Abstract
Previous studies have indicated associations between several OLIG2 gene single-nucleotide polymorphisms (SNPs) and susceptibility to schizophrenia among Caucasians. Consistent with these findings, postmortem brain and diffusion tensor imaging studies have indicated that the schizophrenia-risk-associated allele (A) in the OLIG2 SNP rs1059004 predicts lower OLIG2 gene expression in the dorsolateral prefrontal cortex (DLPFC) of schizophrenia patients and reduced white matter (WM) integrity of the corona radiata in normal brains among Caucasians. In an effort to replicate the association between this variant and WM integrity among healthy Japanese, we found that the number of A alleles was positively correlated with WM integrity in some fiber tracts, including the right posterior limb of the internal capsule, and with mean blood flow in a widespread area, including the inferior frontal operculum, orbital area, and triangular gyrus. Because the A allele affected WM integrity in opposite directions in Japanese and Caucasians, we investigated a possible association between the OLIG2 gene SNPs and the expression level of OLIG2 transcripts in postmortem DLPFCs. We evaluated rs1059004 and additional SNPs in the 5′ upstream and 3′ downstream regions of rs1059004 to cover the broader region of the OLIG2 gene. The 2 SNPs (rs1059004 and rs9653711) had opposite effects on OLIG2 gene expression in the DLPFC in Japanese and Caucasians. These findings suggest ethnicity-dependent opposite effects of OLIG2 gene SNPs on WM integrity and OLIG2 gene expression in the brain, which may partially explain the failures in replicating associations between genetic variants and psychiatric phenotypes among ethnicities.
Keywords: OLIG2 polymorphism, white matter integrity/cerebral blood flow/OLIG2 gene expression/ethnic difference, schizophrenia
Introduction
Several lines of evidence have indicated that oligodendrocyte dysfunction may be involved in the etiology of schizophrenia based on the findings of brain magnetic resonance imaging (MRI) based diffusion tensor imaging (DTI) and postmortem brain studies.1,2 Prior postmortem brain studies have found abnormal expression levels of myelin-related genes in some brain regions in patients with schizophrenia compared with normal brains.3 Consistent with these findings, several genetic association studies have indicated that single-nucleotide polymorphisms (SNPs) in oligodendrocyte-related genes are associated with schizophrenia risk.4–8 Furthermore, multiple previous DTI studies have revealed abnormalities in white matter (WM) integrity in various brain fiber tracts in the brains of patients with schizophrenia.9,10
Oligodendrocyte lineage transcription factor 2 (OLIG2) is an oligodendrocyte-related gene and a basic helix-loop-helix transcription factor mostly expressed in the brain and spinal cord ventricular zone.11 The main function of OLIG2 is to generate motor neurons and oligodendrocytes from a common pool of progenitors termed the pMN domain in the spinal cord.11,12 Although oligodendrocytes are known to form myelin sheaths, which increase impulse speed through saltatory conduction of action potentials in the central nervous system,13 recent studies have provided interesting evidence of the critical role of oligodendrocytes in WM angiogenesis via interaction with the vascular endothelium.14,15 Hypoxia-inducible factor, which is expressed in oligodendrocytes, promotes angiogenesis in the brain, and oligodendrocyte-driven angiogenesis is reportedly critical for axon/WM integrity.16 Recent stereological postmortem brain studies have revealed that the decrease in oligodendrocyte numbers in a part of the anterior and the entire hippocampal subfield are related to cognitive deficits in schizophrenia patients.17 Intravenous transplantation of neural stem cells overexpressing OLIG2 has also reportedly improved memory function via remyelination in the aged hippocampus after transient cerebral ischemia.18 Therefore, as a transcription factor involved in oligodendrocyte generation and differentiation, OLIG2 may play a crucial role in WM integrity, the vascular system, and cognitive functions.
Prior postmortem brain studies have found reduced OLIG2 gene expression in the postmortem brains of patients with schizophrenia, with a few exceptions.19–21 Further, several studies have shown genetic associations between OLIG2 gene SNPs and psychiatric disorders.4,22–24 As shown in table 1, several OLIG2 SNPs, including rs1059004, which is located in the 3′ untranslated region (UTR) of exon 2 of the OLIG2 gene, have been identified as associated with schizophrenia in certain populations.4,22,25 Moreover, consistent with the significant associations between the OLIG2 gene polymorphisms and schizophrenia, the schizophrenia-risk-associated allele (A) of rs1059004 and allele (C) of rs9653711, which are in strong linkage disequilibrium (LD) with each other in Caucasian populations, have been shown to predict reduced OLIG2 mRNA levels in the postmortem dorsolateral prefrontal cortices of Caucasian patients with schizophrenia.22,26
Table 1.
Summary of the Results of Prior Genetic Association Studies Between OLIG2 Gene Polymorphisms and Schizophrenia
| Study | Population | No. of samples | SNP ID (major allele/minor allele) | Position (chr21:_) | Functional consequence | Minor allele frequency | |||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | P-value | |||||
| Georgieva et al.22 | Caucasian | 648 | 712 | rs2834070 (G/T) | 33015144 | None | 0.39 | 0.336 | .002 |
| rs9978551 (C/G) | 33018125 | None | 0.067 | 0.07 | .78 | ||||
| rs11701698 (A/C) | 33021637 | None | 0.215 | 0.189 | .09 | ||||
| rs6517135 (A/G) | 33025263 | 2 kb upstream variant | 0.129 | 0.147 | .17 | ||||
| rs1005573 (T/C) | 33026408 | Intron variant | 0.302 | 0.347 | .012 | ||||
| rs762178 (C/T) | 33027093 | Synonymous variant | 0.391 | 0.461 | .0003 | ||||
| rs1059004 (A/C) | 33028155 | 3-prime UTR variant | 0.425 | 0.5 | .0001 | ||||
| rs6517137 (A/G) | 33028471 | 3-prime UTR variant | 0.1 | 0.099 | .88 | ||||
| rs13046814 (T/G) | 33029069 | 3-prime UTR variant | 0.241 | 0.277 | .03 | ||||
| rs9653711 (G/C) | 33029641 | 500 bp downstream variant | 0.425 | 0.375 | .007 | ||||
| 33322832 C3A (C/A) | 0.054 | 0.051 | .8 | ||||||
| 33322853 G3A (G/A) | 0.197 | 0.192 | .751 | ||||||
| rs11701762 (C/T) | 33030918 | Intron variant | 0.137 | 0.116 | .08 | ||||
| rs881666 (G/C) | 33032958 | Intron variant | 0.43 | 0.392 | .04 | ||||
| rs762237 (T/C) | 33035869 | Intron variant | 0.384 | 0.367 | .35 | ||||
| rs2834072 (A/G) | 33038156 | None | 0.485 | 0.483 | .78 | ||||
| Usui et al.25 | Japanese | 759 | 757 | rs6517135 (A/G) | 33025263 | 2 kb upstream variant | 0.2 | 0.21 | .68 |
| rs1005573 (C/T) | 33026408 | Intron variant | 0.38 | 0.38 | .68 | ||||
| rs762178 (T/C) | 33027093 | Synonymous variant | 0.17 | 0.19 | .29 | ||||
| rs6517137 (A/G) | 33028471 | 3-prime UTR variant | 0.1 | 0.09 | .67 | ||||
| Huang et al.4 | Chinese | 329 | 288 | rs1005573 (C/T) | 33026408 | Intron variant | 0.41 | 0.39 | .47 |
| rs762178 (T/C) | 33027093 | Synonymous variant | 0.09 | 0.15 | .014 | ||||
| rs1059004 (C/A) | 33028155 | 3-prime UTR variant | 0.16 | 0.12 | .2 |
Note: OLIG2, oligodendrocyte lineage transcription factor 2; SNP, single nucleotide polymorphism.
Consistent with the results of gene expression analysis in postmortem brains,26,27 Prata et al27 indicated an association between the schizophrenia-risk-associated A allele of rs1059004 and decreased WM integrity of bilateral corona radiata in Caucasian healthy subjects.
Based on these previously reported findings, we hypothesized that OLIG2 gene polymorphisms affected the vascular system and cognitive functions and that the effect of the OLIG2 SNP rs1059004 on WM integrity would be replicated in the Japanese population. To verify this hypothesis, we investigated the impact of OLIG2 gene SNPs on WM integrity, resting cerebral blood flow, and cognitive functions among the Japanese population, which failed to replicate the association between the SNP rs1059004 and WM integrity; instead, we observed the opposite effect of this variant on WM integrity. The association of the OLIG2 SNPs with OLIG2 gene expression in postmortem brain tissue was further evaluated in both Caucasian and Japanese populations to elucidate the ethnicity-specific differences. Among several SNPs associated with schizophrenia in Caucasian populations, we selected rs1059004 and the additional SNPs rs1005573 and rs9653711 (respectively, an intron variant in the 5′ upstream regions and a downstream variant-500B in the 3′ downstream region of rs1059004) for SNP genotyping to cover a broader region of the OLIG2 gene.
Methods
Methods are described in detail in the supplementary material.
The 765 healthy individuals in cohort A underwent an investigation of the association between OLIG2 SNP rs1059004 and brain imaging data and cognitive data. Cohort B consisted of 244 patients with schizophrenia and 952 healthy controls to assess the genetic association between 3 OLIG2 SNPs (rs1059004, rs9653711, and rs1005573) and cognitive functions. After the study procedures were fully explained, written informed consent was obtained from all participants in accordance with the Declaration of Helsinki (1991). These genetic association studies were approved by the Ethics Committees of Tohoku University and Osaka University.
Genomic DNA was extracted from saliva specimens of cohort A and whole blood samples of cohort B. Genomic DNA and total RNA samples extracted from the dorsolateral prefrontal cortex (DLPFC) from healthy Caucasian controls were obtained from the Array Collection at the Stanley Medical Research Institute. DLPFC tissues of healthy Japanese controls were obtained from Japan Brain Bank Net (JBBN). Genomic DNA and total RNA were isolated from the tissue. All brain tissues used in this study were obtained with written informed consent from the legal next of kin in accordance with the Declaration of Helsinki. This postmortem brain study was approved by the Ethics Committees of Tohoku University, Niigata University, Aichi Medical University, Fukushima Medical University, and National Center of Neurology and Psychiatry.
Genotyping of the OLIG2 gene SNPs was conducted using TaqMan assays. OLIG2 mRNA levels were measured by quantitative reverse transcription PCR. 18S rRNA was measured as an internal reference for normalizing confounding variables among the samples.
Statistical analysis is shown in the statistical group-level analysis of genetic data in the SM.
Results
The successfully genotyped 765 subjects of cohort A showed the following genotype distribution: homozygous A allele (n = 21), heterozygous A/C (n = 224), and homozygous C allele (n = 520); these results were consistent with Hardy-Weinberg equilibrium (HWE). The 3 OLIG2 genotypic groups showed no significant differences with respect to age or gender (supplementary table S1).
Demographic data by genotype in the 952 healthy controls and 244 patients with schizophrenia in cohort B are also shown in supplementary table S1. The genotypic distribution did not deviate from HWE in either healthy controls or patients with schizophrenia. The patients with schizophrenia showed no significant differences in age, gender, education history, antipsychotic dose, or Positive and Negative Syndrome Scale (PANSS) score, according to genotype. The genotypic groups in healthy subjects showed no significant differences in age and education history, but the sex distribution differed significantly among genotypes of rs1059004 and rs9653711 (supplementary table S1).
Associations Between OLIG2 SNPs and Cognitive Performance
Based on the 765 healthy subjects of cohort A, analysis of variance (ANOVA) showed no significant differences among the 3 genotypic groups of rs1059004 in any cognitive function.
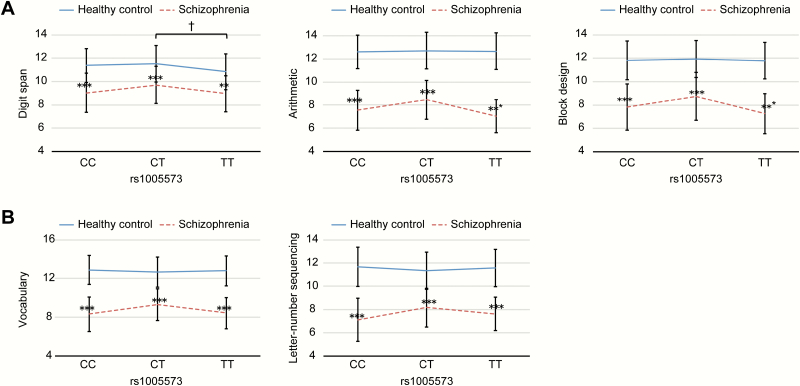
By contrast, in the 952 healthy adults and 244 patients with schizophrenia in cohort B, 2-way ANOVA showed a significant main effect of diagnosis for all subtests and the 4 indices (P < .001). Compared with control subjects, patients with schizophrenia showed significantly lower scores on all subtests and the 4 indices (supplementary table S2). The main effect of genotype in rs1005573 was significant for the arithmetic, digit span, and block design subtests of the Wechsler Adult Intelligence Scale-Third Edition (WAIS-III) (figure 1a). In Bonferroni post hoc analysis, subjects with the TT genotype (TT) displayed significantly lower digit span scores than subjects with the CT genotype (CT) (figure 1a). On the other hand, the genotype-by-diagnosis interaction was significant for the vocabulary and letter-number sequencing subtests (figure 1b).
Fig. 1.
Impact of OLIG2 gene polymorphisms on cognitive function in healthy subjects and patients with schizophrenia. (a) In the SNP rs1005573, 2-way ANOVA indicated that the main effect of genotype was also significant for the digit span, arithmetic, and block design subtests of the WAIS-III (P = .047, P = .028, and P = .031, respectively). Post hoc analysis showed that the subjects with the TT genotype (TT) showed significantly lower scores on the digit span subtest than did the subjects with the CT genotype (CT) (Bonferroni corrected, †P = .018). In each genotype group for rs1005573, there were significantly lower scores on the digit span, arithmetic, and block design subtests in patients with schizophrenia than in healthy controls (Bonferroni corrected, ***P < .001). (b) In the SNP rs1005573, 2-way ANOVA revealed significant genotype-by-diagnosis interactions for the vocabulary and letter-number sequencing subtests (respectively, P = .044and P = .028). In each genotype group for rs1005573, patients with schizophrenia had significantly lower scores on the both subtest than did healthy controls (Bonferroni corrected, ***P < .001 or **P < .01
Effect of the A Allele of OLIG2 rs1059004 on Fractional Anisotropy
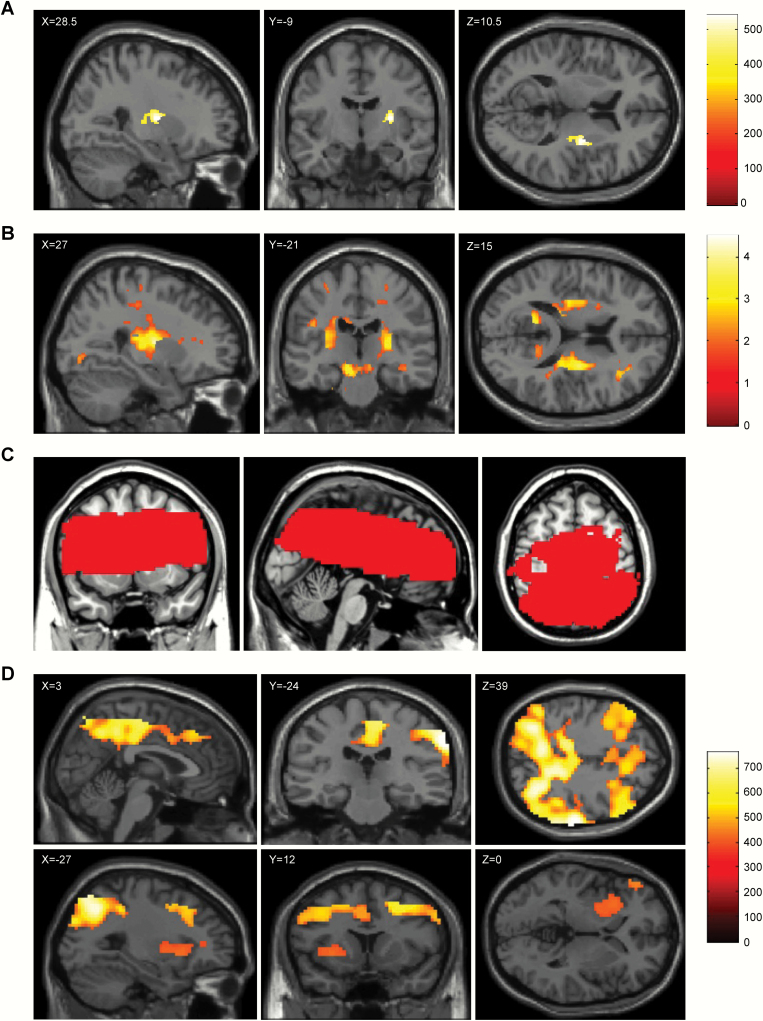
A whole-brain multiple regression analysis corrected for the effects of age and sex showed positive correlations between the number of A alleles and fractional anisotropy (FA) in 3 regions of WM: posterior limb of the internal capsule (PLIC), retrolenticular part of the internal capsule (RPIC), and external capsule (EC). After correction for multiple comparisons, the frequency of the A allele was positively associated with FA in the right PLIC, right RPIC, and right EC (figure 2a: MNI coordinates; x, y, z = 28.5, −9, 10.5, threshold-free cluster enhancement [TFCE] value = 542.08, P = .009, corrected for multiple comparisons: familywise error TFCE). When the associated area was depicted based on the associations without correction for multiple comparisons, the frequencies of the A allele were positively associated with FA in the bilateral PLIC, RPIC, and EC (figure 2b: MNI coordinates; x, y, z = 27, −21, 15, P < .05).
Fig. 2.
Brain regions showing significant associations of the OLIG2 gene polymorphism rs1059004 with fractional anisotropy and regional cerebral blood flow. (a) The white matter region where the positive association between the fractional anisotropy and number of A alleles of the OLIG2 gene polymorphism rs1059004 was observed based on threshold-free cluster enhancement, with the significance level set at P < .05 (corrected for familywise error rate based on 5000 permutations), is shown in yellow-orange. The associated region is overlaid on a single-subject T1 SPM8 image. Significantly associated white matter was widespread in the right posterior limb of the internal capsule, the right retrolenticular part of the internal capsule, and the right external capsule. The anatomical labels and significant clusters of major white matter fibers were determined using the ICBM DTI-81 Atlas (http://www.bmap.ucla.edu/portfolio/atlases/ICBM_DTI-81_Atlas/).28 (b) The white matter region where the positive association between the fractional anisotropy and the number of A alleles of the OLIG2 SNP rs1059004 was observed, with the significance level set at P < .05 (uncorrected for multiple comparisons), is shown in yellow-orange. The associated region was overlaid on a single-subject T1 SPM8 image. Significantly associated white matter was widespread in the bilateral posterior limb of the internal capsule, the bilateral retrolenticular part of the internal capsule, and the bilateral external capsule. (c) Brain regions marked in red indicate the target areas of the brain analyzed with arterial spin labeling analyses. (d) The brain region where the positive association between the mean resting cerebral blood flow and the number of A alleles of the OLIG2 gene polymorphism rs1059004 was observed based on threshold-free cluster enhancement, with the significance level set at P < .05 (corrected for familywise error rate based on 5000 permutations), is shown in yellow-orange. The associated region is overlaid on a single-subject T1 SPM8 image. Significantly associated brain regions included the precuneus, middle and posterior cingulate cortices, putamen, insula, and globus pallidus.
Effect of the A Allele of OLIG2 rs1059004 on Arterial Spin Labeling
The areas of the brain shown in figure 2c were scanned in the arterial spin labeling (ASL) analysis of the subjects. A whole-brain multiple regression analysis, corrected for the effects of age and sex, revealed that A allele frequency was positively associated with cerebral blood flow (CBF) in 2 clustered brain regions: a cluster, including the precuneus, middle, and posterior cingulate cortices, and another cluster, including the putamen, insula, and globus pallidus (figure 2d, table 2).
Table 2.
Brain Regions That Exhibited Significant Positive Correlations Between the Number of A alleles of the OLIG2 SNP rs1059004 and Regional Cerebral Blood Flow
| Included gray matter areas* (number of significant voxels in each anatomical area in the left and right hemispheres) | x | y | z | TFCE value | Corrected P-value (FWE) | Cluster size (voxels) |
|---|---|---|---|---|---|---|
| Angular gyrus (L:183, R:471)/anterior cingulum (L:5, R:8)/middle cingulum (L:251, R:348) | 66 | −24 | 39 | 766.5 | .005 | 6755 |
| Posterior cingulum (L:36, R:8)/cuneus (L:20, R:4)/inferior frontal operculum (L:30, R:41) | ||||||
| Inferior frontal orbital area (L:7)/inferior frontal triangular (L:206, R:46) | ||||||
| Other middle frontal areas (L:365, R:377)/superior frontal medial area (L:105, R:48) | ||||||
| Other superior frontal areas (L:51, R:83)/middle occipital lobe (L:250, R:89) | ||||||
| Superior occipital lobe (L:152, R:79)/paracentral lobule (L:16, R:51) | ||||||
| Inferior parietal lobule (L:251, R:312)/superior parietal lobule (L:292, R:182) | ||||||
| Postcentral gyrus (L:13, R:155)/precentral gyrus (L:70, R:129)/precuneus (L:473, R:384) | ||||||
| Rolandic operculum (R:2)/supplemental motor area (L:39, R:134) | ||||||
| Supramarginal gyrus (R:335)/middle temporal gyrus (R:17)/superior temporal gyrus (R:43) | ||||||
| Inferior frontal triangular (L:18)/insula (L:74)/pallidum (L:11)/putamen (L:97) | −30 | 15 | 0 | 416.48 | .041 | 235 |
Note: TFCE, threshold-free cluster enhancement; OLIG2, oligodendrocyte lineage transcription factor 2; FWE, familywise error. The brain regions and TFCE values of significantly positive associations between the number of A alleles of the OLIG2 SNP rs1059004 and regional cerebral blood flow are shown in the table. The labeling of the anatomical regions of the gray matter was based on the WFU PickAtlas Tool (http://www.fmri.wfubmc.edu/cms/software#PickAtlas/)29,30 and the PickAtlas automated anatomical labeling atlas option.31 Temporal pole areas included all subregions in the areas of this atlas. The table shows the FWE-corrected P values for multiple comparisons in the brain regions that were significantly associated with the number of A alleles.
Impact of OLIG2 rs1059004, rs9653711, and rs1005573 on OLIG2 Gene Expression in Postmortem DLPFC
The postmortem interval (PMI) differed significantly between CC carriers and A allele carriers of rs1059004 among the Caucasian postmortem brains. At the same time, there were no significant differences in age, gender, tissue pH, and RNA integrity number (RIN) between the genotypic groups (supplementary table S3). The Japanese postmortem brains showed a significant difference only in gender and not in the other confounding variables among the 2 genotypes (supplementary table S3).
RIN significantly influenced raw OLIG2 mRNA levels, 18S rRNA levels, and OLIG2 mRNA levels normalized to 18S rRNA levels in the Japanese postmortem brains. There was a significant effect of tissue pH on 18S mRNA levels in both Japanese and Caucasian postmortem brains. PMI had a significant impact only on 18S rRNA levels in the Caucasian postmortem brain (supplementary table S5).
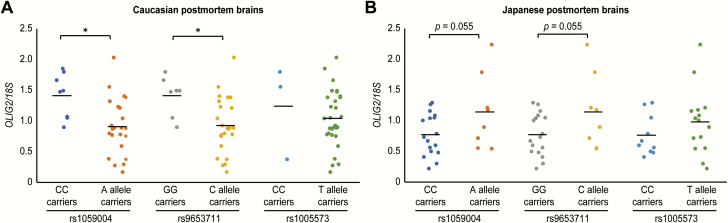
Analysis of covariance (ANCOVA) controlling for PMI revealed that normalized OLIG2 mRNA levels were significantly lower in A allele carriers than in CC carriers of rs1059004 among Caucasian subjects [F(1, 28) = 4.389, P = .045, figure 3a). In addition, the C allele carriers of rs9653711 showed significantly lower OLIG2 transcripts than GG carriers [F(1, 28) = 4.905, P = .035, figure 3a]. Conversely, ANCOVA controlling for RIN displayed normalized OLIG2 mRNA levels that were higher in the A-allele carriers than in the CC carriers of rs1059004 in the Japanese group, although the difference reached a marginal level of statistical significance [F(1, 22)= 4.116, P = .055, figure 3b]. Additionally, the minor C allele carriers at SNP rs9653711 had higher OLIG2 expression levels than the GG carriers [F(1, 22)= 4.116, P = .055, figure 3b]. This SNP rs9653711 was in perfect LD (1000 Genomes Project data: https://www.internationalgenome.org/) with rs1059004 in Japanese subjects, so the results based on rs9653711 were the same as for rs1059004.
Fig. 3.
Impact of the OLIG2 gene SNPs on OLIG2 gene expression among Caucasian and Japanese postmortem DLPFC specimens. (a) Carriers of the A allele of the SNP rs100594 showed significantly lower OLIG2 gene expression in the postmortem DLPFC than CC carriers among Caucasians (*P = .045). In the OLIG2 gene SNP rs9653711, C allele carriers had significantly decreased OLIG2 gene expression compared with GG carriers among Caucasians (*P = .035). (b) In contrast, among the Japanese population, carriers of the A allele of the SNP rs100594 showed higher OLIG2 gene expression in the postmortem DLPFC than CC carriers, although the difference did not reach statistical significance (P = .055). As the SNP rs9653711 is in perfect LD with rs1059004, the results of OLIG2 gene expression analysis of rs9653711 were consistent with the results of rs1059004.
Discussion
The present study, based on a large sample of the Japanese population, showed that the number of A alleles of the OLIG2 SNP rs1059004 was positively correlated with WM integrity in some fiber tracts, including the right PLIC, and with mean CBF over a wide area, including the precuneus and insula in Japanese normal brains, contrary to previous findings indicating a negative correlation between the number of A alleles and WM integrity among Caucasians. Consistent with the converse influence of rs1059004 on WM integrity and CBF between these 2 ethnic groups, these findings also revealed that rs1059004 and rs9653711, which are in strong LD with each other, had opposite effects on OLIG2 gene expression in Caucasian and Japanese postmortem DLPFC tissues.
To our knowledge, 2 previous trials have attempted to elucidate the genetic associations between rs1059004 and the WM integrity of the brain in the Caucasian population. Voineskos et al8 indicated positive correlations between the number of A alleles of rs1059004 and the FA values of multiple brain regions, including the bilateral cingulum bundle, inferior longitudinal fasciculus, arcuate fasciculus, inferior occipitofrontal fasciculus, and corpus callosum, in the region-of-interest analysis but not in whole-brain analysis in healthy Caucasian subjects. Another study based on whole-brain analysis indicated that the A allele was negatively associated with FA values in the bilateral corona radiata of a healthy Caucasian population.27 Our study, which was based on whole-brain DTI analysis, similar to the study of Prata et al,27 also revealed a negative association between the number of A alleles of rs1059004 and the FA values in some fiber tracts, including the PLIC. As OLIG2 is involved in the differentiation of precursor cells into motor neurons as well as oligodendrocytes, this gene polymorphism may underlie the difference in WM integrity around the internal capsule, carrying signals from the primary motor cortex to the lower motor neurons in the spinal cord.
The results of the ASL study revealed that rs1059004 affected resting CBF in broad areas of the normal brain. Based on the results from a postmortem gene expression analysis of Caucasian patients with schizophrenia in which the non-A-allele carriers showed higher expression levels of OLIG2 mRNA than the A-allele carriers, we first assumed that the number of A alleles was significantly and negatively correlated with resting-state CBF as well as FA in healthy Japanese subjects. However, our study using healthy Japanese subjects indicated positive correlations between the number of A alleles and resting CBF or FA in the normal brain.
Because the A allele affected WM integrity in opposite directions in Japanese and Caucasian subjects, we assessed the association between the OLIG2 gene SNPs and OLIG2 gene expression levels in the postmortem brain tissues. We genotyped rs1059004 and 2 SNPs in the 5′ upstream and 3′ downstream regions of rs1059004, one of several SNPs associated with schizophrenia in Caucasians. The SNP rs2834070 in the 5′ upstream region of rs1059004 was excluded from SNP genotyping in this study, as the minor allele frequency (T:0.077) of the SNP was very low (1000 Genomes Project data). The current gene expression analyses in the postmortem brain found that rs1059004 and rs9653711 affected OLIG2 gene expression in opposite directions in the Caucasian and Japanese populations. As rs9653711 is in perfect (r2 = 1) and strong (r2 = .98) LD with rs1059004 in the Japanese and Caucasian groups, respectively (1000 Genomes Project data), the results of gene expression analyses were not independent.
Meanwhile, the results for rs1005573 differed from the results for the other 2 SNPs, as rs1005573 is in moderate (r2 = .39) and very low (r2 = .19) LD with rs1059004 in Caucasian and Japanese populations, respectively (1000 Genomes Project data). The findings suggested that the difference in the influence of rs1059004 on OLIG2 gene expression in the brain may constitute the mechanism of the opposite effects of rs1059004 on WM integrity and resting-state CBF between these 2 ethnic groups. Although the definitive biological mechanism through which rs1059004 affects resting-state CBF and FA remains unclear, the difference in the degree of oligodendrocyte differentiation affected by rs1059004 genotypes may influence postnatal brain angiogenesis and myelination, possibly resulting in differences in resting CBF and FA.
Recent genome-wide quantitative trait locus (eQTL) studies have identified that numerous cis-SNPs significantly associated with gene expression levels in the human brain.32–37 Notably, the cis-effects displayed in the eQTL databases are not consistent among brain regions. The effects of eQTLs on gene expression levels can be observed in a brain region-specific manner, as shown by the data publicly available in BRAINEAC via http://www.braineac.org.38 Gene expression analyses of multiple primary immune cells revealed that the C allele of the SELL gene SNP rs222328 affects SELL gene expression levels in opposite directions between B-cells and monocytes.39 However, ethnicity-specific genetic effects on gene expression levels have rarely been evaluated, with the exception that the effect of the coding SNP (G94 > A) of the GSTL1 gene polymorphism rs7975 on protein expression levels differs between the Caucasian and African American populations, potentially due to the LD between rs7975 and the promoter G > A −1002 SNP rs7160195, which has been shown to be associated with the expression level of this gene.40
Thus, the discrepancies observed between the current MRI studies of the Japanese population and previous studies of the Caucasian population can be explained by region- and ethnicity-dependent opposite cis-effects of this OLIG2 variant, and the current postmortem study has provided the first direct evidence of an ethnicity-dependent opposite cis-effect of this genetic variant, which has previously been overlooked. Notably, similar results have been found with inbred or outbred strains of rodents. Pronounced variations in gene expression, as well as relevant cellular architecture among different strains, have been observed in the central nervous system, which may underlie strain-specific behavioral and physiological responses.41–44 Investigations into strain-specific phenomena utilizing commercially available outbred mice45 along with investigations into human subjects may elucidate the molecular mechanisms underlying ethnicity-specific phenotypes.
Our neurocognitive analysis also showed a significant association of rs1005573 with the scores on the arithmetic, digit span, and block design subtests. These results suggest that this SNP may affect visuospatial ability and working memory. We also revealed a significant genotype-by-diagnosis interaction of rs1005573 on vocabulary and letter-number sequencing subtests. The present results indicate that rs1005573 may affect verbal comprehension and working memory in schizophrenia. The effects of the variant on cognitive function may affect susceptibility to schizophrenia among the Japanese population.
This study has several limitations. First, the associations among the genetic variant, cognitive function, and brain imaging, including CBF and WM integrity, were not assessed using the same subjects. Further association studies are needed to verify the relationships.
Second, the MRI data acquisition procedure and subject demographics in the current Japanese study were not strictly the same as those in the previous Caucasian study.
Third, the impact of rs1059004 on OLIG2 gene expression in the brain regions was not assessed in the postmortem tissues where the variant affected WM integrity of the Japanese or Caucasian population, ie, the corona radiata and right PLIC. However, the DLPFC was found to be connected with the human dorsolateral premotor cortex, where the corticospinal fibers originate and descend through the corona radiata and PLIC to reach the brainstem.46 The biological effect of the genetic variant might be consistent throughout the relevant neuronal networks.
In conclusion, the present study indicated that the schizophrenia risk variant OLIG2 SNP rs1059004 affected both WM integrity and the vascular system in the normal brain. In addition, this study presents an initial example of an ethnicity-dependent opposite effect of a genetic variant on gene transcription and organ structure in the brain, which may be a putative mechanism underlying the failure to replicate the association between a genetic variant and a psychiatric phenotype among different ethnicities. Further investigation is required to reveal the molecular mechanism involved in the ethnicity-dependent effect of a genetic variant on gene transcription and organ structure in the brain.
Funding
This work was partially supported by a Japan Society for the Promotion of Science KAKENHI (Grant Number: JP2411600, JP23700306, JP25700012, JP16H06277) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Strategic Research Program for Brain Sciences of the Japan Agency for Medical Research and Development, (JP18dm0107099, JP18dm0107103, JP18dm0107104, JP18dm0107105, JP18dm0107107, JP18ek0109183). This study was also partially supported by Japan Science and Technology Agency (JST)/Research Institute of Science and Technology for Society; JST/Core Research for Evolutionary Science and Technology; the Collaborative Research Project of the Brain Research Institute, Niigata University (2017-2806 and 2018-2809); and the Core Research Cluster of Disaster Science, Tohoku University, Japan.
Supplementary Material
Acknowledgments
We thank Yuki Yamada for operating the MRI scanner; Haruka Nouchi, for conducting the psychological tests; all other assistants for helping with the experiments and the study; and the study participants and all our colleagues at Tohoku University for their support. The authors have declared that there are no conflicts of interest concerning the subject of the study.
References
- 1. Bartzokis G, Nuechterlein KH, Lu PH, Gitlin M, Rogers S, Mintz J. Dysregulated brain development in adult men with schizophrenia: a magnetic resonance imaging study. Biol Psychiatry. 2003;53(5):412–421. [DOI] [PubMed] [Google Scholar]
- 2. Hof PR, Haroutunian V, Copland C, Davis KL, Buxbaum JD. Molecular and cellular evidence for an oligodendrocyte abnormality in schizophrenia. Neurochem Res. 2002;27(10):1193–1200. [DOI] [PubMed] [Google Scholar]
- 3. Davis KL, Stewart DG, Friedman JI, et al. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60(5):443–456. [DOI] [PubMed] [Google Scholar]
- 4. Huang K, Tang W, Tang R, et al. Positive association between OLIG2 and schizophrenia in the Chinese Han population. Hum Genet. 2008;122(6):659–660. [DOI] [PubMed] [Google Scholar]
- 5. Jitoku D, Hattori E, Iwayama Y, et al. Association study of Nogo-related genes with schizophrenia in a Japanese case-control sample. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(5):581–592. [DOI] [PubMed] [Google Scholar]
- 6. Li D, Feng G, He L. Case-control study of association between the functional candidate gene ERBB3 and schizophrenia in Caucasian population. World J Biol Psychiatry. 2009;10(4 Pt 2):595–598. [DOI] [PubMed] [Google Scholar]
- 7. Maeno N, Takahashi N, Saito S, et al. Association of SOX10 with schizophrenia in the Japanese population. Psychiatr Genet. 2007;17(4):227–231. [DOI] [PubMed] [Google Scholar]
- 8. Voineskos AN, Felsky D, Kovacevic N, et al. Oligodendrocyte genes, white matter tract integrity, and cognition in schizophrenia. Cereb Cortex. 2013;23(9):2044–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanaan RA, Kim JS, Kaufmann WE, Pearlson GD, Barker GJ, McGuire PK. Diffusion tensor imaging in schizophrenia. Biol Psychiatry. 2005;58(12):921–929. [DOI] [PubMed] [Google Scholar]
- 10. Kyriakopoulos M, Bargiotas T, Barker GJ, Frangou S. Diffusion tensor imaging in schizophrenia. Eur Psychiatry. 2008;23(4):255–273. [DOI] [PubMed] [Google Scholar]
- 11. Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia 2006;54(1):1–10. [DOI] [PubMed] [Google Scholar]
- 12. Meijer DH, Kane MF, Mehta S, et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13(12):819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradl M, Lassmann H. Oligodendrocytes: biology and pathology. Acta Neuropathol. 2010;119(1):37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyamoto N, Pham LD, Seo JH, Kim KW, Lo EH, Arai K. Crosstalk between cerebral endothelium and oligodendrocyte. Cell Mol Life Sci. 2014;71(6):1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pham LD, Hayakawa K, Seo JH, et al. Crosstalk between oligodendrocytes and cerebral endothelium contributes to vascular remodeling after white matter injury. Glia 2012;60(6):875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuen TJ, Silbereis JC, Griveau A, et al. Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell 2014;158(2):383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falkai P, Steiner J, Malchow B, et al. Oligodendrocyte and interneuron density in hippocampal subfields in schizophrenia and association of oligodendrocyte number with cognitive deficits. Front Cell Neurosci. 2016;10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahn JH, Chen BH, Shin BN, et al. Intravenously infused F3.Olig2 improves memory deficits via restoring myelination in the aged hippocampus following experimental ischemic stroke. Cell Transplant. 2016;25(12):2129–2144. [DOI] [PubMed] [Google Scholar]
- 19. Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr Res. 2005;79(2–3):157–173. [DOI] [PubMed] [Google Scholar]
- 20. McCullumsmith RE, Gupta D, Beneyto M, et al. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr Res. 2007;90(1–3):15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tkachev D, Mimmack ML, Ryan MM, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362(9386):798–805. [DOI] [PubMed] [Google Scholar]
- 22. Georgieva L, Moskvina V, Peirce T, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2006;103(33):12469–12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stewart SE, Platko J, Fagerness J, et al. A genetic family-based association study of OLIG2 in obsessive-compulsive disorder. Arch Gen Psychiatry. 2007;64(2):209–214. [DOI] [PubMed] [Google Scholar]
- 24. Zhang X, Liu J, Guo Y, Jiang W, Yu J. Association study between oligodendrocyte transcription factor 2 gene and obsessive-compulsive disorder in a Chinese Han population. Depress Anxiety. 2015;32(10):720–727. [DOI] [PubMed] [Google Scholar]
- 25. Usui H, Takahashi N, Saito S, et al. The 2’,3’-cyclic nucleotide 3’-phosphodiesterase and oligodendrocyte lineage transcription factor 2 genes do not appear to be associated with schizophrenia in the Japanese population. Schizophr Res. 2006;88(1–3):245–250. [DOI] [PubMed] [Google Scholar]
- 26. Mitkus SN, Hyde TM, Vakkalanka R, et al. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98(1–3):129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prata DP, Kanaan RA, Barker GJ, et al. Risk variant of oligodendrocyte lineage transcription factor 2 is associated with reduced white matter integrity. Hum Brain Mapp. 2013;34(9):2025–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mori S, Oishi K, Jiang H, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage 2008;40(2):570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maldjian JA, Laurienti PJ, Burdette JH. Precentral gyrus discrepancy in electronic versions of the Talairach atlas. Neuroimage 2004;21(1):450–455. [DOI] [PubMed] [Google Scholar]
- 30. Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage 2003;19(3):1233–1239. [DOI] [PubMed] [Google Scholar]
- 31. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15(1):273–289. [DOI] [PubMed] [Google Scholar]
- 32. Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S, Cho H, Lee D, Webster MJ. Association between SNPs and gene expression in multiple regions of the human brain. Transl Psychiatry. 2012;2:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim Y, Xia K, Tao R, et al. A meta-analysis of gene expression quantitative trait loci in brain. Transl Psychiatry. 2014;4:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu C, Cheng L, Badner JA, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15(8):779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–1499. [DOI] [PubMed] [Google Scholar]
- 37. Zou F, Chai HS, Younkin CS, et al. ; Alzheimer’s Disease Genetics Consortium Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8(6):e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramasamy A, Trabzuni D, Guelfi S, et al. ; UK Brain Expression Consortium; North American Brain Expression Consortium Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fairfax BP, Makino S, Radhakrishnan J, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44(5):502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langaee TY, Zhong G, Li W, et al. The influence of human GSTZ1 gene haplotype variations on GSTZ1 expression. Pharmacogenet Genomics. 2015;25(5):239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gahring LC, Persiyanov K, Dunn D, Weiss R, Meyer EL, Rogers SW. Mouse strain-specific nicotinic acetylcholine receptor expression by inhibitory interneurons and astrocytes in the dorsal hippocampus. J Comp Neurol. 2004;468(3):334–346. [DOI] [PubMed] [Google Scholar]
- 42. Rogers SW, Weis JJ, Ma Y, Teuscher C, Gahring LC. Mouse chromosome 11 harbors genetic determinants of hippocampal strain-specific nicotinic receptor expression. Hippocampus 2008;18(8):750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duyck K, DuTell V, Ma L, Paulson A, Yu CR. Pronounced strain-specific chemosensory receptor gene expression in the mouse vomeronasal organ. BMC Genomics. 2017;18(1):965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim JW, Nam SM, Yoo DY, et al. Strain-specific differential expression of astrocytes and microglia in the mouse hippocampus. Brain Behav. 2018;8(5):e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yalcin B, Nicod J, Bhomra A, et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010;6(9):e1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tomassini V, Jbabdi S, Klein JC, et al. Diffusion-weighted imaging tractography-based parcellation of the human lateral premotor cortex identifies dorsal and ventral subregions with anatomical and functional specializations. J Neurosci. 2007;27(38):10259–10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.