunits is considerably more common than previously thought. A combination of in silico modeling, bacterial two-hybrid system screening, and tandem affinity purification (TAP) of toxin subunits suggests that ArtB and PltB interact in vitro, at least under some growth conditions. While different growth conditions yielded slightly higher transcript abundances of artB and pltB, both genes had their highest relative transcript abundances when Salmonella was grown under low-Mg2+ conditions, suggesting that ArtB and PltB may compete for inclusion in the TT. Together, our results suggest that ArtB likely plays an important and previously underappreciated role in the biology of the TT produced by typhoidal and nontyphoidal Salmonella.
IMPORTANCE While previous reports had suggested that the typhoid toxin (TT) could potentially use ArtB as an alternate binding subunit, this was thought to play a minor role in the evolution and biology of the toxin. In this study, we establish that both TT genes and artB are widespread among Salmonella enterica subsp. enterica, suggesting that TT likely plays a broader role in Salmonella virulence that extends beyond its proposed role in typhoid fever. Furthermore, our data suggest the selective maintenance of both toxin binding subunits, which may compete for inclusion in the holotoxin. Last, our data support the importance of characterizing diverse nontyphoidal Salmonella (NTS) serovars, as the presence of classically defined typhoidal virulence factors among NTS serovars continues to challenge the typhoid-nontyphoid Salmonella paradigm.
INTRODUCTION
Salmonella enterica is an important cause of foodborne illness worldwide, contributing an estimated 88 million (95% uncertainty interval, 35 million to 234 million) foodborne infections each year (1). Given the appreciable diversity of S. enterica with >2,600 known serovars (2), efforts to understand this pathogen have largely focused on categorizing serovars into typhoidal, paratyphoidal, and nontyphoidal types (3, 4) and studying select representatives of each (5). Arguably the two best-characterized serovars are S. enterica serovar Typhi and S. enterica serovar Typhimurium, representing typhoidal and nontyphoidal serovars, respectively. S. Typhi, the causative agent of typhoid fever, is associated with an invasive extraintestinal infection (4), while infection with nontyphoidal serovars usually results in self-limiting gastroenteritis (4), although infection with some serovars is associated with a significantly higher likelihood of invasive disease (6). While these two serovars have been, and continue to be, the proverbial workhorses that have greatly progressed our understanding of Salmonella pathogenesis, the frequency of human infections with other less-studied serovars continues to increase (7–11). Regardless, investigations into Salmonella pathogenesis with S. Typhi and S. Typhimurium have identified an array of virulence factors and adaptations that allow diverse Salmonella serovars to successfully colonize and cause disease in a range of hosts (5, 12, 13).
The typhoid toxin (TT), which was originally discovered and characterized in S. Typhi (14, 15), has been proposed as a key player in the development of typhoid fever, acting at both the single-cell and systemic levels in models of infection (14, 16–19). The TT incorporates the nuclease activity of the CdtB subunit of the cytolethal distending toxin (CDT) (14, 20) with the mono-ADP-ribosyltransferase activity of the pertussis toxin (called PltA), resulting in a hybrid toxin that induces a DNA damage response (DDR) in eukaryotic cells, leading to an accumulation of cells in the G2/M cell cycle phase (17, 18) and subsequent senescence (21, 22). The resulting damaged DNA is proposed to play a role in both disease manifestation (18) and colonization and persistence of the toxin-producing bacteria in the host (19, 23), although its role in acute typhoid fever remains uncertain (24).
The binding subunit of the TT was originally characterized as a pentameric ring of PltB monomers (18). ArtB, a homolog of PltB, was later identified in select S. Typhimurium DT104 isolates (25). As a proof of concept, Gao et al. showed that ArtB from S. Typhimurium DT104 can assemble into a homopentamer in vitro and further confirmed that the ArtB homopentamer can interact with CdtB and PltA from S. Typhi to form a biologically active holotoxin in vitro and in vivo (26). While both ArtB and PltB bind to glycan residues on sialic acids on host cells, ArtB is able to bind both N-acetylneuraminic acid (Neu5Ac)- and N-glycolylneuraminic acid (Neu5Gc)-terminated glycans on sialic acids, whereas PltB preferentially binds Neu5Ac-terminated glycans (18, 26). Gao et al. proposed that the expanded binding repertoire of ArtB could reflect the broad host range of S. Typhimurium DT104, despite the fact that S. Typhimurium isolates do not encode cdtB, pltA, or pltB (26).
We and others have previously established that multiple nontyphoidal serovars encode both artB and pltB (16, 27, 28), including S. enterica serovar Javiana, the 4th most commonly isolated serovar from human clinical cases of salmonellosis in the United States (7). Here, we show that the majority of TT-encoding serovars also encode artB and that PltB and ArtB may compete for inclusion in the binding subunit of the holotoxin.
RESULTS
TT genes are significantly overrepresented among specific clades of S. enterica.
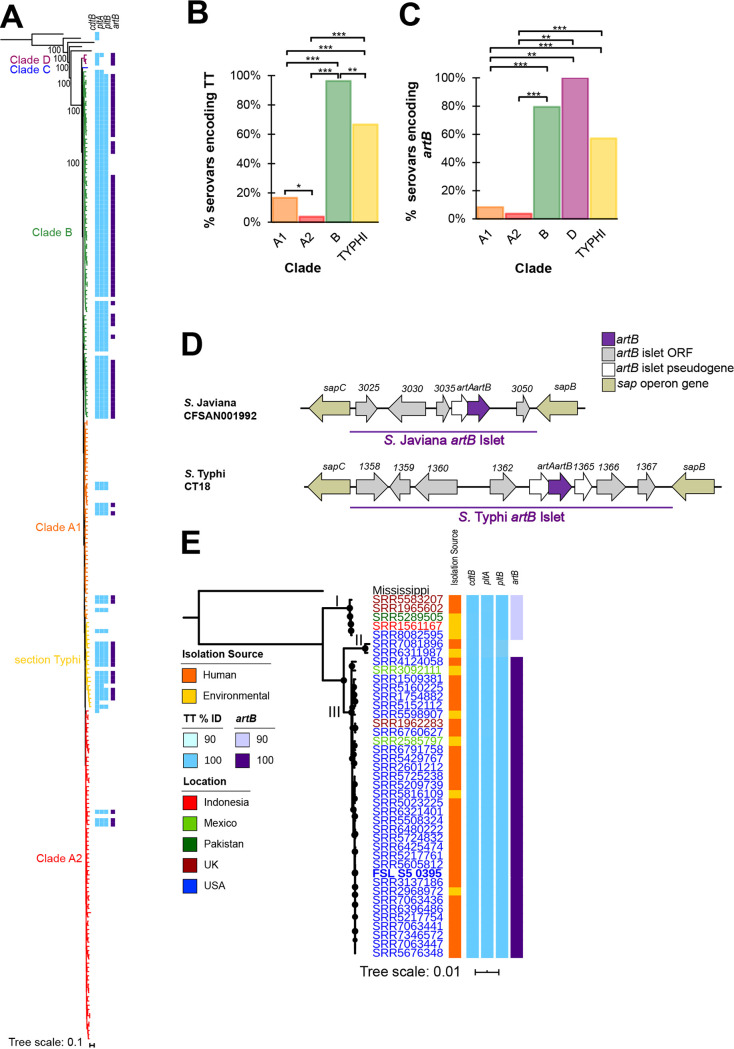
Multiple studies have suggested that genes encoding the TT (cdtB, pltA, and pltB) are distributed across multiple lineages and serovars of Salmonella (27–29). To better understand the evolution of the TT, we queried whole-genome sequence (WGS) data for representative isolates of 235 unique S. enterica subsp. enterica serovars to assess the distribution of TT genes across the main lineages of S. enterica (29). All three TT genes were detected in 105 S. enterica subsp. enterica serovars (45% of serovars examined here) (Fig. 1A). cdtB, encoding one of the active subunits, was detected in an additional seven S. enterica subsp. enterica serovars as well as in representative strains of S. enterica subsp. arizonae and S. enterica subsp. diarizonae (Fig. 1A). While pltA and pltB were not detected in these seven serovars or the representative S. enterica subsp. arizonae and S. enterica subsp. diarizonae strains with the blast settings used, visual examination of these genomes suggested that all except S. Napoli encoded open reading frames (ORFs) that showed nucleotide sequence similarity to S. Javiana pltA (79% to 82% identity) and pltB (65% to 68% identity) (see Table S1 in the supplemental material). Therefore, these serovars/strains were regarded as encoding putative pltA and pltB but were not considered encoding all TT genes for the analyses reported below. Proportions of serovars encoding all TT genes were significantly different among S. enterica phylogenetic clades (P < 0.001; Fisher’s exact test); clade B had the highest proportion of TT-encoding serovars (80 of 83 serovars), followed by section Typhi (14 of 21 serovars), while TT genes were detected in significantly fewer clade A1 and A2 serovars (P < 0.001 for all pairwise comparisons) with just 17% (8 of 40 serovars) and 4% (3 of 76 serovars) of serovars in these clades encoding TT genes, respectively (Fig. 1B). These results demonstrate a strong phylogenetic relationship between the presence of TT genes and Salmonella clades.
FIG 1.
Serovars in some phylogenetic clades are significantly more likely to encode TT genes and artB. (A) Reconstruction of phylogeny of 235 S. enterica subsp. enterica serovars and five additional subspecies based on the comparison of 13,181 core SNPs. Maximum likelihood analysis was computed using a general time-reversible model with gamma distribution in RAxML (66). Bootstrap values for branchpoints between subspecies are shown (based on 100 bootstrap repetitions). Branches are colored to represent the clade of the isolate as reported previously (29) and the detection of TT genes (cdtB, pltA, and pltB; shown in light blue) and artB (shown in purple) among S. enterica isolates using 85% nucleotide identity and 90% query coverage as criteria for detection. S. enterica subsp. arizonae was used to root the phylogeny. Histograms summarizing the proportion of serovars in each clade for which all TT genes (cdtB, pltA, and pltB) (B) or artB (C) was detected with blastn (i.e., the additional 6 S. enterica subsp. enterica serovars having putative pltA and pltB were not included in this analysis); note that clade C is not shown because TT genes and artB were not detected in these serovars. *, P < 0.05; **, P < 0.001; ***, P <0.0001, FDR corrected for pairwise comparisons of Fisher’s exact tests. (D) Comparison of the artB-islet in S. Javiana CFSAN001992 and S. Typhi CT18; ORFs and genes are named according to annotations from NCBI. The artB islet is denoted by the purple horizontal bar. ORFs that are at least 100 nucleotides (nt) in length are shown in gray (those <100-nt long were classified as pseudogenes and are shown in white), while genes shown in gold represent sap operon genes that flank the artB islet; artB is shown in dark purple. Islets are not drawn to scale. (E) Maximum likelihood reconstruction of phylogeny based on 63,994 core SNPs among 41 S. Javiana isolates and a representative isolate of S. Mississippi (used to root the phylogeny). The S. Javiana strain (FSL S5-0395) used in phenotypic experiments is shown in bold. Circles at nodes represent bootstrap values >90 (based on output from 1,000 bootstrap repetitions). Assembly names are colored to reflect the country from which the isolate originated; the isolation sources (human clinical shown in orange, environmental shown in yellow) as well as the nucleotide identity of TT genes (light blue) and artB (purple) are shown as colored bars. Roman numerals are used to depict the clades of the S. Javiana isolates.
Nucleotide lengths and identities of putative pltA and pltB that were not detected with initial blast searches. Download Table S1, PDF file, 0.1 MB (134.7KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we assessed the genomic location of the TT islet using WGS data for representative serovars (see Materials and Methods for a description of how these were selected) from each TT-positive S. enterica subsp. enterica clade that had a closed genome available. The TT islet was previously reported to be located within Salmonella pathogenicity island (SPI) 11 (5, 28); however, the genomic location of SPI-11, and therefore the TT genes, was unknown. While the size of SPI-11 was variable among the serovars examined here, the TT islet was consistently located approximately 3 kb from the terminus of SPI-11. For all genomes evaluated (representing S. enterica serovar Goldcoast [clade A1], S. enterica serovar Milwaukee [clade A2], S. enterica serovar Paratyphi A [section Typhi], S. enterica serovar Indiana [section Typhi], S. enterica serovar Poona [clade B], and S. Javiana [clade B]), except S. Typhi, SPI-11 was within 5 to 7 kb from phoP-phoQ, which encodes a two-component regulatory system that has been shown previously to be the primary system regulating transcription of TT genes (30). However, in S. Typhi CT18, SPI-11 was located >550 kb upstream of phoP-phoQ. These data suggest that, in general, both the location of SPI-11 and the location of the TT islet within SPI-11 are relatively conserved.
While the overall phylogeny suggests multiple acquisition and/or deletion events of TT genes, the most likely scenario appears to be that TT genes were present in the S. enterica ancestor, and subsequent deletion events in major clades as well as possibly additional independent (re)acquisitions of TT genes (e.g., in some serovars in clade A1 and A2 and section Typhi) occurred.
The majority of TT-encoding serovars also encode artB, which is located on an islet within an operon encoding genes associated with resistance to antimicrobial peptides.
Several groups previously established that multiple TT-encoding serovars also encode a homologous binding subunit, artB (28, 31, 32). Therefore, we also assessed the presence of artB to determine the frequency that TT and ArtB are coencoded in a given serovar. Among all 235 serovars examined, artB was detected in a total of 88 serovars (Fig. 1A). Proportions of artB-encoding serovars were significantly different across the phylogenetic clades (P < 0.001; Fisher’s exact test) (Fig. 1C). Serovars in clades B (66 of 83) and D (3 of 3) and section Typhi (12 of 21) had significantly higher proportions of artB-positive serovars than clade A1 (4 of 48) and A2 serovars (3 of 79) (see Fig. 1C for false-discovery rate [FDR] corrected P values for pairwise comparisons). Among the 105 serovars for which a full TT islet was detected (i.e., excluding the three clade D serovars, which encoded putative pltA and pltB), artB was detected in 85 (i.e., 81% of TT-encoding serovars also encoded artB).
Given the relative conservation of artB among TT-positive serovars, we next assessed whether the location of the artB islet was conserved among genomes representing the different S. enterica subsp. enterica phylogenetic clades (A1, A2, and B and section Typhi). Although the genomic locations and the numbers of ORFs identified differed slightly (Fig. 1D; see also Table S2), we found that in all cases, the artB islet was inserted between sapB and sapC (Fig. 1D). The GC content of the artB islet was considerably lower (average, 42.2%; range, 41.2% to 44.7%) than the genome-wide GC content (average, 52.2%; range, 52.1% to 52.3%), suggesting that this islet was likely acquired via horizontal transfer from another source (Table S2). In addition to artB itself, the artB islet includes (i) between 4 to 7 ORFs with no known or predicted functions (Table S2) and (ii) an artA pseudogene, which was identified in all islets examined. The artA pseudogene contains multiple mutations leading to premature stop codons, with the most 5′ premature stop codon leading to a predicted peptide with just 4 amino acid (aa) residues. For one serovar, S. Milwaukee, an additional ORF annotated as an IS3 family transposase, was detected in the middle of the artB islet. Together, these results suggested a common site-specific mode of acquisition of the artB islet as well as a selective advantage for maintenance and/or acquisition of the artB islet among most TT-positive serovars.
Positions of artB islet in closed genomes from representative strains of TT and artB-positive serovars. Download Table S2, PDF file, 0.1 MB (131.5KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For serovar S. Javiana, TT genes are highly conserved, while the presence of intact artB is clade dependent.
Given that 96% of clade B serovars examined here were TT positive and the majority of studies examining the TT had been conducted with S. Typhi, we characterized the role of artB in the clade B nontyphoidal serovar S. Javiana, which is the most prevalent TT-positive serovar among human clinical salmonellosis cases in the United States (33). Among a collection of 41 S. Javiana isolates selected to represent a range of genotypes (different single nucleotide polymorphism [SNP] clusters), geographical sources, and dates of isolation (see Table S3), we found that sequences of cdtB, pltA, and pltB were highly conserved (99% to 100% nucleotide identity). In contrast, while artB was conserved for most of the isolates (100% identity for 34/41 isolates; clade III) (Fig. 1E), we also detected two small clades for which artB was more variable, including (i) five isolates encoding an artB allele containing a 46-nucleotide deletion resulting in a premature stop codon (clade I) (Fig. 1E), and (ii) two isolates for which artB was not detected with blast (clade II) (Fig. 1E); visual inspection of the sap operon further confirmed the absence of the artB islet at this locus as well as a lack of raw sequence reads mapping to artB. These results suggested that even though artB is also relatively conserved among S. Javiana isolates, loss of ArtB function occurred independently in two clades of S. Javiana, due to either artB deletion or artB pseudogene formation.
SRR run identity and metadata for 41 S. Javiana isolates and one S. Mississippi isolate used for phylogenetic and blast analyses. Download Table S3, PDF file, 0.1 MB (111.3KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
artB and pltB are induced when S. Javiana is grown under nutrient-limiting conditions, such as those representing an intracellular host environment.
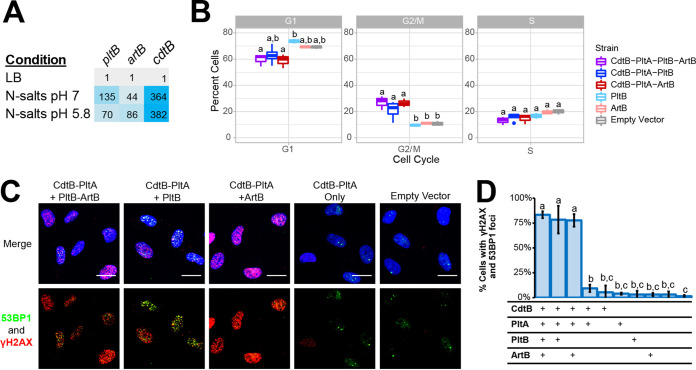
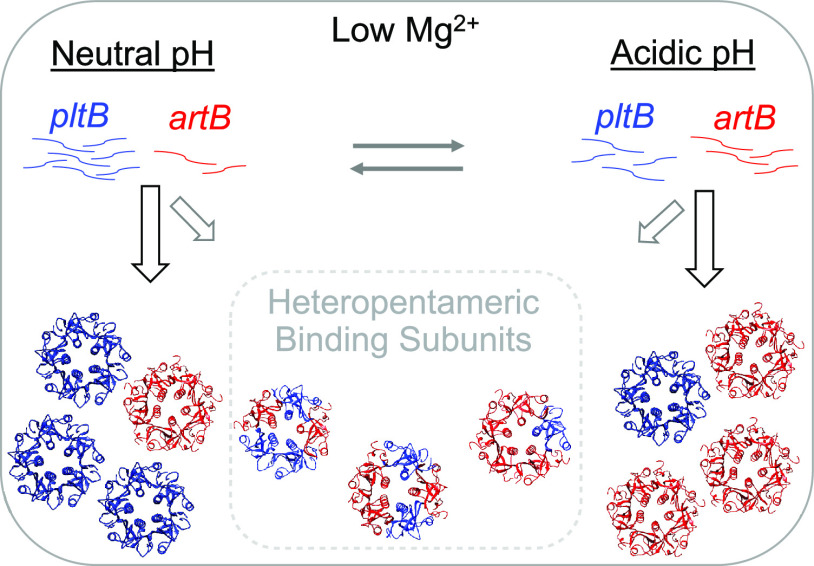
Expression of pltB in S. Typhi was previously shown to occur when cells were grown under conditions that mimic those encountered by intracellular Salmonella (30, 32). Therefore, we compared pltB, cdtB, and artB transcript abundances of S. Javiana grown in Luria-Bertani (LB) broth (pH 7) and N-salts minimal medium containing 8 µM Mg2+ (pH 7). When S. Javiana was grown in N-salts minimal medium at pH 7, transcript abundances of pltB, cdtB, and artB were on average 135-, 364-, and 44-fold higher, respectively, than abundances in S. Javiana grown in LB broth (Fig. 2A). When S. Javiana was grown in N-salts minimal medium acidified to pH 5.8 to simulate intracellular conditions (34), transcript abundances of cdtB and artB were higher (on average 382- and 86-fold compared to growth in LB broth) (Fig. 2A), while pltB transcript abundances were approximately 2-fold lower than abundances obtained during growth in N-minimal medium at pH 7. Collectively, these results suggested that artB and pltB transcription is induced similarly to that of cdtB when S. Javiana is grown in minimal medium where Mg2+ is limiting. However, the relative transcript abundances (relative to growth in LB broth) of pltB were higher at neutral pH than at acidic pH, while both cdtB and artB had higher relative transcript abundances at acidic pH, suggesting that pH may serve as an environmental cue for modulating expression of pltB and artB.
FIG 2.
ArtB or PltB can be used to form an active TT binding subunit. (A) Fold expression (2−ΔΔCT) of pltB, cdtB, and artB in S. Javiana cells grown for 5 h in either N-salts minimal medium at pH 7 or pH 5.8 (representing mid-exponential phase for this condition), normalized to expression of S. Javiana cells grown for 3 h in LB broth, pH 7 (representing mid-exponential phase for this condition). Results are averaged from three independent experiments; cells are shaded to represent high (blue) and low expression (gray) relative to expression in LB broth. (B) Cell cycle analyses of HIEC-6 cells coincubated with lysates of E. coli BTH101 cells expressing different combinations of tagged typhoid toxin subunits: CdtB-His, PltA-Strep, PltB-3×-Flag, and ArtB-c-Myc. HIEC-6 cells were coincubated with toxins for 24 h prior to analyzing DNA content with propidium iodide staining and analysis by flow cytometry. Data represent the averages from three independent experiments; boxplots that do not share lowercase letters are statistically different (Tukey’s adjusted P value < 0.05). (C) Assessment of DNA damage response activation among HIEC-6 cells treated with toxin subunits. Immunofluorescence staining of HIEC-6 cells coincubated for 24 h with total cell lysates containing different combinations of CdtB-His, PltA-Strep, PltB-3×-Flag, and ArtB-c-Myc. DNA damage response proteins are shown in green (53BP1) and red (γH2AX); nucleic acids were stained with DAPI (blue). Scale bars, 20 µm. (D) Quantification of HIEC-6 cell nuclei having ≥4 53BP1-foci as well as γH2AX foci; the table below the graph shows which subunits were present in the supernatant added to the HIEC-6 cells. Two negative controls were included and are represented by the final two bars in the graphic, which represent HIEC-6 cell populations that were treated with supernatants from E. coli BTH101 cells with an empty vector (left) and untreated cell populations (right). Lowercase letters indicate statistically significant differences in the proportions of cells having DDR foci; bars that share lowercase letters are not significantly different at α = 0.05 after correcting for multiple comparisons.
Binding subunits composed of PltB or ArtB result in active TT.
Previous gene knockout experiments suggested that S. Javiana requires either ArtB or PltB to produce active TT, as strains with deletion of either artB or pltB still showed production of active toxin (16). To biochemically confirm this, we expressed various combinations of toxin subunits in a heterologous host and then coincubated lysates containing these toxins with human intestinal epithelial cells (HIEC-6) to assess toxin activity. The accumulation of cells in the G2/M phase continues to be the gold standard for assessing TT activity (14, 17), as cells that have an activated DNA damage response (DDR) will accumulate at this cell cycle checkpoint (35). Therefore, we analyzed the cell cycle progression of HIEC-6 cells treated with lysates containing holotoxins with PltB or ArtB to confirm the activity of both toxin types. Treatment of HIEC-6 cells with lysates containing CdtB, PltA, and either PltB or ArtB resulted in a significantly higher proportion of HIEC-6 cells in the G2/M phase (PltB as binding subunit, P = 0.037, or ArtB as binding subunit, P = 0.006; both compared to cells treated with lysates containing empty vector) (Fig. 2B). There was no significant difference in the proportion of cells in the G2/M phase for HIEC-6 cells treated with TT that used ArtB versus PltB as the binding subunit (P = 0.6983), supporting that holotoxins using either monomer for the binding subunit are active. To confirm that the accumulation of cells in G2/M phase after treatment with these toxins was due to the activation of a DDR, we also used immunofluorescence (IF) staining to detect two proteins involved in the DDR, namely, phosphorylated histone H2AX (γH2AX), and 53 binding protein 1 (53BP1). Treatment with lysates containing CdtB, PltA, and either PltB or ArtB as the binding subunit resulted in DDR activation in ∼78% of HIEC-6 cells (Fig. 2C and D), confirming that both holotoxins are active. Importantly, treatment with individual toxin subunits did not result in DDR activation among treated cells (Fig. 2C and D). Together, these data confirm that the TT encoded by S. Javiana can use either PltB or ArtB as the binding subunit.
In silico modeling suggests that heteropentameric binding subunits of ArtB and PltB may occur.
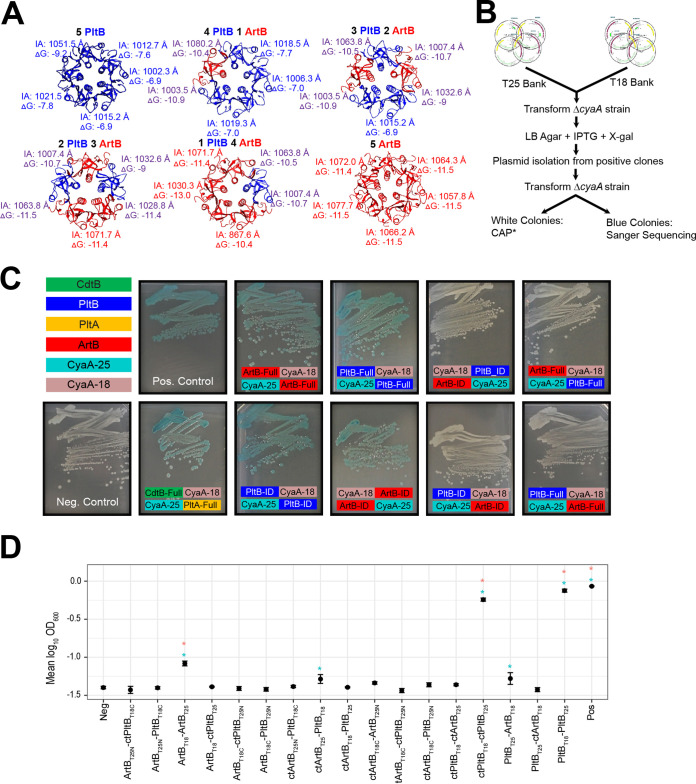
Given that both ArtB and PltB can form active TT and that transcript abundances of artB and pltB are significantly higher when S. Javiana is grown under nutrient-limiting conditions, such as those that might be encountered by intracellular Salmonella, we hypothesized that these subunits likely compete for inclusion in the TT, resulting in toxin binding subunits composed of homo- and heteropentamers. The structures of the TT binding subunits containing homopentamers of PltB (from S. Typhi [18]) or ArtB (from S. Typhimurium DT104 [26]) have been resolved experimentally and were used here to model and predict the energies of interactions between monomers in binding subunits composed of various ratios of ArtB and PltB using the predominant amino acid sequence types for S. Javiana (Fig. 1E and 3A). While alignment of ArtB from S. Typhimurium DT104 with ArtB from clade III S. Javiana (Fig. 1E) showed only 74% amino acid identity, interface areas and the ΔG solvation energy gain of the ArtB complex formation for S. Typhimurium DT104 ArtB and S. Javiana ArtB were similar (see Fig. S1), supporting the modeling strategy used. ArtB was shown to interact with more specificity and stronger hydrophobicity with itself (ArtB-ArtB ΔG average, −11.5 kcal/M) than with PltB (ArtB-PltB ΔG average, −10.5 kcal/M) (Fig. 3A); however, the theoretical interaction of ArtB-PltB was predicted to be more favorable than PltB-PltB (ΔG average, −7.5 kcal/M) (Fig. 3A). These data suggested that ArtB and PltB monomers may interact to form heteropentameric binding subunits, which we next tested experimentally.
FIG 3.
Analysis of PltB-ArtB interactions. (A) Solvation energies of the interface calculated between monomers in the binding subunit are color coded to represent the interaction: PltB-PltB are shown in blue, ArtB-ArtB in red, and PltB-ArtB in purple. IA, interface area in Angstroms (Å); ΔG, solvation free energy gain upon formation of the interface (kcal/M). Negative ΔG values correspond to hydrophobic interfaces, and therefore a positive protein affinity. (B) Schematic of the method used to screen two-hybrid system clones. In the example shown, DNA fragments encoding the PltB polypeptide or C-terminal oligomerization region, as predicted from TT structure, were cloned at the 5′ or 3′ ends of regions encoding adenylate cyclase domain T18 or T25. All T18 or T25 clones were pooled into “banks” that were cotransformed into the cAMP-null (ΔcyaA) E. coli BTH101. Screening was performed by growing the transformants on LB agar supplemented with IPTG and X-Gal. Plasmid DNA was isolated from clones with positive interactions and was retransformed into E. coli BTH101 to exclude constitutively active CAP* mutants. Clones with confirmed interactions were submitted for Sanger sequencing to determine the identity of the interacting domains. (C) E. coli BTH101 two-hybrid system strains grown on LB agar supplemented with IPTG and X-Gal. Clones that showed positive interactions appear blue. Colored boxes show the fusion of either the interacting domain (_ID) or the full-length protein (_Full) for PltB, PltA, CdtB, and ArtB to either the CyaA-25 (shown in teal) or CyaA-18 subunits (shown in mauve) of the two-hybrid system. (D) Detection of interactions of ArtB and PltB in S. Javiana ΔcyaA harboring two-hybrid system plasmids. S. Javiana cells were grown for 24 h in M63 minimal medium with maltose as the sole carbon source, and absorbance was measured at 600 nm. Error bars indicate the standard deviations of the means from three independent experiments. Constructs with the C terminus are denoted with “ct.” *, P < 0.05 versus the negative control before (blue) and after (pink) Dunnett’s test for multiple comparisons adjustment; the PltBT25-ArtBT18 interaction was only marginally significant after multiple comparison corrections (corrected P value = 0.0973).
Results of additional modeling and two-hybrid system constructs in E. coli BTH101 cells. (A) Modeling was performed to compare the calculated energies for S. Typhimurium DT104 ArtB (shown in red font) and S. Javiana CFSAN001992 ArtB (shown in black font). (B) Additional two-hybrid system clones tested as in Fig. 3C. Briefly, E. coli BTH101 cells were transformed with plasmids encoding partial interacting domain (_ID) or full-length proteins (_Full) fused to T18 and T25 subunits of CyaA (50). Blue pigmentation suggests an interaction, while tan colonies are unable to confirm an interaction. Some of these constructs are also shown in Fig. 3C and were included again here for convenience (i.e., showing the entire plate or half of the plate). Download FIG S1, PDF file, 2.8 MB (2.8MB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro interactions between ArtB and PltB monomers and copurification of ArtB and PltB from TTs suggest that heteropentameric binding subunits may occur in vitro.
We first assessed the possibility of ArtB-PltB interactions using an adenylate cyclase two-hybrid system in Escherichia coli BTH101. We specifically used a high-throughput screening approach (Fig. 3B) to test for possible interactions of different domains for each toxin component (CdtB, PltA, PltB, and ArtB) including (i) full-length proteins, (ii) polypeptides without the signal peptide sequence, and (iii) the oligomerization domains predicted from the TT structure (18). Using this technique, we confirmed PltB-PltB, ArtB-ArtB, and CdtB-PltA interactions for both interacting domains and full-length toxin subunits (Fig. 3C), but we were unable to confirm a PltB-ArtB interaction, despite testing interactions for various combinations of interacting domains and full-length PltB and ArtB (Fig. 3C and Fig. S1).
In a heterologous host two-hybrid system assay, issues arising from protein fusion stability and folding as well as a requirement of basal levels of cAMP to initiate the positive-feedback induction loop may interfere with the ability of the system to detect some protein-protein interactions. Hence, we also constructed a cAMP-null S. Javiana strain (ΔcyaA) to assess possible interactions in Salmonella. When ΔcyaA S. Javiana was grown in M63 broth (supplemented with 0.02 mM cAMP, a level that induced basal fusion protein expression but did not support growth of negative controls), we found marginally significant (P < 0.05, uncorrected P value) PltB-PltB, ArtB-ArtB, and ArtB-PltB interactions (Fig. 3D and Table S4); however, upon correcting for multiple comparisons, only one of the ArtB-PltB interactions was considered statistically significant at an α of 0.1 (corrected P value = 0.0973) (Fig. 3D and Table S4). These results suggested that while PltB-PltB and ArtB-ArtB interactions were more readily detected using the two-hybrid system, there was some evidence of ArtB-PltB interactions; we therefore conducted additional experiments to determine whether both ArtB and PltB subunits could be incorporated into a heteropentameric configuration of the TT binding subunit.
Estimates of linear model on log10-transformed absorbance (OD600) data of S. Javiana ΔcyaA mutants. Download Table S4, PDF file, 0.09 MB (95.4KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
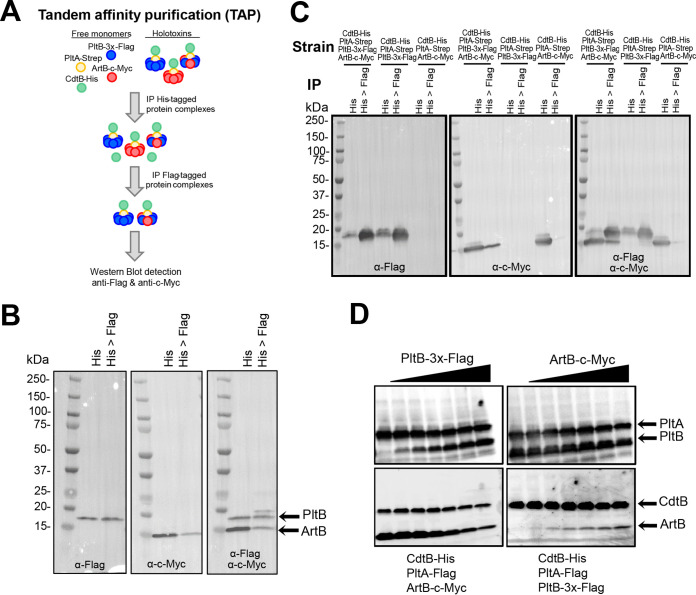
Previous studies have suggested that the TT binding subunit exists as a stable pentamer (18). Therefore, we also used tandem affinity purification (TAP) of different combinations of CdtB-His, PltA-Strep, PltB-3×-Flag, and ArtB-c-Myc overexpressed in E. coli BTH101 (Fig. 4A) to assess whether holotoxins can be formed with heteropentameric binding subunits of PltB and ArtB. Purification of His-tagged CdtB followed by purification of PltB-3×-Flag (Fig. 4B) revealed that ArtB was consistently (results of three independent experiments) (see Fig. S2) copurified with PltB, albeit at lower levels than PltB, in both pulldown steps, supporting that PltB and ArtB can form heteropentameric binding subunits. Additional immunoblotting to control for the possibility of recognition of ArtB-c-Myc by anti-Flag antibodies (used to detect PltB-3×-Flag) confirmed the specificity of this approach (Fig. 4C and Fig. S2). Taken together, both the weak interaction detected using the two-hybrid system and the copurification of ArtB-c-Myc from PltB-3×-Flag-containing toxins suggested that binding subunits composed of heteropentamers of ArtB and PltB may occur, further supporting a scenario in which ArtB and PltB monomers may compete for inclusion in the TT binding subunit.
FIG 4.
PltB replaces ArtB for inclusion in the holotoxin. (A) Schematic for the approach used for TAP and Western blot detection of heteropentameric binding subunits. (B) Detection of PltB-3×-Flag and ArtB-c-Myc using TAP and Western blotting. Holotoxins were pulled down from E. coli BTH101 strains expressing CdtB-His, PltA-Strep, PltB-3×-Flag, and ArtB-c-Myc using α-His antibodies (His) followed by purification of PltB-3×-Flag-containing holotoxins by pulling down with α-Flag magnetic beads (lane His>Flag). Proteins were visualized with antibody staining to detect PltB-3×-Flag (left), ArtB-c-Myc (middle), and both PltB-3×-Flag and ArtB-c-Myc (right). Results of additional replicates can be found in Fig. S2 in the supplemental material. (C) Immunoblots demonstrating the specificity of the pulldown assays and antibody detection used for TAP (additional blots for additional replicates can be found in Fig. S2). (D) Results of the binding subunit exchange assay. PltB-3×-Flag was added at various concentrations to lysates containing CdtB-His, PltA-Flag, and ArtB-c-Myc (left), or ArtB-c-Myc was added to lysates containing CdtB-His, PltA-Flag, and PltB-3×-Flag (right). Holotoxins were purified by pulling down with α-His antibodies to detect CdtB-His, and subunits were detected using tag-specific antibodies (PltA-Flag, CdtB-His, and PltB-3×-Flag). Results from one representative experiment are shown; the assay was performed as three independent experiments. Images for additional replicates can be found in Fig. S2.
Additional biological replicates and controls for experiments shown in Fig. 4. (A) Detection of ArtB-c-Myc with rabbit α-c-Myc antibodies following immunoprecipitation with α-His (lane His) or α-His and α-Flag (lane His>Flag) antibodies from whole-cell lysates containing various combinations of TT proteins and ArtB. (B) Biological replicates 2 and 3 for Fig. 4B in the main text. Immunoprecipitation of lysates from E. coli BTH101 expressing CdtB-His, PltA-Flag, PltB-3×-Flag, and ArtB-c-Myc was performed using either α-His (lane His) or α-His and α-Flag antibodies (lane His>Flag), followed by detection of ArtB-c-Myc (lower band) and PltB-3×-Flag (upper band). (C) Additional replicates 2 and 3 for Fig. 4D in the main text. PltB-3×-Flag was added at various concentrations to lysates containing CdtB-His, PltA-Flag, and ArtB-c-Myc (left), or ArtB-c-Myc was added to lysates containing CdtB-His, PltA-Flag, and PltB-3×-Flag (right). Holotoxins were purified by pulling down with α-His antibodies to detect CdtB-His, and subunits were detected using tag-specific antibodies (PltA-Flag, CdtB-His, PltB-3×-Flag, and ArtB-c-Myc). For replicate number 2, as well as replicate number 1 shown in Fig. 4D in the main text, the eluent from pulldown assays representing each level of added PltB-3×-Flag or ArtB-c-Myc was divided into two aliquots, which were then electrophoresed on separate gels and detected on separate blots to enable better resolution of PltA and PltB; for replicate no. 3, the eluent for each set of pulldowns (i.e., with different amounts of added PltB-3×-Flag [left] or ArtB-c-Myc [right]) was run on a single gel, and all three antibodies (α-His, α-Flag, and α-c-Myc antibodies) were added to the same membrane for detection of CdtB-His, PltA-Flag, PltB-3×-Flag, and ArtB-c-Myc. Download FIG S2, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PltB and ArtB may compete for inclusion in the holotoxin.
Collectively, our data suggested that artB and pltB are both induced when Salmonella is grown in medium mimicking intracellular conditions during Salmonella infection of epithelial cells, and both ArtB and PltB can be used to form biologically active toxins. Therefore, we hypothesized that ArtB and PltB monomers likely compete for inclusion in the holotoxin binding subunit. We thus used a modified competition assay to assess whether addition of PltB to lysates containing CdtB, PltA, and ArtB resulted in replacement of ArtB in the holotoxin, and vice versa. Cell lysate from E. coli BTH101 overexpressing CdtB, PltA, and either ArtB or PltB was challenged with various concentrations of PltB- or ArtB-containing lysate, respectively, prior to TAP and subsequent Western blot detection. Upon challenging cell lysates containing CdtB, PltA, and ArtB with increasing amounts of lysates containing PltB, we found that PltB replaced ArtB, thereby reducing the total amount of ArtB bound in the holotoxin (Fig. 4D and Fig. S2). This result was not reciprocal, however, as addition of ArtB only marginally reduced the amount of PltB in the holotoxin (Fig. 4D and Fig. S2). Together, this suggests that PltB and ArtB may compete for inclusion in the binding subunit.
DISCUSSION
Multiple investigations into the role of ArtB, a homolog of PltB, have suggested that ArtB can form a functional TT binding subunit (16, 26, 32). Our results suggest a role for ArtB in the diversification of the TT in NTS serovars, as (i) both toxin subunits are coencoded by serovars spanning multiple lineages of S. enterica subsp. enterica, (ii) both artB and pltB are expressed under conditions mimicking the intracellular environment, with differences in transcript abundances at neutral and acidic pH potentially favoring assembly of TT with different binding subunits (ArtB or PltB) under different environmental conditions, and (iii) ArtB and PltB may compete for inclusion in the binding subunit.
TT genes are considerably more widespread among S. enterica subsp. enterica serovars than previously thought.
Owing to the initial discovery of cdtB in S. Typhi strain CT18 in 2000, the majority of research efforts have, and continue to, focus primarily on the potential contributions of this toxin to typhoid fever (17, 18, 24, 30, 32, 36–40). Several studies have, however, demonstrated that TT is encoded by many nontyphoidal serovars (27–29, 31), suggesting that this toxin is not the sole contributing factor to the acute onset of typhoid fever, an observation which was recently confirmed in a human challenge study that demonstrated that onset of acute typhoid fever occurred even in subjects challenged with a TT-null strain (24). Importantly, our results support that TT genes are common in clade B serovars and are rare among clade A1 and A2 serovars. As the number of WGS data available for NTS serovars continues to increase, the number of serovars that are known to encode TT will likely increase. Regardless, our results suggest that studies examining the role of the TT in NTS serovars are warranted given the conservation of this toxin in nearly all clade B serovars, as reported here and in previous studies (28, 29).
Maintenance of the artB islet in TT-encoding serovars representing multiple lineages of S. enterica subsp. enterica suggests an important role for ArtB.
Previous studies had suggested that some serovars encode both TT genes and artB (27, 31, 32). Our analyses demonstrate that 81% of TT-positive serovars also encode artB. More importantly, our results indicate that the location of the artB islet within the sap operon is consistent across representative serovars in clades A1, A2, and B and section Typhi. sap operon genes have been shown to play a role in resistance to host-derived antimicrobial peptides for S. Typhimurium (41), uropathogenic E. coli (42), and Haemophilus ducreyi (43). In 2010, Rodas et al. demonstrated that integration of the genomic islet GICT18/1 (putative artB islet) into S. Typhi sapB resulted in an increased sensitivity to the antimicrobial peptide protamine (44). Furthermore, the authors showed that excision of GICT18/1 restored transcription of sapD and sapF and restored resistance to protamine to levels similar to that observed for S. Typhimurium, which lacks the GICT18/1 islet (44). As insertion of the artB islet is expected to result in sensitization to antimicrobial peptides, these results suggest that there is likely an advantage for the selective maintenance of artB by most TT-encoding serovars.
This begs the question of why the majority of TT-positive Salmonella serovars would evolve to encode multiple binding subunits, especially if acquisition of one (ArtB) may result in an increased susceptibility to host-derived antimicrobial peptides. Our current understanding is that PltB and ArtB have adapted to bind to different cell surfaces (26, 32), thereby expanding the variety of cell types that TT can bind to. While PltB has been reported to preferentially bind Neu5Ac-terminated glycans (26), ArtB can bind both Neu5Ac- and Neu5Gc-terminated glycans (26), suggesting that PltB and ArtB allow TT to bind to different cell types. Furthermore, Lee et al. recently established that specific amino acid residues in PltB produced by S. Javiana (called Javiana toxin in that study) were associated with toxin binding to intestinal epithelial cells but not to endothelial cells of brain arterioles (45), therefore supporting the idea that PltB produced by S. Typhi and S. Javiana may have evolved to enable binding to different tissues. Given that single amino acid changes have been found to significantly dampen binding affinity to cell surface glycans (45), collectively, this suggests that encoding two different subunits may provide an evolutionary advantage, as it likely enables TT-positive Salmonella to affect a wider range of cell types, tissues, and possibly hosts. Therefore, future studies examining the sequence variants of ArtB and PltB among a broader range of Salmonella serovars, as well as the binding affinities for these variants for a panel of different cells representing different tissues and hosts, will provide a more complete picture of the potential role of the TT binding subunit in facilitating tissue and or host tropism.
Furthermore, there is also some evidence to support the role of TT as an immunotoxin. Previous reports showed that injection of TT containing PltB or ArtB (called PltC in S. Typhi) into C57BL/6 mice resulted in significantly lower levels of total white blood cells, lymphocytes, and monocytes, than injection with phosphate-buffered saline (PBS) (32). In a human challenge study, subjects that were administered TT-null S. Typhi had higher fold changes of IgA and IgG against O9:lipopolysaccharide (LPS) than subjects that were administered wild-type TT (although not explicitly addressed in the study, this strain would also encode artB) (24). Therefore, TT may play a role in dampening the adaptive immune response, which may have implications for reinfection with TT-producing Salmonella. While these studies suggest that TTs containing ArtB and/or PltB may act as an immunotoxin, additional experimentation to assess the expression of and activity of ArtB- and PltB-containing TTs will be essential for addressing TT’s potential role in modulating the immune response during infection with typhoidal and nontyphoidal TT-producing serovars.
While copurification of ArtB and PltB suggest the formation of heteropentameric forms of the TT binding subunit, homopentamers appear to be the predominant form of the binding subunit.
Overall, our data, combined with previous studies, suggest that multiple mechanisms likely fine-tune ArtB and PltB expression and their assembly into different TT forms (i.e., toxins having binding subunits containing just PltB or ArtB or a mix of both PltB and ArtB). At the transcriptional level, both artB and pltB are induced when cells are grown under low-Mg2+ conditions (this study and reference 30). However, our results also indicate that pH may serve as an additional environmental cue modulating expression of artB and TT genes. For example, artB is more highly induced under acidified conditions (pH 5.8), while induction of pltB is higher at neutral pH. In S. Typhi, artB and pltB are regulated by different two-component systems; artB (called pltC in S. Typhi) expression is primarily regulated by SsrA-SsrB (32), while TT genes are primarily regulated by PhoP-PhoQ (30). However, both SsrA-SsrB and PhoP-PhoQ are induced under low-Mg2+ conditions (34, 46). As SsrB DNA binding is enhanced at low pH (47), while binding of PhoP (predicted regulator of TT genes) is not pH sensitive, our study provides additional support for a role of pH in fine-tuning transcription of TT genes. While this also suggests that regulation of artB and pltB transcription is conserved across S. enterica subspecies additional studies are needed to confirm the regulatory networks that control artB and pltB transcription in a broader range of NTS species.
At the level of toxin assembly, our experimental data suggest that the formation of heteropentameric TT binding subunits may occur, at least under some environmental conditions. The possible occurrence of PltB-ArtB interactions was confirmed by (i) two-hybrid system experiments, which showed evidence for some ArtB-PltB interactions for S. Javiana, and (ii) pulldown experiments, which showed consistent copurification of ArtB with PltB. However, our data also indicate that homopentamers may be the favored configuration of the binding subunit, as (i) PltB-PltB and ArtB-ArtB interactions were readily detected even in a heterologous host, while weak evidence for ArtB-PltB interactions (significant at α = 0.1) was only detected for some constructs in S. Javiana grown in broth, (ii) Western blot detection of PltB and ArtB after TAP suggested that low levels of ArtB and PltB are present when TTs are coimmunoprecipitated in a manner targeting the other binding units (i.e., ArtB or PltB, respectively), and (iii) competition assays demonstrate that PltB efficiently replaces ArtB in the holotoxin at neutral pH, which was in contrast to the results of the in silico modeling suggesting that ArtB-PltB interactions were more favorable than PltB-PltB interactions. Similar to our experimental findings, a recent study suggested that TT produced by S. Typhi is predominantly composed of either PltB or ArtB homopentamers, although the possibility of heteropentameric configurations could not be excluded in that study (32). As both the study described here and a previous study (32) overexpressed toxin subunits in a heterologous host, future studies examining toxin expression and assembly using untagged subunits (as tagged subunits were used in this study) under native conditions in Salmonella will be important for understanding the putative role of heteropentameric configurations of the TT binding subunit. Furthermore, differences in the amino acid sequences of PltA and ArtB encoded by different serovars (such as S. Typhi and S. Javiana [33]) may also need to be considered for studying the assembly of this toxin among a broader range of S. enterica serovars.
It is important to note that the experimental methods used here cannot distinguish configurations of the TT binding subunit with differing ratios of PltB and ArtB; additional structural evidence will be necessary to confirm the formation of heteropentameric configurations of the binding subunit and to assess the ratios of holotoxins with homo- and heteropentameric configurations of the binding subunit. In addition, whether the formation of ArtB-PltB heteropentameric binding subunits represents transitional forms of the TT or whether they have specific functional or virulence benefits also remains to be elucidated. One possibility is that differences in transcriptional regulation of artB and TT genes (cdtB, pltA, and pltB) resulting from changes in environmental cues (e.g., acidification of the Salmonella-containing vacuole [SCV]) could lead to a transitory state in which TTs with heteropentameric binding subunits represent an intermediate step that occurs as the TT transitions into a different homopentameric binding subunit (i.e., ArtB only or PltB only) (Fig. 5). Assessment of TT assembly under different conditions such as those mimicking intracellular conditions (e.g., acidified SCV) will therefore be important for procuring a more complete understanding of the potential complementary roles that ArtB and PltB play in the function of the TT. Additional experiments examining the delicate balance of transcriptional, as well as possible translational and posttranslational, regulatory mechanisms driving the formation of TT will be important for understanding the in vivo occurrence and any potential biological relevance of TTs with different binding subunits.
FIG 5.
Proposed relationship between environmental cues and TT formation. Based on our data and what has been shown previously for the regulation of TT in S. Typhi, at neutral pH, transcription of pltB is induced by PhoP (30), which may lead to the preferential inclusion of PltB in the TT binding subunit. Conversely, at slightly acidic pH, SsrB induces transcription of artB (32) in a pH-sensitive manner (47), which may favor preferential inclusion of ArtB in the TT binding subunit. The copurification of ArtB and PltB suggests the potential formation of heteropentameric forms of the binding subunit, although a role for these heteropentameric subunits will require further investigation.
Overall, our data contribute to a growing body of evidence that suggests that ArtB plays an important and currently underappreciated role in the activity of the TT. As additional serovars are found to encode both ArtB and PltB, a more complete understanding of the TT will require a diversified approach to better assess the role that TT plays in infection with nontyphoidal serovars.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
Bacterial strains used in this study are listed in Table S5 in the supplemental material, vectors and recombinant constructs are listed in Table S6, and primers are listed in Table S7. Bacterial strains were routinely grown in Difco Luria-Bertani (LB) broth, pH 7, containing 5 g/liter NaCl (Becton, Dickinson [BD], Franklin Lakes, NJ). For two-hybrid system interactions, E. coli and S. Javiana strains were grown in M63 medium with maltose as the sole carbon source (48). N-salts minimal medium (pH 5.8 or 7) containing 8 µM MgSO4 (34) was used for experiments assessing transcript abundances of TT genes and artB. Unless otherwise indicated, ampicillin and kanamycin were used at 100 µg/ml and 50 µg/ml, respectively, in complex medium (LB broth) and 50 µg/ml and 25 µg/ml, respectively, in chemically defined media (M9 or N-salts minimal medium).
Strains used in this study. Download Table S5, PDF file, 0.1 MB (142.9KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Vectors and constructs used in this study. Download Table S6, PDF file, 0.1 MB (146.1KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S7, PDF file, 0.1 MB (128.1KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Human intestinal epithelial cells (HIEC-6 cells; ATCC, Manassas, VA) (49) were cultured in Opti-MEM (Gibco-Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco-Invitrogen) and recombinant epidermal growth factor (10 ng/ml; Gibco-Invitrogen) at 37°C with 5% CO2. Cell supernatants were routinely tested to confirm the absence of Mycoplasma and Acholeplasma species infection using the VenorGEM Mycoplasma detection kit (Sigma-Aldrich, St. Louis, MO).
Cloning and expression of toxin proteins in E. coli.
Toxin subunits were expressed in E. coli BTH101 cells (50) using (i) pltB-3×-Flag-artB-c-Myc cloned into the high-copy-number pUT18 vector or (ii) cdtB-His-pltA-Flag and cdtB-His-pltA-Strep, each cloned into the low-copy-number pKNT25 plasmid. All PCRs were performed using the high-fidelity polymerase Q5 (New England Biolabs [NEB], Ipswich, MA) and the primers listed in Table S7. All constructs were designed to contain identical ribosome binding sites (AGGAGG) 5 to 7 bases upstream of the ATG start codons. The DNA sequences of all constructs were confirmed with Sanger sequencing.
For construction of pUT18::pltB-3×-Flag-artB-c-Myc, PCR products representing (i) pltB-3×-Flag and (ii) artB-c-Myc were digested using (i) XbaI and KpnI and (ii) KpnI and EcoRI, respectively, and were ligated into XbaI-EcoRI-digested pUT18 using T4 DNA ligase (NEB). For construction of pKNT25::cdtB-His-pltA-Flag and pKNT25::cdtB-His-pltA-Strep, PCR products (i) cdtB-His and (ii) pltA-Flag or pltA-Strep were digested with (i) XbaI and KpnI and (ii) KpnI and SacI, respectively, followed by ligation into XbaI-SacI-digested pKNT25. To construct pKNT25 containing cdtB-His only, the pltA-Flag region was deleted from pKNT25::cdtB-His-pltA-Flag by digesting plasmid DNA with KpnI and SacI. The resulting 3′ overhang was removed, and the 5′ overhang was filled in with T4 DNA polymerase (NEB). Following purification, the plasmid was self-ligated with T4 DNA ligase. pKNT25::pltA-Flag, pUT18::pltB-3×-Flag and pUT18::artB-c-Myc were constructed in a similar fashion.
Expression of these constructs was carried out in an E. coli BTH101 cAMP-null strain (ΔcyaA) due to the observed toxicity of TT components in E. coli NEB5α cells (NEB). Sequence-confirmed clones were cotransformed (pUT18 and pKNT25) into E. coli BTH101 cells and were selected for ampicillin and kanamycin resistance on LB agar plus 1% (wt/vol) glucose, incubated at 37°C.
Assessment of two-hybrid system interactions.
Interactions of CdtB, PltA, PltB, and ArtB subunits were first assessed using the bacterial adenylate cyclase two-hybrid (BACTH) system in E. coli BTH101 cells (50). The full-length polypeptides of the toxin subunits (including signal peptides) were fused to the N termini of T25 or T18 subunits in pKNT25 and pUT18, respectively (Fig. 3C). To ensure cytoplasmic CyaA activity, we also cloned the coding regions of CdtB, PltA, PltB, and ArtB lacking the signal peptide into BACTH plasmids for fusion at either the N terminus (pKNT25 and pUT18) or C terminus (pKT25 and pUT18C). Regions encoding the C-terminal interacting domains of PltA, PltB, and ArtB, as predicted from the TT structure (18, 26), were also cloned into pKNT25, pUT18, pKT25, and pUT18C plasmids (Tables S6 and S7). As 324 theoretical combinations exist, we used a high-throughput screening method. Screening was performed by cotransforming T18 and T25 vectors into E. coli BTH101 cells, selecting on LB agar plus 1% (wt/vol) glucose with ampicillin and kanamycin, and subsequently substreaking colonies onto LB agar with ampicillin and kanamycin plates containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 40 µg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) for blue-white screening. To ensure that positive interactions were not false positives arising from cAMP-independent CAP* spontaneous mutations, plasmid DNA was purified from blue colonies and retransformed into E. coli BTH101. Plasmids were isolated from clones that resulted in blue colonies after the second transformation and were analyzed by Sanger sequencing to determine the identity of the interacting domains.
Construction of ΔcyaA S. Javiana.
Deletion of cyaA was performed to enable screening for maltose utilization resulting from an interaction of T18- and T25-fused proteins. Whole-genome sequence analysis identified a single full-length cyaA in the S. Javiana strain used in this study (FSL S5-0395). Construction of a ΔcyaA S. Javiana strain was performed using the λ-Red recombinase system as described previously (16). Plasmids and primers used to generate the ΔcyaA strain are listed in Tables S6 and S7, respectively. The in-frame deletion was confirmed by Sanger sequencing. The mutant strain was also phenotypically confirmed by a lack of growth in M63 medium containing maltose as the sole carbon source; growth was restored upon addition of 0.05 mM cAMP.
Expression of the two-hybrid system constructs in M63 minimal medium.
Plasmids containing ArtB and PltB constructs were transformed into the ΔcyaA S. Javiana strain via electroporation. Transformed cells were selected on LB agar plates supplemented with ampicillin and kanamycin. Overnight cultures (12 to 14 h) were grown shaking at 30°C in LB broth with ampicillin and kanamycin. Subsequently, M63 broth containing ampicillin, kanamycin, and 0.02 mM cAMP was inoculated with an overnight culture (diluted 1:100), followed by incubation with shaking at 30°C for 24 h. The addition of 0.02 mM cAMP (a concentration that does not support the growth of negative controls) (Fig. 3D) was necessary for the basal expression of the fusion proteins in S. Javiana. Growth was assessed as optical density at 600 nm (OD600), which was measured after 24 h with a BioTek synergy plate reader (BioTek Instruments, Inc., Winooski, VT). Each growth assay was performed as three independent experiments.
Molecular modeling of different forms of the TT binding subunit.
In silico modeling of TT subunits was done using the Phyre2 server (51). Construction of the TT with different ratios of PltB and ArtB was performed using the MatchMaker tool in Chimera software (52). The stability and biological relevance of homo- and heteropentamer formation were assessed by calculating free energies from individual subunit-subunit interactions within the pentamer, using the PDBe Protein Interfaces, Surfaces and Assemblies (PDBe PISA) server (53).
Tandem affinity purification.
Beveled flasks containing 250 ml of LB broth with ampicillin and kanamycin were inoculated with overnight cultures (12 to 14 h, 1:500 dilution) of E. coli BTH101 harboring plasmids pAG37 and pAG43, which express PltB-3×-Flag ArtB-c-Myc and CdtB-His PltA-Strep, respectively. Cells were grown shaking at 200 rpm for 4.5 h at 37°C, followed by induction with IPTG and cAMP (both 1 mM final concentrations) for an additional 1.5 h. Cells were collected by centrifugation and stored at −80°C. Cells were thawed on ice, resuspended in 2 ml of NTA buffer (50 mM NaH2PO4 [pH 8.0], 0.3 M NaCl) containing 2 mg lysozyme, and incubated at 37°C for 30 min. Cell lysis was achieved with three rounds of freeze-thaw cycles (submersion in liquid nitrogen and incubation at 37°C) followed by sonication using a Branson Sonifier 250 sonicator (80% duty cycle, 7 output control) for 30 s on ice (performed twice). Cell debris was removed by centrifugation at 15,000 × g for 10 min. For CdtB-His purification, 50 µl of Dynabeads (Thermo Fisher Scientific; Waltham, MA) was washed twice with 500 µl of cold NTA buffer in preparation for TAP. Cell lysates were added to beads, followed by incubation overnight (12 to 14 h) at 4°C in a tube rotator. Beads were collected on a magnetic stand and were washed three times at 4°C with 500 µl of cold NTA buffer with incubation periods of 10 min. Proteins were eluted using 300 µl of NTA buffer containing 250 mM imidazole. Eluted proteins were then dialyzed against TBS buffer (50 mM Tris-Cl, 150 mM NaCl, pH 7.5) containing 10% (vol/vol) glycerol. PltB-3×-Flag was pulled down using Flag magnetic beads (EMD Millipore) according to the manufacturer’s instructions. Proteins were eluted from beads by boiling in the presence of 100 µl of 1× SDS-loading dye. PltB-3×-Flag and ArtB-c-Myc were detected in subfractions using Western blot analyses performed with rabbit anti-Flag (Sigma number [no.] F7425, diluted 1:500) and rabbit anti-c-Myc (Sigma no. SAB4301136, diluted 1:4,000) antibodies with goat anti-rabbit-horseradish peroxidase (HRP) antibodies (Thermo Fisher Scientific no. 65-6120, diluted 1:3,000). Horseradish peroxidase-conjugated secondary antibodies were detected using Clarity Western ECL substrate (Bio-Rad). Blots were visualized using the Bio-Rad ChemiDoc MP imaging system.
Western blot detection of proteins.
Protein samples were resolved on 4% to 20% Mini-Protean TGX precast protein SDS-PAGE gels (Bio-Rad Laboratories; Hercules, CA) and blotted on polyvinylidene difluoride (PVDF) membranes using the Trans-Blot Turbo transfer system (Bio-Rad). Membranes were incubated in TBS buffer containing 0.1% (vol/vol) Tween 20 (TTBS) and 5% (wt/vol) blocking reagent (Bio-Rad) with gentle shaking at room temperature for 30 min. Primary antibodies were added as described above with mouse anti-strep (Millipore Sigma no. 71590-M, 1:1,000 dilution) and mouse anti-His antibodies (Santa Cruz Biotechnology, Dallas, TX; no. SC-53073, 1:400 dilution) diluted in TTBS with 0.5% (wt/vol) blocking reagent, followed by incubation with gentle shaking at room temperature for 12 to 14 h. Membranes were washed three times with TTBS, followed by incubation (2 h at room temperature with gentle shaking) with secondary antibodies (goat anti-rabbit-HRP [Thermo Fisher Scientific no. 65-6120, diluted 1:3,000] and donkey anti-mouse-Alexa 647 [Thermo Fisher Scientific no. A-31571, diluted 1:1,000]) diluted in TTBS with 0.5% (vol/vol) blocking reagent. Membranes were washed twice in TTBS and once in TBS before Western blot detection. Blots were visualized using the Bio-Rad ChemiDoc MP imaging system.
Binding subunit exchange assay.
Protein expression and cell lysis of E. coli BTH101 strains (FSL G4-0035 to G4-0038) were performed as described above for TAP with cell pellets resuspended in PBS containing 10% (vol/vol) glycerol. Final total protein concentrations were determined spectrophotometrically (Nanodrop 2000c) (54, 55). The PltB-3×-Flag-containing lysate was added at various protein concentrations to 100 µg of total cell lysate containing CdtB-His, PltA-Flag, and ArtB-c-Myc for a final reaction volume of 50 µl of PBS with 10% (vol/vol) glycerol. The reaction mixture was incubated at 37°C for 30 min, followed by 2 h of incubation at 4°C to ensure equilibrium. A 200-µl aliquot of NTA buffer was added, and protein complexes were purified using Ni-nitrilotriacetic acid (NTA) beads, as described above. The assay was also performed with lysates containing CdtB-His, PltA-Flag, and PltB-3×-Flag as target and ArtB-c-Myc as the competitor. All proteins were detected by Western blotting using antibodies that recognize the corresponding protein tag (i.e., Flag, anti-His, and anti-c-Myc); three independent experiments were performed.
Intoxication of HIEC-6 cells with crude lysates of toxin components expressed in E. coli BTH101 cells.
Fresh Opti-MEM (containing 10 ng/ml recombinant epidermal growth factor [r-EGF] and 10% [vol/vol] FBS) medium supplemented with 10 µg/ml gentamicin was added to HIEC-6 cells grown to confluence on 12-mm coverslips (Thermo Fisher Scientific) in 24-well plates (Corning Inc., Corning, NY). Lysates containing various combinations of toxin subunits were then added to HIEC-6 cells at a final concentration of 400 µg total protein per ml, and the HIEC-6 cells were subsequently incubated at 37°C with 4.5% CO2. Immunofluorescence (IF) detection of DDR foci and cell cycle analyses were performed at 24 ± 2 h after inoculation. Three independent experiments were performed for both cell cycle and IF detection experiments.
IF detection of DDR proteins γH2AX and 53BP1.
IF staining for γH2AX and 53BP1 foci was performed as described previously (16, 33). The following antibodies were used: mouse anti-γH2AX (diluted 1:500, no. 05-636; Millipore Sigma, Billerica, MA), rabbit anti-53BP1 (diluted 1:500, no. NB 100–304; Novus Biologicals, Littleton, CO), donkey anti-rabbit conjugated to Alexa 555 (diluted 1:500, no. A-31572; Thermo Fisher Scientific), and donkey anti-mouse conjugated to Alexa 647 (diluted 1:200, no. A-31571; Thermo Fisher Scientific). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) for 5 min at room temperature. Microscopic observation was performed using a Zeiss 710 confocal microscope. FIJI software was used for image processing (56). Cells (at least 50 were scored per treatment) were considered to have an activated DDR if their nuclei had at least four 53BP1-positive foci and also contained γH2AX foci.
Cell cycle analyses.
Staining with propidium iodide for cell cycle analysis determination was performed as described previously (16, 33). DNA content (for cell cycle analysis) was assessed using the FACSAria flow cytometer (BD). Gating was performed to exclude multiplets as described previously (16, 57).
Whole-genome sequence data analyses for 235 Salmonella serovars.
Assemblies for the 235 S. enterica subsp. enterica serovars and five other S. enterica subspecies were downloaded from NCBI (see Data Set S1 for S. enterica data set). For the S. enterica data set, isolates were selected to include serovars commonly associated with human clinical disease (7) and those representing novel clades of S. enterica (29). Sequences from a convenience sample of 40 S. Javiana isolates, representing 28 unique SNP clusters were downloaded from the NCBI Pathogen Detection browser (https://www.ncbi.nlm.nih.gov/pathogens/isolates/#/search/) (see Table S3 for details); WGS data for S. Javiana FSL S5-0395 was also included. S. Mississippi isolate SRR1960042 was included as an outgroup for phylogenetic analyses (Table S3). For these data, Illumina adapters from sequence reads were trimmed, and low-quality bases were removed using Trimmomatic 0.33 with default settings (58). Determination of the quality of trimmed reads was performed using FastQC v0.11.7 (59). De novo assembly of all genomes was performed using SPAdes 3.6.0 (60). To assess the qualities of draft genomes, QUAST 3.2 (61) was used, followed by BBmap 35.49 (62) and SAMtools 1.3.1 (63) to calculate average coverage. Serotypes were confirmed using SISTR (64). kSNP3 was used to identify core SNPs in (i) the S. enterica data set and (ii) 40 S. Javiana genomes and strain FSL S5-0395; a k-mer size of 19 was used for both data sets, as determined using kSNP3’s Kchooser function (65). Phylogenetic trees were constructed with RAxML (66) using a general time-reversible model with gamma-distributed sites constructed from either 100 (S. enterica data set) or 1,000 (S. Javiana data set) bootstrap repetitions. iTOL v4 was used for editing of phylogenetic trees (67, 68).
Predicted serotype data for whole-genome assemblies used in analyses included in Fig. 1. Download Data Set S1, XLSX file, 0.1 MB (36.1KB, xlsx) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection of TT genes and artAB.
The presence of artA, artB, and TT genes was queried for all isolates using nucleotide BLAST (blastn) version 2.3.0 with default settings to query artA, artB, pltA, cdtB, and pltB sequences from S. Javiana strain CFSAN001992 (69); an E value of 1e−20 was used for querying S. Javiana genomes, and an E value of 1e−10 was used for querying the 235 serovars. In addition, closed genomes were downloaded to visually assess the location of TT genes and artB in serovars representing different phylogenetic clades (see Table S2 for assembly and strain information). Genomes were selected if they (i) represented commonly used strains from serovars associated previously with outbreaks of human clinical illness (e.g., CT18 for S. Typhi, AKU_12601 for S. Paratyphi A, CFSAN001992 for S. Javiana, and ATCC BAA-1673 for S. Poona) or (ii) were among the few closed genomes available for serovars that were positive for both TT and artB (serovars S. Goldcoast [clade A1] and S. Milwaukee [clade A2]); serovar S. Indiana was selected as an additional serovar for assessing the genomic location of toxin genes in section Typhi serovars, as S. Paratyphi A and S. Typhi represented a unique phylogenetic clade in our core genome analysis (Fig. 1A). In addition, to confirm the absence of artB in select S. Javiana assemblies, mapping of raw reads to artB from CFSAN001992 was performed using BWA version 0.7.17.
RT-qPCR quantification of artB and pltB transcript levels.
Overnight cultures (16 to 18 h) of FSL S5-0395 grown in LB broth were subcultured 1:1,000 into LB or N-salts minimal medium (either pH 7 or pH 5.8), and subcultured samples were grown at 37°C with shaking at 200 rpm until cells reached mid-exponential phase (3 h for LB broth, 5 h for N-salts minimal medium). RNA was stabilized with RNA protect (Qiagen) and was collected using the RNeasy kit (Qiagen). DNA was depleted with Ambion DNase I (Life Technologies), and DNA depletion was confirmed with quantitative PCR (qPCR); a cycle threshold (CT) of 34 for rpoB in DNase-treated RNA samples was used for judging successful DNA depletion. cDNA libraries were prepared with the Superscript reverse transcription kit (Thermo Fisher), according to manufacturer’s instructions. qPCR was performed with SYBR green 2× master mix (Applied Biosystems) in a reaction mixture containing 0.4 μM each primer (see Table S7) and 1 µl of cDNA (approximately 15 ng of cDNA) as the template. Fold expression was calculated by raising the ΔΔCT to the power of the efficiency calculated for each primer pair (70). Results are the averages from three independent experiments, each performed in technical duplicates.
Statistical analyses.
Statistical differences were assessed using R studio version 3.4.2. using packages lme4 (71) 1.1-14, emmeans version 1.3.3 (72), lmerTest version 2.0-33 (73), and multicomp version 1.4-8 (74) for Dunnett’s test for multiple comparisons adjustment.
Data availability.
Scripts and data sets are available online at https://github.com/ram524/2019_ArtB.
ACKNOWLEDGMENTS
We thank Pete Chandrangsu for his guidance with the in silico modeling and the staff at the Cornell Statistical Consulting Unit and the Biotechnology Resource Center Imaging Facility for their guidance with statistical analyses and flow cytometry data collection, respectively.
A.G., A.S.H., M.W., and R.A.C. designed the study. A.G., A.S.H., A.R.C., and R.A.C. performed experimental, bioinformatical, and/or statistical analyses. All authors wrote and assisted with the revision of the manuscript.
Confocal microscopy data were acquired through the Cornell University Biotechnology Resource Center, with NIH 1S10RR025502 funding for the shared Zeiss LSM 710 Confocal Microscope. R.A.C. was partially supported by USDA 2020-67034-31905.
We declare that the submitted work was carried out in the absence of any personal, professional, or financial relationships that could potentially impact the outcomes of this research.
REFERENCES
- 1.Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference Group. 2015. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, Fields PI, Weill F-X. 2014. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res Microbiol 165:526–530. doi: 10.1016/j.resmic.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Crump JA, Mintz ED. 2010. Global trends in typhoid and paratyphoid fever. Clin Infect Dis 50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 6.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. 2008. Salmonellosis outcomes differ substantially by serotype. J Infect Dis 198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- 7.CDC. 2016. National enteric disease surveillance: Salmonella annual report, 2016. CDC, Atlanta, GA. [Google Scholar]
- 8.Boore AL, Hoekstra RM, Iwamoto M, Fields PI, Bishop RD, Swerdlow DL. 2015. Salmonella enterica infections in the United States and assessment of coefficients of variation: a novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS One 10:e0145416. doi: 10.1371/journal.pone.0145416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson DA, Lane CR, Easton M, Valcanis M, Strachan J, Veitch MG, Kirk MD, Howden BP. 2018. Increasing antimicrobial resistance in nontyphoidal Salmonella isolates in Australia from 1979 to 2015. Antimicrob Agent Chemother 62:e02012-17. doi: 10.1128/AAC.02012-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). 2019. The European Union one health 2018 zoonoses report. EFSA J 17:e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 12.Fàbrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivera-Chavez F, Baumler AJ. 2015. The pyromaniac inside you: Salmonella metabolism in the host gut. Annu Rev Microbiol 69:31–48. doi: 10.1146/annurev-micro-091014-104108. [DOI] [PubMed] [Google Scholar]
- 14.Haghjoo E, Galán JE. 2004. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 16.Miller RA, Betteken MI, Guo X, Altier C, Duhamel GE, Wiedmann M. 2018. The typhoid toxin produced by the nontyphoidal Salmonella enterica serotype Javiana is required for induction of a DNA damage response in vitro and systemic spread in vivo. mBio 9:e00467-18. doi: 10.1128/mBio.00467-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spanò S, Ugalde JE, Galán JE. 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Song J, Gao X, Galán JE. 2013. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Bel Belluz L, Guidi R, Pateras IS, Levi L, Mihaljevic B, Rouf SF, Wrande M, Candela M, Turroni S, Nastasi C, Consolandi C, Peano C, Tebaldi T, Viero G, Gorgoulis VG, Krejsgaard T, Rhen M, Frisan T. 2016. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog 12:e1005528. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nešić D, Hsu Y, Stebbins CE. 2004. Assembly and function of a bacterial genotoxin. Nature 429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 21.Ibler AEM, ElGhazaly M, Naylor KL, Bulgakova NA, F El-Khamisy S, Humphreys D. 2019. Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection. Nat Commun 10:4040. doi: 10.1038/s41467-019-12064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphreys D, ElGhazaly M, Frisan T. 2020. Senescence and host–pathogen interactions. Cells 9:1747. doi: 10.3390/cells9071747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge Z, Feng Y, Whary MT, Nambiar PR, Xu S, Ng V, Taylor NS, Fox JG. 2005. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect Immun 73:3559–3567. doi: 10.1128/IAI.73.6.3559-3567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibani MM, Jones E, Barton A, Jin C, Meek J, Camara S, Galal U, Heinz E, Rosenberg-Hasson Y, Obermoser G, Jones C, Campbell D, Black C, Thomaides-Brears H, Darlow C, Dold C, Silva-Reyes L, Blackwell L, Lara-Tejero M, Jiao X, Stack G, Blohmke CJ, Hill J, Angus B, Dougan G, Galan J, Pollard AJ. 2019. Investigation of the role of typhoid toxin in acute typhoid fever in a human challenge model. Nat Med 25:1082–1088. doi: 10.1038/s41591-019-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh M, Tanaka K, Nishimori K, Makino S-I, Kanno T, Ishihara R, Hatama S, Kitano R, Kishima M, Sameshima T, Akiba M, Nakazawa M, Yokomizo Y, Uchida I. 2005. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology (Reading) 151:3089–3096. doi: 10.1099/mic.0.27933-0. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Deng L, Stack G, Yu H, Chen X, Naito-Matsui Y, Varki A, Galán JE. 2017. Evolution of host adaptation in the Salmonella typhoid toxin. Nat Microbiol 2:1592–1599. doi: 10.1038/s41564-017-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Rivera LD, Bowen BM, den Bakker HC, Duhamel GE, Wiedmann M. 2015. Characterization of the cytolethal distending toxin (typhoid toxin) in non-typhoidal Salmonella serovars. Gut Pathog 7:19. doi: 10.1186/s13099-015-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova E, Furtado MR, Wiedmann M. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425. doi: 10.1186/1471-2164-12-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worley J, Meng J, Allard MW, Brown EW, Timme RE. 2018. Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. mBio 9:e2303-18. doi: 10.1128/mBio.02303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowler CC, Galan JE. 2018. Decoding a Salmonella Typhi regulatory network that controls typhoid toxin expression within human cells. Cell Host Microbe 23:65.e6–76.e6. doi: 10.1016/j.chom.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamamura Y, Tanaka K, Uchida I. 2017. Characterization of pertussis-like toxin from Salmonella spp. that catalyzes ADP-ribosylation of G proteins. Sci Rep 7:2653–2653. doi: 10.1038/s41598-017-02517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowler CC, Stack G, Jiao X, Lara-Tejero M, Galán JE. 2019. Alternate subunit assembly diversifies the function of a bacterial toxin. Nat Comm 10:3684. doi: 10.1038/s41467-019-11592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller RA, Wiedmann M. 2016. The cytolethal distending toxin produced by nontyphoidal Salmonella serotypes Javiana, Montevideo, Oranienburg, and Mississippi induces DNA damage in a manner similar to that of serotype Typhi. mBio 7:e02109-16. doi: 10.1128/mBio.02109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 35.Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. 2011. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology (Reading) 157:1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SJ, Jin SC, Jiao X, Galan JE. 2019. Unique features in the intracellular transport of typhoid toxin revealed by a genome-wide screen. PLoS Pathog 15:e1007704. doi: 10.1371/journal.ppat.1007704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang S-J, Song J, Galán JE. 2016. Receptor-mediated sorting of typhoid toxin during its export from Salmonella Typhi-infected cells. Cell Host Microbe 20:682–689. doi: 10.1016/j.chom.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galán JE, Varki A. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodak H, Galan JE. 2013. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep 14:95–102. doi: 10.1038/embor.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang YA, Lee S, Zhao J, Thompson AJ, McBride R, Tsogtbaatar B, Paulson JC, Nussinov R, Deng L, Song J. 2018. In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat Microbiol 3:155–163. doi: 10.1038/s41564-017-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parra‐Lopez C, Baer MT, Groisman EA. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J 12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mount KL, Townsend CA, Rinker SD, Gu X, Fortney KR, Zwickl BW, Janowicz DM, Spinola SM, Katz BP, Bauer ME. 2010. Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect Immun 78:1176–1184. doi: 10.1128/IAI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodas PI, Contreras I, Mora GC. 2010. Salmonella enterica serovar Typhi has a 4.1 kb genetic island inserted within the sapABCDF operon that causes loss of resistance to the antimicrobial peptide protamine. J Antimicrob Chemother 65:1624–1630. doi: 10.1093/jac/dkq197. [DOI] [PubMed] [Google Scholar]
- 45.Lee S, Yang Y-A, Milano SK, Nguyen T, Ahn C, Sim JH, Thompson AJ, Hillpot EC, Yoo G, Paulson JC, Song J. 2020. Salmonella typhoid toxin PltB subunit and its non-typhoidal Salmonella ortholog confer differential host adaptation and virulence. Cell Host Microbe 27:937–949. doi: 10.1016/j.chom.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi E, Groisman EA, Shin D. 2009. Activated by different signals, the PhoP/PhoQ two-component system differentially regulates metal uptake. J Bacteriol 191:7174–7181. doi: 10.1128/JB.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liew ATF, Foo YH, Gao Y, Zangoui P, Singh MK, Gulvady R, Kenney LJ. 2019. Single cell, super-resolution imaging reveals an acid pH-dependent conformational switch in SsrB regulates SPI-2. Elife 8:e45311. doi: 10.7554/eLife.45311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Perreault N, Beaulieu JF. 1996. Use of the dissociating enzyme thermolysin to generate viable human normal intestinal epithelial cell cultures. Exp Cell Res 224:354–364. doi: 10.1006/excr.1996.0145. [DOI] [PubMed] [Google Scholar]
- 50.Karimova G, Ullmann A, Ladant D. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J Mol Microbiol Biotechnol 3:73–82. [PubMed] [Google Scholar]
- 51.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Layne E. 1957. Spectrophotometric and turbidimetric methods for measuring proteins. Methods Enzymol 3:447–454. doi: 10.1016/S0076-6879(57)03413-8. [DOI] [Google Scholar]
- 55.Stoscheck CM. 1990. Quantitation of protein. Methods Enzymol 182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 56.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler‐Stevenson M, Gabrielson E. 2001. Doublet discrimination in DNA cell‐cycle analysis. Cytometry 46:296–306. doi: 10.1002/cyto.1171. [DOI] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bushnell B. 2015. BBMap v. 35.49. https://sourceforge.net/projects/bbmap/.
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 66.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young VB, Knox KA, Schauer DB. 2000. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect Immun 68:184–191. doi: 10.1128/IAI.68.1.184-191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allard MW, Muruvanda T, Strain E, Timme R, Luo Y, Wang C, Keys CE, Payne J, Cooper T, Luong K, Song Y, Chin CS, Korlach J, Roberts RJ, Evans P, Musser SM, Brown EW. 2013. Fully assembled genome sequence for Salmonella enterica subsp. enterica serovar Javiana CFSAN001992. Genome Announc 1:e0008113. doi: 10.1128/genomeA.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 71.Bates D, Maechler M, Bolker B, Walker S, Christensen R, Singmann H, Dai B, Grothendieck G, Green P. 2017. lme4: linear mixed-effects models using Eigen and S4, R package version 1.1-14. https://CRAN.R-project.org/package=lme4.
- 72.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 73.Kuznetsova A, Brockhoff PB, Christensen RHB. 2015. Package ‘lmerTest’. R package version 2. https://CRAN.R-project.org/package=lmerTest.
- 74.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom J 50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide lengths and identities of putative pltA and pltB that were not detected with initial blast searches. Download Table S1, PDF file, 0.1 MB (134.7KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Positions of artB islet in closed genomes from representative strains of TT and artB-positive serovars. Download Table S2, PDF file, 0.1 MB (131.5KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SRR run identity and metadata for 41 S. Javiana isolates and one S. Mississippi isolate used for phylogenetic and blast analyses. Download Table S3, PDF file, 0.1 MB (111.3KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of additional modeling and two-hybrid system constructs in E. coli BTH101 cells. (A) Modeling was performed to compare the calculated energies for S. Typhimurium DT104 ArtB (shown in red font) and S. Javiana CFSAN001992 ArtB (shown in black font). (B) Additional two-hybrid system clones tested as in Fig. 3C. Briefly, E. coli BTH101 cells were transformed with plasmids encoding partial interacting domain (_ID) or full-length proteins (_Full) fused to T18 and T25 subunits of CyaA (50). Blue pigmentation suggests an interaction, while tan colonies are unable to confirm an interaction. Some of these constructs are also shown in Fig. 3C and were included again here for convenience (i.e., showing the entire plate or half of the plate). Download FIG S1, PDF file, 2.8 MB (2.8MB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Estimates of linear model on log10-transformed absorbance (OD600) data of S. Javiana ΔcyaA mutants. Download Table S4, PDF file, 0.09 MB (95.4KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional biological replicates and controls for experiments shown in Fig. 4. (A) Detection of ArtB-c-Myc with rabbit α-c-Myc antibodies following immunoprecipitation with α-His (lane His) or α-His and α-Flag (lane His>Flag) antibodies from whole-cell lysates containing various combinations of TT proteins and ArtB. (B) Biological replicates 2 and 3 for Fig. 4B in the main text. Immunoprecipitation of lysates from E. coli BTH101 expressing CdtB-His, PltA-Flag, PltB-3×-Flag, and ArtB-c-Myc was performed using either α-His (lane His) or α-His and α-Flag antibodies (lane His>Flag), followed by detection of ArtB-c-Myc (lower band) and PltB-3×-Flag (upper band). (C) Additional replicates 2 and 3 for Fig. 4D in the main text. PltB-3×-Flag was added at various concentrations to lysates containing CdtB-His, PltA-Flag, and ArtB-c-Myc (left), or ArtB-c-Myc was added to lysates containing CdtB-His, PltA-Flag, and PltB-3×-Flag (right). Holotoxins were purified by pulling down with α-His antibodies to detect CdtB-His, and subunits were detected using tag-specific antibodies (PltA-Flag, CdtB-His, PltB-3×-Flag, and ArtB-c-Myc). For replicate number 2, as well as replicate number 1 shown in Fig. 4D in the main text, the eluent from pulldown assays representing each level of added PltB-3×-Flag or ArtB-c-Myc was divided into two aliquots, which were then electrophoresed on separate gels and detected on separate blots to enable better resolution of PltA and PltB; for replicate no. 3, the eluent for each set of pulldowns (i.e., with different amounts of added PltB-3×-Flag [left] or ArtB-c-Myc [right]) was run on a single gel, and all three antibodies (α-His, α-Flag, and α-c-Myc antibodies) were added to the same membrane for detection of CdtB-His, PltA-Flag, PltB-3×-Flag, and ArtB-c-Myc. Download FIG S2, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download Table S5, PDF file, 0.1 MB (142.9KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Vectors and constructs used in this study. Download Table S6, PDF file, 0.1 MB (146.1KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S7, PDF file, 0.1 MB (128.1KB, pdf) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted serotype data for whole-genome assemblies used in analyses included in Fig. 1. Download Data Set S1, XLSX file, 0.1 MB (36.1KB, xlsx) .
Copyright © 2021 Gaballa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Scripts and data sets are available online at https://github.com/ram524/2019_ArtB.