Abstract
Chemicals in commerce or under development must be assessed for genotoxicity; assessment is generally conducted using validated assays (e.g. Tk mouse lymphoma assay) as part of a regulatory process. Currently, the MutaMouse FE1 cell mutagenicity assay is undergoing validation for eventual use as a standard in vitro mammalian mutagenicity assay. FE1 cells have been shown to be metabolically competent with respect to some cytochrome P450 (CYP) isozymes; for instance, they can convert the human carcinogen benzo[a]pyrene into its proximate mutagenic metabolite. However, some contradictory results have been noted for other genotoxic carcinogens that require two-step metabolic activation (e.g. 2-acetylaminofluorene and 2-amino-3-methylimidazo[4,5-f]quinoxaline). Here, we examined three known or suspected human carcinogens, namely acrylamide, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 4-aminobiphenyl (4-ABP), together with their proximate metabolites (i.e. glycidamide, N-OH-PhIP and N-OH-4-ABP), to aid in the validation of the FE1 cell mutagenicity assay. Assessments of the parent compounds were conducted both in the presence and absence of an exogenous metabolic activation mixture S9; assessments of the metabolites were in the absence of S9. The most potent compound was N-OH-PhIP -S9, which elicited a mutant frequency (MF) level 5.3-fold over background at 5 µM. There was a 4.3-fold increase for PhIP +S9 at 5 µM, a 1.7-fold increase for glycidamide −S9 at 3.5 mM and a 1.5-fold increase for acrylamide +S9 at 4 mM. Acrylamide −S9 elicited a marginal 1.4-fold MF increase at 8 mM. Treatment with PhIP −S9, 4-ABP ±S9 and N-OH-4-ABP −S9 failed to elicit significant increases in lacZ MF with any of the treatment conditions tested. Gene expression of key CYP isozymes was quantified by RT-qPCR. Cyp1a1, 1a2 and 1b1 are required to metabolise PhIP and 4-ABP. Results showed that treatment with both compounds induced expression of Cyp1a1 and Cyp1b1 but not Cyp1a2. Cyp2e1, which catalyses the bioactivation of acrylamide to glycidamide, was not induced after acrylamide treatment. Overall, our results confirm that the FE1 cell mutagenicity assay has the potential for use alongside other, more traditional in vitro mutagenicity assays.
Introduction
Prior to being introduced into the marketplace, regulatory requirements ensure that the safety of new substances must be adequately evaluated. In order to test the genotoxic potential of chemicals, many in vitro and in vivo assays, as well as in silico tools, have been developed. Available and commonly used assays collectively assess a variety of endpoints (e.g. mutagenicity and clastogenicity) (1); the Organisation for Economic Co-operation and Development (OECD) publishes Guidelines for the Testing of Chemicals that detail internationally accepted assessment protocols. Typically, three in vitro endpoints (i.e. gene mutations, structural and numerical chromosomal aberrations) are evaluated for a satisfactory assessment of genotoxicity. The tests commonly used for the in vitro assessments are the Ames test (OECD 471), the in vitro mammalian chromosomal aberration test (OECD 473) and the micronucleus test (OECD 487) (2–5). The two most commonly used OECD-validated in vitro mammalian mutagenicity assessment assays are the Hprt (OECD 476) and Tk (OECD 490) locus mutation assays (1,6,7). However, the cell lines used for these assays (e.g. human TK6, L5178Y mouse lymphoma and CHO) have been criticised for their lack of metabolic capacity and impaired p53 function (8). Although various cell lines and primary cells from transgenic rodents are available for in vitro mutagenicity assessment, none are currently accepted for regulatory purposes since they have not yet been appropriately validated.
A working group at the 7th International Workshop on Genotoxicity Testing reviewed all available transgenic rodent in vitro assays, concluding that assays utilising cells from the MutaMouse and lacZ plasmid mouse are the most promising for validation and use in routine mutagenicity testing (1). Therefore, the MutaMouse FE1 cell mutagenicity assay is currently in early stages of validation at Health Canada (Ottawa, ON) and collaborating laboratories. FE1 cells are an epithelial cell line isolated from the MutaMouse lung that are cytogenetically stable and show normal p53 functionality (9). As FE1 cells are derived from MutaMouse tissue, they contain the λgtlacZ shuttle vector, which can be excised from the DNA of FE1 cells and packaged into λ phage heads. The phage is then absorbed into Escherichia coli, which, in turn, are plated onto a titre plate and a mutant plate containing P-galactosidase. The lacZ mutant frequency (MF) is determined as the ratio of the number of mutant colonies to the number of colonies on the titre plate (10). As outlined in White et al. (1), to date, the reproducibility and reliability of the FE1 cell mutagenicity assay across different operators have shown a concordance of ~90% and a total of 25 reference compounds have been assessed previously in this assay (9,11–14). More details on experimental results can be found in Table 4 of White et al. (1). Of the 14 known in vivo mutagens tested, 10 elicited positive responses, including benzo[a]pyrene (BaP), aflatoxin B1 and 3-nitrobenzanthrone; two known in vivo non-mutagens ampicillin trihydrate and D-mannitol were negative (11,12). Substances often referred to as false or misleading in vitro positives (15), meaning that they are negative in in vivo mutagenicity assays but are frequently positive in other in vitro mutagenicity tests were also examined. Nine such substances (e.g. curcumin, eugenol and p-nitrophenol) were evaluated but they did not elicit positive responses in the FE1 cell mutagenicity assay (12). Additionally, the FE1 cell line has been characterised for its enzymatic capabilities. Using quantitative reverse transcription polymerase chain reaction (RT-qPCR), it has been shown that mRNA expression is inducible in FE1 cells for important Phase I [e.g. cytochrome P450 (CYP) 1A2, 1A1, 1B1] and Phase II enzymes [e.g. sulfotransferases (SULTs), N-acetyltransferases (NATs)], following chemical treatment (11,16). Thus, it is not surprising that FE1 cells can activate mutagenic carcinogens, such as BaP, without an exogenous metabolic activation system (S9). This constitutes a noteworthy benefit in comparison with the aforementioned commonly used mammalian cell mutagenicity assays. Nevertheless, although important xenobiotic-metabolising enzymes (XMEs) are expressed in MutaMouse FE1 cells, some contradictory results have been obtained. For instance, the aromatic amines 2-acetylaminofluorene (2-AAF) and 2-aminoanthracene (2-AA), which both require two-step metabolic activations to exert their genotoxic effects, elicited positive responses without S9, leading to the assumption that NATs and SULTs are expressed in FE1 cells (11,13). However, when assessing the heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), which requires SULT-catalysed sulfonation of N-OH-PhIP for bioactivation, a positive result was only observed in the presence of S9 (9). In contrast, the heterocyclic amine 2-amino-3-methylimidazo[4,5-f]quinoxaline (IQx), despite having a similar bioactivation pathway, was not mutagenic in FE1 cells even when S9 was added (13). In light of the positive results for some (heterocyclic) aromatic amines (e.g. 2-AAF ±S9 or PhIP +S9), and negative results for others (e.g. Iqx ±S9), the metabolic activation capabilities of FE1 cells seems to be complex and compound specific, even across substances that are all known to require a similar two-step bioactivation. Furthermore, some compounds (e.g. 1-methylpyrene) that require Cyp2e1 metabolism yielded negative responses in the FE1 cell mutagenicity assay (11).
No guidelines are available regarding the number of compounds that should be tested for the evaluation and validation of in vitro mutagenicity assays; however, e.g., 43 chemicals were evaluated to validate the mouse lymphoma assay (17). To contribute to the continued validation of the FE1 cell mutagenicity assay, the aim of this study was to assess the mutagenicity of three known or suspected human carcinogens, namely acrylamide, PhIP, and 4-aminobiphenyl (4-ABP), and together with their proximate metabolites glycidamide, N-OH-PhIP and N-OH-4-ABP, via which three carcinogens form pre-mutagenic DNA adducts; if not repaired, the adducts may contribute to the establishment of mutations (Supplementary Figures S1–3).
PhIP is a pyrolysis product formed during the cooking of meat and fish (18) that has been classified by the International Agency for the Research on Cancer (IARC) as a Group 2B human carcinogen (‘possibly carcinogenic’) inducing guanine mutations in vivo and in vitro (19). As noted, it has been previously evaluated in FE1 cells and has been used as a positive control to ensure that the metabolic activation is functioning as expected (9,12). As stated, PhIP has previously failed to elicit a positive response in the absence of S9; it was hypothesised that this indicates a lack of SULT activity. By directly assessing the PhIP metabolite N-OH-PhIP, this hypothesis can be evaluated.
4-ABP is an aromatic amine found in tobacco smoke and cooking fumes, which induces gene mutations in in vitro and in vivo systems. It is the only compound examined herein that is classified as Group 1 human carcinogen by IARC (20). PhIP and 4-ABP follow similar pathways of metabolism, requiring initial bioactivation predominantly by Cyp1a1, 1a2 and 1b1, followed by conversion by NATs and SULTs to unstable esters, which can undergo heterolytic cleavage and form an electrophilic nitrenium ion, that can bind to DNA (18). By including both compounds in the current study, the aforementioned two-step metabolic activation pathway, which requires both Phase I and II enzymes, can be further evaluated. Recent in vivo studies have further shown that Cyp2e1 expression is as important as Cyp1a2 for N-hydroxylation of 4-ABP (21).
Acrylamide has been classified by IARC as probably carcinogenic to humans (Group 2A) (22) and is found in cooked starch-rich foods (23); it is metabolised by CYP2E1 to the known mutagen glycidamide (24). These compounds were included in the current study to further test the hypothesis that compounds requiring Cyp2e1 metabolism are unable to elicit positive responses in the FE1 cell mutagenicity assay, i.e. they are false negatives. Glycidamide has been shown to be mutagenic in various in vivo and in vitro systems (25–29). In contrast, acrylamide induced mutations in vivo but in vitro results have been mixed (25–29). To optimise treatment conditions, crystal violet staining and Western blotting were used to examine cell viability and DNA damage response (DDR) in exposed cells, respectively. Gene expression analysis by RT-qPCR was employed to measure the induction of CYP enzyme transcript levels (i.e. Cyp1a1, 1a2, 1b1, 2e1) in mutagen-treated FE1 cells. Here, we present an investigation of the mutagenic potential of acrylamide, PhIP and 4-ABP, as well as their proximate metabolites glycidamide, N-OH-PhIP and N-OH-4-ABP to aid in the validation of the FE1 cell mutagenicity assay.
Material and methods
Test compounds
Acrylamide, glycidamide and BaP were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Syntheses of PhIP and N-OH-PhIP were performed at the Biochemical Institute for Environmental Carcinogens (Großhansdorf, Germany). 4-ABP was purchased from Honeywell Riedel-de Haën (Charlotte, North Carolina, USA), and N-OH-4-ABP from Toronto Research Chemicals (Toronto, Ontario, Canada). Acrylamide and glycidamide stocks were prepared in water at 2 M, sterile filtered (0.22 µm) and aliquots stored at −20°C. The remainder of stock solutions were prepared at 50 mM (PhIP, N-OH-PhIP), 100 mM (4-ABP, N-OH-4-ABP) or 250 mM (BaP) by dissolving the compounds in water-free DMSO. Aliquots of PhIP, 4-ABP and BaP were stored at −20°C; aliquots of N-OH-PhIP and N-OH-4-ABP were stored under nitrogen gas at −80°C.
Culture of FE1 cells
FE1 cells were isolated at Health Canada (Ottawa, Ontario, Canada) and cultured at 37°C and 5% CO2 in growth medium (DMEM:Nutrient Mixture F-12; Thermo Fisher Scientific, Waltham, Massachusetts, USA) supplemented with 2% (v/v) foetal bovine serum (Thermo Fisher Scientific), 100 U/ml penicillin, 100 µg/ml streptomycin (Thermo Fisher Scientific) and 1 ng/ml human epidermal growth factor (Thermo Fisher Scientific). For passaging, cells were detached using 0.25% trypsin-EDTA (Thermo Fisher Scientific) for 1–3 min and then suspended in growth medium; cells were reseeded at the desired cell number. Cells were counted using a Countess FL2 automated haemocytometer (Life Technologies).
Crystal violet staining assay for cell viability
Crystal violet stains adherent cells by binding to DNA and proteins and depends on the detachment of dead cells from the plate. Consequently, staining is indicative of cell viability in the wells. FE1 cells were seeded onto six-well plates at 20 000 cells/well and incubated overnight. The following day, the growth medium was removed, and cells washed with phosphate buffered saline (PBS). Serum-free treatment medium with test compounds was added. For acrylamide, PhIP and 4-ABP, a metabolic activation system was added to the exposure medium, i.e. 0.2% (v/v) Aroclor-1254-induced rat liver S9 from male Sprague–Dawley rats (Molecular Toxicology Inc.), 1.26 mM MgCl2 (Thermo Fisher Scientific), 8.3 mM KCl (Sigma-Aldrich), 1.26 mM glucose-6-phosphate (Sigma-Aldrich) and 1 mM NADP (Sigma-Aldrich). For controls, DMSO (0.04% for N-OH-PhIP or 0.1% for PhIP, 4-ABP, N-OH-4-ABP) or water (for acrylamide and glycidamide) was added to the serum-free treatment medium. After 6 h, the treatment medium was removed, cells were rinsed with PBS and then incubated for a further 72 h in fresh growth medium. Treatment was performed in duplicate wells at 37°C and 5% CO2. At 72 h after treatment was initiated, cells were washed with PBS followed by staining with 0.1% (w/v) crystal violet dye (Sigma-Aldrich) in 10% ethanol for at least 10 min. To remove excess crystal violet, cells were washed twice with PBS and air-dried in the dark at room temperature. Crystal violet was solubilised in 1 ml 50% ethanol/well to quantify the amount of staining of attached cells. Absorbance was determined at 595 nm using a plate reader (ELx800, Bio-Tek, Winooski, Vermont, USA). Data shown are mean absorbance values relative to control and are the results of at least three independent experiments. As suggested previously (12), cell viability at the beginning and end of the experiment was considered by staining a well with crystal violet prior to treating the cells. To normalise cytotoxicity with the pre-exposure cytotoxicity values, this initial absorbance value was subtracted from the final one.
Western blotting
For the Western blot experiment, cell numbers were adjusted to recover enough protein at the end of a 6-h exposure period. Thus, cells were seeded at 520 000 cells/well in six-well plates and treated the following day with subcytotoxic and cytotoxic concentrations of the test compounds in serum-free treatment medium for 6 h that resulted in 80%, 60% and 30% cell viability. For acrylamide, PhIP and 4-ABP, treatments with a metabolic activation (S9) system were included (see above). Treated cells were washed with PBS and lysed in 62.5 mM Tris (pH 6.8), 1 mM EDTA (pH 8.0), 2% (w/v) sodium dodecyl sulphate, 10% glycerol supplemented with 1X HaltTM Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Expression of phospho-p53 (Ser15), p21, γ-H2ax (Ser139), phospho-Chk1 (Ser345) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was assessed. Western blotting was performed as described previously (30). After blocking with 3% milk in TBS-T for 1 h at room temperature, blots were incubated overnight at 4°C with the following primary antibodies: anti-phospho-p53 (1:2000; Cell Signalling, Danvers, Massachusetts, USA), anti-p21 (1:2000; BD Biosciences, Franklin Lakes, New Jersey, USA), anti-γ-H2ax (1:1000; Cell Signalling) and anti-phospho-Chk1 (1:1,000; Cell Signalling). Incubation with the loading control anti-Gapdh (1:25,000; Chemicon International, Temecula, California, USA) was performed for 30 min at room temperature. After incubations with primary antibodies, blots were incubated with species-specific horse radish peroxidase-conjugated secondary antibodies (anti-mouse or anti-rabbit; Bio-Rad) for 1 h at room temperature and proteins detected by chemiluminescence.
Gene expression analysis by RT-qPCR
For the gene expression analysis, cell numbers were adjusted to recover enough RNA at the end of a 6-h period. Cells were seeded at 1.3 × 106/25-cm2 flasks and incubated overnight. The next day, the growth medium was removed, and cells washed with PBS. Serum-free treatment medium with acrylamide, PhIP or 4-ABP was added to lead to 80%, 60% and 30% cell viability. After 6 h, pellets were prepared, and RNA was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. After establishing the concentration and quality parameters of the RNA using a NanoDrop spectrophotometer, RNA was reverse transcribed into cDNA using a High-Capacity RNA-to-cDNA™ Kit (Thermo Fisher Scientific). RT-qPCR was performed using a 2X TaqMan™ Gene Expression Master Mix (Thermo Scientific) and the Roche Universal Probe Library designed intron-spanning assays (i.e. primers and matching probe) for the following NCBI sequences: NM_009992.4 (Cyp1a1), NM_009993.3 (Cyp1a2), NM_009994.1 (Cyp1b1) and NM_021282.2 (Cyp2e1). Gene expression was analysed using a 7500 Fast Real-Time PCR System (Applied Biosystems). Relative gene expression was normalised to the housekeeping gene Gapdh (NM_001289726.1) and analysed by the comparative threshold cycle (Ct) method. Results were reported as the fold change in gene expression between the treated and solvent control samples (2−ΔΔCt method).
FE1 cell mutagenicity assay
FE1 cells were seeded onto 10-cm plates at 300 000 cells/plate. The following day, cells were rinsed with PBS and serum-free treatment medium containing the test compound was added to the cells. For acrylamide, PhIP and 4-ABP S9 was added to the exposure medium (see above). After 6 h, the treatment medium was removed and the cells were washed with PBS and incubated for a further 72 h in growth medium. Cells were exposed to a range of subcytotoxic and cytotoxic concentrations of the test compounds (see Table 1) and solvent control (0.1% DMSO: PhIP, 4-ABP and N-OH-4-ABP; 0.04% DMSO: N-OH-PhIP; water: acrylamide and glycidamide) in triplicate plates. Positive controls (0.4 μM BaP −S9; 4.5 μM PhIP +S9) were included. Following the 72-h sampling period, cells were harvested, and pellets stored at −20°C until DNA was isolated using a standard phenol–chloroform extraction method. The phenyl-β-D galactosidase (P-gal) positive selection assay was performed as described previously (9,12). Briefly, the λgt10lacZ shuttle vector was recovered from the DNA and packaged into bacteriophage particles using Transpack reagent (Agilent Technologies, Santa Clara, California, USA). After incubating the phage particles with Escherichia coli (E. coli C ΔlacZ, galE-, recA-, Kanr, pAA119), they were plated onto non-selective titre plates and the remainder on mutant selective plates containing P-Gal. Following an incubation overnight at 37°C, plaques were manually scored and MF calculated as the ratio of the mutant plaque forming units (pfu) on the selective plates to the total number of pfu calculated from the non-selective titre plates.
Table 1.
Carcinogen concentrations used to treat FE1 cells for the FE1 cell mutagenicity assay based on cell viability (% of control) determined by crystal violet staining
| Cell viability (% of control) | Acrylamide (mM) | Glycidamide −S9 (mM) | PhIP (μM) | N-OH-PhIP −S9 (μM) | 4-ABP (μM) | N-OH-4-ABP −S9 (μM) | |||
|---|---|---|---|---|---|---|---|---|---|
| −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | ||||
| 100 | 2 | 0.5 | 1 | 25 | 0.625 | 1.25 | 100 | 100 | 1 |
| 80 | 3 | 1 | 1.5 | 50 | 1.25 | 2 | 200 | 300 | 1.5 |
| 60 | 4 | 2 | 2 | 100 | 2.5 | 2.5 | 300 | 400 | 3 |
| 40 | 8 | 3 | 2.5 | 200 | 2.75 | 4 | 500 | 500 | 3.5 |
| 20 | 10 | 4 | 3.5 | 250 | 5 | 5 | 550 | 520 | 4.5 |
Statistics
Results are shown as mean values ± standard deviation (SD). The sample size is indicated in each section. Statistical analysis for the FE1 cell mutagenicity assay was performed in SAS v.9.1 (SAS Institute, Cary, NC) using Poisson regression. A compound was categorised as genotoxic if the analyses revealed a significant treatment-related effect at P < 0.05, and at least one concentration elicited a significant increase in MF relative to the solvent control (P < 0.05). GraphPad Prism version 8.2.0 (GraphPad Software Inc., La Jolla, California, USA) was used for the remaining statistical analysis. Cell viability was expressed as the percentage of control (untreated) cells, and the area under the curve (AUC) and half-maximal inhibitory concentration (IC50) values were calculated. AUCs and IC50 values were compared by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. Relative mRNA expression data (2−ΔΔCt) were log2 transformed and analysed using a single-sample t-test with Bonferroni correction against the population control mean of 0. Significance levels are *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
Cell viability in FE1 cells
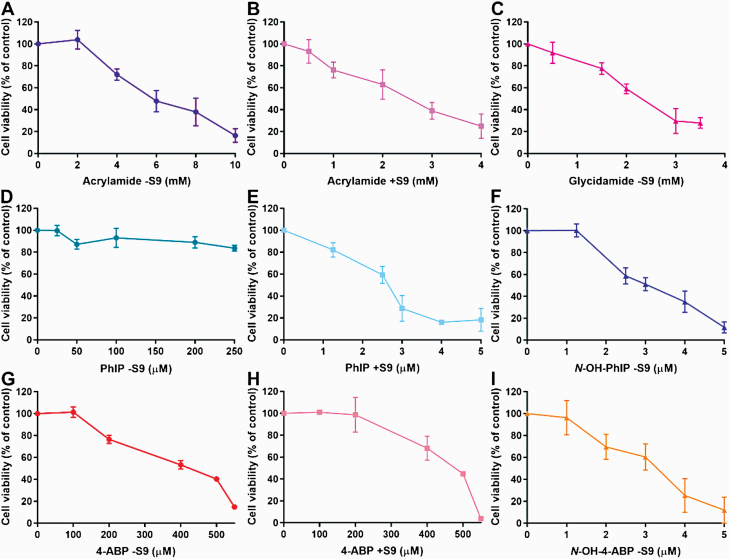
Cell viability after 6-h treatment with glycidamide (0–4 mM), acrylamide (0–10 mM), PhIP (0–250 µM), N-OH-PhIP (0–5 µM), 4-ABP (0–550 µM) or N-OH-4-ABP (0–5 µM), followed by a 72-h sampling period, was established by crystal violet staining. For acrylamide, PhIP and 4-ABP cell viability was assessed in the presence and absence of S9.
All treatments except PhIP −S9 decreased cell viability in a concentration-dependent manner. Glycidamide and acrylamide +S9 led to very similar (P > 0.05) levels of cell viability with virtually the same IC50 values of 2.4 mM (acrylamide +S9) and 2.3 mM (glycidamide −S9), whereas the calculated IC50 value for acrylamide −S9 of 6.1 mM was significantly higher (P < 0.001; Figure 1A–C). Similar effects were observed for PhIP +S9 and N-OH-PhIP −S9 (Figure 1D–F). For N-OH-PhIP −S9 or PhIP +S9, IC50 values were very similar (P > 0.05), calculated as 2.5 and 3.0 µM, respectively. In contrast, cell viability was unaffected at PhIP −S9 concentrations up to 250 µM and the calculated IC50 value of 5.6 mM was significantly different to the one of PhIP +S9 and N-OH-PhIP −S9 (P < 0.01). Interestingly, these effects were not seen for 4-ABP ±S9 and N-OH-4-ABP −S9 (Figure 1G–I). Levels of cell viability were highly similar (P > 0.05) for 4-ABP ±S9 with IC50 values of 391 µM (−S9) and 455 µM (+S9); N-OH-4-ABP −S9 was much more cytotoxic at 100-fold lower concentrations with an IC50 value of 2.9 µM (P < 0.001). A summary of all IC50 values can be found in Supplementary Table S1.
Fig. 1.
Cell viability assessment in FE1 cells. Cells were treated with various concentrations of acrylamide −S9 (A), acrylamide +S9 (B), glycidamide −S9 (C), PhIP −S9 (D), PhIP +S9 (E), N-OH-PhIP −S9 (F), 4-ABP −S9 (G), 4-ABP +S9 (H) or N-OH-4-ABP −S9 (I) for 6 h followed by a 72-h sampling time. Cell viability (% control) was assessed by staining with crystal violet. Cells treated with water (A–C) or DMSO (D–I) served as controls. Shown are mean values ± SD (n > 3).
AUCs for each treatment condition were compared (Supplementary Table S2). No significant difference was observed between glycidamide and acrylamide +S9 treatment. Results were the same when comparing N-OH-PHIP −S9 and PhIP +S9 treatment (P > 0.05). In contrast, AUCs for treatment with both acrylamide +S9 and glycidamide, as well as PhIP +S9 and N-OH-PhIP −S9, were significantly different from the acrylamide −S9 and PhIP −S9 treatments, respectively (P < 0.001). Also, treatment with N-OH-4-ABP −S9 was significantly different from the 4-ABP ±S9 treatment (P < 0.001), but only minor differences were found between 4-ABP with and without S9 treatments (P < 0.05).
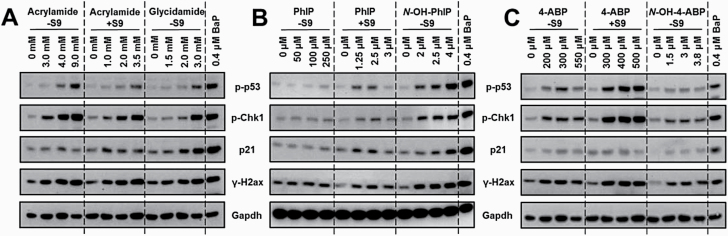
Induction of DDR proteins in FE1 cells
Based on the cell viability assessment, three concentrations of each treatment condition that induce approximately 80%, 60% and 30% cell viability, compared to untreated controls, were chosen for Western blot analyses as indicated in Figure 2. Induction of DDR proteins phospho-p53, phospho-Chk1, p21 and γ-H2ax was assessed after 6 h exposure.
Fig. 2.
DDR in FE1 cells. Western blot analysis of various DDR proteins (phospho-p53, p21, phospho-Chk1 and γ-H2ax) in FE1 cells exposed to indicated concentrations of acrylamide ±S9 or glycidamide −S9 (A), PhIP ±S9 or N-OH-PhIP −S9 (B) and 4-ABP ±S9 or N-OH-4-ABP −S9 (C) for 6 h. GAPDH was used as loading control, and cells treated with 0.4 µM BaP −S9 as positive control. Representative images of Western blot analysis are shown. Analysis was performed in duplicate from independent experiments.
For acrylamide ±S9 and glycidamide −S9 (Figure 2A), most of the examined DDR proteins were induced in a concentration-dependent manner, with the highest concentration tested leading to a similar expression of DDR proteins as had been observed in BaP-treated cells (i.e. positive control). Overall, p21 was induced after glycidamide treatment, but not after acrylamide treatment. After exposure to PhIP −S9, all examined DDR proteins were weakly expressed but not induced in response to PhIP exposure (Figure 2B). All DDR protein levels were lower than that observed for BaP-treated cells. With the addition of S9, all DDR proteins were induced with increasing PhIP +S9 concentrations but still lower than in BaP-treated cells. However, expression levels were the greatest at 2.5 µM, and then decreased again at the highest concentration tested (i.e. 3 µM), which could be due to increased cell death. In contrast, all examined DDR proteins were induced after N-OH-PhIP −S9 treatment in a concentration-dependent manner. The highest N-OH-PhIP −S9 concentration tested (4 µM) induced similar levels of DDR protein expression as seen with the positive control BaP. Most DDR proteins were induced after 4-ABP ±S9 and N-OH-4-ABP −S9 exposure (Figure 2C). However, strongest overall induction was found after 4-ABP +S9 treatment, with similar or even higher levels induced compared to treatment with BaP. The expression of phospho-p53 was very low after N-OH-4-ABP −S9 exposure, whereas, after 4-ABP +S9 treatment, it was substantially induced. For 4-ABP −S9, phospo-p53 first increased at lower concentrations but then decreased at the highest concentration tested (550 µM). phospho-Chk1 and γ-H2ax were both induced at all treatment conditions, with the strongest induction observed again after exposure to 4-ABP +S9, followed by 4-ABP −S9 and N-OH-4-ABP −S9. Interestingly, p21 expression levels were quite low with only minor induction being observed.
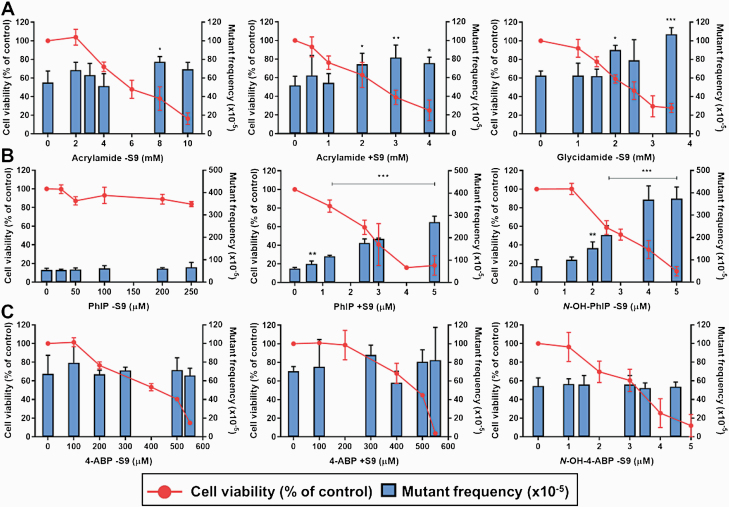
Determination of lacZ mutant frequency
Based on the crystal violet staining assay, four subcytotoxic and cytotoxic concentrations of each compound (i.e. parent compound ±S9 and proximate metabolite −S9) were chosen to evaluate lacZ mutagenicity in FE1 cells (Table 1). Cells treated with 0.4 µM BaP −S9 and 4.5 µM PhIP +S9 were included as positive controls; they both elicited strong positive responses (Supplementary Figure S4). In addition, it is noteworthy that all solvent-exposed controls were within the 5th and 95th percentile of the historical control data, i.e. 24–95 × 10–5 with a mean of ~44 × 10–5 (n = 653) (11).
Acrylamide ±S9 and glycidamide −S9 (P < 0.001) both elicited a significant treatment-related effect (P < 0.05 and P < 0.001, respectively), and at least one test concentration showed a statistically significant increase in MF as compared with the solvent control (Figure 3A). More precisely, acrylamide +S9 yielded a significant induction of MF at the top three concentrations tested with a 1.5-fold increase at 4 mM and glycidamide −S9 yielded a significant increase of MF at two tested concentrations that almost doubled at 3.5 mM in comparison to the negative control. Thus, both acrylamide +S9 and glycidamide −S9 were categorised as positive responses. Only one concentration of acrylamide −S9 (8 mM) yielded a significant 1.4-fold increase of MF compared to the negative control. However, as a lack of a dose-related increase in MF indicates that the response is marginal, acrylamide −S9 was categorised as equivocal. Treatment with PhIP −S9 failed to elicit a significant increase in MF over background (P > 0.05). In contrast, N-OH-PhIP -S9 and PhIP +S9 treatment both induced significant (P < 0.001) lacZ MF treatment effects (Figure 3B). PhIP +S9 yielded a significant increase in MF at all concentrations tested with a 4.3-fold increase at 5 µM. N-OH-PhIP −S9 significantly increased MF at the top four concentrations tested, with a maximum increase of 5.3-fold at 5 μM. N-OH-4-ABP −S9 and 4-ABP ±S9 failed to elicit significant increases in lacZ MF at any treatment condition (P > 0.05; Figure 3C).
Fig. 3.
Induction of lacZ mutant frequency in FE1 cells. Cells were treated as indicated with acrylamide ±S9 or glycidamide −S9 (A), PhIP ±S9 or N-OH-PhIP −S9 (B) and 4-ABP ±S9 or N-OH-4-ABP −S9 (C) for 6 h followed by a 72-h sampling period. Control cells were treated with water (A) or DMSO (B and C) only. The lacZ mutant frequencies were calculated as the number of lacZ mutants per total number of recovered lacZ copies. Statistical analysis was performed in SAS v.9.1 using Poisson regression values (*P < 0.05; **P < 0.01; ***P < 0.001 compared with control). Values shown are the means of three replicate experiments + SD.
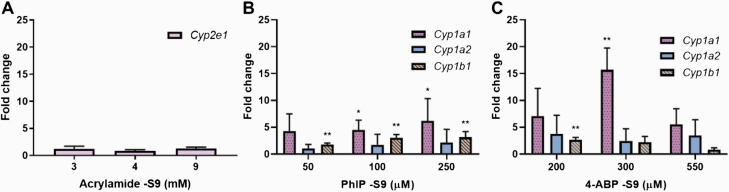
Gene expression in FE1 cells
RT-qPCR was employed to assess mRNA expression of the main metabolising enzymes known to bioactivate the parent compounds investigated; levels were examined after 6 h treatment with cytotoxic and subcytotoxic concentrations of acrylamide −S9, PhIP −S9 or 4-ABP −S9 as indicated in Figure 4. Acrylamide is metabolised by CYP2E1 to glycidamide and, although low levels of Cyp2e1 were detected in FE1 cells, no induction of Cyp2e1 was found at any acrylamide concentration tested (Figure 4A). PhIP and 4-ABP are both metabolised by CYP1A1, CYP1A2 and CYP1B1. As shown in Figure 4B, Cyp1a1 and Cyp1b1 gene expression increased significantly after PhIP treatment. Cyp1a2 expression was not affected in a statistically significant manner by PhIP treatment. The highest PhIP concentration (250 µM) elicited a six-fold induction of Cyp1a1 expression, and four-fold induction of Cyp1b1 expression; Cyp1a2 mRNA expression only doubled. Cyp1a1 was expressed and significantly induced (~15-fold) after treatment with 300-µM 4-ABP (Figure 4C). This corresponds to the Western blot results, whereby the expression of DDR proteins was greatest at this concentration rather than at the highest tested concentration (550 µM). However, the main 4-ABP metabolising enzyme Cyp1a2 was not significantly induced. In addition, a weak but significant induction of Cyp1b1 was observed after treatment with 200 µM 4-ABP but not at other tested concentrations. In general, Cyp1b1 gene expression changes were minimal.
Fig. 4.
Relative gene expression of relevant Cyp enzymes in response to carcinogen treatment. FE1 cells were treated with indicated concentrations of acrylamide −S9 (A), PhIP −S9 (B) and 4-ABP −S9 (C) for 6 h. Controls were exposed to water (acrylamide) or 0.1% DMSO (PhIP and 4-ABP) only. Cyp1a1, 1a2, 1b1 and 2e1 expression was determined by RT-qPCR and the 2−ΔΔCt method. Values are normalised to mRNA expression of the housekeeping gene GAPDH and are relative to the water (A) or DMSO (B and C) control (n > 3). Statistical analysis was performed by log2 transforming the data and analysis using a single-sample t-test with Bonferroni correction against the population control mean of 0 (*P < 0.05, **P < 0.01).
Discussion
To date, more than 30 compounds, including 25 reference compounds (e.g. BaP), nanomaterials (e.g. nanoparticulate quartz) and complex mixtures (e.g. diesel exhaust) have been studied in the FE1 cell mutagenicity assay (1). The reference compounds included 14 in vivo mutagens (‘true positives’) that should be detected as positive in in vitro assays, 9 in vivo non-mutagens (‘true negatives’) that should fail to elicit mutagenicity in vitro and 9 compounds that are often detected as positive in in vitro assays, although they are non-genotoxic in vivo (‘false positives’). For the reference compounds, an overall concordance of 88% was achieved with 10/14 true positives (e.g. BaP, PhIP and 2-AAF) (1,11), 9/9 false positives (e.g. p-Nitrophenol, curcumin and eugenol) (12) and 2/2 true negatives (ampicillin trihydrate and D-mannitol) leading to the expected genotoxicity readout (11). The four true positives that did not show the expected positive response were 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 1-methylpyrene, dimethylnitrosamine and IQx (11,13). Although the negative response for 1-methylpyrene, dimethylnitrosamine and NNK are most likely due to a lack of Phase I enzymes (i.e. Cyp2e1 and Cyp2a6), it remains unclear why IQx did not elicit a positive response (1). Here, we studied an additional three known or suspected human carcinogens (i.e. acrylamide, PhIP and 4-ABP), and their proximate metabolites (i.e. glycidamide, N-OH-PhIP, and N-OH-4-ABP), to better understand misleading results previously observed in the FE1 cell mutagenicity assay. A positive genotoxic response was observed for acrylamide +S9, glycidamide −S9, N-OH-PhIP −S9 and PhIP +S9, while PhIP −S9, N-OH-4-ABP −S9 and 4-ABP ±S9 did not elicit significant positive responses. Acrylamide −S9 only elicited an MF value significantly above control at a single concentration, so it was categorised as an equivocal response. When comparing the mutagenic potency at concentrations that led to similar levels of cytotoxicity (i.e. 20% cell viability), acrylamide +S9 was the least mutagenic compound yielding a 1.5-fold increase after treatment with 4 mM (+S9). This was followed by glycidamide −S9 at 3.5 mM, PhIP +S9 at 5 µM and N-OH-PhIP −S9 at 5 µM resulting in a 1.7-, 4.5- and 5.3-fold increase of MF, respectively. To put this into context, treatment with 0.4 µM BaP −S9 yields a ~16-fold increase of MF level at a similar levels of cytotoxicity (12). This indicates that each compound tested in the present study, albeit mutagenic in FE1 cells, is far less mutagenic than BaP. Nevertheless, numerous responses were statistically elevated relative to concurrent controls. The results of this study in relation to OECD-validated assays are discussed below.
Glycidamide −S9 and acrylamide +S9 elicited positive responses, with the treatment-related effect generally being much more pronounced for glycidamide −S9 (P < 0.001) than acrylamide +S9 (P < 0.05). Due to a lack of concentration-related increase of MF, acrylamide −S9 gave equivocal mutagenic responses in the FE1 cell mutagenicity assay. Previous studies have also reported both positive and negative results for acrylamide −S9 (25–27). Both the significant positive response for glycidamide −S9 and the positive response for acrylamide +S9 are in agreement with results obtained for these compounds in other OECD-validated in vitro assays (i.e. Tk and Hprt; Table 2) (25–27). In the Tk in vitro mammalian cell mutagenicity test, acrylamide −S9 (mouse lymphoma cells) and acrylamide ±S9 (human Tk6 cells) elicited a positive response at concentrations albeit only at concentrations over 12 mM (25,26); in the Hprt in vitro mutagenicity assay, acrylamide −S9 yielded a negative result (27). As acrylamide is metabolised by CYP2E1 to glycidamide, these results are most likely due to a lack of bioactivation in the test system. Indeed, gene expression analysis in FE1 cells did not reveal a significant change in Cyp2e1 mRNA expression after acrylamide treatment. This was expected as FE1 cells are isolated from mouse lung and a study of Cyp expression profiling in various mouse tissues has noted only minimum levels of Cyp2e1 in the lung (31). In addition, Aroclor-1254 is a poor inducer of Cyp2e1 and has even been shown to suppress Cyp2e1 expression (32). Consequently, it is not surprising that results for acrylamide are negative in many screening tests for genotoxicity even when an exogenous metabolic activation system is added. Furthermore, previous studies have found a lack of FE1 cell mutagenicity for other procarcinogens that require Cyp2e1 bioactivation, such as 1-methylpyrene and N-nitrosodimethylamine (1). The modest fold change in MF for acrylamide +S9 in the present study suggest that the observed genotoxicity may be the result of Cyp2e1-independent mechanisms previously observed for acrylamide (e.g. induction of reactive oxygen species and oxidative damage to DNA or Michael-type nucleophilic addition reactions) (33–35). However, further tests beyond the scope of this study are required to make a clear statement.
Table 2.
Comparisons of the test-compound mutagenicity observed herein with previously published results for other in vitro mammalian cell mutagenicity assays
| Compound | lacZ | Tk | Hprt | ||
|---|---|---|---|---|---|
| MutaMouse FE1 cells | Mouse lymphoma L5178Y cells | Human TK6 cells | Chinese hamster V79 cells | Chinese hamster ovary cells | |
| Acrylamide −S9 | Equivocal | Positivea (25) | Positivea (26) | Negative (27) | No data |
| Acrylamide +S9 | Positive | No data | Positivea (26) | No data | No data |
| Glycidamide −S9 | Positive | Positive (25) | Positive (26) | Positive (27) | No data |
| PhIP −S9 | Negative | No data | No data | Negative (36) | No data |
| PhIP +S9 | Positive | No data | Positive (37) | Positiveb (36) | No data |
| N-OH-PhIP −S9 | Positive | No data | No data | No data | No data |
| 4-ABP −S9 | Negative | No data | No data | No data | No data |
| 4-ABP +S9 | Negative | Equivocalc (38) | No data | No data | Positive (39) |
| N-OH-4-ABP −S9 | Negative | No data | No data | No data | No data |
aOnly weakly genotoxic at concentrations above 10 mM.
bCells expressed human CYP1A2.
cOnly positive at one criteria; large variability.
A strong positive response in the FE1 cell mutagenicity assay was observed after N-OH-PhIP -S9 and PhIP +S9 treatment; PhIP −S9 did not elicit a significant response increase at any of the concentrations tested. This is in agreement with previous results in FE1 cells (9). N-OH-PhIP has not been assessed in OECD-validated assays, but PhIP ±S9 is mutagenic in the Tk and Hprt mutation assay (Table 2) (36,37). However, PhIP −S9 only resulted in positive results in V79 cells that expressed human CYP1A2 (36). Thus, it is not possible to directly compare the three assays (i.e. Tk, Hprt and lacZ FE1) for these compounds. Previously, it has been hypothesised that PhIP −S9 most likely does not elicit a positive response in the FE1 cell mutagenicity assay because the cells do not possess SULT activity, which is necessary to convert N-OH-PhIP to its ultimate mutagenic carcinogen (e.g. N-sulfoxy-PhIP) (1). However, the present study examined N-OH-PhIP; since the results revealed a positive response in the absence of S9, the hypothesis regarding lack of Phase II capacity is not supported. It is important to note that, although SULTs have been shown to enhance bioactivation of N-OH-PhIP leading to DNA adduct formation, non-enzymatic covalent binding of N-OH-PhIP has also been demonstrated in vitro (40). In the present study, it seems that rather than a lack of Phase II enzymes, FE1 cells lack Phase I enzymes necessary to sufficiently metabolise PhIP to N-OH-PhIP. It has been shown previously that, although Cyp1a2 is important in PhIP metabolism in humans (N2-hydroxylation), in rats and mice, this Cyp enzyme most likely results in the detoxication of PhIP (4’-hydroxylation) (41,42). In contrast, Cyp1a1 and 1b1 have been shown to be extremely important for PhIP metabolism and activation in extrahepatic tissues of mice. For instance, levels of PhIP-DNA adducts in lungs of Cyp1a2−/− mice were comparable to those observed in wild-type mice, whereas levels in Cyp1a1−/− mice were approximately 50% lower, highlighting the importance of Cyp1a1 for PhIP activation in murine lung (43). Furthermore, Cyp1a1 is expressed more predominantly in mouse lung than in other tissues (31). The Cyp1 gene expression data agree with the suggestion that PhIP is not efficiently metabolised to N-OH-PhIP in FE1 cells. Cyp1a2 was not significantly induced after PhIP treatment, whereas both Cyp1a1 and 1b1 were induced at low levels with the highest induction seen for Cyp1a1 (six-fold). The conversion of PhIP to N-OH-PhIP may have been insufficient to elicit lacZ mutants or a DDR response. Future work could investigate the level of N-OH-PhIP following exposure of FE1 cells to the parent compound. Overall, there seems to be a species- and compound-specific catalytic activity of CYP enzymes that can explain the negative result for PhIP-S9 in the FE1 cell mutagenicity assay. More heterocyclic aromatic amines [e.g. 2-amino-3-methylimidazo[4,5-f]quinoline (IQ)] need to be tested as enzyme kinetic parameters for N-oxidation catalysed by CYPs are likely to be different for different heterocyclic aromatic amines. In another study, using primary hepatocytes of the MutaMouse PhIP elicited a positive response without the addition of S9 (44) because liver cells express all XMEs required for PhIP metabolism.
Although very low concentrations of N-OH-4-ABP −S9 were cytotoxic to FE1 cells, a negative response was obtained in the FE1 cell mutagenicity assay. As shown previously in a variety of other in vitro models, cytotoxicity is not necessarily indicative of genotoxicity (45–47). Similarly, 4-ABP ±S9 treatment failed to elicit a significant increase in lacZ MF. Others have shown that 4-ABP ±S9 and N-OH-4-ABP −S9 are mutagenic in mammalian cells, including human uroepithelial and mouse lymphoma cell lines (38,48). With respect to the OECD-validated Tk and Hprt mutagenicity assays, 4-ABP +S9 elicited significant positive responses indicating that the response profile of these assays is distinct from that observed to date for the FE1 cell mutagenicity assay (Table 2) (38,39). N-OH-4-ABP has not been tested in the Tk and Hprt mutagenicity assays; therefore, a cross-assay comparison is not possible. The observed negative response for 4-ABP −S9 could be explained by low expression of CYP enzymes responsible for the bioactivation of 4-ABP. The negative response obtained for 4-ABP +S9 could be due to the fact that Cyp2e1 is important for the bioactivation for 4-ABP and is inhibited by DMSO, which was used as solvent for 4-ABP (21,49). It is surprising, however, that N-OH-4-ABP failed to elicit a positive response in the FE1 cell mutagenicity assay. This may be explainable by the rapid repair of 4-ABP-DNA adducts as shown previously in urinary bladder transitional cell carcinoma cell lines (50,51). Thus, it is possible that 4-ABP-DNA adducts were formed in FE1 cells but then efficiently repaired prior to mutation induction. Studies investigating 4-ABP-DNA adduct formation in FE1 cells would be required to test this hypothesis; however, this is beyond the scope of the present study.
In conclusion, as expected, we observed that acrylamide +S9, glycidamide −S9, PhIP +S9 and N-OH-PhIP −S9 elicited significant increases in lacZ MF, whereas the negative responses for 4-ABP ±S9 and N-OH-4-ABP −S9 were unexpected. A perplexing problem with the FE1 mutagenicity assay remains the observed inconsistent results for heterocyclic aromatic amines. This issue should be addressed by testing additional heterocyclic aromatic amines (e.g. IQ) by expanding the compound specificity assessment of the assay and via further quantification of the induced expression of XME genes. Acrylamide +S9 did elicit a positive mutagenic response, but results indicate that alternative mechanisms, such as induction of reactive oxygen species and oxidative damage to DNA, appear to be more important than the formation of glycidamide-DNA adducts. The lack of Cyp2e1 expression in FE1 cells remains a limitation of the FE1 cell mutagenicity assay; this might be solved by creating human CYP2E1 knock-in cells, achieved with other mammalian cell systems (e.g. human CYP2E1-expressing V79 cells) (36,52). This type of enhancement of metabolic capacity would likely improve the sensitivity of the FE1 cell mutagenicity assay, particularly for the assessment of compound metabolism/activation by CYP2E1. Such enhancements would likely reduce the frequency of false-negative responses (e.g. for NNK, 1-methylpyrene). Despite the limitations of the FE1 cell mutagenicity assay noted here and elsewhere (1), and the unexpected results that need to be further clarified, the FE1 cell mutagenicity assay shows great potential for routine chemical safety assessment.
Funding
Lisa Hölzl-Armstrong was supported by a PhD studentship from the MRC Centre for Environment and Health and a UKEMS Technology Transfer and Training Bursary from the United Kingdom Environmental Mutagen Society (UKEMS). This work was supported by Cancer Grand Challenges Mutographs team award funded by Cancer Research UK [C98/A24032]. Work at Health Canada was partially supported by the Government of Canada’s CMP (Chemicals Management Plan) research fund.
Conflict of interest statement: None declared.
Supplementary Material
References
- 1. White P. A., Luijten M., Mishima M., Cox J. A., Hanna J. N., Maertens R. M., and Zwart E. P (2019) In vitro mammalian cell mutation assays based on transgenic reporters: a report of the International Workshop on Genotoxicity Testing (IWGT). Mutat. Res. Genet. Toxicol. Environ. Mutagen., 847, 403039. [DOI] [PubMed] [Google Scholar]
- 2. Corvi R. and Madia F (2017) In vitro genotoxicity testing—can the performance be enhanced? Food Chem. Toxicol., 106, 600–608. [DOI] [PubMed] [Google Scholar]
- 3. OECD (2016) Test guideline: 473. In Vitro Mammalian Chromosomal Aberration Test. OECD Guidelines for Testing of Chemicals. Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 4. OECD (2016) Test fuideline 487. In Vitro Mammalian Cell Micronucleus Test. OECD Guidelines for Testing of Chemicals. Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 5. OECD (2020) Test guideline 471. In Bacterial Reverse Mutation Test. OECD Guidelines for Testing of Chemicals. Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 6. OECD (2015) Test guideline 490. In Vitro Mammalian Cell Gene Mutation Tests Using the Thymidine Kinase Gene OECD Guidelines for Testing of Chemicals. Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 7. OECD (2016) Test guideline 476. In Vitro Mammalian Cell Gene Mutation Tests Using the Hprt and xprt Genes. OECD Guidelines for Testing of Chemicals. Organization for Economic Co-operation and Development, Paris. [Google Scholar]
- 8. Kirkland D., Pfuhler S., Tweats D., et al. (2007) How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: report of an ECVAM Workshop. Mutat. Res., 628, 31–55. [DOI] [PubMed] [Google Scholar]
- 9. White P. A., Douglas G. R., Gingerich J., et al. (2003) Development and characterization of a stable epithelial cell line from Muta Mouse lung. Environ. Mol. Mutagen., 42, 166–184. [DOI] [PubMed] [Google Scholar]
- 10. Lambert I. B., Singer T. M., Boucher S. E. and Douglas G. R (2005) Detailed review of transgenic rodent mutation assays. Mutat. Res., 590, 1–280. [DOI] [PubMed] [Google Scholar]
- 11. Hanna J. (2018) Validation of an In Vitro Mutagenicity Assay Based on Pulmonary Epithelial Cells from the Transgenic MutaMouse: Intra-Laboratory Variability and Metabolic Competence. University of Ottawa, Ottawa: https://ruor.uottawa.ca/handle/10393/37312. [Google Scholar]
- 12. Maertens R. M., Long A. S. and White P. A (2017) Performance of the in vitro transgene mutation assay in MutaMouse FE1 cells: Evaluation of nine misleading (“False”) positive chemicals. Environ. Mol. Mutagen., 58, 582–591. [DOI] [PubMed] [Google Scholar]
- 13. Villeneuve N. (2018) Validation of an In Vitro Mutagenicity Assay Based on Immortalized Pulmonary Flat Epithelium Isolate 1 (FE1) Cells From the Transgenic MutaMouse. University of Ottawa, Ottawa. [Google Scholar]
- 14. Arlt V. M., Gingerich J., Schmeiser H. H., Phillips D. H., Douglas G. R. and White P. A (2008) Genotoxicity of 3-nitrobenzanthrone and 3-aminobenzanthrone in MutaMouse and lung epithelial cells derived from MutaMouse. Mutagenesis, 23, 483–490. [DOI] [PubMed] [Google Scholar]
- 15. Kirkland D., Kasper P., Martus H. J., Müller L., van Benthem J., Madia F. and Corvi R (2016) Updated recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 795, 7–30. [DOI] [PubMed] [Google Scholar]
- 16. Berndt-Weis M. L., Kauri L. M., Williams A., White P., Douglas G. and Yauk C (2009) Global transcriptional characterization of a mouse pulmonary epithelial cell line for use in genetic toxicology. Toxicol. In Vitro, 23, 816–833. [DOI] [PubMed] [Google Scholar]
- 17. Clive D., Johnson K. O., Spector J. F., Batson A. G. and Brown M. M (1979) Validation and characterization of the L5178Y/TK+/- mouse lymphoma mutagen assay system. Mutat. Res., 59, 61–108. [DOI] [PubMed] [Google Scholar]
- 18. Turesky R. J. and Le Marchand L (2011) Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem. Res. Toxicol., 24, 1169–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. IARC (1993) Some Naturally Occuring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. Vol. 56 IARC, Lyon. [Google Scholar]
- 20. IARC (2010) Some Aromatic Amines, Organic Dyes, and Related Exposures. Vol. 99 IARC Press, Lyon. [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S., Bott D., Tung A., Sugamori K. S. and Grant D. M (2015) Relative contributions of CYP1A2 and CYP2E1 to the bioactivation and clearance of 4-aminobiphenyl in adult mice. Drug Metab. Dispos., 43, 916–921. [DOI] [PubMed] [Google Scholar]
- 22. IARC (1994) Some Industrial Chemicals. Vol. 60 IARC Press, Lyon. [Google Scholar]
- 23. Tareke E., Rydberg P., Karlsson P., Eriksson S. and Törnqvist M (2002) Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J. Agric. Food Chem., 50, 4998–5006. [DOI] [PubMed] [Google Scholar]
- 24. Kraus D., Rokitta D., Fuhr U. and Tomalik-Scharte D (2013) The role of human cytochrome P450 enzymes in metabolism of acrylamide in vitro. Toxicol. Mech. Methods, 23, 346–351. [DOI] [PubMed] [Google Scholar]
- 25. Mei N., Hu J., Churchwell M. I., Guo L., Moore M. M., Doerge D. R. and Chen T (2008) Genotoxic effects of acrylamide and glycidamide in mouse lymphoma cells. Food Chem. Toxicol., 46, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koyama N., Yasui M., Oda Y., et al. (2011) Genotoxicity of acrylamide in vitro: Acrylamide is not metabolically activated in standard in vitro systems. Environ. Mol. Mutagen., 52, 11–19. [DOI] [PubMed] [Google Scholar]
- 27. Baum M., Fauth E., Fritzen S., Herrmann A., Mertes P., Merz K., Rudolphi M., Zankl H. and Eisenbrand G (2005) Acrylamide and glycidamide: genotoxic effects in V79-cells and human blood. Mutat. Res., 580, 61–69. [DOI] [PubMed] [Google Scholar]
- 28. Manjanatha M. G., Aidoo A., Shelton S. D., Bishop M. E., McDaniel L. P., Lyn-Cook L. E. and Doerge D. R (2006) Genotoxicity of acrylamide and its metabolite glycidamide administered in drinking water to male and female Big Blue mice. Environ. Mol. Mutagen., 47, 6–17. [DOI] [PubMed] [Google Scholar]
- 29. Manjanatha M. G., Guo L. W., Shelton S. D. and Doerge D. R (2015) Acrylamide-induced carcinogenicity in mouse lung involves mutagenicity: cII gene mutations in the lung of big blue mice exposed to acrylamide and glycidamide for up to 4 weeks. Environ. Mol. Mutagen., 56, 446–456. [DOI] [PubMed] [Google Scholar]
- 30. Hölzl-Armstrong L., Kucab J. E., Moody S., et al. (2020) Mutagenicity of acrylamide and glycidamide in human TP53 knock-in (Hupki) mouse embryo fibroblasts. Arch. Toxicol., 94, 4173–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Renaud H. J., Cui J. Y., Khan M. and Klaassen C. D (2011) Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol. Sci., 124, 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Escobar-Garcia D., Camacho-Carranza R., Pérez I., Dorado V., Arriaga-Alba M. and Espinosa-Aguirre J. J (2001) S9 induction by the combined treatment with cyclohexanol and albendazole. Mutagenesis, 16, 523–528. [DOI] [PubMed] [Google Scholar]
- 33. Blasiak J., Gloc E., Wozniak K. and Czechowska A (2004) Genotoxicity of acrylamide in human lymphocytes. Chem. Biol. Interact., 149, 137–149. [DOI] [PubMed] [Google Scholar]
- 34. Jiang L., Cao J., An Y., Geng C., Qu S., Jiang L. and Zhong L (2007) Genotoxicity of acrylamide in human hepatoma G2 (HepG2) cells. Toxicol. In Vitro, 21, 1486–1492. [DOI] [PubMed] [Google Scholar]
- 35. Friedman M. (2003) Chemistry, biochemistry, and safety of acrylamide. A review. J. Agric. Food Chem., 51, 4504–4526. [DOI] [PubMed] [Google Scholar]
- 36. Yadollahi-Farsani M., Gooderham N. J., Davies D. S. and Boobis A. R (1996) Mutational spectra of the dietary carcinogen 2-amino-1-methyl-6- phenylimidazo[4,5-b]pyridine(PhIP) at the Chinese hamsters hprt locus. Carcinogenesis, 17, 617–624. [DOI] [PubMed] [Google Scholar]
- 37. Morgenthaler P. M. and Holzhäuser D (1995) Analysis of mutations induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in human lymphoblastoid cells. Carcinogenesis, 16, 713–718. [DOI] [PubMed] [Google Scholar]
- 38. Guo X., Heflich R. H., Dial S. L., Richter P. A., Moore M. M. and Mei N (2016) Quantitative analysis of the relative mutagenicity of five chemical constituents of tobacco smoke in the mouse lymphoma assay. Mutagenesis, 31, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oberly T. J., Rexroat M. A., Bewsey B. J., Richardson K. K. and Michaelis K. C (1990) An evaluation of the CHO/HGPRT mutation assay involving suspension cultures and soft agar cloning: results for 33 chemicals. Environ. Mol. Mutagen., 16, 260–271. [DOI] [PubMed] [Google Scholar]
- 40. Turesky R. J., Lang N. P., Butler M. A., Teitel C. H. and Kadlubar F. F (1991) Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis, 12, 1839–1845. [DOI] [PubMed] [Google Scholar]
- 41. Cheung C., Ma X., Krausz K. W., Kimura S., Feigenbaum L., Dalton T. P., Nebert D. W., Idle J. R. and Gonzalez F. J (2005) Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem. Res. Toxicol., 18, 1471–1478. [DOI] [PubMed] [Google Scholar]
- 42. Langouët S., Paehler A., Welti D. H., Kerriguy N., Guillouzo A. and Turesky R. J (2002) Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rat and human hepatocytes. Carcinogenesis, 23, 115–122. [DOI] [PubMed] [Google Scholar]
- 43. Ma X., Idle J. R., Malfatti M. A., Krausz K. W., Nebert D. W., Chen C. S., Felton J. S., Waxman D. J. and Gonzalez F. J (2007) Mouse lung CYP1A1 catalyzes the metabolic activation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis, 28, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cox J. A., Zwart E. P., Luijten M. and White P. A (2019) The development and prevalidation of an in vitro mutagenicity assay based on MutaMouse primary hepatocytes, Part II: Assay performance for the identification of mutagenic chemicals. Environ. Mol. Mutagen., 60, 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kucab J. E., Phillips D. H. and Arlt V. M (2012) Metabolic activation of diesel exhaust carcinogens in primary and immortalized human TP53 knock-in (Hupki) mouse embryo fibroblasts. Environ. Mol. Mutagen., 53, 207–217. [DOI] [PubMed] [Google Scholar]
- 46. Kucab J. E., Zou X., Morganella S., et al. (2019) A compendium of mutational signatures of environmental agents. Cell, 177, 821–836.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piberger A. L., Krüger C. T., Strauch B. M., Schneider B. and Hartwig A (2018) BPDE-induced genotoxicity: relationship between DNA adducts, mutagenicity in the in vitro PIG-A assay, and the transcriptional response to DNA damage in TK6 cells. Arch. Toxicol., 92, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bookland E. A., Reznikoff C. A., Lindstrom M. and Swaminathan S (1992) Induction of thioguanine-resistant mutations in human uroepithelial cells by 4-aminobiphenyl and its N-hydroxy derivatives. Cancer Res., 52, 1615–1621. [PubMed] [Google Scholar]
- 49. Hickman D., Wang J. P., Wang Y. and Unadkat J. D (1998) Evaluation of the selectivity of In vitro probes and suitability of organic solvents for the measurement of human cytochrome P450 monooxygenase activities. Drug Metab. Dispos., 26, 207–215. [PubMed] [Google Scholar]
- 50. Torino J. L., Burger M. S., Reznikoff C. A. and Swaminathan S (2001) Role of TP53 in repair of N-(deoxyguanosin-8-yl)-4-aminobiphenyl adducts in human transitional cell carcinoma of the urinary bladder. Carcinogenesis, 22, 147–154. [DOI] [PubMed] [Google Scholar]
- 51. Swaminathan S., Torino J. L. and Burger M. S (2002) Human urinary bladder epithelial cells lacking wild-type p53 function are deficient in the repair of 4-aminobiphenyl-DNA adducts in genomic DNA. Mutat. Res., 499, 103–117. [DOI] [PubMed] [Google Scholar]
- 52. Jin G., Cai L., Hu K., Luo Y., Chen Y., Glatt H. and Liu Y (2019) Mutagenic activity of N-nitrosodiethylamine in cell lines expressing human CYP2E1-adequacy of dimethylsulfoxide as solvent. Environ. Mol. Mutagen., 60, 214–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.