Abstract
Dinoflagellates possess many cellular characteristics with unresolved evolutionary histories. These include nuclei with greatly expanded genomes and chromatin packaged using histone-like proteins and dinoflagellate-viral nucleoproteins instead of histones, highly reduced mitochondrial genomes with extensive RNA editing, a mix of photosynthetic and cryptic secondary plastids, and tertiary plastids. Resolving the evolutionary origin of these traits requires understanding their ancestral states and early intermediates. Several early-branching dinoflagellate lineages are good candidates for such reconstruction, however these cells tend to be delicate and environmentally sparse, complicating such analyses. Here, we employ transcriptome sequencing from manually isolated and microscopically documented cells to resolve the placement of two cells of one such genus, Abedinium, collected by remotely operated vehicle in deep waters off the coast of Monterey Bay, CA. One cell corresponds to the only described species, Abedinium dasypus, whereas the second cell is distinct and formally described as Abedinium folium, sp. nov. Abedinium has classically been assigned to the early-branching dinoflagellate subgroup Noctilucales, which is weakly supported by phylogenetic analyses of small subunit ribosomal RNA, the single characterized gene from any member of the order. However, an analysis based on 221 proteins from the transcriptome places Abedinium as a distinct lineage, separate from and basal to Noctilucales and the rest of the core dinoflagellates. The transcriptome also contains evidence of a cryptic plastid functioning in the biosynthesis of isoprenoids, iron–sulfur clusters, and heme, a mitochondrial genome with all three expected protein-coding genes (cob, cox1, and cox3), and the presence of some but not all dinoflagellate-specific chromatin packaging proteins.
Keywords: Dinoflagellate evolution, noctilucoid, cryptic plastid, single-cell transcriptomics
Introduction
Dinoflagellates are some of the most abundant and diverse eukaryotes in the ocean, assuming a wide range of morphologies and ecological roles (Taylor et al. 2008; Keeling and del Campo 2017). Of the free-living species, most consume prey, whereas many photosynthesize. Many of the latter are mixotrophs, exhibiting facultative phagotrophy (Stoecker 1999; Jeong et al. 2005; Seong et al. 2006). Symbiotic lineages form associations ranging from mutualism, as in many members of the family Symbiodiniaceae (Trench 1993; LaJeunesse et al. 2018), to parasitism of other eukaryotes (Coats 1999). Dinoflagellates are of great ecological and economic importance in coastal ecosystems, facilitating primary production and nutrient cycling that support the greater food web (Jeong et al. 2010). Many species form dense seasonal blooms that often dominate plankton assemblages, and some species produce toxic metabolites that bioaccumulate in filter feeders, threatening shellfisheries and human health (Shumway 1990).
Despite this diversity, the members of the “core” dinoflagellates (or Dinokaryota) share many complex, distinctive traits. These include highly reduced mitochondrial genomes with extensive mRNA editing (Zhang and Lin 2008), nonpolycistronic tandem array mRNA expression and transsplicing (Zhang et al. 2007; Beauchemin et al. 2012), the nonnucleosomal packaging of chromatin using histone-like proteins (HLPs) and dinoflagellate-viral nucleoproteins (DVNPs) (Lin 2011; Gornik et al. 2012), and extremely large genomes, which can be as large as 245 Gb in some taxa (Veldhuis et al. 1997). Dinoflagellates also seem to be particularly amenable to plastid acquisition, giving rise to the only clearly documented cases of tertiary and serial secondary endosymbiosis (Keeling 2013).
Because they are so unusual, reconstructing the origins and evolutionary interrelationships between these traits has been a subject of some interest (Lukeš et al. 2009; Wisecaver and Hackett 2011; Janouškovec et al. 2017). This task has been significantly informed by investigations into early diverging dinoflagellate lineages (i.e., relatives closely related to but not within the core dinoflagellates), the phylogenetic position and biology of which help to infer ancestral states. However, although many core dinoflagellates are readily found in nature and cultured for detailed study, many of these early diverging species are more elusive. None as of yet is known to be photosynthetic, many of the most common are probably parasites and mostly only known from environmental surveys of small subunit (SSU, 18S) rRNA (Chambouvet et al. 2008; Guillou et al. 2008), and many others are widespread but numerically rare heterotrophs (Le Bescot et al. 2016). Noctilucales or “noctilucoid” dinoflagellates are one such lineage; with the exception of the bloom-forming Noctiluca scintillans, these large, heterotrophic grazers tend to be environmentally sparse and physically delicate, making them challenging subjects for culture and in-depth molecular analysis. The only molecular phylogeny of noctilucoids to date (Gómez et al. 2010), established that Spatulodinium, Kofoidinium, and Abedinium (=Leptophyllus) are members of the Noctilucales, but none of these genera is well studied. Indeed, Abedinium dasypus (Cachon & Cachon-Enjumet) Loeblich & Loeblich III, 1966 is arguably the least-studied of the noctilucoids. This monotypic genus has been described only a few times in the literature, in reports from the Mediterranean Sea, and Indian and Atlantic Oceans, indicating a cosmopolitan distribution (Cachon and Cachon-Enjumet 1964; Margalef 1973; Gómez et al. 2010; Saburova et al. 2013). By all accounts, A. dasypus cells are rare, fragile, and prone to disintegration, all of which may also contribute to their underrepresentation in environmental samples.
Here, we apply a novel approach using single-cell transcriptome sequencing to analyze cells identified as Abedinium including a new species we describe as Abedinium folium sp. nov., collected near Monterey Bay, CA. A phylogenomic analysis resolves the placement of this rare genus, revealing that Abedinium is not a noctilucoid, but instead represents a new lineage sister to the clade consisting of N. scintillans and the core dinoflagellates. The transcriptome data also show Abedinium retains a functioning reduced plastid, and reveals its full mitochondrial genome as well as the presence of some, but not all, chromatin-binding proteins found in core dinoflagellates.
Materials and Methods
Sample Collection and Imaging
Sample collection focused on rare or unidentifiable taxa and was performed on two separate cruises as part of a survey of the diversity of marine heterotrophic flagellates in the mesopelagic to bathypelagic ocean. In all, 177 cells were collected from 23 samples (250–500 ml/sample). Cells were chosen for sequencing if they possessed features distinguishing them from common and easily identifiable taxa. Water samples were not assessed for general community composition and the focus was on rapidly surveying the contents of samples to document and isolate cells before they began to deteriorate or lyse. The cell “DICHO17-03” (hereafter referred to as A. folium) was sampled during a Monterey Bay Aquarium Research Institute (MBARI) cruise on September 7, 2017. The cell was isolated from water collected at 600 m using a Niskin Bottle mounted on the remotely operated vehicle Ventana deployed from the research vessel Rachel Carson at mid-Monterey Canyon in Monterey Bay, California. The water was dark incubated in a cooler on ice before being concentrated ∼100-fold by gravity filtration on a 0.8-µm filter (Pall) prior to cell isolation. Cell “DSEL18-54” (hereafter referred to as A. dasypus) was collected using a Niskin rosette from the research vessel Western Flyer at 160 m on February 3, 2018. Additional images and video were taken of an apparently identical specimen in the same sample (“DSEL18-59”) that perished during imaging. All cells were isolated with a microcapillary tube under a Leica DM IL LED microscope, imaged with a Sony Alpha 6000 camera, transferred to a 0.2-ml PCR tube containing lysis buffer (Picelli et al. 2014), and stored at −80 °C.
Single-Cell Transcriptome Assembly
An Illumina library was prepared from isolated cells (Kolisko et al. 2014; Picelli et al. 2014), then sequenced on MiSeq and HiSeq platforms (A. folium and A. dasypus, respectively) at the Sequencing and Bioinformatics Consortium at University of British Columbia. Two separate assemblies were performed for each transcriptome using different methods for trimming and assembly to maximize capture of the most complete data set. First, forward and reverse reads were trimmed using Trimmomatic v0.36 to remove Nextera adapters and Illumina-specific primers (bases below a threshold quality of 5 cut from leading and trailing ends of reads; Bolger et al. 2014), combined using PEAR (Zhang et al. 2014), and assembled with Trinity v2.4.0 (Grabherr et al. 2011). In parallel, each set of raw reads were also trimmed with Cutadapt (Martin 2011) and then assembled using rnaSPAdes v3.13.2 (Bankevich et al. 2012). Reads were inspected using FASTQC (Andrews 2010) to confirm the removal of adapters after each trimming step. Raw reads and assemblies were submitted to the National Center for Biotechnology Information Sequence Read Archive (accession: PRJNA608604) and Dryad (https://doi.org/10.5061/dryad.pg4f4qrk0), respectively. After assembly, contaminants were identified for removal using megaBLAST to search against Uniprot reference proteomes for divergent contigs. TransDecoder v5.1.0 (Haas et al. 2013) was used to identify open reading frames and annotation was performed using BlastP (Altschul et al. 1990) against the SwissProt database (Poux et al. 2017) with an e-value threshold ≤1e10−5.
Phylogenetic Analyses
Curated protein alignments of 263 genes (Burki et al. 2016) were used as queries for BlastP searches through all peptides predicted in the transcriptome. After resulting hits were aligned with respective queries, maximum likelihood (ML) trees were generated from each alignment using RAxML HPC-PTHREADS-SSE3 (model: PROTGAMMALG; Stamatakis 2006). Contaminants, paralogs, and isoforms were identified via visual inspection of the trees and removed from parent alignments. Resulting clean alignments were processed using SCaFoS v4.55 (Roure et al. 2007) to select relevant taxa and genes with ≥ 60% presence across selected operational taxonomic units. This yielded a total concatenated alignment of 221 genes, from which an ML tree was generated using IQ-TREE, employing ProtTest to determine the best fit model: LG+F + I+G4 (Abascal et al. 2005; Nguyen et al. 2015). Bayesian analysis was performed on the same alignment using Phylobayes-MPI in four independent runs (Lartillot et al. 2009). For this analysis, the model CAT+GTR+G4 was used, as CAT+GTR is considered the best fit model for data sets with >400 aligned positions (according to the Phylobayes manual v4.1). After surpassing 10,000 generations per run and removing the first 10% of trees, we checked for convergence. Out of four runs, three converged to an identical phylogeny and the difference in the fourth run did not affect the placement of Abedinium. Every two trees were sampled to construct a consensus tree from the converged runs. Several topology tests were performed to rule out alternative tree topologies (supplementary table S1, Supplementary Material online).
A BlastN search was performed against the assembled transcriptome data sets using the A. dasypus SSU rRNA gene sequence as a query (Gómez et al. 2010; accession number GU355678.1). Hits were added to the multiple sequence alignment from Gómez et al. (2010), aligned with MAFFT v7.212 (Katoh and Standley 2013), and trimmed with a 10% gap threshold in trimAl v3 (Capella-Gutiérrez et al. 2009). An ML tree was constructed using IQ-TREE with jModelTest (Posada 2008) to determine the best fit model (TIM2+I+G4) and BlastN was used to calculate sequence similarity between SSU rDNA sequences of both cells (Zhang et al. 2000). SSU rDNA sequences from A. folium and A. dasypus were submitted to GenBank under the accession numbers MT191358 and MT191359.
Organellar Protein Characterization
To look for the mitochondrial genes cob, cox1, and cox3, a BlastP search was performed against the translated data sets of both cells using select dinoflagellate sequences as queries (accession numbers AAK67258.2, ACR45472.1, and ABR15109.1, respectively). The resulting mtDNA transcript for cox3 was aligned with orthologs belonging to core dinoflagellates known to transsplice their cox3 mRNA together to look for evidence of a poly-adenylated tail remnant at the splice site (Jackson et al. 2007; Jackson and Waller 2013). For the nuclear packaging proteins DVNP, HLPs I and II, and core histones, a BlastP search (e-value threshold ≤1e−5) was conducted and hit identities were confirmed with BlastP in GenBank. For proteins that were not recovered, additional searches were performed using Hidden Markov Model profiles hand-curated from known homologs and built with hmmbuild (HMMER v2.4; http://hmmer.org/) to confirm their absence from transcriptome data.
Dinoflagellate homologs for enzymes involved in plastid-associated heme, isoprenoid, and iron–sulfur cluster biosynthesis pathways were used as queries in a BlastP search (e-value threshold ≤1e−25) against assemblies from both cells. The identity of each hit was initially confirmed by BlastP and then added to alignments including homologs from a wide range of eukaryotes and prokaryotes (Hehenberger et al. 2019). Alignments were trimmed with trimAl to remove weakly aligned regions (gap threshold: 80%) before ML tree construction with IQ-TREE (supplementary trees, Supplementary Material online; alignments available upon request). Signal sequence prediction was performed using SignalP v3.0 (to view the cleavage site prediction in cases of low signal probability) and v5.0 (default settings; Nielsen et al. 1997) and transmembrane regions (TMRs) were predicted using TMHMM v2.0 (Sonnhammer et al. 1998) after visual inspection of alignments for intact N-termini and the presence of N-terminal extensions. To search for evidence of the continued existence of a plastid organelle, queries for proteins associated with plastid division and transport were used to perform a broad BlastP search (supplementary table S2, Supplementary Material online).
Geographic Metadata Search
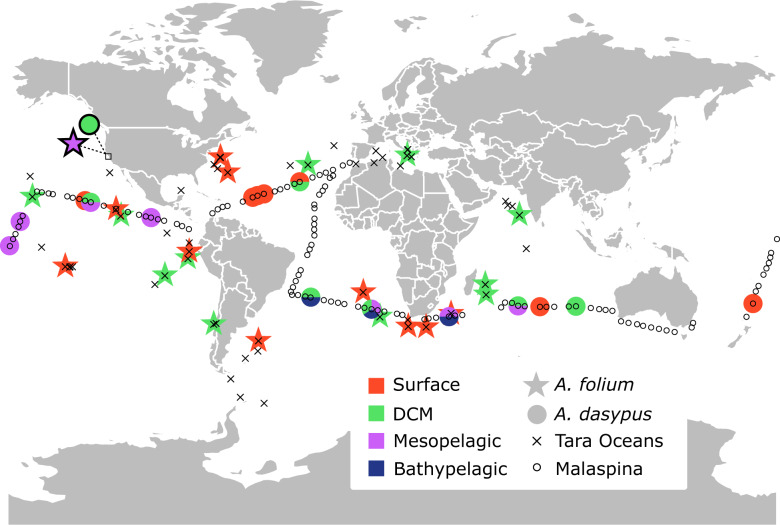
To assess the global distribution of each Abedinium species, SSU rDNA sequences were used to search data from two global-sequencing expeditions: V4 metabarcoding of the picoplanktonic size fraction (0.2–3 µm) at 13 stations and seven depths taken on the Malaspina 2010 expedition (Duarte 2015; Giner et al. 2020), and the Marine Atlas of Tara Oceans Unigenes (MATOU) from the Tara Oceans expeditions (Bork et al. 2015; Carradec et al. 2017). Hits of ≥99% similarity with a minimum of length of 230 bp were considered the same species. MATOU data were searched and visualized using the Ocean Gene Atlas (Villar et al. 2018).
Results and Discussion
Isolation and Identification of A. dasypus and A. folium sp. nov.
Immediately after collection, seawater was concentrated to improve the rate of cell discovery immediately after sampling and observed by microscopy on the ship immediately after they were collected to explore the diversity of marine microorganisms that were not easily identifiable as common and well-studied species. Three cells matching the overall description of Abedinium were documented from two sites where they were collected at 160 m in 2018 (two cells) and 600 m in 2017 (one cell) and all were observed and documented by photo- or video-microscopy to provide information about their morphology and behavior prior to processing for transcriptomics. Cell DSEL18-54 (subsequently identified as A. dasypus, see below) was observed in a nonmotile “balled up” state with one posterior flagellum visible (fig. 1A, supplementary video 1 and all videos available at https://doi.org/10.5061/dryad.pg4f4qrk0), and in the same sample a second cell (DSEL18-59) with the same overall appearance was also recorded to show active curling behavior (supplementary video 2). In previous records of A. dasypus, orange or red pigment was observed at the distal end of its tentacle, as well as a single reddish pigmented body within or near the nucleus (Cachon and Cachon-Enjumet 1964; Gómez et al. 2010; Saburova et al. 2013). No pigment was visible in the A. dasypus cells observed here, however, it may have been obscured in the balled-up cell, and the curling cell lacked the bulbous tip typically associated with pigment on its tentacle. In the other sample, cell DICHO17-03 (subsequently described as A. folium, see below) was shown to have a posterior flagellum that was observed just before it degraded away from the cell (fig. 1B and C) and an obovate leaf-shaped body plan with undulated margins that appeared to be in a semicontracted state. In this posture, the cell body was ∼75 µm in length. However, in a relaxed state, it would likely extend, perhaps to 100–150 µm. Abedinium folium showed no visible pigment, although a colorless body is visible where the nucleus-associated pigment has been observed in A. dasypus (fig. 1B and C). No tentacle was visible in A. folium, but this might have been due to its position during viewing—the contractile behavior that A. dasypus is known to exhibit involves tucking its tentacle into its body before curling into a ball (Cachon and Cachon-Enjumet 1964; Gómez et al. 2010; Saburova et al. 2013; supplementary video 2). Alternatively, if the cell lacks a tentacle, this specimen may simply have undergone recent binary fission—in A. dasypus, this process yields one daughter cell that retains the mother’s tentacle, whereas the other must grow a new one (Cachon and Cachon-Enjumet 1964). Although Abedinium has never been observed in the act of feeding, they are thought to be heterotrophic predators, as they lack chlorophyll. The absence of large visible food vacuoles is likely explained by a specialization in picoplanktonic prey, consistent with recent findings using isotope probe analysis that A. dasypus consumes Micromonas pusilla (Orsi et al. 2018). Abedinium folium was collected from the Monterey Canyon waters, a system where Micromonas is frequently observed (Simmons et al. 2016; Limardo et al. 2017).
Fig. 1.
Light micrographs of two species of Abedinium collected in Monterey Bay, CA. After imaging, each cell was prepared for RNA sequencing and analysis. (A) Abedinium dasypus is shown in a fully rolled up state with one posterior flagellum visible. (B and C) The posterior flagellum of Abedinium folium sp. nov. is shown degrading away during imaging. The cell has a bilaterally symmetrical leaf-shaped body plan with undulated cell margins in a semicontracted state. Arrows indicate a colorless body where pigment is often seen in A. dasypus but appears here to lack pigment in A. folium. The scale bar depicts 50 µm. Images and video of an open cell isolated from the same sample as A. dasypus are available in Supplementary Material online.
The SSU rDNA sequences from DSEL18-54 and DICHO17-03 were added to the data set of Gómez et al. (2010), and ML phylogenetic reconstruction placed them in a clade with A. dasypus with full bootstrap support (fig. 2). DSEL18-54 branched closely with A. dasypus, whereas DICHO17-03 sister to both. Consistent with this, SSU sequences DSEL18-54 and A. dasypus isolate 239 (GU355678.1) shared 99% identity, whereas DICHO17-03 shared only 95% identity. We conclude that DSEL18-54 is a representative of A. dasypus, whereas DICHO17-03 is a representative of a distinct species, which we describe as A. folium, the second member described in the Abedinium genus.
Fig. 2.
Maximum likelihood phylogeny of dinoflagellate SSU rRNA gene sequences adapted from Gómez et al. (2010). Bold IDs indicate sequences of interest in the present study. Node numbers represent bootstrap values; values of 100 are indicated with a black dot. The scale bar provides reference for the estimated number of nucleic acid substitutions per site.
We identified both A. folium and A. dasypus SSU rRNA in global environmental surveys of eukaryotic diversity. Both sequence types were globally widespread (fig. 3), but typically appeared in low abundance at the locations where they were found. The best hit for A. dasypus (99.7% similar) occurred at the surface, deep chlorophyll maximum (DCM), mesopelagic, and bathypelagic depths at various sites sampled on the Malaspina expedition. The best hit for A. folium (99.16% similar) was found at the surface and DCM in Tara Oceans sampling, the only depths sampled on this expedition. Hits with ≥99% similarity for A. dasypus were not found in Tara Oceans sampling and hits meeting this threshold for A. folium were similarly not found in the Malaspina data set. These surveys reveal a rare but widespread distribution of Abedinium globally, but also a broad distribution across water column depth. This is consistent with our observations, in that cell density of Abedinium spp. was very low. For instance, of the 177 cells collected from 23 water samples during the two cruises, we found only a single cell of A. folium, two cells of A. dasypus, and no other noctilucoid cells. This is a substantially lower occurrence than that of eupelagonemid-like cells (unpublished data), one of the most prevalent protist group of the marine environment during 2017–2018 campaign as well as the previous survey (Gawryluk et al. 2016; Okamoto et al. 2019). This distribution pattern (i.e., low density and wide vertical distribution range) seems to be shared among other phylogenetically unresolved noctilucoid species (Gómez and Furuya 2005; Gómez 2010; Saburova et al. 2013). The ability to survive in sunlit surface waters as well as in the sunless bathypelagic may suggest a broad diet not restricted to photosynthetic prey. However, since no information exists on the life cycles of Abedinium, the possibility remains that these sequences represent errant cysts or cells displaced by animal movement or currents.
Fig. 3.
Schematic depiction of diagnostic sequence hits with >99% similarity to SSU rRNA gene sequences of Abedinium folium and Abedinium dasypus from two global expeditions. Sampling sites are indicated for Tara Oceans and Malaspina expeditions, as well as which sites yielded hits (see legend). Colors signify the depth zone at which a hit was sampled. Shapes depicted with a black border were sampled in the present study. DCM, deep chlorophyll maximum.
Phylogenomic Analysis Reveals Abedinium Is a Distinct, Early-Branching Lineage of Dinoflagellates
We constructed and sequenced single-cell transcriptome libraries for both A. dasypus (DSEL18-54) and A. folium (DICHO17-03) to see if Abedinium lends any insights to dinoflagellate character evolution based on phylogenomics. The highest quality assemblies for A. folium and A. dasypus included 62% and 20% of conserved eukaryotic single-copy genes in BUSCO coverage estimates, respectively, and in total 52,052 and 15,743 predicted peptides, respectively. It is not clear why A. dasypus showed such low coverage, but it may have been caused by deterioration of the cell during sampling or RNA degradation postsampling, as these are factors that can affect the success of the single-cell sequencing approach. We performed parallel searches for genes of interest in these as well as the lower quality assemblies produced from alternative trimming and assembly methods to maximize detection of the most complete sequences.
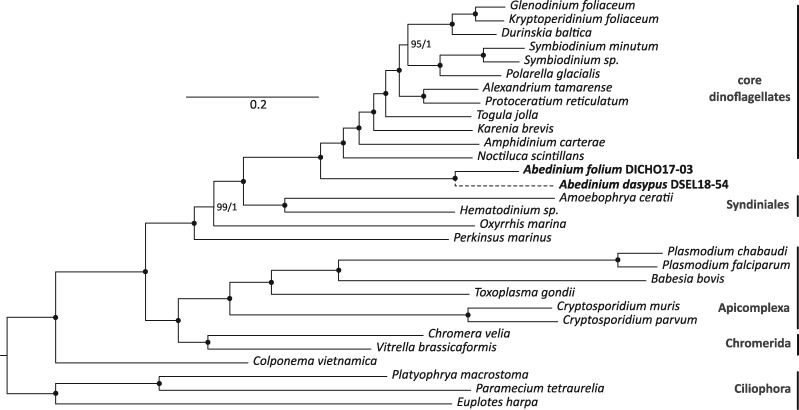
The inclusion of Abedinium in the family Leptodiscaceae within the noctilucoids was based on aspects of its body plan and SSU rRNA phylogeny, which was not strongly supported (Gómez et al. 2010). Using transcriptome data, we were able to expand the number of genes analyzed many-fold, which resulted in consistent strong support for an alternative topology: in ML analysis of 221 genes, A. folium (represented by 79% of the analyzed genes) branched as the sister lineage to a clade consisting of N. scintillans and core dinoflagellates with full bootstrap support (fig. 4). About 76 of the single gene trees making up this analysis showed this placement, and no other position was consistently found in any large number of genes (most other single gene trees showed Abedinium branching in the dinoflagellates or some position basally within the myzozoa—the group containing dinoflagellates, perkinsids, chromerids, and apicomplexans). The inclusion of A. dasypus (possessing only 35% of the analyzed genes) yielded the same topology with both Abedinium species forming a fully supported clade, and with complete support for the position of Abedinium relative to other dinoflagellates. Bayesian analyses and topology tests both confirmed the position of Abedinium, with and without A. dasypus (supplementary table S1, Supplementary Material online). Based on this new phylogenetic insight, we conclude that Abedinium represents an independent lineage sister to Noctilucales, giving rise to the newly described order, Abediniales (taxonomic summary available below). It is unclear if the other members of Leptodiscacea would also be included in Abediniales, as this family was defined primarily on morphology, which is challenging as they are known to exhibit polymorphism during their lifecycle (Gómez and Furuya 2005).
Fig. 4.
Maximum likelihood (ML) phylogeny of select alveolates based on 221 protein orthologs (all found in >60% of analyzed taxa). The dotted branch shows the placement of Abedinium dasypus in an alternate analysis including this taxon, which was excluded here due to relatively poor coverage of its transcriptome. Numbers at nodes represent bootstrap values over posterior probabilities; values of 100 and 1 are indicated with a black dot (all Bayesian posterior probability values are 1). In three out of four independent runs, Bayesian analysis predicted that Noctiluca scintillans branches with Amphidinium carterae (leaving the placement of Abedinium unaffected), but the topology was otherwise identical to that shown. The scale bar provides reference for the estimated number of amino acid substitutions per site.
Abedinium and Early Dinoflagellate Character Evolution
As a new lineage branching near the origin of core dinoflagellates, Abedinium may help reconstruct the evolutionary history of important traits that appeared around this time. Several unique characters that have attracted considerable speculation are proteins involved in the formation of chromatin. As expected, both species of Abedinium expressed the packaging protein DVNP, characteristic of all dinoflagellates including early-branching taxa (Janouškovec et al. 2017). The presence of a lysine-rich N-terminal extension confirmed that these transcripts belonged to a dinoflagellate and not a virus (supplementary fig. S1, Supplementary Material online; Gornik et al. 2019). The advent of liquid crystalline chromosome condensation in dinoflagellates occurred after DVNP acquisition and is thought to coincide with the appearance of dinoflagellate HLPs. HLPs were acquired at least twice through the horizontal transfer of bacterial HU-like proteins—HLP II appearing in early-branching lineages starting with Noctiluca, and HLP I in core taxa—in contrast to the archaeal origins of canonical histones (Sandman and Reeve 2000; Wong et al. 2003; Janouškovec et al. 2017). We found no evidence of either type of HLP in Abedinium, but a search for other chromatin-binding proteins uncovered canonical H3 and H4 core histone homologs in A. folium, and H2A in A. dasypus. Although further study is needed to confirm or refute the absence of HLPs, we will note that they are consistently highly represented in transcriptomic data from other dinoflagellates, and much more highly represented than canonical histones (Riaz et al. 2019). An absence of HLPs in this lineage would significantly narrow the timeframe for the origin of HLPs, suggesting that they were acquired after the divergence of Abedinium, but before noctilucoids. Currently, N. scintillans is the earliest-branching dinoflagellate known to exhibit liquid crystalline chromatin packaging (and possess HLPs), but do so only in their haploid gametic life stage (Fukuda and Endoh 2008). Interestingly, Noctiluca is also the only core dinoflagellate known to have a diploid trophont, whereas those of later branching taxa are haploid with permanently condensed chromosomes throughout the life cycle (Pfiester 1984). This observation has led to the hypothesis that taxa with haploid trophonts descend from a neotenous haploid zoospore similar to that of Noctiluca (Fukuda and Endoh 2008). Further exploration into the life cycle of Abedinium is needed to reveal whether these taxa possess condensed chromatin at any life stage, and whether haploid trophonts are indeed monophyletic in the core dinoflagellates, or if Noctiluca is simply an outlier.
Another character that has been well studied is the plastid, which has had a complex history in dinoflagellates, including multiple losses of photosynthesis (Saldarriaga et al. 2001). Plastid-derived enzymes for heme, isoprenoid, and iron–sulfur cluster assembly biosynthetic pathways have been found to persist in the nucleus of several nonphotosynthetic dinoflagellates and other myzozoans studied to date, and are generally concluded to encode plastid-targeted proteins whose functions explain the persistence of an organelle (Hehenberger et al. 2014; Janouškovec et al. 2017; Mathur et al. 2019). Highly similar, but not identical patterns of loss and retention of these pathways have been observed across many heterotrophic myzozoans with nonphotosynthetic plastids (biosynthesis in other lineages with cryptic plastids is more diverse), but in general, these pathways are indicative of plastid retention (Janouškovec et al. 2017; Mathur et al. 2019). In A. folium, we recovered transcripts for all but four of the 24 expected genes associated with these pathways (fig. 5), but found none in A. dasypus, which is most likely a reflection of the fact that the coverage of the A. folium transcriptome is generally much deeper.
Fig. 5.
Schematic depiction of iron–sulfur cluster, isoprenoid, and heme biosynthesis enzymes in Abedinium folium with N-terminal targeting information. N-terminal extensions are depicted as being plastid-targeting signals, undetermined extensions, incomplete, or absent (see legend). Although most proteins indicated as having plastid-targeting signals contain predicted signals, proteins with truncated signals that still contain identifiable transit domains are included in this designation.
Phylogenies were inferred for all these genes, and A. folium always clustered with the peridinin plastid-derived homologs of other dinoflagellates, with only one exception: the complete sequence for ispF in A. folium was highly divergent, and clustered with Perkinsus outside of the dinoflagellates. Two sufS homologs were recovered, both clustering in dinoflagellate clades and both with incomplete N-termini (supplementary trees, Supplementary Material online). The four enzymes that were not found (sufD, petF, ispD, and hemH) may be absent because of sampling, but it is unclear. Many transcripts were truncated, but seven were determined to be full-length when compared with other full-length homologs within the alignment, and these were found to encode an N-terminus extension with characteristics expected of a dinoflagellate plastid-targeting leader. Specifically, these proteins were predicted to encode signal peptides followed by sequences with characteristics expected of a dinoflagellate transit peptide (fig. 5 and supplementary fig. S2 and alignments, Supplementary Material online). Dinoflagellate transit peptides come in two forms: Class I transit peptides contain a TMR ending with a negatively charged, arginine-rich domain upstream of the mature protein, whereas Class II peptides lack this TMR (Patron et al. 2005). Most intact A. folium plastid sequences had Class I transit peptides, whereas ispF bore an ambiguous signal-peptide-encoding extension and nifU had a presequence devoid of recognizable signal peptides or TMRs (supplementary spreadsheet: https://doi.org/10.5061/dryad.pg4f4qrk0). N-terminus extensions for dxr and hemB were truncated and missing a signal peptide domain, however both contained the Class I TMR and were thus inferred to have plastid-targeting leaders. Three proteins, dxs, hemC, and hemE, contained TMR with “FVAP” motifs, a characteristic of plastid-targeting sequences that is not associated with other endomembrane targeting (Patron et al. 2005; supplementary spreadsheet: https://doi.org/10.5061/dryad.pg4f4qrk0).
To provide more compelling evidence for the retention of a plastid organelle, an additional search was performed for protein homologs associated with plastid import and division. Unlike metabolic enzymes, which theoretically could perform their function in another cell compartment, plastid translocators or division machinery have no conceivable function outside the presence of a plastid. Of the orthologs queried (supplementary table S2, Supplementary Material online), only one returned a hit of plausible plastidial origin: A homolog of “Group V” TIC62 (Balsera et al. 2007) was recovered from the A. folium transcriptome (supplementary spreadsheet: https://doi.org/10.5061/dryad.pg4f4qrk0). TIC62 is part of the translocation complex that transports proteins across the inner-most membrane of the plastid (Küchler et al. 2002). “Group V” TIC62 proteins in particular are exclusive to eukaryotes (Balsera et al. 2007). The homolog recovered from Abedinium grouped at the base of a dinoflagellate clade within a TIC62 phylogeny (at the same position inferred for Abedinium: supplementary fig. S3, Supplementary Material online), suggesting it is not the byproduct of food or contamination. Unfortunately, the sequence lacked a complete N-terminus, leaving no information about targeting.
At a minimum, the presence of peridinin plastid proteins indicates that Abedinium is yet another parallel example of plastid reduction in dinoflagellates. Although transcriptome data alone cannot definitively confirm the existence of a nonphotosynthetic organelle, the presence of near-complete peridinin plastid-derived pathways and plastid-targeting signals as well as evidence of plastid transport machinery all strongly suggests that these proteins are indeed transported to an extant plastid, as no organism has ever been found to possess these characteristics in the absence of a plastid.
Like their apicomplexan sisters, dinoflagellates possess a highly reduced mitochondrial genome containing only three protein-coding genes, cob, cox1, and cox3 (Jackson et al. 2007; Slamovits et al. 2007). All three genes were found in both A. folium and A. dasypus. Many dinoflagellate cox3 transcripts are the result of a unique trans-splicing event that incorporates bases from the poly-A tail of the 5′ transcript into the coding sequence (Jackson et al. 2007). The Abedinium cox3 cDNA sequences spanned this trans-splice site but showed no evidence of a poly-A tract. A genomic survey would be required to confidently conclude there was no trans-splicing (e.g., that does not incorporate any of the poly-A tail), however, we feel this is nevertheless the likely explanation, given that other deep-branching taxa Oxyrrhis and Hematodinium have also been shown to lack cox3 trans-splicing (Slamovits et al. 2007; Jackson et al. 2012; Jackson and Waller 2013), and Abedinium sharing this ancestral state is compatible with its phylogenetic position. Nuclear trans-splicing was evident in several cDNA sequences that encoded the canonical 22 nucleotide spliced leader motif common to other dinoflagellates and perkinsids (Zhang et al. 2007), but we note that relatively few transcripts with complete spliced leaders were identified (12 in total).
Concluding Remarks
Understanding eukaryotic diversity and evolution requires more information from taxa that are rare, ephemeral, and unculturable. In the case of dinoflagellates, additional challenges, like their well-known nuclear complexity, pose hurdles due to the limits of current genome-sequencing technology. Here, we show for Abedinium that single-cell transcriptomics can be linked to morphological identification and achieve adequate sampling from very limited material to address several questions and establish a phylogenetic position with much greater certainty than was possible with single-gene sequencing. Obviously, more information can be gleaned from finding more cells and the use of other methods for a truly comprehensive species understanding, but for some kinds of protists, single-cell transcriptomics can provide information from thousands of genes where previously we had only one. In the case of Abedinium, procuring cells poses a significant challenge for further study as geographic surveys reveal that they are globally distributed, but only sparsely so. For these and other rare or intractable species, single-cell sequencing offers a level of detailed study and description that might not otherwise be possible, and linking these data with pictures and videos provides a solid connection between morphology and microscopical identification, and molecular phylogeny and character evolution.
Taxonomic Summary
Class DINOPHYCEAE West & Fritch, 1927
Order ABEDINIALES ord. nov. Cooney, Okamoto & Keeling, 2020
Diagnosis. Cell dorso-ventrally flattened and bilaterally symmetrical with a posterior flagellum, undulated cell margin, and hyaline appearance.
Type genus. Abedinium Loeblich & Loeblich III, 1966
Etymology. The order name is derived from the type genus, Abedinium.
Family ABEDINIACEAE fam. nov. Cooney, Okamoto & Keeling, 2020
Diagnosis. (See above)
Type genus. Abedinium Loeblich & Loeblich III, 1966
Etymology. The family name is derived from the type genus, Abedinium.
Genus Abedinium Loeblich & Loeblich III, 1966
Abedinium folium sp. nov. Cooney, Okamoto & Keeling, 2020
Diagnosis. Unicellular heterotroph with a dorso-ventrally flattened, bilaterally symmetrical cell body. Cell is hyaline in appearance with no pigmentation. Possesses an undulated cell margin and a posterior flagellum. Found in the mesopelagic zone within Monterey Bay, California, but diagnostic sequences were also reported from environmental samples in the Mediterranean, Atlantic, Pacific, and Indian open oceans in both northern and southern hemispheres.
Holotype. Specimen pictured in figure 1B and C.
Etymology. Folium (fo’li.um), the Latin word for “leaf,” refers to the flat body plan of the cell, which resembles an obovate leaf or leaflet.
Type locality. Obtained from mid Monterey Canyon in Monterey Bay, CA at 600 m. Collector: N. Okamoto.
Sequence data. Raw reads: SRA (accession number SRR11184859). Single-cell transcriptome: https://doi.org/10.5061/dryad.pg4f4qrk0. SSU rRNA gene: GenBank accession number MT191358.
Supplementary Material
Acknowledgments
We would like to thank Ramon Massana for his help with metadata searches. Additional thanks to Varsha Mathur, Nick Irwin, Filip Husnik, Juan Saldarriaga, and Vittorio Boscaro for their advice and assistance with data analysis. This work was supported by a grant from the Gordon and Betty Moore Foundation (GBMF3307 to P.J.K., T.A.R., A.Z.W., and A.E.S.) and the National Science and Engineering Research Council of Canada (2019 03994 and 03986 to P.J.K. and B.S.L.).
Data Availability
The data underlying this article are available in the article and in its Supplementary Material online.
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21(9):2104–2105. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc.
- Balsera M, Stengel A, Soll J, Bölter B. 2007. Tic62: a protein family from metabolism to protein translocation. BMC Evol Biol. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin M, et al. 2012. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc Natl Acad Sci U S A. 109(39):15793–15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Genome analysis Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, et al. 2015. Tara Oceans studies plankton at planetary scale. Science 348(6237):873–873. [DOI] [PubMed] [Google Scholar]
- Burki F, et al. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc R Soc B Biol Sci. 283: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon J, Cachon-Enjumet M. 1964. Leptospathium navicula nov. gen. nov. sp. et Leptophyllus dasypus nov. gen. nov. sp., Peridiniens Noctilucidae (Hertwig) du plancton neritique de Villefranche-sur-Mer. Bull l’Institut Oceanogr Monaco. 62:1–12. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carradec Q, et al. 2017. A global oceans atlas of eukaryotic genes. Nat Commun. 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambouvet A, Morin P, Marie D, Guillou L. 2008. Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science 322(5905):1254–1258. [DOI] [PubMed] [Google Scholar]
- Coats DW. 1999. Parasitic life styles of marine dinoflagellates. J Eukaryot Microbiol. 46(4):402–409. [Google Scholar]
- Duarte CM. 2015. Seafaring in the 21st century: the Malaspina 2010 circumnavigation expedition. Limnol Oceanogr Bull. 24(1):11–14. [Google Scholar]
- Fukuda Y, Endoh H. 2008. Phylogenetic analyses of the dinoflagellate Noctiluca scintillans based on β-tubulin and Hsp90 genes. Eur J Protistol. 44(1):27–33. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, et al. 2016. Morphological identification and single-cell genomics of marine diplonemids. Curr Biol. 26(22):3053–3059. [DOI] [PubMed] [Google Scholar]
- Giner CR, et al. 2020. Marked changes in diversity and relative activity of picoeukaryotes with depth in the world ocean. ISME J. 14(2):437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez F. 2010. Diversity and distribution of noctilucoid dinoflagellates (Noctilucales, Dinophyceae) in the open Mediterranean Sea. Acta Protozool. 49:365–372. [Google Scholar]
- Gómez F, Furuya K. 2005. Leptodiscaceans (Noctilucales, Dinophyceae) from the Pacific Ocean: first records of Petalodinium and Leptodiscus beyond the Mediterranean Sea. Eur J Protistol. 41(3):231–239. [Google Scholar]
- Gómez F, Moreira D, López-García P. 2010. Molecular phylogeny of noctilucoid dinoflagellates (Noctilucales, Dinophyceae). Protist 161(3):466–478. [DOI] [PubMed] [Google Scholar]
- Gornik SG, et al. 2012. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr Biol. 22(24):2303–2312. [DOI] [PubMed] [Google Scholar]
- Gornik SG, Hu I, Lassadi I, Waller RF. 2019. The biochemistry and evolution of the dinoflagellate nucleus. Microorganisms 7(8):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, et al. 2008. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ Microbiol. 10(12):3349–3365. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Gast RJ, Keeling PJ. 2019. A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc Natl Acad Sci U S A. 116(36):17934–17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Imanian B, Burki F, Keeling PJ. 2014. Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts. Genome Biol Evol. 6(9):2321–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, et al. 2007. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 5(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Gornik SG, Waller RF. 2012. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol Evol. 4(1):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Waller RF. 2013. A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLoS One 8(2):e56777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2017. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc Natl Acad Sci U S A. 114(2):E171–E180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, et al. 2005. Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic. Aquat Microb Ecol. 40:133–150. [Google Scholar]
- Jeong HJ, et al. 2010. Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci J. 45(2):65–91. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. 2013. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 64(1):583–607. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, del Campo J. 2017. Marine protists are not just big bacteria. Curr Biol. 27(11):R541–R549. [DOI] [PubMed] [Google Scholar]
- Kolisko M, Boscaro V, Burki F, Lynn DH, Keeling PJ. 2014. Single-cell transcriptomics for microbial eukaryotes. Curr Biol. 24(22):R1081–R1082. [DOI] [PubMed] [Google Scholar]
- Küchler M, Decker S, Hörmann F, Soll J, Heins L. 2002. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21(22):6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC, et al. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 28(16):2570–2580. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Le Bescot N, et al. 2016. Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ Microbiol. 18(2):609–626. [DOI] [PubMed] [Google Scholar]
- Limardo AJ, et al. 2017. Quantitative biogeography of picoprasinophytes establishes ecotype distributions and significant contributions to marine phytoplankton. Environ Microbiol. 19(8):3219–3234. [DOI] [PubMed] [Google Scholar]
- Lin S. 2011. Genomic understanding of dinoflagellates. Res Microbiol. 162(6):551–569. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Leander BS, Keeling PJ. 2009. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Light Evol. 3:65–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalef R. 1973. Fitoplancton marino de la region de afloramiento del NW de Africa. Result. Exped Cient del B/O Cornide. 2:65–94. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Mathur V, et al. 2019. Multiple independent origins of apicomplexan-like parasites. Curr Biol. 29(17):2936–2941. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10(1):1–6. [DOI] [PubMed] [Google Scholar]
- Okamoto N, et al. 2019. A revised taxonomy of diplonemids including the Eupelagonemidae n. fam. and a type species, Eupelagonema oceanica n. gen. & sp. J Eukaryot Microbiol. 66(3):519–524. [DOI] [PubMed] [Google Scholar]
- Orsi WD, et al. 2018. Identifying protist consumers of photosynthetic picoeukaryotes in the surface ocean using stable isotope probing. Environ Microbiol. 20(2):815–827. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Archibald JM, Keeling PJ. 2005. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 348(4):1015–1024. [DOI] [PubMed] [Google Scholar]
- Pfiester LA. 1984. Dinoflagellate nuclei In: Spector DL, editor. Dinoflagellates. Orlando: Academic Press; p. 181–199. [Google Scholar]
- Picelli S, et al. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 9(1):171–181. [DOI] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256. [DOI] [PubMed] [Google Scholar]
- Poux S, et al. 2017. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 33(21):3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz S, Niaz Z, Khan S, Liu Y, Sui Z. 2019. Detection, characterization and expression dynamics of histone proteins in the dinoflagellate Alexandrium pacificum during growth regulation. Harmful Algae. 87:101630. [DOI] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. 2007. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburova M, Polikarpov I, Al-Yamani F. 2013. First records of noctilucoid dinoflagellates Abedinium dasypus and Scaphodinium mirabile (Dinophyceae) from the Indian Ocean. Mar Biodivers Rec. 6:1–7. [Google Scholar]
- Saldarriaga JF, Taylor FJR, Keeling PJ, Cavalier-Smith T. 2001. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 53(3):204–213. [DOI] [PubMed] [Google Scholar]
- Sandman K, Reeve JN. 2000. Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch Microbiol. 173(3):165–169. [DOI] [PubMed] [Google Scholar]
- Seong KA, Jeong HJ, Kim S, Kim GH, Kang JH. 2006. Bacterivory by co-occurring red-tide algae, heterotrophic nanoflagellates, and ciliates. Mar Ecol Prog Ser. 322:85–97. [Google Scholar]
- Shumway SE. 1990. A review of the effects of algal blooms on shellfish and aquaculture. J World Aquaculture Soc. 21(2):65–104. [Google Scholar]
- Simmons MP, et al. 2016. Abundance and biogeography of picoprasinophyte ecotypes and other phytoplankton in the eastern North Pacific Ocean. Appl Environ Microbiol. 82(6):1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. 2007. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 372(2):356–368. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 6:175–182. [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stoecker DK. 1999. Mixotrophy among dinoflagellates. J Eukaryot Microbiol. 46(4):397–401. [Google Scholar]
- Taylor FJR, Hoppenrath M, Saldarriaga JF. 2008. Dinoflagellate diversity and distribution. Biodivers Conserv. 17(2):407–418. [Google Scholar]
- Trench RK. 1993. Microalgal-invertebrate symbioses: a review. Endocytobiosis Cell Res. 9:135–175. [Google Scholar]
- Veldhuis MJW, Cucci TL, Sieracki ME. 1997. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J Phycol. 33(3):527–541. [Google Scholar]
- Villar E, et al. 2018. The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Nucleic Acids Res. 46(W1):W289–W295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. 2011. Dinoflagellate genome evolution. Annu Rev Microbiol. 65(1):369–387. [DOI] [PubMed] [Google Scholar]
- Wong JTY, New DC, Wong JCW, Hung VKL. 2003. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot Cell. 2(3):646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. 2007. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci U S A. 104(11):4618–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin S. 2008. mRNA editing and spliced-leader RNA trans-splicing groups Oxyrrhis, Noctiluca, Heterocapsa, and Amphidinium as basal lineages of dinoflagellates. J Phycol. 44(3):703–711. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7(1–2):203–214. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, et al. 2014. A comparative study of techniques for differential expression analysis on RNA-seq data. PLoS One 9(8):e103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Material online.