Abstract
Although cyanobacteria and algae represent a small fraction of the biomass of all primary producers, their photosynthetic activity accounts for roughly half of the daily CO2 fixation that occurs on Earth. These microorganisms are able to accomplish this feat by enhancing the activity of the CO2-fixing enzyme Rubisco using biophysical CO2 concentrating mechanisms (CCMs). Biophysical CCMs operate by concentrating bicarbonate and converting it into CO2 in a compartment that houses Rubisco (in contrast with other CCMs which concentrate CO2 via an organic intermediate, such as malate in the case of C4 CCMs). This activity provides Rubisco with a high concentration of its substrate, thereby increasing its reaction rate. The genetic engineering of a biophysical CCM into land plants is being pursued as a strategy to increase crop yields. This review focuses on the progress toward understanding the molecular components of cyanobacterial and algal CCMs, as well as recent advances toward engineering these components into land plants.
Keywords: carboxysome, pyrenoid, CO2 concentrating mechanism, Rubisco, synthetic biology, crop yields
INTRODUCTION
The CO2-Fixing Activity of the Enzyme Rubisco Is Limited by Slow Catalytic Rate and a Competing Reaction with O2
Carbon is an essential building block for life on Earth, and nearly all the organic carbon that is accessible to living organisms is thought to have passed through the enzyme Rubisco at some point in time. Rubisco, short for d-ribulose-1,5-bisphosphate carboxylase/oxygenase, captures inorganic CO2 and catalyzes its addition to ribulose-1,5-bisphosphate (RuBP), generating two molecules of 3-phosphoglycerate (3-PGA) that continue through the Calvin Benson Bassham (CBB) cycle to produce a sugar precursor.
Rubisco is highly productive on a global scale, collectively fixing roughly 1011 tons of carbon per year (22). However, the CO2-fixing activity of Rubisco is limited by a slow catalytic rate and a competing reaction with O2. Whereas the median turnover rate (kcat) of central carbon metabolism enzymes is approximately 79 reactions per second (6a), more than 95% of characterized Rubiscos catalyze only 1 to 10 carboxylation reactions per second (24). Furthermore, when Rubisco binds to O2 instead of CO2, it catalyzes a counterproductive oxygenation reaction that generates 2-phosphoglycolate from RuBP, which results in the removal of carbon from the CBB cycle. 2-phosphoglycolate must be recycled through a process called photorespiration that wastes energy and liberates fixed CO2 (68).
According to molecular dynamics simulations, Rubisco has an obvious preference for CO2 over O2 when both gases are present at equal concentrations (91). However, at atmospheric concentrations of 21% O2 and 0.04% CO2, Rubisco’s oxygenase activity occurs frequently, leading to substantial photorespiration. In C3 plants, which include trees and many crops such as rice and wheat, photorespiration results in the loss of about a quarter of fixed carbon (94).
Some organisms have evolved Rubisco with a higher specificity for CO2 (SCO2/O2), but this seems to come at the expense of the enzyme’s turnover rate for CO2 (kcat,C). The examination of Rubisco orthologs from a variety of species has revealed an inverse correlation between Rubisco’s kcat,C and its SCO2/O2, suggesting a trade-off between the two parameters (81). A proposed explanation for this trade-off is that because CO2 is a fairly featureless molecule, improving Rubisco’s SCO2/O2 may only be possible through stabilizing the carboxyketone intermediate that forms while CO2 is being added to RuBP (89). Tighter binding to the carboxyketone intermediate would slow conversion to the final product, resulting in a slower kcat,C.
C3 plants usually contain Rubiscos with a higher SCO2/O2 but lower kcat,C, and to compensate for Rubisco’s slow activity, these plants express abundant amounts of the enzyme, dedicating up to 25% of leaf nitrogen to do so (78).
CO2 Concentrating Mechanisms Promote Rubisco’s Carboxylase Activity
Many organisms, including C4 plants, cyanobacteria, and algae, overcome Rubisco’s limitations by using CO2 concentrating mechanisms (CCMs). CCMs increase the CO2/O2 ratio around Rubisco, thereby speeding carboxylation while limiting the occurrence of oxygenation.
By creating an environment around Rubisco that is enriched with CO2, organisms with CCMs are able to use a Rubisco with a lower SCO2/O2 and higher kcat,C. With a faster Rubisco, the same rate of carbon fixation can be achieved using less of the enzyme, which allows the organism to allocate fewer resources to producing Rubisco, improving the growth rate and amount of biomass produced per unit of nitrogen (28). Additionally, for land plants, CCMs allow capture of a greater fraction of the CO2 that diffuses into leaves through stomata, allowing plants to have fewer and/or smaller stomata, which decreases water losses due to transpiration and thus improves growth in arid environments. These benefits come at an energetic cost, but this is a small price to pay in many environments where photosynthetic energy is available in excess.
CCMs increase the concentration of CO2 in a compartment that houses Rubisco, but CO2 cannot be directly transported because it easily diffuses across membranes. Therefore, CCMs operate by converting CO2 to a charged intermediate that can be concentrated before being converted back to CO2 at a site near Rubisco.
C4 plants have biochemical CCMs, which rely on an enzyme temporarily fixing CO2 into a four-carbon organic molecule that is shuttled to a compartment containing Rubisco, where the molecule is decarboxylated to produce concentrated CO2. In most C4 plants, the initial fixation and decarboxylation occur in different tissues, the mesophyll and the bundle sheath, respectively. These two tissues are arranged in characteristic concentric rings, known as Kranz anatomy (Figure 1a).
Figure 1.
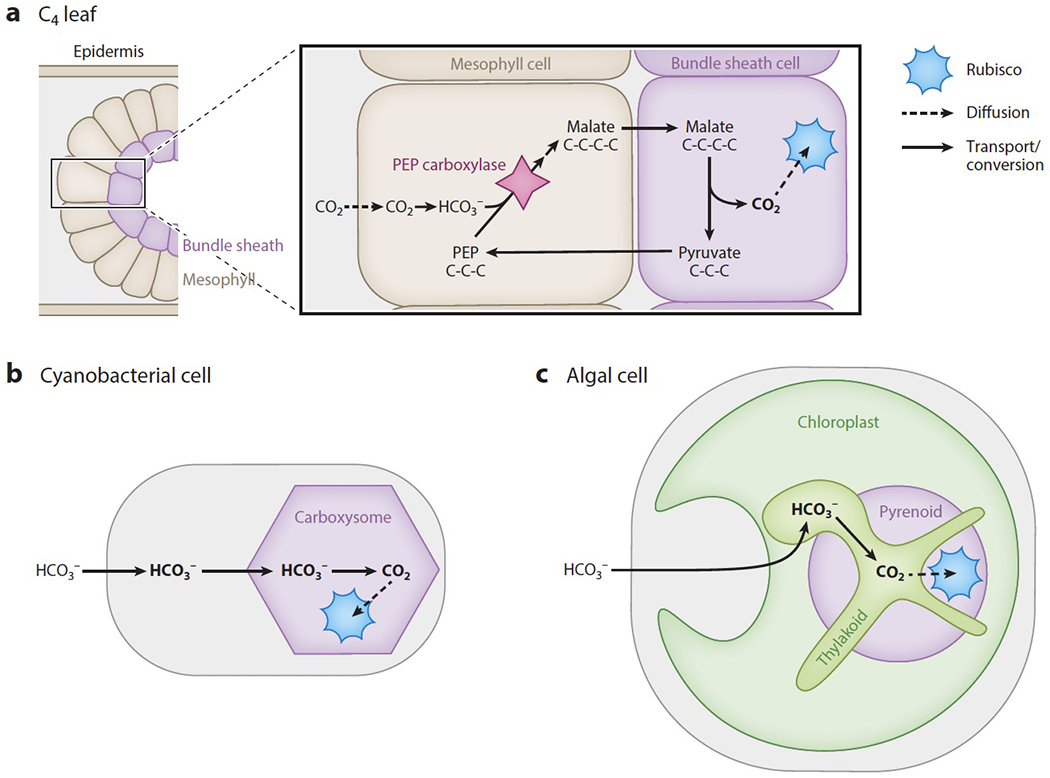
C4 plants, cyanobacteria, and algae concentrate CO2 around the enzyme Rubisco using different strategies. (a) In the mesophyll cells of C4 plants, HCO3− is added to the three-carbon molecule phosphoenolpyruvate (PEP) by the enzyme PEP carboxylase, generating the four-carbon molecule malate or aspartate. The malate or aspartate is then decarboxylated to form the three-carbon molecule pyruvate and release CO2 in bundle sheath cells, where Rubisco is expressed. The biophysical CCMs of (b) cyanobacteria and (c) algae actively transport HCO3− into the cell, then convert it to CO2 at a site of clustered Rubisco, which is either the (b) carboxysome or (c) pyrenoid
There are three known subtypes of the C4 mechanism, but in general they operate by using the enzyme phosphoenolpyruvate (PEP) carboxylase to temporarily fix CO2 in mesophyll cells. Dissolved CO2 is converted into bicarbonate (HCO3− hereafter), and PEP carboxylase adds HCO3− to the three-carbon molecule PEP, generating the four-carbon product oxaloacetate. Oxaloacetate is eventually converted to malate or aspartate, which is transported into bundle sheath cells that surround the vascular tissues. Within the bundle sheath cells, the four-carbon malate or aspartate is decarboxylated to the three-carbon molecule pyruvate, releasing CO2 in the vicinity of Rubisco. It is worth noting that some plants (18a, 82a) and some diatoms (78a) operate a single-celled C4 mechanism where the metabolic pathways are compartmentalized within a single cell rather than partitioned between two cell types. A more detailed discussion of the C4 pathway and efforts to engineer it into C3 plants is beyond the scope of this review; we refer the reader to expert reviews by Schlüter and Weber in this volume (81a) and by others (5, 26, 82).
Cyanobacteria and algae have biophysical CCMs that operate by transporting HCO3− and converting it to CO2 near a site of clustered Rubisco (Figure 1b,c). The negative charge on HCO3− impedes its diffusion across membranes, preventing it from escaping and allowing high concentrations of HCO3− to be maintained within the cell. HCO3− is then converted to CO2 in the proximity of Rubisco.
Cyanobacteria and algae leverage the properties of HCO3− and CO2 to enhance their CCMs. In this field, HCO3− and CO2 are referred to as inorganic carbon (Ci) because they lack C–H bonds. These Ci species equilibrate with each other, and their equilibrium concentrations depend on pH. HCO3− is the most abundant Ci species between pH 6 and 9, while CO2 is the most abundant below pH 6. Both algal (64) and cyanobacterial (54) CCMs leverage a high pH to capture CO2 into HCO3−. Algae appear to additionally convert HCO3− into CO2 at a low pH, which favors the reaction and uses protons concentrated by the photosynthetic light reactions to create a sink for HCO3−. Spontaneous equilibration between HCO3− and CO2 occurs slowly, on the timescale of 10 seconds (32), but is accelerated dramatically by carbonic anhydrase enzymes, which are among the world’s fastest enzymes, able to perform up to 106 reactions per second (38). Thus, by localizing the carbonic anhydrases to specific subcellular compartments, the cell is able to control where the conversion happens.
Engineering a CO2 Concentrating Mechanism into Land Plants Has the Potential to Improve Yields
Genetically engineering a CCM into C3 crops could improve yields by both increasing Rubisco’s carboxylation rate and reducing the energy lost to photorespiration. Free-air CO2 enrichment experiments have shown that crops produce higher yields when grown with elevated levels of CO2 (35). This supports the idea that engineering plants to concentrate CO2 at the subcellular level would also lead to increased yields. The percentage increase in yield observed in free-air CO2 enrichment experiments varies depending on the plant species. For example, cotton has an average of 42% increase in yield, whereas wheat and rice have a more modest yield increase of 15% (1). This discrepancy is attributed to differences in physiology; cotton leaves develop more rapidly, which may allow the plant to take advantage of enhanced photosynthesis starting at an earlier stage of its growth.
Limiting the occurrence of photorespiration is one of the key ways in which a CCM could improve plant growth. By elevating the CO2/O2 ratio around Rubisco, CCMs decrease both the energy and fixed CO2 lost to photorespiration. Significant progress has already been made toward engineering an alternative pathway that reduces the energetic cost of processing 2-phosphoglycolate, although it still liberates fixed CO2 (86). Transgenic tobacco plants engineered with the alternative pathway display a 24% increase in seasonal biomass as compared to wild-type plants. C3 plants engineered to contain a CCM would be expected to achieve the same benefits and additional yield increases by not liberating fixed CO2. Furthermore, plants with an engineered CCM are expected to have improved nitrogen and water use efficiency because they will require less Rubisco and less leaf gas exchange, as is the case for existing C4 plants (28).
This review focuses on our current understanding of the molecular components of biophysical CCMs as well as recent progress toward engineering these components into other organisms. The first section of this review covers progress that has been made toward characterizing the cyanobacterial CCM. The second section of this review discusses what is known about the algal CCM from studies of the freshwater alga Chlamydomonas reinhardtii. Within each section, progress toward engineering these CCMs into land plants and important future directions are discussed.
THE CYANOBACTERIAL CO2 CONCENTRATING MECHANISM
The concentration of Ci within a cyanobacterial cell begins with the active transport of HCO3− across the plasma membrane. Once concentrated within the cytosol, HCO3− diffuses into the carboxysome, which is composed of an icosahedral protein shell wrapped around Rubisco and a carbonic anhydrase. This carbonic anhydrase catalyzes the conversion of HCO3− into CO2, feeding Rubisco’s carboxylase activity (Figure 1b).
Two types of cyanobacteria, α-cyanobacteria, which are found predominately in marine environments, and β-cyanobacteria, which favor freshwater, appear to have convergently evolved carboxysomes. Components of α-carboxysomes are encoded by the cso operon in α-cyanobacteria, and components of β-carboxysomes are encoded either by the ccm operon or other operons at separate genomic loci. This review focuses on the β-carboxysome, but many of the molecular components that make up the α-cyanobacterial CCM are conceptually similar and are described briefly. Table 1 lists likely functionally analogous proteins between the CCMs of α-cyanobacteria, β-cyanobacteria, and the eukaryotic alga C. reinhardtii. Several outstanding recent reviews provide more detailed comparisons of α- and β-carboxysomes (37, 75, 101).
Table 1.
The biophysical CO2 concentrating mechanisms (CCMs) of α-cyanobacteria, β-cyanobacteria, and the alga Chlamydomonas reinhardtii have many analogous core components, although they evolved independently
| Type of protein | α-Cyanobacteria | β-Cyanobacteria | Chlamydomonas reinhardtii |
|---|---|---|---|
| Rubisco large subunit | CbbL | RbcL | RbcL |
| Rubisco small subunit | CbbS | RbcS | RbcS |
| Rubisco linker | CsoS2 (CsoS2A and CsoS2B isoforms) | CcmM (M35 and M58 isoforms) | EPYC1 |
| Carbonic anhydrases near the site of Rubisco | CsoS3 (CsoSCA) | CcmM58 N terminus, CcaA | CAH3 |
| Shell proteins: hexameric, pentameric, pore-forming | Hexameric: CsoS1A, CsoS1B, CsoS1C Pentameric: CsoS4A, CsoS4B Pore-forming: CsoS1D, CsoS1E |
Hexameric: CcmK2, CcmK3, CcmK4, CcmO Pentameric: CcmL Pore-forming: CcmP |
The Chlamydomonas pyrenoid does not appear to have a shell. |
| Bicarbonate transporters | Plasma membrane: BicA, SbtA | Plasma membrane: BicA, SbtA, BCT1 | Plasma membrane: HLA3 and LCI1 Chloroplast envelope: LCIA and CIA8 Thylakoid membrane: BST1, BST2, BST3 |
| CO2 recapture | NDH-I4 | NDH-I3, NDH-I4 | LCIB/LCIC complex |
Inorganic Carbon Is Concentrated in the Cell by HCO3− Transporters and Converted to CO2 by a Carbonic Anhydrase
At the plasma membrane of β-cyanobacteria, the heteromultimeric ATP-binding cassette transporter BCT1 and the homomultimeric Na+/HCO3− symporters BicA and SbtA actively transport HCO3− into the cytosol (Figure 2) (69). Both BCT1 and SbtA have a high affinity for HCO3−, while BicA has a low affinity. A previously published review has covered these Ci transporters in more detail (e.g., 69).
Figure 2.
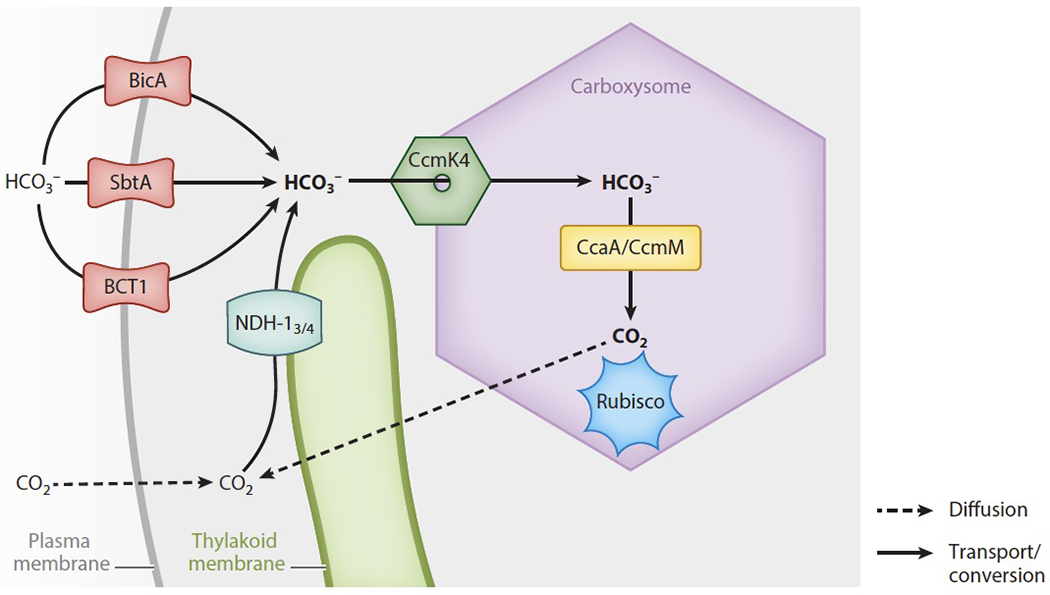
HCO3− transport in a β-cyanobacterial cell starts at the plasma membrane, where the transporters BicA, SbtA, and BCT1 are located. After being concentrated in the cytoplasm, HCO3− diffuses into the carboxysome, likely via pores in the shell protein CcmK4. Inside the carboxysome, carbonic anhydrases convert HCO3− into CO2, which can then be fixed by Rubisco clustered there. CO2 that diffuses into the cytoplasm or leaks out of the carboxysome is converted to HCO3− by the NDH-13 and NDH-14 complexes.
CO2 that diffuses across the plasma membrane or that leaks out of the carboxysome is captured in the cytosol and converted into HCO3− by the NDH-13 and NDH-14 complexes (Figure 2). β-cyanobacteria express both complexes, whereas α-cyanobacteria only encode NDH-14 (6). In β-cyanobacteria, the expression of the higher-affinity NDH-13 is induced when Ci is limited, whereas the lower-affinity NDH-14 is constitutively expressed (69).
Both of the NDH-1 complexes are membrane-bound, but whether they reside on the plasma or thylakoid membrane is controversial and may vary between species (6, 8). Little is known about the precise mechanism through which the NDH-13 and NDH-14 complexes facilitate CO2 uptake, but it is thought that they operate by using energy from electron transport to drive the unidirectional hydration of CO2 into HCO3− (5a).
Ultimately, the coordinated action of these Ci uptake systems leads to a state where the cytosolic HCO3− concentration is about 30 times the concentration found in water with pH 7 at 25°C (93). Maintaining this high concentration of HCO3− within the cytosol is important for the CCM, as it drives HCO3− diffusion into the carboxysome where HCO3− is converted into concentrated CO2 to feed Rubisco (72). The absence of carbonic anhydrase activity in the cytosol is critical for maintaining CCM function. When a carbonic anhydrase from humans was heterologously expressed in the cyanobacterial cytosol, cells required high CO2 to grow, indicating that their CCM was impaired (70). This is presumably because the presence of a carbonic anhydrase in the cytosol drives the conversion of HCO3− into CO2, allowing CO2 to diffuse out of the cell before it reaches Rubisco within the carboxysome.
Engineering a HCO3− Transport System Is an Important Step in Transferring a Cyanobacterial CO2 Concentrating Mechanism into Land Plants
Installing a cyanobacterial HCO3− transporter at the chloroplast inner envelope membrane (IEM) of a land plant has been suggested as a simple strategy for modestly boosting CO2 flux to Rubisco in the chloroplast without needing to build a carboxysome (71). The imported HCO3− could be converted to CO2 by β-carbonic anhydrases that are natively expressed in plant chloroplasts (16). One computational model predicted that incorporating a single HCO3− transporter could increase CO2 uptake by 9% and that expressing multiple transporters could lead to a 16% increase (58). However, this model had assumed ideal growth conditions, and a more detailed model using field data for crop plants (102) predicts that a full cyanobacterial CCM is needed to see crop improvement.
Because cyanobacteria are prokaryotes, they have a different cellular organization from plants, which adds a challenge for properly localizing engineered components. Cyanobacteria are the evolutionary relative and topological equivalent of plant chloroplasts. Therefore, to be targeted to their site of function, components from cyanobacteria need to either be directly transformed into the chloroplast genome or transformed into the nuclear genome fused to an exogenous chloroplast targeting sequence (also called transit sequence). Chloroplast transformation is straightforward in the dicotyledonous reference plant tobacco, which facilitates proof-of-concept studies. However, chloroplast genome transformation is ineffective in monocotyledonous plants, which include major CCM target crops such as rice and wheat, because a lack of selectable markers makes it difficult to obtain plants where each chloroplast expresses the transgenes (30). Therefore, practical efforts toward achieving a CCM in these target crops are likely to ultimately require expressing components from the nuclear genome with targeting sequences to localize them to the desired chloroplast sub-compartment.
An initial attempt was made to express BicA in tobacco leaf chloroplasts using biolistic transformation to insert the foreign DNA directly into the chloroplast genome (67). While BicA could be expressed without hindering the plant’s growth, only about 25% of the protein localized to the chloroplast envelope while 75% was found in the thylakoid membranes instead.
Experiments performed using nuclear transformation of BicA and SbtA fused with chloroplast transit peptides were more successful, both in Nicotiana benthamiana (79) and in Arabidopsis (90). Rolland et al. (79) identified N-terminal sequences in large Arabidopsis transmembrane proteins that could reliably redirect BicA and SbtA to the chloroplast envelope of N. benthamiana. Uehara et al. (90) were able to specifically direct BicA and SbtA to the IEM of Arabidopsis by fusing these HCO3− transporters to both a chloroplast transit peptide and a mature portion of a protein that is natively located in the IEM. In their study, BicA could stay embedded in the IEM even after removal of the transit peptide. This represents an encouraging step toward engineering functional prokaryotic transporters into the chloroplast membrane.
To identify the HCO3− transporter homologs that are most likely to function in other organisms, researchers expressed HCO3− transporters from various cyanobacterial species in Escherichia coli and tested them for functionality through examining cellular uptake of NaH14CO3 (17). This screen identified six active SbtA homologs, but none of the BicA or BCT1 homologs tested were able to transport HCO3−. Identifying additional factors necessary for BicA or BCT1 functionality could be important for reconstituting a cyanobacterial CCM in a land plant.
Going forward, a major goal for the field will be demonstrating that HCO3− is concentrated in the chloroplasts of engineered plants.
Shell Proteins Allow Selective Diffusion of Charged Molecules Between the Cytosol and the Carboxysome Interior
The protein shell of the carboxysome encapsulates densely packed Rubisco and is thought to selectively allow the channeling of HCO3− and RuBP into the carboxysome and 3-PGA out. Just three monomeric proteins are sufficient for building the icosahedral shell structure; these are CcmK2, CcmO, and CcmL (Figure 3a,b) (74). CcmK2 forms a hexamer that takes on a flattened hexagonal shape and forms the large sheets that make up the faces of the carboxysome icosahedron. CcmO forms trimers that have a hexagonal shape, and it likely bends to form the edges of the icosahedron. Finally, CcmL is a pentamer that creates the vertices of the shell. In α-cyanobacteria, analogous roles are performed by the hexameric proteins CsoS1A, CsoS1B, and CsoS1C, which make up the bulk of the carboxysome shell faces, and the pentameric proteins CsoS4A and CsoS4B, which form the vertices (Table 1) (40, 88).
Figure 3.
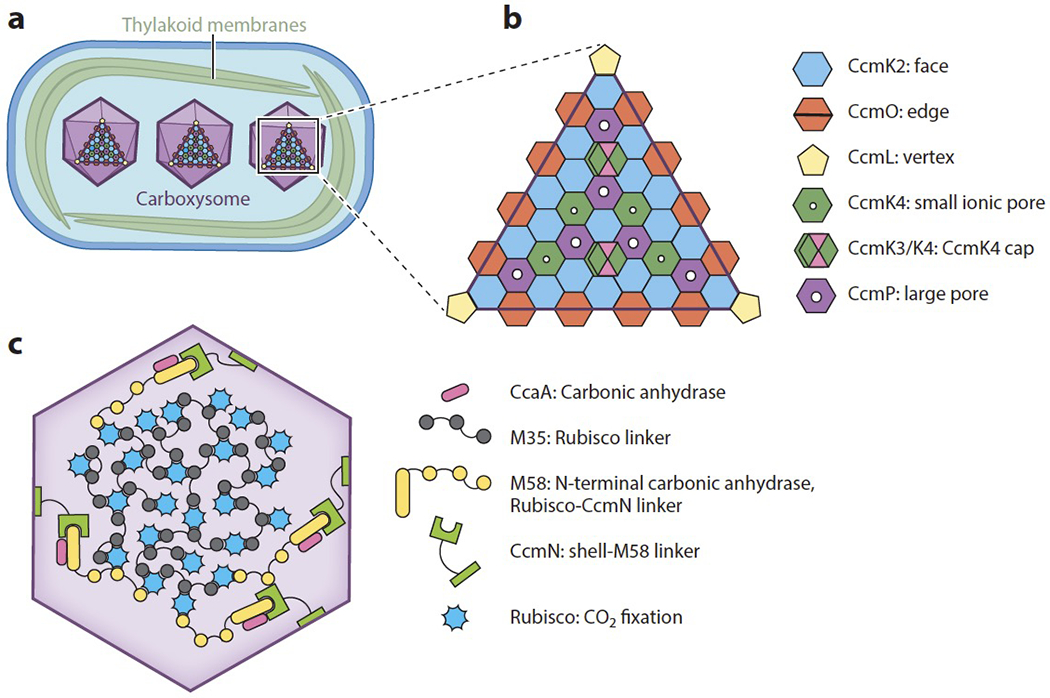
Carboxysomes are made up of an icosahedral protein shell that surrounds clustered Rubisco. (a) There are several carboxysomes per cyanobacterial cell, and they are evenly distributed across the longitudinal axis. (b) Assembly of the icosahedral carboxysome shell relies on the hexameric CcmK2 that makes up the faces of the icosahedron, the trimeric CcmO at the edges, and pentameric CcmL at the vertices. The hexameric protein CcmK4 has a small ionic pore through which HCO3− could diffuse, and CcmP has a larger pore that could allow an exchange of Calvin cycle intermediates. The CcmK3/K4 hexamer may act as a cap to block the flow of HCO3− through CcmK4 when needed. (c) The interior of the carboxysome is made up of Rubisco that is clustered by the CcmM isoform M35. The CcmM isoform M58 also binds Rubisco and is thought to reside at the edges, where CcmN links it to the shell.
Although CcmK2, CcmO, and CcmL are sufficient to build the shell’s structure, the oligomers CcmP, CcmK3, and CcmK4 are thought to be necessary for mediating diffusion of metabolites across the shell, thereby enabling the CCM to function properly (Figure 3b). CcmP is a hexamer and forms a double layer of hexagonal shapes (42). The double layer allows CcmP to have an open and a closed conformation. In its open conformation, CcmP has a central pore large enough to allow the diffusion of CBB cycle metabolites such as the Rubisco substrate RuBP and product 3-PGA.
CcmK4 can independently form a hexamer containing a central pore that is predicted to be permeable to anions such as HCO3− (53). The CcmK4 hexamer is thought to be dispersed throughout the faces of the carboxysome and to control the influx of HCO3−.
CcmK3 cannot form large oligomers on its own, but it is able to form a heterohexamer with CcmK4 that has a ratio of four CcmK4 monomers to two CcmK3 monomers (85). Certain residues on CcmK3 make it unlikely to fit favorably alongside CcmK2 in the carboxysome shell. However, the CcmK3/K4 heterohexamer can form a dodecamer by fitting on the top of CcmK4, suggesting a model in which the heterohexamer may be used as a cap to limit the flux of HCO3− under certain conditions that would induce CcmK3 expression (Figure 3b).
CcmK3 and CcmK4 are encoded at a separate locus from the rest of the ccm operon, indicating that they could be under different transcriptional control (84). While CcmK4 is necessary for growth, CcmK3 is not (85).
Linker Proteins Bind and Cluster Rubisco Inside the Carboxysome
In β-cyanobacteria, carboxysome assembly begins with the clustering of Rubisco into an electron-dense body called the procarboxysome (13). This clustering is carried out by M35, a truncated isoform of the protein CcmM that arises by translation initiated at an internal ribosome entry site in the ccmM transcript (Figure 3c) (48). M35 is necessary for procarboxysome formation (49) and sufficient for clustering Rubisco into a procarboxysome-like structure in cyanobacterial mutants lacking the ccm operon (13).
The amino acid sequence of M35 consists of three repeated Rubisco small subunit-like (SSUL) domains, each of which binds to the Rubisco large subunit, allowing its clustering to form a matrix within the carboxysome. The sequence similarity between the SSUL domains of CcmM and the Rubisco small subunit led Price et al. (72a) to propose a model where each SSUL domain of CcmM binds to the Rubisco large subunit by displacing the Rubisco small subunit. Intriguingly, recent work has shown that in vitro the SSUL domain can bind at an equatorial region between the Rubisco large subunit dimers, suggesting that CcmM could link Rubiscos without displacing the Rubisco small subunit (80, 95). However, it remains possible that the original hypothesis is correct, and the interactions observed in vitro are representative of an intermediate complex that forms before a chaperone replaces the Rubisco small subunit with the SSUL domain.
The full-length 58-kDa isoform of CcmM, M58, has an N-terminal γ-carbonic anhydrase domain in addition to the three SSUL domain repeats (49). In some species, this N-terminal domain functions as a carbonic anhydrase to convert HCO3− into CO2 within the carboxysome, and in other species, there is a separate carbonic anhydrase called CcaA that is recruited to the carboxysome by the M58 N-terminal domain (46, 59).
M58 is expressed less abundantly than M35, supporting a model in which M35 clusters Rubisco into a matrix at the carboxysome center while M58 resides at the periphery and mediates interactions between the Rubisco matrix and the shell (Figure 3c) (48). Interactions between the shell and the carboxysome interior are thought to be facilitated by the protein CcmN. The N terminus of CcmN interacts with M58, while the C terminus interacts with the abundant hexagonal shell protein CcmK2 (41). CcmK2 fails to localize to the procarboxysome in mutants lacking CcmN, demonstrating that CcmN is necessary to recruit CcmK2 to the procarboxysome (13).
In α-cyanobacteria, Rubisco clustering and carboxysome shell assembly are thought to occur simultaneously (33). The protein CsoS2 plays a similar role to the β-cyanobacterial protein CcmM, acting as a Rubisco linker (12). Like CcmM, CsoS2 has two isoforms that arise through posttranscriptional mechanisms: the full-length isoform CsoS2B, and a truncated isoform, CsoS2A (15). However, while the truncated M35 form of CcmM is produced through an internal ribosomal entry site, CsoS2A is produced from programmed ribosomal frameshifting.
While both CcmM and CsoS2 cluster Rubisco into a dense matrix, they are different in several structural and functional aspects. For example, CcmM contains SSUL domains and binds to the Rubisco large subunit, but CsoS2 is intrinsically disordered and interacts with the Rubisco small subunit (45). While both isoforms of CcmM are necessary to assemble a β-carboxysome, only the full-length CsoS2B isoform is necessary to assemble an α-carboxysome, and the role of CsoS2A is unknown (15).
Steps Have Been Taken to Build Synthetic Carboxysomes
To achieve the goal of engineering a cyanobacterial CCM into land plants, researchers are pursuing efforts to reconstitute carboxysomes in heterologous systems in parallel with efforts to target cyanobacterial HCO3− transporters to plant membranes (Figure 4). Both β- and α-carboxysome-like structures have been reconstituted in heterologous systems including E. coli and tobacco. While tobacco is a good reference for the crop plants that will ultimately be the target for engineering, E. coli is a useful chassis organism for rapidly identifying the minimal gene set for assembling a carboxysome.
Figure 4.
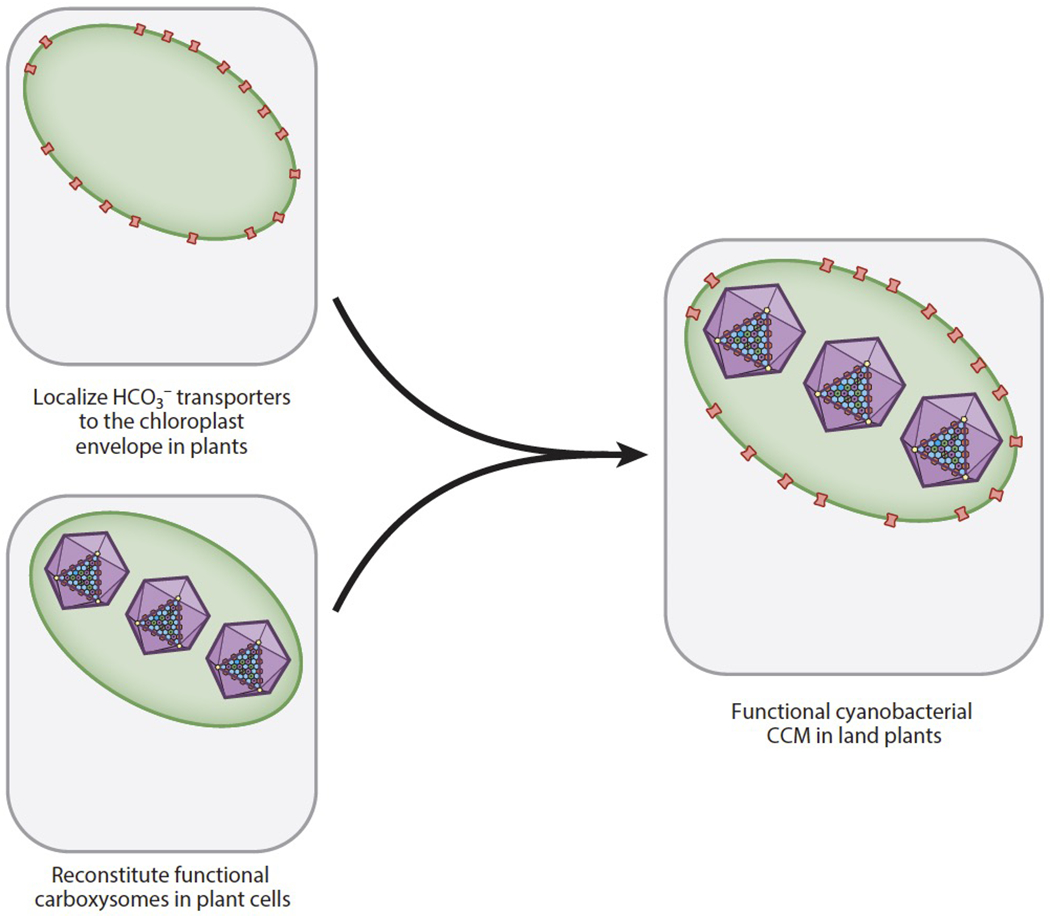
The goals of localizing HCO3− transporters to the proper plant membranes and building a carboxysome have been pursued in parallel. Once HCO3− transporters and carboxysomes can independently function in a plant cell, combining them into a single plant will be the next step toward reconstituting a cyanobacterial CO2 concentrating mechanism (CCM) in land plants.
Simple β-carboxysome-like shells could be assembled in E. coli using a synthetic operon consisting of only four genes from the cyanobacterium Halothece sp. PCC 7418 (11). These genes encode the hexameric proteins that make up the faces of the carboxysome icosahedron, CcmK2 and K1; a K2 homolog found in a subset of β-cyanobacteria; the trimeric protein CcmO that forms the edges; and the pentameric vertex CcmL.
Fang et al. (20) expanded on this work by creating and expressing an IPTG-inducible synthetic ccm operon using genes found in the β-cyanobacterial reference organism Synechococcus elongatus PCC 7942. This synthetic operon combined 12 genes normally found at five different chromosomal loci into a single operon. These genes encode the shell-associated proteins CcmK2, CcmO, CcmL, CcmK3, CcmK4, CcmP, and CcmN, as well as the Rubisco linker CcmM, the Rubisco large and small subunits, the Rubisco chaperone RbcX, and the carbonic anhydrase CcaA. When expressed in E. coli, the synthetic operon generated carboxysome-like structures that were slightly larger and more irregularly shaped than native carboxysomes, indicating that the ratio of these carboxysome components may need to be optimized in future experiments. Nevertheless, the packing of Rubisco within the synthetic carboxysome lumen was similar to the native packing density. Furthermore, 14C could be fixed by isolated carboxysomes treated with NaH14CO3, indicating that the carboxysomes had an active carbonic anhydrase and Rubisco.
An α-carboxysome has been assembled in E. coli using the cso operon from Halothiobacillus neapolitanus, a chemolithotrophic proteobacteria that also packages Rubisco into a microcompartment, demonstrating that all the proteins necessary to build an α-carboxysome are encoded in the cso operon (9). Furthermore, α-carboxysome-like structures could be observed in tobacco chloroplasts expressing simply the α-cyanobacterial Rubisco large and small subunits alongside the Rubisco linker CsoS2 and the shell protein CsoS1A (47).
Given the promising results in E. coli, a major near-term frontier is the production of carboxysomes in plant chloroplasts. Replacing a plant’s endogenous Rubisco with the ortholog from cyanobacteria is a critical step toward ensuring that the enzyme can be packaged into the carboxysome, as the linker protein interacts with specific sites in the cyanobacterial ortholog. Lin et al. (44) have been able to replace the native Rubisco large subunit gene in tobacco transplastomic lines with the β-cyanobacterial large and small subunits, and Long et al. (47) accomplished this with the α-cyanobacterial Rubisco. In both cases, the transgenic plants are able to grow autotrophically in high CO2, indicating that the cyanobacterial Rubisco is functional in the plant.
Excitingly, minimal carboxysome-like structures have been reconstituted in tobacco using either β- or α-carboxysome components. Lin et al. (44) observed procarboxysome-like structures when the β-cyanobacterial Rubisco was expressed along with the M35 isoform of CcmM in tobacco. Long et al. (47) reconstituted minimal α-carboxysome-like structures in tobacco using a minimal gene set containing Rubisco, the linker CsoS2, and the shell component CsoS1A. Going forward, the next challenge for both systems is incorporating the remaining components necessary for a fully functional carboxysome, including other shell proteins and a carbonic anhydrase. Once this is accomplished, a functional carboxysome can be combined with a HCO3− transport system to reconstitute a full CCM (Figure 4).
THE CO2 CONCENTRATING MECHANISM IN EUKARYOTIC ALGAE
Much of our molecular understanding of the algal CCM has come from studies of the reference freshwater alga Chlamydomonas reinhardtii (Chlamydomonas hereafter), which is the focus of this section of the review. The CCM in Chlamydomonas relies on HCO3− transport across several different membranes to concentrate Ci near Rubisco, which is housed in a non-membrane-bound organelle within the chloroplast called the pyrenoid (Figure 5). The core of the pyrenoid is a region known as the matrix, which consists of densely packed Rubisco. Starch granules form a sheath around the matrix. Membrane tubules traverse the matrix, exit through gaps in the starch sheath, and connect to the photosynthetic thylakoid membranes in the stroma. The core model of the algal CCM is as follows: HCO3− is concentrated in the lumen of the pyrenoid tubules through the action of HCO3− transporters. There, an acidic environment and a carbonic anhydrase cause the conversion of HCO3− to CO2. This CO2 is then able to diffuse across the pyrenoid tubule membranes out to the Rubisco in the matrix.
Figure 5.
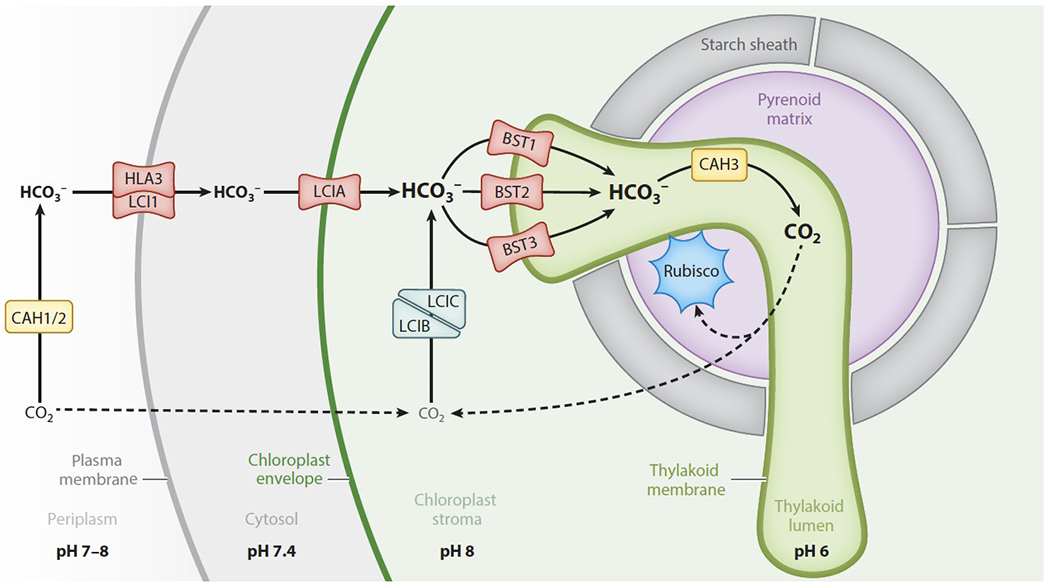
In the green alga Chlamydomonas reinhardtii, HCO3− transport across a series of membranes leads to concentrated HCO3− in the pyrenoid tubules, which are continuous with the thylakoid membranes. In the tubules, a carbonic anhydrase (CAH3) converts HCO3− to CO2, which then diffuses out to reach Rubisco in the matrix. CO2 that leaks out of the pyrenoid is converted back into HCO3− by the LCIB/LCIC complex and then recycled through the pyrenoid. The pH of various subcellular compartments is important for determining the net direction of CO2/HCO3− conversion. The neutral to slightly basic pH of the periplasm and chloroplast stroma supports conversion of CO2 into HCO3−, and the acidic pH of the thylakoid lumen supports conversion into CO2.
HCO3− Transport Must Occur at the Plasma Membrane, Chloroplast Envelope, and Thylakoid Membrane
The flux of Ci through a Chlamydomonas cell begins in the periplasm, where the carbonic anhydrases CAH1 (Cre04.g223100) and CAH2 (Cre04.g223050) are located (Figure 5, Table S1). The pH of the periplasm is thought to match the pH of the extracellular environment. Active transport at the plasma membrane removes HCO3− from the periplasm, so CAH1 and CAH2 likely function to replenish HCO3− by converting CO2 into HCO3− (21).
At the plasma membrane, HCO3− uptake is mediated by LCI1 (low CO2 inducible protein 1, Cre03.g162800) (66) and HLA3 (high light activated protein 3, Cre02.g097800) (18), which form a complex (51). Overexpression of LCI1 in high CO2 conditions, when the rest of the CCM is not induced, leads to higher accumulation of Ci within the cell (66). The sequence of LCI1 does not have any recognizable structural domains, so its specific role in Ci uptake is still unknown (99), and HLA3 is a putative ATP-binding cassette HCO3− transporter (18). The ATP-dependent HCO3− transport activity of HLA3 has been demonstrated in a heterologous expression system, indicating that it can function independently from other Chlamydomonas-specific factors to actively pump HCO3− into the cell (57).
Once in the cytoplasm, HCO3− must then pass through the chloroplast envelope to become concentrated in the chloroplast stroma. LCIA (limiting CO2 inducible A, Cre06.g309000), a formate/nitrite transporter homolog, localizes to the chloroplast envelope and has been implicated in bicarbonate transport there (99), although it is unclear whether LCIA mediates active or passive transport. Photosynthesis is inhibited in lcia mutants at pH 9, when most Ci is expected to be HCO3−, demonstrating that LCIA likely has a role in HCO3− uptake (97). Furthermore, heterologous expression of LCIA in Xenopus oocytes led to a twofold increase in HCO3− accumulation (55).
Inside the stroma, according to the prevailing model, HCO3− crosses one final membrane to enter the thylakoid lumen (63). The genes BST1 (Cre16.g662600), BST2 (Cre16.gg663400), and BST3 (Cre16.g663450) each encode putative bestrophins (65), which are a family of chloride channels that are in some cases permeable to HCO3− (73). BST1, BST2, and BST3 each localize to the thylakoid membrane, suggesting that one or more of these proteins could facilitate the shuttling of HCO3− into the thylakoid lumen (51). The hypothesis that BST1-3 play an important role in the CCM is further supported by the observation that a bst1-3 triple knockdown generated by RNAi grows more slowly at air levels of CO2 than wildtype cells (65).
Proton pumping during the light reactions of photosynthesis creates an acidic environment within the thylakoid lumen, which promotes the conversion of HCO3− to CO2 (2, 77). This conversion is likely catalyzed by the carbonic anhydrase CAH3 (Cre09.g415700), which localizes to the thylakoid lumen (63). Concentrated CO2 produced in the thylakoid lumen is thought to diffuse across the pyrenoid tubules to feed the Rubisco in the matrix.
The proteins LCIB (Cre10.g452800) and LCIC (Cre06.g307500) form a complex that is thought to recapture unfixed CO2 that leaks out of the matrix, converting this CO2 back into HCO3− (100). The structure of LCIB resembles functional β-carbonic anhydrases, and homologs of LCIB have carbonic anhydrase activity (36). LCIB and LCIC bind to some of the bestrophins on the thylakoid membrane (51), suggesting that the HCO3− recaptured by LCIB and LCIC can rapidly access the thylakoid lumen for another opportunity to be fixed by Rubisco.
CIA8 (Ci accumulation 8, Cre09.g395700), a putative member of the sodium bile acid symporter family, has recently been implicated in Ci uptake, although both its function and localization within the chloroplast are yet unresolved (50). cia8 mutants have impaired growth at air levels of CO2 and reduced Ci accumulation both at pH 7.3 and at pH 9. This is notable because many of the proteins involved in Ci uptake have overlapping functions, so a growth defect is usually not observed from a single gene knockout. Continuing to study the function of CIA8 will lead to a more complete picture of Ci uptake in Chlamydomonas.
Rubisco, Linked by EPYC1, Is Clustered in the Liquid-Like Pyrenoid Matrix
When the CCM is induced, an estimated 90% of the cell’s Rubisco clusters in the pyrenoid matrix (10). This clustering of Rubisco ensures that the enzyme is supplied with concentrated CO2, which is thought to arrive via the membranous pyrenoid tubules that traverse the matrix.
Assembly of the Chlamydomonas pyrenoid matrix relies on the Rubisco linker EPYC1 (essential pyrenoid component 1, Cre10.g436550) (Figure 6a), which is one of the most abundant proteins in the matrix (52). The amino acid sequence of EPYC1 consists primarily of four nearly identical repeats, each with a predicted α-helical region followed by a region predicted to be highly disordered, suggesting that EPYC1 has at least four binding sites for Rubisco. Aggregation of Rubisco into the pyrenoid requires two solvent-facing α-helices found on the algal Rubisco small subunit, making this region a potential site for EPYC1 binding (60, 4a).
Figure 6.
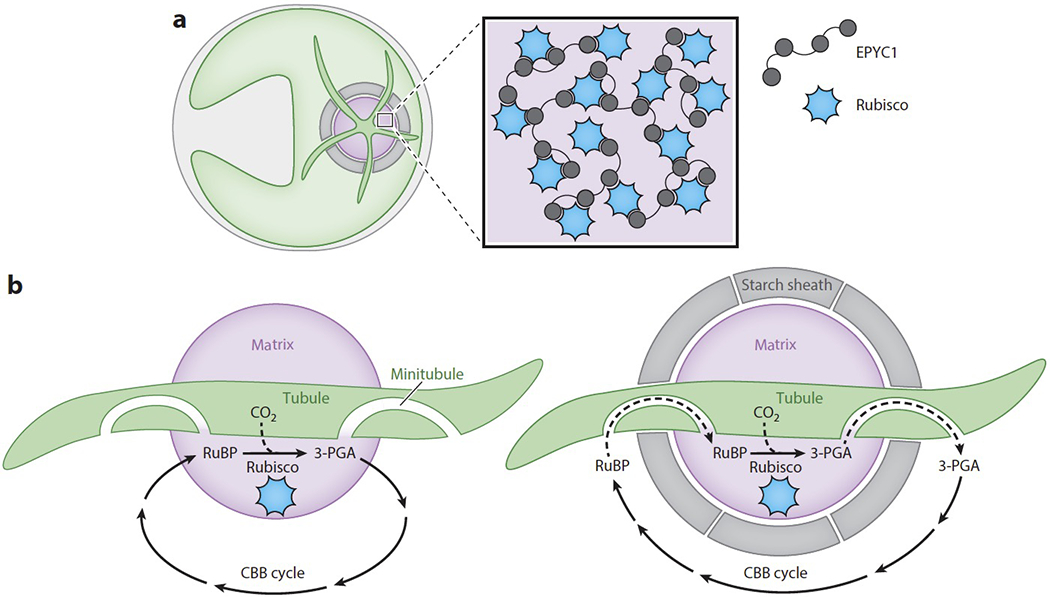
A pyrenoid forms at the center of the Chlamydomonas cell’s cup-shaped chloroplast. (a) Rubisco is clustered within the pyrenoid matrix by the linker protein EPYC1. (b) Before the Rubisco matrix is fully encapsulated by a starch sheath, Rubisco can directly exchange its substrate ribulose-1,5-bisphosphate (RuBP) and its product 3-phosphoglycerate (3-PGA) with the other Calvin Benson Bassham (CBB) cycle enzymes in the chloroplast stroma. However, when a starch sheath is in place, this exchange may depend on diffusion through minitubules embedded within the pyrenoid tubules. These minitubules are continuous with the stroma and pyrenoid matrix.
Observation of fluorescently tagged matrix components within Chlamydomonas has revealed that the matrix displays liquid-like properties, such as internal mixing and formation of spherical droplets that can fuse (25). Pyrenoids are also inherited by daughter cells through fission of the mother cell’s pyrenoid, although a portion of the matrix rapidly disperses into the stroma at the end of each cell division cycle. The rapid dispersal of the pyrenoid during cell division could be important for ensuring that both daughter cells have enough starting material to form a pyrenoid de novo in case fission fails. In addition, the phase-separated nature of the pyrenoid matrix could allow it to form rapidly as a response to conditions that induce the CCM, such as low CO2 (10).
Purified Rubisco does not form liquid-like droplets on its own in vitro, but the addition of EPYC1 is sufficient to drive demixing into liquid droplets (98). The internal mixing dynamics of these in vitro droplets occur on a similar time scale to the pyrenoid, suggesting that EPYC1 and Rubisco are sufficient to reconstitute the Chlamydomonas pyrenoid matrix.
The Pyrenoid Starch Sheath May Serve as a Diffusion Barrier to Slow CO2 Escape
Both green algae and plants store some of the energy they capture from photosynthesis as starch, a polymer of glucose that forms large lens-shaped granules in the chloroplast stroma. Many green algal species also assemble a subset of their starch granules in a shell around the pyrenoid, forming a structure called the starch sheath. Unlike globular stromal starch, this pyrenoid starch has a curved morphology, and is made up of distinct plates that wrap around the pyrenoid and appear to form a seal interrupted by gaps to allow the passage of pyrenoid tubules (61).
In Chlamydomonas, the conditions that induce the CCM also induce the starch sheath to form (76). The total amount of starch accumulated is coordinated with the diurnal cycle, but whether starch is primarily in the stroma or surrounding the pyrenoid depends on whether the CCM is active. Nitrogen deprivation, which slows down photosynthetic activity, causes cells to accumulate more stromal starch rather than pyrenoid starch (23). The apparent coordination between CCM induction and pyrenoid starch formation suggests that the starch sheath may have a role in the CCM.
It has been suggested that the starch sheath could act as a barrier to slow the CO2 efflux from the pyrenoid (76). Starch is composed of alternating amorphous layers of the unbranched polymer amylose and crystalline layers of the branched polymer amylopectin (103), the latter of which is thought to be impermeable to gases, including CO2 and O2 (27). Furthermore, the curved morphology of pyrenoid starch granules and the seal they form around the pyrenoid would appear to support this function.
Despite the proposed role of the starch sheath in preventing CO2 efflux, mutants that are unable to synthesize starch still appear to have a fully functional CCM at air levels of CO2, suggesting that the starch sheath may not actually be necessary for the CCM after all (92). This conundrum could be resolved if the starch sheath were found to be important under slightly different growth conditions than those tested in the laboratory, or if it were found to confer only a small advantage that is still relevant evolutionarily.
Several proteins that contain starch-binding domains localize to the periphery of the pyrenoid and form different localization patterns there (51). The granule-bound starch synthase STA2 (Cre17.g721500) and the starch branching enzyme SBE3 (Cre10.g444700) appear to localize to the starch plates, whereas the protein LCI9 (Cre02.g130700) localizes to a mesh pattern, possibly filling the gaps between starch plates. LCI9 contains two starch-binding domains and is homologous to glucan 1,4-α-glucosidases, enzymes that typically function to liberate glucose monomers from glucan chains. This homology suggests that LCI9 could degrade starch at the gaps between starch plates, possibly ensuring a close fit for adjacent starch plates.
Two mutants have been described with interesting phenotypes related to the pyrenoid starch. Cells lacking a protein that localizes to the starch sheath, named SAGA1 (starch granules abnormal 1, Cre11.g467712), have thin and elongated pyrenoid starch granules (34). Cells lacking another protein called BSG1 (bimodal starch granule 1, Cre02.g091750) have enlarged pyrenoid starch granules under nitrogen-limiting conditions that would normally favor the accumulation of stromal starch, indicating that BSG1 may be involved in controlling the transition from pyrenoid to stroma starch (23). Continued studies of the pyrenoid starch sheath could provide insights into why starch granules across the green lineage have specific morphologies and how these morphologies are produced.
Pyrenoid Tubules Are Thought to Deliver Concentrated CO2 and May Provide a Path for Diffusion of Calvin Benson Bassham Cycle Metabolites
The pyrenoid tubules, which are continuous with the thylakoid membranes, penetrate into the pyrenoid matrix through distinct gaps in the starch sheath and fuse into a reticulated network at the center (19). When a starch sheath is present, the tubules are thought to be the primary route for entry of inorganic carbon into the pyrenoid: HCO3− enters the thylakoid lumen via transporters outside the pyrenoid, and diffuses along the inside of the tubules before being converted to CO2 in the portion of the tubules that traverses the matrix (Figure 5).
Formation of the starch sheath occurs more slowly than formation of the other components of the pyrenoid, so for a short period of time after the CCM is induced, the starch sheath does not fully enclose the pyrenoid matrix (76). During this time, Rubisco’s substrate RuBP and its product 3-PGA can directly diffuse between Rubisco in the pyrenoid and the other CBB cycle enzymes, which are located in the chloroplast stroma (87) (Figure 6b).
Cryo-electron tomography images of the pyrenoid tubules have shown that there are several minitubules embedded within each tubule (Figure 6b) (19). The lumens of these minitubules are continuous with both the pyrenoid and the chloroplast stroma, and are wide enough for the passage of RuBP and 3-PGA. Therefore, the minitubules may serve as a conduit for the diffusion of metabolites between Rubisco in the matrix and other CBB cycle enzymes in the stroma along their respective concentration gradients when the starch sheath is fully formed.
Mutants that lack a pyrenoid matrix still form a pyrenoid tubule network at the canonical location within the chloroplast (14). This observation suggests that the process of building the pyrenoid tubule network occurs separately from the assembly of the rest of the pyrenoid, and that these tubules contain the information for where a pyrenoid should be placed. Therefore, the pyrenoid tubules may actually localize the matrix and the starch sheath.
Although the pyrenoid tubules play a crucial role in the algal CCM, not much is known about their biogenesis. The topology of the transition zone between the thylakoid membranes that exist as stacked sheets in the chloroplast and the more cylindrical tubules that traverse the pyrenoid is complex (19), and how its formation is mediated molecularly is unknown. The protein PSAH (Cre07.g330250), associated with photosystem I, is enriched in the pyrenoid tubules (51) and may have a function there. Identifying and characterizing more proteins involved in constructing the pyrenoid tubules will be important for future attempts to install an algal CCM in land plants.
Early Progress Suggests Promising Prospects of Transferring an Algal CO2 Concentrating Mechanism into Land Plants
Several important components of the Chlamydomonas CCM have been fused to green fluorescent protein (GFP) and expressed in Arabidopsis and tobacco leaves to examine their localization (3). Most of these algal proteins localized to the correct subcellular compartment in the plant cell without requiring changes to their protein sequence. For example, the periplasmic carbonic anhydrase CAH1 and the plasma membrane HCO3− transporters LCI1 and HLA3 all localized to the cell periphery. The putative CO2 recapture complex proteins LCIB and LCIC localized to the chloroplast stroma, while the putative HCO3− channel LCIA was targeted to the chloroplast envelope. These results are encouraging because they suggest that expressing algal CCM components from the nucleus of a land plant will require very few modifications at the protein sequence level, and the key challenge is determining which genes need to be transferred.
As in the case of cyanobacterial transporters, it is not clear whether algal HCO3− transporters are active, and an important goal for the future will be to test their activity. Furthermore, the full algal CCM likely requires HCO3− transport across the thylakoid membranes. A voltage-dependent chloride channel that has homology to the bestrophin-like proteins on the Chlamydomonas thylakoids already resides on Arabidopsis thaliana thylakoids (31). It will be interesting to learn whether this channel is sufficient for CCM activity, or whether it is necessary to express the putative algal HCO3− transporters BST1, BST2 and BST3.
Another important step toward reconstituting a pyrenoid in a plant is to engineer the plant Rubisco small subunit to contain specific residues on the two solvent-facing α-helices present on the Chlamydomonas ortholog, which may be required for binding to the Rubisco linker EPYC1 (60). Atkinson et al. (4) were able to construct a functional hybrid small subunit which replaced the native Arabidopsis small subunit α-helices with those of Chlamydomonas. This hybrid small subunit was functional, as demonstrated by its ability to rescue an Arabidopsis double mutant that grew slowly due to impairment of two out of four homologous small subunit genes. Modifying these α-helices did not appear to significantly impact the catalytic properties of the Rubisco holoenzyme.
Several stages for engineering an algal system into plants can be pursued in parallel; these include reconstituting a pyrenoid matrix in a land plant, continuing to identify and transfer HCO3− transporters to plants, and studying the mechanisms of pyrenoid tubule biogenesis. After these stages are achieved, an important goal will be to orient a pyrenoid matrix around pyrenoid tubules and finally to combine this matrix/tubule system with the HCO3− transporters (Figure 7).
Figure 7.
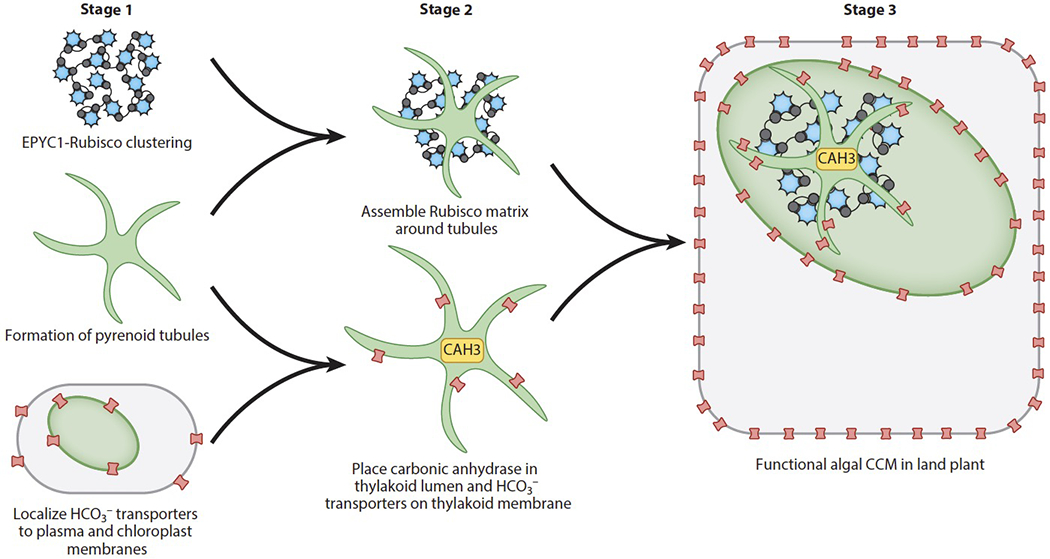
Several milestones toward engineering an algal CO2 concentrating mechanism (CCM) into land plants can be pursued in parallel.
Characterizing and Engineering the Algal CO2 Concentrating Mechanism Will Benefit from New Resources
Much of the progress that has been made toward understanding the genes involved in the algal CCM has relied on forward genetics studies, wherein mutants generated by chemical mutagenesis or random insertion mutagenesis are screened for a CCM phenotype (86a, 96). The classic CCM phenotype is impaired photoautotrophic growth at air levels of CO2 that is rescued by elevated CO2. It will be important to be able to approach saturation in such screens to produce a comprehensive list of factors that need to be transferred to plants. To this end, an insertional mutant library has recently been created, covering 83% of Chlamydomonas nuclear genes (43). Many new genes with roles in photosynthesis have been identified through pooled screening of this library, and some may include novel CCM factors. Additionally, this library now enables the reverse genetic characterization of mutants in genes that become CCM candidates as a result of their sequence or presence in other high-throughput data sets. Furthermore, recent advances in RNA interference (39) and CRISPR-Cas9 gene editing (29) in Chlamydomonas provide the complementary ability to study genes that are not represented in the library or homologous genes that may be partially functionally redundant and thus may not show a phenotype in a single-gene knockout mutant.
Systematic characterization of protein localization and protein-protein interactions is synergistic with studies of mutant phenotypes because it identifies new genes of interest and greatly accelerates the process of understanding gene function. A high-throughput fluorescence protein-tagging effort in Chlamydomonas has provided data about the localizations of 135 candidate CCM proteins, 89 of which localize to at least six distinct patterns within the pyrenoid (51). Moreover, the interactions of 38 core CCM proteins were identified through affinity purification and mass spectrometry. A parallel effort characterized the Chlamydomonas pyrenoid proteome (104), providing independent evidence of pyrenoid localization of proteins that were also identified in the protein-tagging effort and additionally identifying other candidate pyrenoid components.
CONCLUSIONS AND OUTLOOK
Genetically engineered crops that are able to grow more quickly while using fewer resources will be important tools for meeting global agricultural demands, which are expected to rise significantly in the near future (89a). One promising target for engineering is the improvement of the function of the carbon-fixing enzyme Rubisco through installing a CCM, as this would promote Rubisco’s productive carboxylase activity while minimizing its unwanted oxygenase activity. Progress is being made toward understanding a variety of types of CCMs, such as the C4 pathway found in many plant species and the biophysical CCMs found in single-celled cyanobacteria and algae. Efforts to reconstitute these CCMs into land plants are yielding encouraging results. Each system has its own set of opportunities and challenges, some of which remain unknown (Table 2), so continuing the pursuit of characterizing and engineering C4, α-cyanobacterial, β-cyanobacterial, and algal CCMs in parallel will maximize the chance of at least one approach being successful. Exploring hybrid approaches that combine elements from different CCMs could also be advantageous. The availability of genetic tools that can now be used to both study the components of CCMs and begin to engineer them into plants is expected to lead to exciting advances in the near future.
Table 2.
C4, cyanobacterial, and algal CO2 concentrating mechanisms (CCMs) each provide different opportunities and challenges for engineering into land plants
| Type of CCM | Advantages | Challenges |
|---|---|---|
| C4 plant | Most closely related evolutionarily to C3 plants | Requires engineering tissue development to give leaves Kranz anatomy |
| Both cyanobacteria and algae | Operates within a single cell, therefore no need to engineer cell differentiation | Requires replacing Rubisco, because Rubisco linkers appear to bind only Rubisco from their host organisms |
| Cyanobacteria | Components are well characterized | Most evolutionarily distant from plants Components are not natively encoded in a eukaryotic nucleus and targeted to a chloroplast |
| Algae | Components are natively encoded in a eukaryotic nucleus and targeted to a chloroplast | Components are poorly characterized |
Supplementary Material
SUMMARY POINTS.
The enzyme Rubisco is crucial for converting CO2 into biomass but has limitations, including a relatively slow reaction rate and a competing reaction with O2.
CO2 concentrating mechanisms (CCMs) enhance Rubisco’s CO2-fixing activity by feeding the enzyme with concentrated CO2 and increasing the CO2/O2 ratio so that carboxylation is favored over oxygenation.
The CCMs in both cyanobacteria and green algae operate by concentrating HCO3− through transport and converting the molecule into CO2 at a site near clustered Rubisco.
Rubisco is clustered by linker proteins to form a subcellular structure: the carboxysome in cyanobacteria and the pyrenoid in algae.
In recent progress toward the goal of engineering functional cyanobacterial and algal CCMs into plants, researchers have targeted HCO3− transporters from both types of organisms to plant membranes.
Carboxysome-like structures have been synthesized in chassis organisms and land plants, and a pyrenoid matrix has been reconstituted in vitro.
ACKNOWLEDGMENTS
We would like to thank Moritz Meyer, Eric Franklin, Alexandra Wilson, Shan He, Lianyong Wang, Michael Bender, Cornelia Spetea Wiklund, and the anonymous reviewer for their helpful input on the manuscript. Recent research by J.H.H. in this area has been supported by the National Institute of General Medical Science of the National Institutes of Health (T32GM007388). The research of M.C.J. is funded by the National Institutes of Health (DP2-GM-119137), the Simons Foundation and Howard Hughes Medical Institute (55108535), the Department of Energy (DE-SC0020195) and the National Science Foundation (MCB-1914989 and MCB-1935444).
DISCLOSURE STATEMENT
M.C.J. is funded by National Science Foundation grant MCB-1935444, which aims to engineer an algal CO2 concentrating mechanism into land plants; and is a co-inventor on a patent application titled “Rubisco-Binding Protein Motifs and Uses Thereof”, which offers tools relevant to efforts to engineer CO2 concentrating mechanisms into land plants.
Glossary
- Rubisco
d-ribulose-1,5-bisphosphate carboxylase/oxygenase
- Ribulose-1,5-bisphosphate (RuBP)
Rubisco’s substrate
- 3-Phosphoglycerate (3-PGA)
Rubisco’s product
- CBB cycle
Calvin Benson Bassham cycle
- kcat
catalytic rate
- SCO2/O2
Rubisco’s specificity for CO2 over O2
- kcat,C
Rubisco’s turnover rate for CO2
- CCM
CO2 concentrating mechanism
- Biochemical CCM
A CO2 concentrating mechanism that uses intermediate organic molecule that is decarboxylated to release CO2 near Rubisco.
- Kranz anatomy
the arrangement of mesophyll and bundle sheath tissues in a C4 leaf
- Biophysical CCM
A CO2 concentrating mechanism that operates by directly transporting inorganic carbon (in the form of bicarbonate) and dehydrating it to CO2 near Rubisco
- Inorganic carbon (Ci)
Defined in this field as carbon that is not bound directly to hydrogen
- IEM
chloroplast inner envelope membrane
LITERATURE CITED
- 1.Ainsworth EA, Long SP. 2005. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 165:351–71 [DOI] [PubMed] [Google Scholar]
- 2.Antal TK, Kovalenko IB, Rubin AB, Tyystjarvi E. 2013. Photosynthesis-related quantities for education and modeling. Photosynth. Res 117:1–30 [DOI] [PubMed] [Google Scholar]
- 3.Atkinson N, Feike D, Mackinder LCM, Meyer MT, Griffiths H, et al. 2016. Introducing an algal carbon-concentrating mechanism into land plants: location and incorporation of key components. Plant Biotechnol. J 14:1302–15 [DOI] [PMC free article] [PubMed] [Google Scholar]; 3. Several algal CCM components could localize to their corresponding subcellular compartments in tobacco and Arabidopsis.
- 4.Atkinson N, Leitão N, Orr DJ, Meyer MT, Carmo-Silva E, et al. 2017. Rubisco small subunits from the unicellular green alga Chlamydomonas complement Rubisco-deficient mutants of Arabidopsis. New Phytol. 214(2):655–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Atkinson N, Velanis CN, Wunder T, Clarke DJ, Mueller-Cajar O, McCormick AJ. 2019. The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis–Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit. Journal of Experimental Botany 70:5271–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot 62:3049–59 [DOI] [PubMed] [Google Scholar]
- 5a.Badger MR, Price GD. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot 54:609–22 [DOI] [PubMed] [Google Scholar]
- 6.Badger MR, Price GD, Long BM, Woodger FJ. 2006. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot 57(2):249–65 [DOI] [PubMed] [Google Scholar]
- 6a.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, et al. 2011. The Moderately Efficient Enzyme: Evolutionary and Physicochemical Trends Shaping Enzyme Parameters. Biochemistry 50: 4402–10. [DOI] [PubMed] [Google Scholar]
- 7.Deleted in proof.
- 8.Battchikova N, Eisenhut M, Aro E-M. 2011. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta Bioenerget 1807:935–44 [DOI] [PubMed] [Google Scholar]
- 9.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, et al. 2012. Modularity of a carbon-fixing protein organelle. PNAS 109:478–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borkhsenious ON, Mason CB, Moroney JV. 1998. The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 116:1585–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai F, Bernstein SL, Wilson SC, Kerfeld CA. 2016. Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170:1868–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, et al. 2015. Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life 5:1141–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155:1131–40 [DOI] [PubMed] [Google Scholar]; 13. Observing β-carboxysome formation in Δccm cells expressing truncated ccm operons provided key insights into the order of assembly of β-carboxysomes.
- 14.Caspari OD, Meyer MT, Tolleter D, Wittkopp TM, Cunniffe NJ, et al. 2017. Pyrenoid loss in Chlamydomonas reinhardtii causes limitations in CO2 supply, but not thylakoid operating efficiency. J. Exp. Bot 68:3903–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaijarasphong T, Nichols RJ, Kortright KE, Nixon CF, Teng PK, et al. 2016. Programmed ribosomal frameshifting mediates expression of the α-carboxysome. J. Mol. Biol 428:153–64 [DOI] [PubMed] [Google Scholar]
- 16.DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV. 2017. Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol. Plant 10:30–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du J, Förster B, Rourke L, Howitt SM, Price GD. 2014. Characterisation of cyanobacterial bicarbonate transporters in E. coli shows that SbtA homologs are functional in this heterologous expression system. PLOS ONE 9:e115905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duanmu D, Miller AR, Horken KM, Weeks DP, Spalding MH. 2009. Knockdown of limiting-CO2–induced gene HLA3 decreases HCO3− transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. PNAS 106:5990–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Edwards GE, Franceschi VR, Voznesenskaya Elena V. 2004. Single-Cell C4 Photosynthesis Versus The Dual-Cell (Kranz) Paradigm. Annual Review of Plant Biology 55:173–96 [DOI] [PubMed] [Google Scholar]
- 19.Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W. 2015. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4:e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Huang F, Faulkner M, Jiang Q, Dykes GF, et al. 2018. Engineering and modulating functional cyanobacterial CO2-fixing organelles. Front. Plant Sci. 9:739. [DOI] [PMC free article] [PubMed] [Google Scholar]; 20. A synthetic β-carboxysome with functional Rubisco was assembled in E. coli using 12 genes from the β-cyanobacterium Synechococcus elongatus PCC7942.
- 21.Fett JP, Coleman JR. 1994. Regulation of periplasmic carbonic anhydrase expression in Chlamydomonas reinhardtii by acetate and pH. Plant Physiol. 106(1):103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–40 [DOI] [PubMed] [Google Scholar]
- 23.Findinier J, Laurent S, Duchêne T, Roussel X, Lancelon-Pin C, et al. 2019. Deletion of BSG1 in Chlamydomonas reinhardtii leads to abnormal starch granule size and morphology. Sci. Rep. 9:1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flamholz AI, Prywes N, Moran U, Davidi D, Bar-On YM, et al. 2019. Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry 58:3365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, et al. 2017. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171:148–62.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]; 25. The Chlamydomonas pyrenoid matrix has liquid-like properties, and a portion disperses at the end of each cell cycle.
- 26.Furbank RT. 2016. Walking the C4 pathway: past, present, and future. J. Exp. Bot. 67:4057–66 [DOI] [PubMed] [Google Scholar]
- 27.García MA, Martino MN, Zaritzky NE. 1999. Edible starch films and coatings characterization: scanning electron microscopy, water vapor, and gas permeabilities. Scanning 21:348–53 [Google Scholar]
- 28.Ghannoum O, Evans JR, von Caemmerer S. 2010. Chapter 8 Nitrogen and Water Use Efficiency of C4 Plants In C4 Photosynthesis and Related CO2 Concentrating Mechanisms., ed. R A., S R., 32. Advances in Photosynthesis and Respiration: Springer; Number of. [Google Scholar]
- 29.Guzmán-Zapata D, Sandoval-Vargas JM, Macedo-Osorio KS, Salgado-Manjarrez E, Castrejón-Flores JL, et al. 2019. Efficient editing of the nuclear APT reporter gene in Chlamydomonas reinhardtii via expression of a CRISPR-Cas9 module. Int. J. Mol. Sci 20:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson MR, Gray BN, Ahner BA. 2013. Chloroplast transformation for engineering of photosynthesis. J. Exp. Bot. 64:731–42 [DOI] [PubMed] [Google Scholar]
- 31.Herdean A, Teardo E, Nilsson AK, Pfeil BE, Johansson ON, et al. 2016. A voltage-dependent chloride channel fine-tunes photosynthesis in plants. Nat. Commun. 7:11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho C, Sturtevant JM. 1963. The kinetics of the hydration of carbon dioxide at 25°. J. Biol. Chem. 238:3499–501 [PubMed] [Google Scholar]
- 33.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. 2010. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itakura AK, Chan KX, Atkinson N, Pallesen L, Wang L, et al. 2019. A Rubisco-binding protein is required for normal pyrenoid number and starch sheath morphology in Chlamydomonas reinhardtii. PNAS 116:18445–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jablonski LM, Wang X, Curtis PS. 2002. Plant reproduction under elevated CO2 conditions: a meta‐analysis of reports on 79 crop and wild species. New Phytol. 156:9–26 [Google Scholar]
- 36.Jin S, Sun J, Wunder T, Tang D, Cousins AB, et al. 2016. Structural insights into the LCIB protein family reveals a new group of β-carbonic anhydrases. PNAS 113:14716–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerfeld CA, Melnicki M. 2016. Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31:66–75 [DOI] [PubMed] [Google Scholar]
- 38.Khalifah RG. 1971. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 246:2561–73 [PubMed] [Google Scholar]
- 39.Kim E-J, Cerutti H. 2009. Targeted gene silencing by RNA interference in Chlamydomonas. Methods Cell Biol. 93:99–110 [DOI] [PubMed] [Google Scholar]
- 40.Kinney JN, Axen SD, Kerfeld CA. 2011. Comparative analysis of carboxysome shell proteins. Photosynth. Res. 109:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287:17729–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsson AM, Hasse D, Valegård K, Andersson I. 2017. Crystal structures of β-carboxysome shell protein CcmP: Ligand binding correlates with the closed or open central pore. J. Exp. Bot. 68:3857–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, et al. 2019. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51:627–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR. 2014. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513:547–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, He X, Lim W, Mueller J, Lawrie J, et al. 2018. Deciphering molecular details in the assembly of alpha-type carboxysome. Sci. Rep. 8:15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long BM, Badger MR, Whitney SM, Price GD. 2007. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282:29323–35 [DOI] [PubMed] [Google Scholar]
- 47.Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, et al. 2018. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9:3570. [DOI] [PMC free article] [PubMed] [Google Scholar]; 47. α-Carboxysome-like structures were formed in tobacco chloroplasts through transformation of a minimal gene set.
- 48.Long BM, Rae BD, Badger MR, Price GD. 2011. Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth. Res. 109:33–45 [DOI] [PubMed] [Google Scholar]
- 49.Long BM, Tucker L, Badger MR, Price GD. 2010. Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 153:285–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machingura MC, Bajsa-Hirschel J, Laborde SM, Schwartzenburg JB, Mukherjee B, et al. 2017. Identification and characterization of a solute carrier, CIA8, involved in inorganic carbon acclimation in Chlamydomonas reinhardtii. J. Exp. Bot. 68:3879–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, et al. 2017. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171:133–47. e14 [DOI] [PMC free article] [PubMed] [Google Scholar]; 51. This protein–protein interactome study in Chlamydomonas has illuminated multiple novel interactions in the algal CCM.
- 52.Mackinder LCM, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, et al. 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. PNAS 113:5958–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahinthichaichan P, Morris DM, Wang Y, Jensen GJ, Tajkhorshid E. 2018. Selective permeability of carboxysome shell pores to anionic molecules. J. Phys. Chem. B 122:9110–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangan NM, Flamholz A, Hood RD, Milo R, Savage DF. 2016. pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. PNAS 113:E5354–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mariscal V, Moulin P, Orsel M, Miller AJ, Fernández E, Galván A. 2006. Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 157:421–33 [DOI] [PubMed] [Google Scholar]
- 56.Deleted in proof.
- 57.McCloskey MA, Duanmu D, Benge N, Spalding MH. 2017. The HLA3 protein of C. reinhardtii enhances HCO3-transport activity of mammalian cells. Biophys. J. 112:571A [Google Scholar]
- 58.McGrath JM, Long SP. 2014. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 164:2247–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGurn LD, Moazami-Goudarzi M, White SA, Suwal T, Brar B, et al. 2016. The structure, kinetics and interactions of the β-carboxysomal β-carbonic anhydrase, CcaA. Biochem. J. 473:4559–72 [DOI] [PubMed] [Google Scholar]
- 60.Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, et al. 2012. Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. PNAS 109:19474–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer MT, Whittaker C, Griffiths H. 2017. The algal pyrenoid: key unanswered questions. J. Exp. Bot. 68:3739–49 [DOI] [PubMed] [Google Scholar]
- 62.Deleted in proof.
- 63.Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, et al. 2011. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth. Res. 109:133–49 [DOI] [PubMed] [Google Scholar]
- 64.Moroney JV, Ynalvez RA. 2007. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6:1251–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukherjee A, Lau CS, Walker CE, Rai AK, Prejean CI, et al. 2019. Thylakoid localized bestrophin-like proteins are essential for the CO2 concentrating mechanism of Chlamydomonas reinhardtii. PNAS 116:16915–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohnishi N, Mukherjee B, Tsujikawa T, Yanase M, Nakano H, et al. 2010. Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell 22:3105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pengelly JJL, Förster B, von Caemmerer S, Badger MR, Price GD, Whitney SM. 2014. Transplastomic integration of a cyanobacterial bicarbonate transporter into tobacco chloroplasts. J. Exp. Bot. 65:3071–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, et al. 2010. Photorespiration. Arabidopsis Book 8:e0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price GD. 2011. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 109:47–57 [DOI] [PubMed] [Google Scholar]
- 70.Price GD, Badger MR. 1989. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype: evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 91:505–13 [DOI] [PMC free article] [PubMed] [Google Scholar]; 70. Expressing a carbonic anhydrase in the cyanobacterial cytosol impairs the CCM, demonstrating that HCO3− is maintained at high concentrations in the cytosol.
- 71.Price GD, Badger MR, von Caemmerer S. 2011. The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol. 155:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price GD, Coleman JR, Badger MR. 1992. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:784–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72a.Price GD, Howitt SM, Harrison K, Badger MR. 1993. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. Journal of Bacteriology 175:2871–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qu Z, Hartzell HC. 2008. Bestrophin Cl− channels are highly permeable to HCO3−. Am. J. Physiol. Cell Physiol. 294:C1371–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rae BD, Long BM, Badger MR, Price GD. 2012. Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLOS ONE 7:e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77:357–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramazanov Z, Rawat M, Henk MC, Mason CB, Matthews SW, Moroney JV. 1994. The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195:210–16 [Google Scholar]
- 77.Raven JA. 1997. CO2-concentrating mechanisms: a direct role for thylakoid lumen acidification? Plant Cell Environ. 20:147–54 [Google Scholar]
- 78.Raven JA. 2013. Rubisco: still the most abundant protein of Earth? New Phytol. 198:1–3 [DOI] [PubMed] [Google Scholar]
- 78a.Reinfelder JR, Milligan AJ, Morel FMM. 2004. The Role of the C4 Pathway in Carbon Accumulation and Fixation in a Marine Diatom. Plant Physiology 135:2106–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rolland V, Badger MR, Price GD. 2016. Redirecting the cyanobacterial bicarbonate transporters BicA and SbtA to the chloroplast envelope: Soluble and membrane cargos need different chloroplast targeting signals in plants. Front. Plant Sci. 7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]; 79. With a chloroplast transit peptide from Arabidopsis, cyanobacterial HCO3− transporters could be targeted to the Nicotiana benthamiana chloroplast envelope.
- 80.Ryan P, Forrester TJB, Wroblewski C, Kenney TMG, Kitova EN, et al. 2019. The small RbcS-like domains of the β-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit. J. Biol. Chem. 294:2593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Savir Y, Noor E, Milo R, Tlusty T. 2010. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. PNAS 107:3475–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81a.Schlüter U, Weber APM. 2020. Regulation of C4 Photosynthesis. Annu. Rev. Plant. Biol. 71:X-X [DOI] [PubMed] [Google Scholar]
- 82.Schuler ML, Mantegazza O, Weber APM. 2016. Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87:51–65 [DOI] [PubMed] [Google Scholar]
- 82a.Sharpe RM, Offermann S. 2014. One decade after the discovery of single-cell C4 species in terrestrial plants: what did we learn about the minimal requirements of C4 photosynthesis? Photosynth Res 119:169–80 [DOI] [PubMed] [Google Scholar]
- 83.Deleted in proof.
- 84.Sommer M, Cai F, Melnicki M, Kerfeld CA. 2017. β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J. Exp. Bot. 68:3841–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sommer M, Sutter M, Gupta S, Kirst H, Turmo A, et al. 2019. Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol. 179:156–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.South PF, Cavanagh AP, Liu HW, Ort DR. 2019. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363:eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86a.Spalding MH, Spreitzer RJ, Ogren WL. 1983. Carbonic anhydrase-deficient mutant of Chlamydomonas reinhardtii requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol. 73:268–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suss KH, Prokhorenko I, Adler K. 1995. In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, ferredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy. Plant Physiol. 107:1387–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, et al. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083–86 [DOI] [PubMed] [Google Scholar]
- 89.Tcherkez GGB, Farquhar GD, Andrews TJ. 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. PNAS 103:7246–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89a.Tilman D, Balzer C, Hill J, Befort BL. 2011. Global food demand and the sustainable intensification of agriculture. PNAS 108: 20260–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uehara S, Adachi F, Ito-Inaba Y, Inaba T. 2016. Specific and efficient targeting of cyanobacterial bicarbonate transporters to the inner envelope membrane of chloroplasts in Arabidopsis. Front. Plant Sci 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]; 90. The cyanobacterial HCO3− transporters BicA and SbtA were targeted to the Arabidopsis chloroplast envelope inner membrane.
- 91.van Lun M, Hub JS, van der Spoel D, Andersson I. 2014. CO2 and O2 distribution in Rubisco suggests the small subunit functions as a CO2 reservoir. J. Am. Chem. Soc. 136:3165–71 [DOI] [PubMed] [Google Scholar]
- 92.Villarejo A, Martinez F, del Pino Plumed M, Ramazanov Z. 1996. The induction of the CO2 concentrating mechanism in a starch-less mutant of Chlamydomonas reinhardtii. Physiol. Plant. 98:798–802 [Google Scholar]
- 93.Volokita M, Zenvirth D, Kaplan A, Reinhold L. 1984. Nature of the inorganic carbon species actively taken up by the cyanobacterium Anabaena variabilis. Plant Physiol. 76:599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. 2016. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 67:107–29 [DOI] [PubMed] [Google Scholar]
- 95.Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, et al. 2019. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566:131–35 [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Yamano T, Kajikawa M, Hirono M, Fukuzawa H. 2014. Isolation and characterization of novel high-CO2-requiring mutants of Chlamydomonas reinhardtii. Photosynth. Res. 121:175–84 [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Spalding MH. 2014. Acclimation to very low CO2: contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant Physiol. 166:2040–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wunder T, Cheng SLH, Lai S-K, Li H-Y, Mueller-Cajar O. 2018. The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 9:507698. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rubisco and its linker EPYC1 were sufficient to form pyrenoid matrix-like droplets in vitro.
- 99.Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. 2015. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. PNAS 112:7315–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamano T, Tsujikawa T, Hatano K, Ozawa S-I, Takahashi Y, Fukuzawa H. 2010. Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51:1453–68 [DOI] [PubMed] [Google Scholar]
- 101.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681–91 [DOI] [PubMed] [Google Scholar]
- 102.Yin X, Struik PC. 2017. Can increased leaf photosynthesis be converted into higher crop mass production? A simulation study for rice using the crop model GECROS. J. Exp. Bot. 68:2345–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeeman SC, Kossmann J, Smith AM. 2010. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61:209–34 [DOI] [PubMed] [Google Scholar]
- 104.Zhan Y, Marchand CH, Maes A, Mauries A, Sun Y, et al. 2018. Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLOS ONE 13:e0185039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


