Abstract
Accurate taxonomic identification of alien species is crucial to detect new incursions, prevent or reduce the arrival of new invaders and implement management options such as biological control. Globally, the taxonomy of non-native Prosopis species is problematic due to misidentification and extensive hybridization. We performed a genetic analysis on several Prosopis species, and their putative hybrids, including both native and non-native populations, with a special focus on Prosopis invasions in Eastern Africa (Ethiopia, Kenya and Tanzania). We aimed to clarify the taxonomic placement of non-native populations and to infer the introduction histories of Prosopis in Eastern Africa. DNA sequencing data from nuclear and chloroplast markers showed high homology (almost 100 %) between most species analysed. Analyses based on seven nuclear microsatellites confirmed weak population genetic structure among Prosopis species. Hybrids and polyploid individuals were recorded in both native and non-native populations. Invasive genotypes of Prosopis juliflora in Kenya and Ethiopia could have a similar native Mexican origin, while Tanzanian genotypes likely are from a different source. Native Peruvian Prosopis pallida genotypes showed high similarity with non-invasive genotypes from Kenya. Levels of introduced genetic diversity, relative to native populations, suggest that multiple introductions of P. juliflora and P. pallida occurred in Eastern Africa. Polyploidy may explain the successful invasion of P. juliflora in Eastern Africa. The polyploid P. juliflora was highly differentiated from the rest of the (diploid) species within the genus. The lack of genetic differentiation between most diploid species in their native ranges supports the notion that hybridization between allopatric species may occur frequently when they are co-introduced into non-native areas. For regulatory purposes, we propose to treat diploid Prosopis taxa from the Americas as a single taxonomic unit in non-native ranges.
Keywords: Eastern Africa, genetic diversity, hybridization, invasive alien species, mesquite, microsatellites, polyploidy, taxonomic uncertainty, tree invasions
Uncertain taxonomy can complicate the management of invasive species. Using population genetic data Castillo et al. aimed to clarify the taxonomic placement of selected invasive Prosopis populations. They found tetraploid P. juliflora to be highly differentiated from all other diploid species of the genus. Diploid species, on the other hand, had low genetic differentiation, supporting anecdotal evidence that suggests these species often form hybrid swarms in their non-native ranges. For regulatory purposes all invasive diploid Prosopis taxa should be treated as a single taxonomic unit.
Introduction
Biological invasions are a major threat to biodiversity, ecosystem services and human well-being (Pimentel et al. 2005; van Wilgen et al. 2011; Shackleton et al. 2014). With globalization, the number of species being translocated, intentionally or accidently, is ever increasing as part of socio-economic development (Seebens et al. 2020).
Sound taxonomic knowledge of invasive populations is crucial to detect new invasions, to determine the potential sources and pathways of introduction(s), to prevent or reduce the arrival of new invaders, to accurately model potential ecological niches and to implement management options such as biological control (Le Roux and Wieczorek 2009; Ensing et al. 2013). However, the taxonomy of many alien taxa remains problematic due to unresolved phylogenetic relationships, uncertain native-range geographic distributions and interspecific hybridization, among other factors (Pyšek et al. 2013). For example, invasive Heracleum species belong to a taxonomically complex group, making identification of several invasive taxa difficult (Jahodová et al. 2007). In the USA, large areas of riparian and wetland habitats have been invaded by Eurasian saltcedar (Tamarix) species. Gaskin and Schaal (2002) found hybridization among Tamarix species to be widespread in the invaded range, while levels of hybridization in the native range appear to be low. Under such complex scenarios, complementing ecomorphological approaches (i.e. morphological data and environmental requirements) with genetic information may be critical to delimit species boundaries (Le Roux and Wieczorek 2009).
Comparative ecological and genetic studies between conspecific invasive alien species from different parts of the world provide opportunities to clarify genetic relationships and provide insights into taxonomy and invasion history, i.e. knowing which taxa are invasive and where (Gaskin and Schaal 2002; Gallego-Tévar et al. 2019). Research examining genetic diversity and differentiation within and among invasive populations as well as between invasive and native populations, is commonly conducted to unravel introduction histories (Lavergne and Molofsky 2007; Le Roux et al. 2010; Hirsch et al. 2019), dispersal routes within non-native areas (Lachmuth et al. 2010) and the role that genetic constraints play in invasive performance (Dlugosch and Parker 2008). Historical range expansions and past demographic processes may also affect levels of genetic diversity present in invasive populations (Taylor and Keller 2007; Le Roux et al. 2011). Lastly, intra- or interspecific hybridization following introduction may replenish species genetic diversity (Ellstrand and Schierenbeck 2000), mask deleterious alleles and cause fixed heterosis (te Beest et al. 2012).
The genus Prosopis (Leguminosae), commonly known as mesquite, includes some of the world’s worst woody invasive species (Shackleton et al. 2014). The taxonomy of Prosopis species is problematic because diagnostic morphological traits are often lacking and because the native distributions of many species remain contentious (Pasiecznik et al. 2001). Following Burkart (1976), the genus comprises 44 species from the Americas, South West Asia and North Africa, which are mostly found in arid and semiarid regions. Prosopis species have been grouped into five sections, from these, the section Algarobia is divided into six series based on leaf morphological traits (Burkart 1976). The validity of these series has been questioned due to taxonomic uncertainty, interspecific hybridization and a probable polyphyletic origin (Bessega et al. 2006; Burghardt and Espert 2007; Sherry et al. 2011).
Prosopis species have been intentionally moved around the globe for many reasons, including for soil stabilization and to provide fuel and livestock fodder. These movements have been characterized by multiple introductions, often of multiple species from various sources, to different localities (Pasiecznik et al. 2001). Alien Prosopis species are now present in 103 countries and are considered invasive in 49 of these (Shackleton et al. 2014). Given the problematic taxonomy of Prosopis species many studies simply refer to the taxon as Prosopis in their non-native ranges. Taxonomic uncertainty is further exacerbated due to frequent hybridization between different species. For example, in South Africa, numerous species were introduced and became invasive, including Prosopis glandulosa, Prosopis velutina and Prosopis laevigata (Poynton 2009). Here, DNA sequencing data showed that extensive hybridization is occurring and were unable to identify ‘pure’ parental species (Mazibuko 2012). In Australia, introduced populations have been morphologically identified as Prosopis juliflora, P. glandulosa, Prosopis pallida, P. velutina, and their hybrids (van Klinken and Campbell 2001), with the most severe infestation being represented by a hybrid swarm between P. pallida × P. velutina × P. glandulosa var. glandulosa (van Klinken 2012). In Hawaii, morphological hybrids between P. juliflora and the invasive P. pallida seem to be present in several locations (Gallaher and Merlin 2010).
In Eastern Africa, various Prosopis species were introduced and some became invasive. Importantly, there is no credible information available on the origin(s) of Prosopis individuals, their introduction histories or their taxonomic classification in this region (Choge et al. 2011). In Kenya, P. juliflora and P. pallida were first introduced in 1973 to Mombasa (Johansson 1990) with later introductions of various Prosopis species during the 1970s and 1980s to different parts of Kenya, including Baringo County, Tana River and Taveta (Johansson 1990; Otsamo and Maua 1993; Choge et al. 2002; Little 2019). The aggressive spread of P. juliflora has been documented (Choge et al. 2002; Mbaabu et al. 2019; M. L. Castillo et al., unpubl. data), while P. pallida has seemingly not become invasive (M. L. Castillo et al., unpubl. data). The overlapping morphological traits and native-range distributions of these two species (Burkart 1976; Díaz Celis 1995) have often led to misidentifications in both native and non-native areas, with some suggesting that they should be treated as a species complex (Pasiecznik et al. 2001). Intermediate morphotypes between P. juliflora and P. pallida have been observed in Kenya, i.e. putative hybrids (W. Okellu, CABI, unpubl. data). In Ethiopia and Tanzania, morphological identification of invasive trees remains unclear and studies only refer to the taxon as Prosopis or P. juliflora (Wakie et al. 2014; Kilawe et al. 2017; Shiferaw et al. 2019). Hybrids between P. juliflora and P. pallida are assumed to be absent or rare (Wakie et al. 2014; Kilawe et al. 2017; Shiferaw et al. 2019). In Ethiopia, Prosopis was first introduced in the early 1980s into the Afar Region, with additional introductions between the 1980s and 1990s (Admasu 2008; Kebede and Coppock 2015). Prosopis is now considered one of the country’s worst invasives. In Tanzania, Prosopis was thought to have been first introduced in 1953 to Mombo Arboretum and Tanga region (J. R. Mbwambo, Tanzania Forestry Research Institute, pers. comm.), with later introductions between 1988 and 1995 (Kilawe et al. 2017). Prosopis is considered to be at an early stage of invasion in Tanzania.
Studies at large biogeographic scales, including both native and non-native ranges, may provide valuable information about the genetic diversity and differentiation of invasive Prosopis species, the occurrence of hybridization, and may help clarify taxonomic uncertainties. In this study, we assessed the genetic diversity, differentiation and structure, and evaluated the occurrence of interspecific hybridization in native and non-native populations of several Prosopis species, with a special focus on non-native populations of P. juliflora and P. pallida in Eastern Africa (Ethiopia, Kenya and Tanzania). For the latter region, we also wanted to indirectly infer the introduction histories of both Prosopis species by comparing levels of genetic diversity and differentiation between native and Eastern African populations.
Materials and Methods
Sampling and DNA extraction
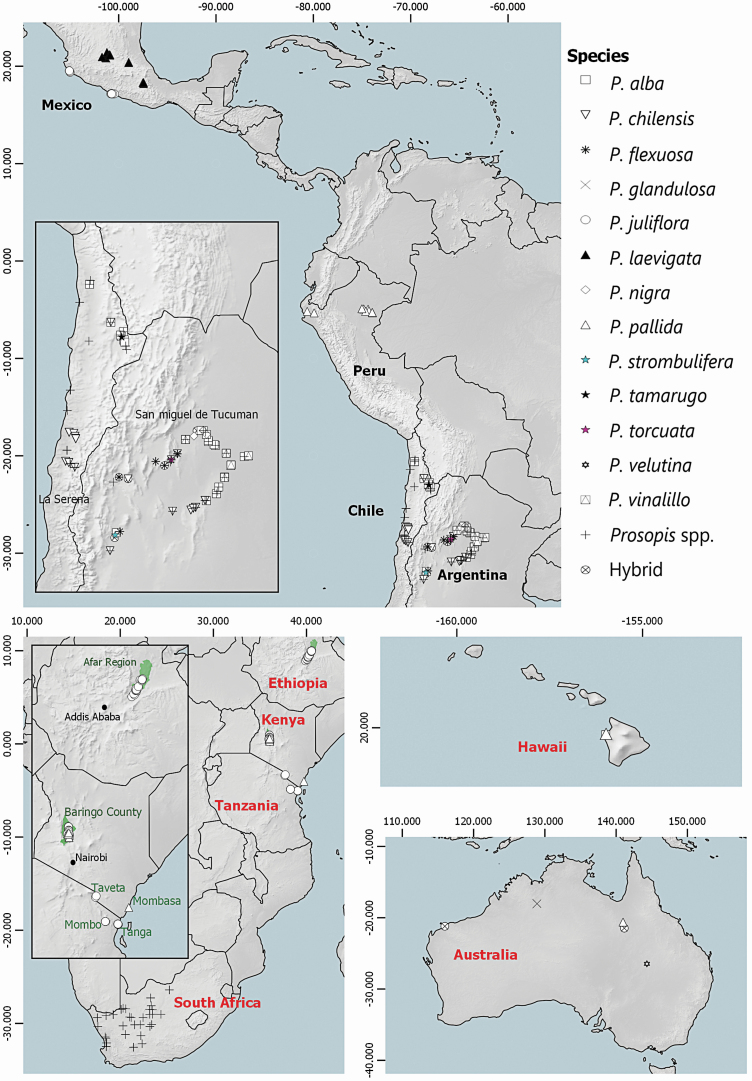
Leaf material of different Prosopis species was collected from various native and non-native areas worldwide in 2016 (Table 1; Fig. 1). For this, sampling in the native areas was done within the two known centres of diversification of the genus, the Argentine–Paraguayan–Chilean region and the Texan–Mexican region. We aimed to include a high number of species, rather than sampling comprehensively across the distributions of only a few species. From the native range, we sampled individuals of P. juliflora and P. laevigata from Mexico; P. pallida from Peru; Prosopis alba, Prosopis chilensis, Prosopis flexuosa, Prosopis nigra, Prosopis strombulifera, Prosopis torcuata and Prosopis vinalillo and putative hybrids from Argentina, as well as Prosopis tamarugo, P. chilensis and P. alba from Chile. A total of 67 sampled individuals from native areas in Argentina and Chile could not be morphologically identified to species level (hereafter referred to only as Prosopis spp.). The native ranges of sampled species are provided in Supporting Information—Appendix S1.
Table 1.
Prosopis species included in the study from various native, introduced and invasive populations. For each species, the number of individuals (N) sampled in each country, its status in each country (native, introduced or invasive) and the section/series where it belongs to, are shown. The series Chilenses, Pallidae and Ruscifoliae are part of the section Algarobia.
| Species | Country | N | Status | Section/series |
|---|---|---|---|---|
| P. alba | Argentina | 29 | Native | Series Chilenses |
| Chile | 11 | Native | ||
| P. chilensis | Argentina | 19 | Native | Series Chilenses |
| Chile | 25 | Native | ||
| P. flexuosa | Argentina | 9 | Native | Series Chilenses |
| P. glandulosa | Australia | 2 | Invasive | Series Chilenses |
| P. juliflora | Mexico | 20 | Native | Series Chilenses |
| Ethiopia | 202 | Invasive | ||
| Kenya | 470 | Invasive | ||
| Tanzania | 50 | Invasive | ||
| P. laevigata | Mexico | 25 | Native | Series Chilenses |
| P. nigra | Argentina | 6 | Native | Series Chilenses |
| P. pallida | Peru | 14 | Native | Series Pallidae |
| Australia | 2 | Invasive | ||
| Hawaii | 15 | Invasive | ||
| Kenya | 57 | Introduced | ||
| P. strombulifera | Argentina | 1 | Native | Section Strombocarpa |
| P. tamarugo | Chile | 1 | Native | Section Strombocarpa |
| P. torquata | Argentina | 1 | Native | Section Strombocarpa |
| P. velutina | Australia | 1 | Invasive | Series Chilenses |
| P. vinalillo | Argentina | 7 | Native | Series Ruscifoliae |
| P. alba × P. chilensis | Argentina | 2 | Native | |
| P. alba × P. nigra | Argentina | 3 | Native | |
| P. alba × P. rustifolia | Argentina | 1 | Native | |
| P. alba × P. vinalillo | Argentina | 1 | Native | |
| P. chilensis × P. flexuosa | Argentina | 4 | Native | |
| Hybrids | Australia | 4 | Invasive | |
| Prosopis spp. | South Africa | 58 | Invasive | |
| Prosopis spp. | Argentina | 35 | Native | |
| Prosopis spp. | Chile | 32 | Native | |
| Total | 1107 |
Figure 1.
Sampling sites of various Prosopis taxa and putative hybrids from native (black labels) and non-native areas (red labels). Inset maps indicate sampling sites in Argentina, Chile, Ethiopia, Kenya and Tanzania.
From non-native areas, we sampled individuals of P. glandulosa, P. pallida, P. velutina and putative hybrids from Australia, P. pallida from Hawaii and various Prosopis species and putative hybrids from South Africa that could not be identified to species level (hereafter referred to only as Prosopis spp.). From Eastern Africa, we included areas where P. juliflora and P. pallida individuals were first introduced; we sampled P. juliflora from the Afar Region, Ethiopia, P. juliflora and P. pallida from Baringo County, Mombasa and Taveta, Kenya, and P. juliflora from Mombo and Tanga arboreta, Tanzania. All sampled species belong to the sections Algarobia and Strombocarpa, and from the section Algarobia, the species belong to the series Chilenses, Pallidae and Ruscifoliae (Table 1). One to 35 sampling sites were included per country for each species and 1–474 adult trees were sampled per species per country (Table 1; Fig. 1; ntotal = 1107 individuals). This uneven sampling reflects the availability of individuals at each location, i.e. areas where only one tree was located versus areas with dense invasive populations. We sampled trees that were separated by at least 30 m to avoid collecting genetically related material (Vilardi et al. 1988). In the case of Kenya, Ethiopia and Tanzania, some sampled trees were separated by less than 30 m. Throughout this manuscript we use the term ‘population’ to refer to a group of individuals of the same species that were sampled in a specific native or non-native area in each country. Leaf material was air-dried and stored on silica gel until further use for DNA extraction. Because P. juliflora is the only polyploid member of the genus (2n = 4x), we performed flow cytometry analysis on a subset of P. juliflora individuals (n = 75). Further details on DNA extraction, morphological classification of individuals and flow cytometry analyses are provided in Supporting Information—Appendix S1.
Nuclear and chloroplast DNA sequencing
To assess evolutionary history (i.e. phylogeny) of our study species, we optimized and sequenced the nuclear external transcribed spacer region (ETS) and two chloroplast intergenic spacers (rpl32-trnL and psbA-trnH) for 12 Prosopis individuals initially to check for genetic variability at these gene regions prior to sequencing a more representative sample of individuals. ETS and rpl32-trnL have previously been successfully employed at the intraspecific level for Leguminosae species (e.g. Australian Acacia species; Le Roux et al. 2011) and the psbA-trnH marker is generally highly variable across angiosperms (Shaw et al. 2007). Details of the Prosopis individuals selected for initial screening, primers used for PCR amplification are provided in Supporting Information—Appendix S1. DNA sequence data gene were aligned and edited for each gene region separately using BioEdit version 7.0.5.3 (Hall 1999).
Microsatellite genotyping
We selected 11 nuclear microsatellite markers considering their levels of polymorphism across different Prosopis species, functional annotations in some instances and similar annealing temperatures [see Supporting Information—Table S1]. Details of marker amplification and genotyping are provided in Supporting Information—Table S2 and Appendix S1. From these markers, the following four loci were excluded from subsequent analyses: I-P00930c as it was monomorphic; I-P07653, GL23 and Prb8 as these showed extensive patterns of non-specific binding in numerous samples. Samples that failed to amplify at more than five loci were removed from all subsequent analyses (including the single P. tamarugo individual), leaving a total of 1072 individuals. Individuals not identified as P. juliflora and that had more than two alleles at least one locus were excluded from subsequent analyses because their ploidy could not be reliably determined (n = 14; see Supporting Information—Table S3).
Genetic diversity and differentiation between native and non-native populations
Departures from Hardy–Weinberg equilibrium (HWE) were tested for all loci for all diploid species (i.e. excluding P. juliflora) using the R packages adegenet version 2.0.1 (Jombart 2008) and pegas version 0.11 (Paradis 2010) and significance was tested using a permutation test (10 000 permutations). We calculated various statistics of genetic diversity separately for all native and non-native populations of various Prosopis species, putative hybrids from Australia and Prosopis spp. individuals from South Africa. For these analyses, all putative hybrids from Argentina were analyzed as a single taxon, and species for which we were only able to collect one or two individuals at a particular location were not included in the analysis. A total of 18 populations were included in the analyses. In addition, to evaluate whether genetic diversity differs between native and non-native populations of Prosopis, we grouped all native-range individuals of all Prosopis species, putative hybrids and Prosopis spp. individuals, referred to hereafter as ‘Native Prosopis’. Separately, we grouped all non-native-range individuals (i.e. introduced and invasive) of all Prosopis species, putative hybrids and Prosopis spp. individuals, referred to hereafter as ‘Non-native Prosopis’. Then, we estimated genetic diversity indexes separately for each group and compared them. To assess levels of genetic diversity, numbers of alleles per locus, observed heterozygosity (HO), expected heterozygosity (HE) and inbreeding coefficients (FIS) were estimated using the SPAGeDi version 1.5 software for polyploid P. julifora (Hardy and Vekemans 2002). For all diploid individuals, HO and HE were estimated with the software GenoDive version 3.0 (Meirmans 2020), and numbers of alleles per locus and FIS values were estimated with the diveRsity R package version 1.9.90 (Keenan et al. 2013). Since there is a positive correlation between population size and HE (Nybom 2004), GenoDive and SPAGeDi analyses included corrections for sample sizes for HE calculations. Lastly, allelic richness (AR) and the number of private alleles were calculated with the software ADZE version 1.0 (Szpiech et al. 2008). ADZE uses a rarefaction approach to calculate sample size-corrected estimates for these metrics. The number of individuals included in these analyses per population is reported in Table 2.
Table 2.
Population genetic diversity indices for native, and non-native (introduced and invasive) populations of various Prosopis species, putative hybrids and Prosopis spp. individuals from South Africa. Native Prosopis and non-native Prosopis groups (i.e. all native and non-native Prosopis individuals, respectively) were analysed as well. Statistics were calculated as mean values of each index over the seven loci analysed. N = number of samples; HE = expected heterozygosity expected; HO = observed heterozygosity observed; FIS = inbreeding coefficient.
| Species | Country | Category | N | H E | H O | F IS |
|---|---|---|---|---|---|---|
| P. alba | Argentina | Native | 28 | 0.71 | 0.65 | 0.05 |
| P. alba | Chile | Native | 11 | 0.68 | 0.49 | 0.22 |
| P. alba | All | 39 | 0.70 | 0.57 | 0.13 | |
| P. chilensis | Argentina | Native | 9 | 0.69 | 0.57 | 0.09 |
| P. chilensis | Chile | Native | 24 | 0.66 | 0.55 | 0.15 |
| P. chilensis | All | 43 | 0.68 | 0.56 | 0.16 | |
| P. flexuosa | Argentina | Native | 8 | 0.70 | 0.60 | 0.02 |
| P. juliflora | Mexico | Native | 20 | 0.31 | 0.44 | -0.20 |
| P. juliflora | Ethiopia | Invasive | 200 | 0.35 | 0.42 | 0.08 |
| P. juliflora | Kenya | Invasive | 457 | 0.42 | 0.46 | 0.18 |
| P. juliflora | Tanzania | Invasive | 46 | 0.28 | 0.29 | 0.10 |
| P. juliflora | All | 723 | 0.41 | 0.44 | 0.19 | |
| P. laevigata | Mexico | Native | 24 | 0.48 | 0.47 | 0.33 |
| P. nigra | Argentina | Native | 6 | 0.57 | 0.47 | 0.01 |
| P. pallida | Peru | Native | 12 | 0.42 | 0.30 | 0.30 |
| P. pallida | Hawaii | Invasive | 14 | 0.29 | 0.20 | 0.24 |
| P. pallida | Kenya | Introduced | 57 | 0.39 | 0.28 | 0.21 |
| P. pallida | All | 83 | 0.37 | 0.26 | 0.22 | |
| P. vinalillo | Argentina | Native | 7 | 0.68 | 0.58 | 0.09 |
| Hybrids | Argentina | Native | 10 | 0.71 | 0.68 | -0.08 |
| Hybrids | Australia | Invasive | 3 | 0.66 | 0.33 | 0.41 |
| Prosopis spp. | South Africa | Invasive | 48 | 0.69 | 0.58 | 0.14 |
| Native Prosopis | 229 | 0.62 | 0.49 | 0.26 | ||
| Non-native Prosopis | 829 | 0.45 | 0.36 | 0.32 |
We also estimated genetic differentiation between native and non-native populations of various Prosopis species, putative hybrids from Argentina and Australia, and Prosopis spp. individuals from South Africa as described above (i.e. 18 populations and the same number of individuals per population as detailed in Table 2). For P. juliflora, a matrix of pairwise genetic distances (FST) was calculated using the R package PolySat with 95 % confidence intervals calculated via bootstrapping across loci. For diploid individuals, pairwise FST values were calculated following Weir (1996). For this, the FreeNA software (Chapuis and Estoup 2007) was used to calculate corrected and uncorrected FST estimates since it applies an ‘excluding null alleles’ (ENA) correction to account for the presence of null alleles. The 95 % confidence intervals for FST values were obtained by 10 000 simulations. FST estimates depend on within-population genetic diversity and therefore, on sample sizes (Meirmans and Hedrick 2011). Therefore, we also calculated pairwise G″ST estimates, which includes a correction for sampling bias, using the GenoDive software (Meirmans and Hedrick 2011). In addition, a hierarchical analysis of molecular variance (AMOVA) was performed including native and non-native P. juliflora and P. pallida populations of Ethiopia, Kenya and Tanzania and using the pegas R package (Paradis 2010). For P. juliflora, a matrix of pairwise distances between individuals was generated using Bruvo distances (Bruvo et al. 2004), while for P. pallida, Euclidian distance based on the allele frequencies was used to generate pairwise distances between individuals.
Genetic structure and hybridization
To identify the number of genetic clusters present in the overall data set, Bayesian assignment tests were used as implemented in the software STRUCTURE version 2.3.4 (Pritchard et al. 2000). A hierarchical clustering approach (Le Roux et al. 2010) was applied including native and non-native populations of all investigated Prosopis species, putative hybrids and Prosopis spp. individuals. Details of model parameters and settings are provided in Supporting Information—Appendix S1.
Principal component analyses (PCAs) were also performed. A first PCA included native and non-native populations of all Prosopis species, their putative hybrids and Prosopis spp. individuals. We used the PolySat R package (Clark and Jasieniuk 2011) to generate a matrix of pairwise distances between individuals using Bruvo distances since this method can incorporate distances between microsatellite alleles without information on allele copy number (Bruvo et al. 2004). In a second ‘diploid-only’ PCA (i.e. excluding P. juliflora individuals), we generated a matrix of Euclidian distances between individuals considering allele frequencies.
We tested the morphological assignment of diploid individuals to pure species and putative hybrids using microsatellite data and the NewHybrids version 1.1beta software (Anderson and Thompson 2002). This software identifies six genotype classes (i.e. pure species 1, pure species 2, F1 hybrids, F2 hybrids, species 1 backcrosses and species 2 backcrosses) without information on the allele frequency of the parental species. The program provides probabilities of an individual belonging to any of the genotype classes and therefore how well our a priori morphological assignment of individuals aligned with the genetic data. An analysis was done between all possible pairs of species from Argentina: P. alba, P. chilensis, P. flexuosa, P. nigra, P. vinalillo and their putative hybrids. In the case of Chile, the analysis was done between P. alba and P. chilensis individuals. A last analysis was done between Peruvian P. pallida individuals and Hawaiian and Kenyan P. pallida individuals. A burn-in period of 30 000 generations and 50 000 MCMC iterations was used. We used ‘Jeffrey’s like priors’ and a posterior probability of 0.8 was used to assign individuals to the six genotype classes. Individuals that could not be assigned to genotype classes were considered of ‘mixed’ ancestry.
Results
Nuclear and chloroplast DNA sequencing
DNA sequencing data for the ETS, psbA-trnH and rpl32-trnL gene regions indicate extremely low sequence variability across our initial subset of Prosopis species from native and non-native populations. The exception was P. tamarugo, where we found 15 substitutions for the psbA-trnH region, 17 substitutions for the rpl32-trnL region and ~240 substitutions for the ETS region. When excluding P. tamarugo, we found only one substitution in the psbA-trnH region between P. pallida from Peru and all other species, and one substitution in the rpl32-trnL region between P. nigra and P. flexuosa and all other species. For the ETS region, we found only four substitutions between P. juliflora from Kenya and all other studied species. Prosopis spp. from South Africa differed by three substitutions with all other species, while P. glandulosa and the putative hybrids from Australia, and P. pallida from Peru had one substitution each when compared with the rest of the studied species. Given this low differentiation between species in the Algarobia section we did not sequence additional individuals.
Genetic diversity and differentiation between native and non-native populations
For the microsatellite data and for diploid Prosopis species, 21 loci for each species by country combination (27.3 %) did not meet HWE expectations. All seven loci were polymorphic in the overall data set. Four markers were not polymorphic for some native and non-native populations of Prosopis species and putative hybrids from Australia [see Supporting Information—Table S4]. The average number of alleles per locus was 17.4 (range 6–33 alleles).
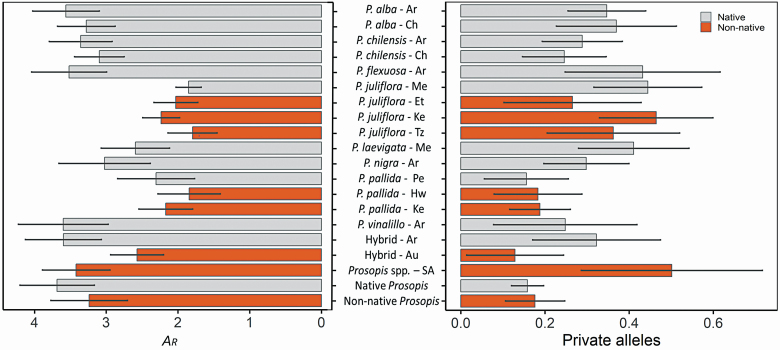
Overall, native populations of pure Prosopis species, putative hybrids from Argentina and Prosopis spp. individuals from South Africa had higher numbers of alleles per locus, levels of AR, HE and HO than native and non-native populations of P. juliflora and P. pallida and Australian putative hybrids (Table 2; Fig. 2). Inbreeding coefficients (FIS) were high in most native and non-native populations of Prosopis species. The number of private alleles was similar among native and non-native populations of Prosopis species and putative hybrids from Argentina (Table 2).
Figure 2.
Allelic richness (AR) and number of private alleles. (±1 SE) for native and non-native populations of various Prosopis species, putative hybrids and Prosopis spp. individuals from South Africa. Native Prosopis and non-native Prosopis groups (i.e. all native and non-native Prosopis individuals, respectively) were analyzed as well. Country codes are: Argentina (Ar), Australia (Au), Chile (Ch), Ethiopia (Et), Hawaii (Hw), Kenya (Ke), Mexico (Me), South Africa (SA) and Tanzania (Tz).
In the case of P. juliflora, native Mexican populations had lower HE compared to invasive population from Kenyan, but higher HE and HO than invasive Tanzanian populations. Native and non-native populations of P. pallida had similar levels of AR. In the case of P. pallida, native Peruvian populations had similar HE and HO than introduced Kenyan populations. Invasive Hawaiian populations had lower HE and HO compared to native and other non-native populations of the species. Lastly, native Prosopis populations (i.e. Native Prosopis) had higher levels of AR, HE and HO than the non-native populations (i.e. non-native Prosopis), but lower levels of FIS. The number of private alleles was similar between these groups (Fig. 2).
When estimating genetic differentiation, similar results were obtained with uncorrected and ENA-corrected pairwise FST estimates (Kruskal–Wallis chi-square = 0.03, P = 0.85); therefore, uncorrected pairwise FST values with 95 % confidence intervals are presented [see Supporting Information—Tables S5 and S6]. Overall, we found low genetic differentiation between some Prosopis species and hybrids in spite of their allopatric distributions. Levels of differentiation based on pairwise G″ST estimates were similarly low [see Supporting Information—Table S7]. For example, genetic distances (i.e. pairwise FST and G″ST values) between some sympatric Prosopis species from Chile and Argentina were similar to those between these species and P. laevigata from Mexico, Prosopis spp. individuals from South Africa and putative Australian hybrids.
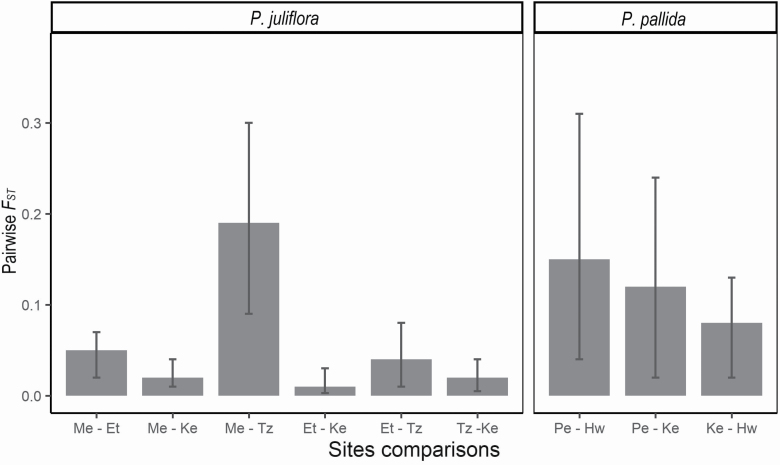
Regarding P. juliflora, FST-based genetic differentiation between invasive Tanzanian and native Mexican populations was higher than between the latter and invasive populations from Kenya and Ethiopia, and was also higher than the differentiation between invasive Kenyan and Ethiopian populations (Fig. 3). These results were also supported by pairwise G″ST estimates (see Supporting Information—Table S7). The hierarchical AMOVA indicated considerable, but not significant, genetic variation between native and non-native P. juliflora populations (71.36 %), while significant, and similar, genetic variation was found among invasive populations (12.32 %) and within invasive populations (16.32%; Table 3). In the case of P. pallida, levels of genetic differentiation (based on FST and G″ST) were similar between native Peruvian populations and non-native (both introduced and invasive) populations from Hawaii and Kenya (Fig. 3; see Supporting Information—Table S7). There was also some, but not significant, genetic variation between native and non-native populations of P. pallida (37.29 %), while the genetic variation between native, introduced and invasive populations (28.66 %) was significant and slightly lower than the variation within populations (34.05 %; Table 3).
Figure 3.
Pairwise FST (± 95 % confidence interval) between native Mexican (Me) and invasive populations of P. juliflora in Ethiopia (Et), Kenya (Ke) and Tanzania (Tz); between invasive populations of P. juliflora; between native populations from Peru (Pe) and invasive populations from Hawaii (Hw) and introduced populations from Kenya (Ke) and between Ke and Hw populations of P. pallida.
Table 3.
Hierarchical AMOVA partitioning of genetic variation for various native, introduced and invasive populations of P. juliflora and P. pallida. * Significant fixation indices, tested using 10 000 random permutations. d.f. = degrees of freedom.
| Source of variation | d.f. | Sum of squares | Variance | Percent variation (%) | Fixation index |
|---|---|---|---|---|---|
| P. juliflora | |||||
| Native versus non-native populations | 1 | 1.28 | 170.02 | 71.36 | 0.45 |
| Among native and invasive populations | 2 | 1.58 | 29.36 | 12.32 | 0.13* |
| Within populations | 710 | 21.87 | 38.88 | 16.32 | 0.52 |
| P. pallida | |||||
| Native versus non-native populations | 1 | 37.71 | 22.48 | 37.29 | 0.18 |
| Among native, introduced and invasive populations | 1 | 13.53 | 17.28 | 28.66 | 0.23* |
| Within populations | 80 | 432.18 | 20.53 | 34.05 | 0.06 |
Genetic structure and hybridization
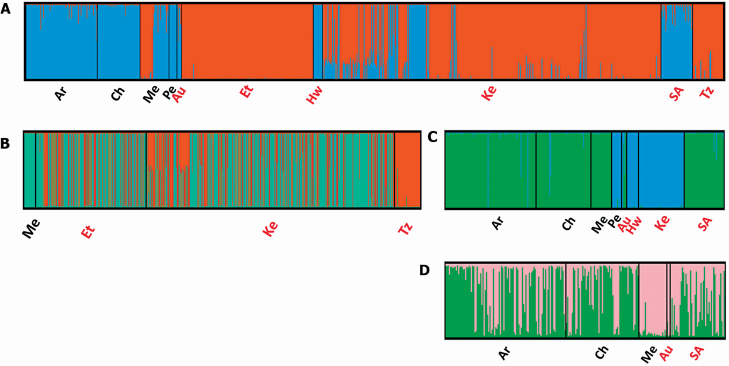
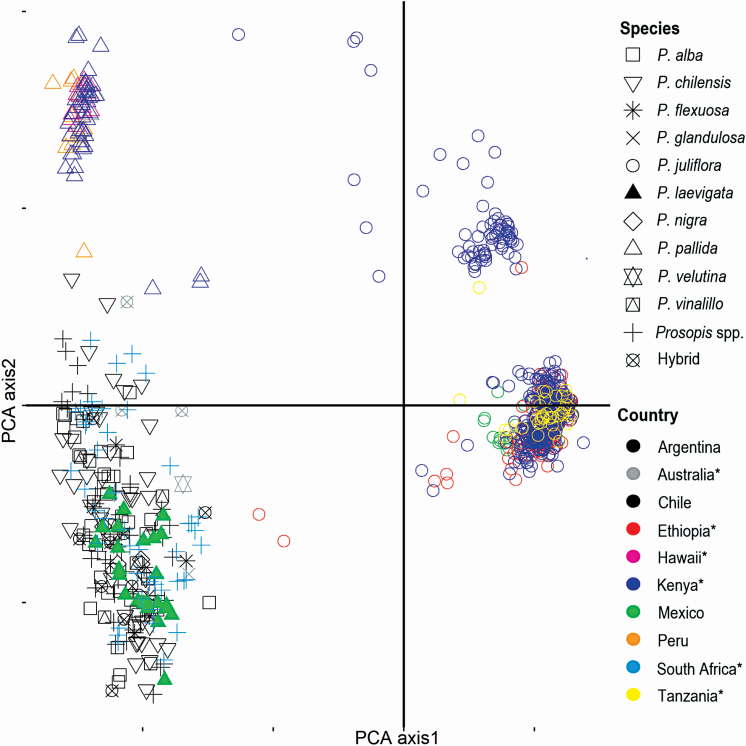
Both Bayesian assignment tests and PCAs indicated that overall genetic structure largely reflected ploidal variation, with polyploid P. juliflora being highly differentiated from the rest of the diploid Prosopis species included here. A second level of hierarchical structure largely reflected series-level relationships, showing genetic differentiation between P. pallida and the remaining diploid species, while there was not a clear genetic structure among Prosopis species from Argentina, Australia, Chile, Mexico and Prosopis spp. from South Africa. Interestingly, both analyses confirmed the presence of P. juliflora in Ethiopia, Kenya and Tanzania, and also identified some admixed individuals, i.e. hybrids, in Kenya. Lastly, STRUCTURE, but not PCA, showed low genetic differentiation between native Mexican P. juliflora populations and invasive populations from Ethiopia and Kenya (Figs 4 and 5; see Supporting Information—Figs S1 and S2).
Figure 4.
Hierarchical Bayesian clustering analyses of individuals of native (black labels) and non-native (red labels) populations of various Prosopis species and putative hybrids: Argentina (Ar) = P. alba, P. chilensis, P. flexuosa, P. strombulifera, P. nigra, P. torcuata and P. vinalillo, putative hybrids and Prosopis spp. individuals; Chile (Ch) = P. alba, P. chilensis, and Prosopis spp. individuals; Mexico (Me) = P. juliflora and P. laevigata; Peru (Pe) = P. pallida; Australia (Au) = P. glandulosa, P. pallida, P. velutina and putative hybrids; Ethiopia (Et) = P. juliflora; Hawaii (Hw) = P. pallida; Kenya (Ke) = P. juliflora and P. pallida; South Africa (SA) = Prosopis spp. individuals; Tanzania (Tz) = P. juliflora. Individuals were genotyped using seven nuclear microsatellite loci and clustered at three levels. (A) Level 1: ‘P. juliflora’ cluster in orange and ‘other Prosopis species’ cluster in blue; (B) Level 2: only P. juliflora individuals and (C) Level 2: ‘P. pallida’ cluster in blue ‘other Prosopis species’ cluster in green; (D) Level 3: individuals of ‘other Prosopis species’ cluster from Argentina, Chile, Mexico, Australia and South Africa. Vertical axes represent the assignment (qik values) of individual genomes to the inferred number of genetic clusters, in all cases K = 2 [see Supporting Information—Fig. S1].
Figure 5.
Principal component analysis (PCA) showing genetic structure among native and non-native populations of different Prosopis taxa and their putative hybrids. Countries from which non-native populations originated are indicated by asterisks (*). PCA was performed using Bruvo distances calculated in PolySat (Bruvo et al. 2004). PCA 1 and PCA 2 captured 63.6 % and 11.0 % of the variation, respectively.
Assignment tests in NewHybrids were done between pairs of species per site [see Supporting Information—Fig. S3]. These analyses were able to identify only three genotype classes: pure parental species and their hybrids. For most comparisons including species from Argentina and Chile, individuals morphologically identified as one of the two species, aligned with the genetic data and were assigned as pure genotypes of the same species between 57.1–100 % of assignments. In contrast, individuals were also assigned as pure genotypes of the other species (3.7–21.1 % of assignments); or as having mixed ancestry (5.3–42.9 % of assignments). Only when including pairs of the species P. flexuosa–P. nigra and P. flexuosa–P. vinalillo from Argentina, were the models unable to assign individuals to any class. Interestingly, for two of the three putative Argentinean hybrids, morphological identification did not align well with the genotype classification. That is, morphological hybrids were genetically mainly classified as being one of the pure parental species (66.7 %). Lastly, an analysis between P. pallida individuals from Peru and P. pallida individuals from Hawaii and Kenya classified all Peruvian individuals as pure parental genotypes. For P. pallida from Hawaii, half of the individuals represented pure genotypes that differ from Peruvian genotypes, few of them were classified as pure P. pallida genotypes from Peru (7.14 % of assignments) and the rest were found to have mixed ancestry (42.9 %). In contrast, almost all P. pallida individuals from Kenya were classified as pure Peruvian P. pallida genotypes (92.0 %) and a few as having mixed ancestry (8.0 %).
Discussion
While numerous studies have reported on the genetic relationships among Prosopis species and population-level genetic variation (e.g. Ramírez et al. 1999; Saidman et al. 2000; Bessega et al. 2006; Catalano et al. 2008; Moncada et al. 2019; Aguilar et al. 2020), ours is the first to provide genetic insights on the taxonomic uncertainty of non-native Prosopis species. We found low genetic differentiation between most diploid Prosopis species, suggesting that hybridization between previously allopatric species may occur frequently when they are co-introduced into new ranges. Polyploid individuals were detected in both native and non-native areas, with tetraploid P. juliflora being highly differentiated from the rest of the diploid species in the genus. Levels of genetic diversity suggest that invasive populations in Eastern Africa (Kenya and Ethiopia) resulted from multiple introductions of both P. juliflora and P. pallida. While hybridization is thought to promote invasiveness of Prosopis in countries like Australia and South Africa, this seems not be the case in Eastern Africa. Here polyploidy appears to benefit invasion success.
Uncertain taxonomy of diploid Prosopis taxa
The taxonomy of Prosopis has been much debated (Saidman and Vilardi 1987; Saidman et al. 2000; Pasiecznik et al. 2001). Low genetic variability among diploid taxa has been postulated to blur species boundaries, with some authors considering Algarobia species to constitute a so-called ‘syngameon’, i.e. a hybrid swarm (Palacios and Bravo 1981). Pre-zygotic reproductive barriers (e.g. differences in phenology or the use of different pollinators) are thought to be weak in Prosopis, while post-zygotic reproductive barriers (i.e. pollen inviability) may be more important (Palacios and Bravo 1981; Naranjo et al. 1984). Our DNA sequencing data indicated that many Algarobia species shared almost 100 % genetic similarity. These results suggest a recent radiation of these species and possibly incomplete reproductive isolation between them (also see Catalano et al. 2008). This may lead to frequent hybridization and introgression between species in this section (Hunziker et al. 1986), especially when they are co-introduced into new ranges (e.g. van Klinken et al. 2006; Mazibuko 2012). Only P. juliflora was found to be highly differentiated from the rest of Algarobia species included in our analyses.
We found genetic diversity in Prosopis populations to be high and similar among species and native and non-native regions, with the exception of P. pallida and P. juliflora (also see Juárez-Muñoz et al. 2006; Sherry et al. 2011). We also identified Prosopis individuals that had more than two alleles at some loci in both native and non-native areas. These individuals were not initially classified as tetraploid P. julifora based on morphology, but rather as hybrids from Australia, P. flexuosa from Argentina, P. laevigata from Mexico and individuals from South Africa that could not be identified to species level but are presumed to be hybrids. While polyploidy has been reported in Prosopis (Burkart 1976; Hunziker et al. 1986; Fontana et al. 2018), Trenchard et al. (2008) proposed that P. juliflora is the only polyploid species in the genus. Ploidal variation is an important mechanism that underlies reproductive isolation, and thus could be promoting genetic differentiation between P. juliflora and its congeners.
Taxonomic uncertainty in Prosopis was further illustrated by our genetic analysis of hybridization. We found consistent disagreement between taxonomic classification of Prosopis species based on morphological versus on genetic data. One possible explanation for this is that hybridization, followed by extensive backcrossing, can lead to individuals expressing the morphological traits of one parental species while retaining genetic information of the other (e.g. see Gaskin and Kazmer 2009; Boswell et al. 2016). We also used leaf morphological traits (see Burkart 1976) to classify our species; however, these can be highly plastic (Bessega et al. 2006, 2009; Verga et al. 2009). Future research should focus on identifying diagnostic traits, and their heritability, for different Prosopis taxa.
Our study also provides clarity on the identity of Prosopis species in Eastern Africa. Firstly, our genetic results confirmed the presence of P. juliflora in Ethiopia, Kenya and Tanzania. Our results using native and non-native genetic material, together with previous work from the native range (Catalano et al. 2008; Palacios et al. 2012), also indicate important genetic differences between P. pallida and P. juliflora, confirming that they are indeed distinct taxa. Secondly, we identified a few instances of hybridization between these two species. These hybrids are likely to be triploid and sterile and therefore unlikely to increase invasiveness.
Prosopis invasion in Eastern Africa
Our study provides the first genetic analysis of the origins of P. juliflora and P. pallida in Eastern Africa. Importantly, in Kenya, Ethiopia and Tanzania, we included comprehensive sampling from areas where P. juliflora and P. pallida individuals were first introduced, and became invasive, in the case of P. juliflora. We found that genetic material of P. juliflora appears to be similar for most Kenyan and Ethiopian individuals, and closely related to native Mexican ones. These results suggest that invasive Kenyan and Ethiopian genotypes could have a similar Mexican origin. In the case of P. pallida in Kenya, individuals were genetically similar to Peruvian individuals, indicating a South American origin. Additionally, similar levels of genetic diversity were observed between Mexican P. juliflora and invasive Ethiopian populations and Peruvian P. pallida and introduced Kenyan individuals. In contrast, invasive Kenyan individuals of P. juliflora had higher heterozygosity than native individuals from Mexico. It is surprising that is not the case in Ethiopia given similar introduction histories shared by these two countries. Similar or higher levels of genetic diversity in native and non-native populations may be indicative of multiple introductions or it may simply reflect a unique introduction from a source generated by admixture of multiple populations (Le Roux et al. 2011). Higher levels of genetic diversity than native individuals can also occur due to cultivation, and can generate genetic novelties (Thompson et al. 2012). Compared to traditional statements that multiple introductions characterize the introduction of Prosopis species to non-native areas globally (Pasiecznik et al. 2001), our study is the first to provide support to this hypothesis for P. juliflora and P. pallida in Kenya and Ethiopia.
For Tanzania, the origin and identity of Prosopis is more complicated and the available evidence limited. The source(s) and species identity of Prosopis individuals originally introduced to two arboreta in the 1960s remains unknown and have been speculated to include P. juliflora from other non-native regions like India, Israel and/or South Africa (C. J. Kilawe and J. R. Mbwambo, Tanzania Forestry Research Institute, pers. comm.). However, it is thought that P. chilensis and P. pallida have also been introduced to Tanzania (C. J. Kilawe, Sokoine University of Agriculture, Tanzania, and J. R. Mbwambo, Tanzania Forestry Research Institute, pers. comm.). Our genetic analyses showed that the trees we collected from two arboreta were P. juliflora, similar to some genotypes from Ethiopia and Kenya but not closely related to native Mexican ones (Fig. 4B). In other areas of Tanzania, not included in this study, where Prosopis is invasive (i.e. Kahe, Mwanga and Simajiro), repeated introductions would have been made from Taita Taveta (C. J. Kilawe, Sokoine University of Agriculture, Tanzania, pers. comm.). It is therefore likely that these invasive populations are also P. juliflora since trees collected in Taita Taveta were identified as this species and most individuals were assigned to the same genetic cluster than Tanzanian individuals (results not shown). Therefore, our genetic results showed that, unlike in Kenya and Ethiopia, Tanzanian P. juliflora genotypes in arboreta are not closely related to the Mexican individuals, supporting the notion of additional and unknown sources for Tanzanian plantings.
Hybridization, polyploidy and invasiveness in Prosopis
In agreement with previous studies, we identified instances of hybridization between Prosopis species in both native (e.g. see Saidman et al. 2000) and non-native ranges (e.g. see Zimmermann 1991; van Klinken et al. 2006; Mazibuko 2012; Muturi 2012). The success of many plant invasions has been attributed to hybridization (Schierenbeck and Ellstrand 2009; Zalapa et al. 2010; Gaskin et al. 2012) and this may also be the case for some Prosopis invasions such as those in Australia and South Africa (van Klinken et al. 2006; Mazibuko 2012), but not in Eastern Africa. In the native range, hybridization between Prosopis species seems to be promoted by certain environmental conditions (Vega and Hernández 2005), with hybrids frequently found in disturbed areas (Verga 2005). Considering this, interspecific hybridization between Prosopis species in the invaded range may not only be dependent on the genetic relatedness of species, but also on whether certain habitat features facilitate co-occurrence of, and interbreeding between, them. It may also be that only certain Prosopis genotypes, or hybrid combinations, are successful under particular environmental conditions, or that only hybrid genotypes are able to spread extensively in new environments. In Australia, P. pallida occurs widely in the north of the country, from the east coast of Queensland through the Northern territory, to the west coast of Western Australia (van Klinken and Campbell 2001; CRC Weed Management Guide 2003; van Klinken 2012). However, this species is not found in the cooler southern states of Australia, where P. velutina and hybrids between this species and P. glandulosa var. torreyana seem to dominate (van Klinken and Campbell 2001; CRC Weed Management Guide 2003; van Klinken 2012). While these biogeographic patterns may reflect the initial introduction of only certain species to certain areas (van Klinken and Campbell 2001), they may also be indicative of variation in soil or climate preferences of these species and their hybrids.
Our results also show that polyploidization facilitates immediate reproductive isolation between Prosopis species. Polyploidy often also leads to higher levels of stress tolerance, growth vigour through increased plant size, seed size, flower size, niche breadth and phenotypic plasticity, among others, traits that will benefit invasive species (for a review, see te Beest et al. 2012). This may well explain why only tetraploid P. juliflora, and not diploid P. pallida, became invasive in Eastern Africa, despite the similar introduction histories of the two species to the region.
Our findings may also have implications for the management of Prosopis invasions. For example, the fact that P. juliflora is genetically highly differentiated from other Prosopis species raises the question whether biological control agents that have been tested against (mostly diploid) invasive Prosopis species in Australia and South Africa could perform differently on invasive P. juliflora in Eastern Africa. Moreover, hybridization between Prosopis species may also reduce the likelihood of finding effective biological control agents against any particular taxon (Goolsby et al. 2006). Lastly, given extensive hybridization between Prosopis species, it may be prudent to treat diploid species from the Americas as a single unit when developing regulations to govern management, and not to regulate individual species. This not only has obvious management advantages, but also circumvents potential legal challenges to such regulations (e.g. see Little 2019). However, even under such a classification scheme we think that future research should still aim to determine whether different taxa and their hybrids differ in invasiveness and their responses to different management practices.
Supporting Information
The following additional information is available in the online version of this article—
Appendix S1. Supporting Information—materials and methods.
Table S1. Details of the 51 microsatellites loci tested for amplification.
Table S2. Volume of the 11 microsatellites primers included in one multiplex PCR assay.
Table S3. List of Prosopis individuals, from native and non-native populations, that presented more than two alleles in at least one locus.
Table S4. Number of alleles per microsatellites locus for native and non-native populations of different Prosopis species, putative hybrids and Prosopis spp. individuals.
Table S5. Pairwise FST values calculated for various native and non-native populations of Prosopis species, putative hybrids and Prosopis spp. individuals.
Table S6. 95 % confidence interval of pairwise FST values (calculated on bootstrap resampling over loci) for various native and non-native populations of Prosopis species, putative hybrids and Prosopis spp. individuals.
Table S7. Pairwise G’’ST values calculated for various native and non-native populations of Prosopis species, putative hybrids and Prosopis spp. individuals.
Figure S1. Identification of the optimal number clusters (K) inferred by an hierarchical Bayesian clustering analyses with the software STRUCTURE.
Figure S2. Percentage of individuals assigned to genotype classes by NewHybrids software using seven nuclear microsatellites loci.
Data Availability
The genotype data generated in the study are available on the DRYAD online repository (doi:10.5061/dryad.zgmsbcc97).
Acknowledgements
We thank M. Sedutla, J. Foster, A. Malan, J. Sergon, P. Coech, L. Campos, R. Zuñiga, C. Basualto-Castillo, S. Choge, Mr. Mburu, A. Ramírez-Martínez, E. Chimal-Sánchez, E. Blancas, F. Pérez-Baleón, and staff of the Kenya Forestry Research Institute for assistance; A. Verga from the Instituto de Fisiología y Recursos Genéticos Vegetales (IFRGV), CIAP, INTA, Argentina, for species identification; C. Kilawe from Sokoine University of Agriculture, Tanzania, J. R. Mbwambo from Tanzania Forestry Research Institute, G. C. Alcedo from Universidad de Piura, Peru and J. Delatorre-Herrera from Universidad Arturo Prat, Chile, for providing Prosopis samples. We also thank the editor and the two anonymous reviewers whose suggestions greatly helped to clarify and improve this manuscript.
Populations & Communities. Chief Editor: Jean Burns
Sources of Funding
Funding for this study was provided by the Swiss Programme for Research on Global Issues for Development (r4d), funded by the Swiss National Science Foundation (SNSF) and the Swiss Agency for Development and Cooperation (SDC), for the project ‘Woody invasive alien species in East Africa: Assessing and mitigating their negative impact on ecosystem services and rural livelihood’ (grant number: 400440_152085). U.S. was supported by CABI with core financial support from its member countries (see http://www.cabi.org/about-cabi/who-we-work-with/key-donors/).
Conflict of Interest
None declared.
Contributions by the Authors
M.L.C., J.J.L.R. and U.S. designed the study. M.L.C. and N.M.M. did field collection. A.C. and R.O.B. provided assistant in collection of samples. B.vW. provided management information. M.L.C. and M.J.M generated the genetic data. M.L.C. performed the data analyses. M.L.C. and J.J.L.R. led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Literature Cited
- Admasu D 2008. Invasive plants and food security: the case of Prosopis juliflora in the Afar Region of Ethiopia. Technical Report. Ethiopia. FARM-Africa, IUCN. [Google Scholar]
- Aguilar DL, Acosta MC, Baranzelli MC, Sérsic AN, Delatorre-Herrera J, Verga A, Cosacov A. 2020. Ecophylogeography of the disjunct South American xerophytic tree species Prosopis chilensis (Fabaceae). Biological Journal of the Linnean Society 129:793–809. [Google Scholar]
- Anderson EC, Thompson EA. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessega C, Saidman BO, Darquier MR, Ewens M, Sánchez L, Rozenberg P, Vilardi JC. 2009. Consistency between marker- and genealogy-based heritability estimates in an experimental stand of Prosopis alba (Leguminosae). American Journal of Botany 96:458–465. [DOI] [PubMed] [Google Scholar]
- Bessega C, Vilardi JC, Saidman BO. 2006. Genetic relationships among American species of the genus Prosopis (Mimosoideae, Leguminosae) inferred from ITS sequences: evidence for long-distance dispersal. Journal of Biogeography 33:1905–1915. [Google Scholar]
- Boswell A, Sing SE, Ward SM. 2016. Plastid DNA analysis reveals cryptic hybridization in invasive Dalmatian toadflax (Linaria dalmatica) populations. Invasive Plant Science and Management 9:112–120. [Google Scholar]
- Bruvo R, Michiels NK, D’Souza TG, Schulenburg H. 2004. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molecular Ecology 13:2101–2106. [DOI] [PubMed] [Google Scholar]
- Burghardt AD, Espert SM. 2007. Phylogeny of Prosopis (Leguminosae) as shown by morphological and biochemical evidence. Australian Systematic Botany 20:332–332. [Google Scholar]
- Burkart A 1976. A monograph of the genus Prosopis (Leguminosae subfam. Mimosoideae). Journal of the Arnold Arboretum 57:219–525. [Google Scholar]
- Catalano SA, Vilardi JC, Tosto D, Saidman BO. 2008. Molecular phylogeny and diversification history of Prosopis (Fabaceae: Mimosoideae). Biological Journal of the Linnean Society 93:621–640. [Google Scholar]
- Chapuis MP, Estoup A. 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24:621–631. [DOI] [PubMed] [Google Scholar]
- Choge SK, Ngunjiri FD, Kuria MW, Basaka EA, Muthondeki JK. 2002. Status and impact of Prosopis in Kenya. Unpublished Technical Report. Nairobi, Kenya. KEFRI. [Google Scholar]
- Choge S, Pasiecznik N, Awan S, Harris P, Wright J, Harvey M. 2011. Prosopis pods as human food, with special reference to Kenya. Water SA 33:419–424. [Google Scholar]
- Clark LV, Jasieniuk M. 2011. POLYSAT: an R package for polyploid microsatellite analysis. Molecular Ecology Resources 11:562–566. [DOI] [PubMed] [Google Scholar]
- CRC Weed Management Guide 2003. Mesquite - Prosopis species. https://www.environment.gov.au/biodiversity/invasive/weeds/publications/guidelines/wons/pubs/prosopis.pdf (16 November 2019). [Google Scholar]
- Díaz Celis A 1995. Los algarrobos. Lima, Peru: Concytec. [Google Scholar]
- Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology 17:431–449. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America 97:7043–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensing DJ, Moffat CE, Pither J. 2013. Taxonomic identification errors generate misleading ecological niche model predictions of an invasive hawkweed. Botany 91:137–147. [Google Scholar]
- Fontana ML, Pérez VR, Luna CV. 2018. Características evolutivas en Prosopis spp.: citogenética, genética e hibridaciones. Rodriguésia 69:409–421. [Google Scholar]
- Gallaher T, Merlin M. 2010. Biology and impacts of Pacific island invasive species. 6. Prosopis pallida and Prosopis juliflora (Algarroba, Mesquite, kiawe) (Fabaceae). Pacific Science 64:489–526. [Google Scholar]
- Gallego-Tévar B, Grewell BJ, Rousseau H, Keller J, Ainouche A, Lima O, Dréano S, Salmon A, Figueroa E, Aïnouche M, Castillo JM. 2019. Genetic structure of Spartina hybrids between native Spartina maritima and invasive Spartina densiflora in Southwest Europe. Perspectives in Plant Ecology, Evolution and Systematics 37:26–38. [Google Scholar]
- Gaskin JF, Birken AS, Cooper DJ. 2012. Levels of novel hybridization in the saltcedar invasion compared over seven decades. Biological Invasions 14:693–699. [Google Scholar]
- Gaskin JF, Kazmer DJ. 2009. Introgression between invasive saltcedars (Tamarix chinensis and T. ramosissima) in the USA. Biological Invasions 11:1121–1130. [Google Scholar]
- Gaskin JF, Schaal BA. 2002. Hybrid Tamarix widespread in U. S. invasion and undetected in native Asian range. Proceedings of the National Academy of Sciences of the United States of America 99:11256–11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby JA, DE Barro PJ, Makinson JR, Pemberton RW, Hartley DM, Frohlich DR. 2006. Matching the origin of an invasive weed for selection of a herbivore haplotype for a biological control programme. Molecular Ecology 15:287–297. [DOI] [PubMed] [Google Scholar]
- Hall TA 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98. [Google Scholar]
- Hardy OJ, Vekemans X. 2002. SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2:618–620. [Google Scholar]
- Hirsch H, Castillo ML, Impson FAC, Kleinjan C, Richardson DM, Le Roux JJ. 2019. Ghosts from the past: even comprehensive sampling of the native range may not be enough to unravel the introduction history of invasive species-the case of Acacia dealbata invasions in South Africa. American Journal of Botany 106:352–362. [DOI] [PubMed] [Google Scholar]
- Hunziker JH, Saidman BO, Naranjo CA, Palacios RA, Poggio L, Burghardt AD. 1986. Hybridization and genetic variation of Argentine species of Prosopis. Forest Ecology and Management 16:301–315. [Google Scholar]
- Jahodová Š, Trybush S, Pyšek P, Wade M, Karp A. 2007. Invasive species of Heracleum in Europe: an insight into genetic relationships and invasion history. Diversity and Distributions 13:99–114. [Google Scholar]
- Johansson S 1990. Controlling and containing the spreading of Prosopis spp. at Bura. An outline of options and required actions. Research component in Bura fuelwood project. Technical Report. Kenya. KEFRI. [Google Scholar]
- Jombart T 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Juárez-Muñoz J, Carrillo-Castañeda GM, Rubluo A. 2006. Polymorphism determination in two natural mesquite (Prosopis laevigata) populations using RAPD. Biotecnología Aplicada 23:229–235. [Google Scholar]
- Kebede AT, Coppock DL. 2015. Livestock-mediated dispersal of Prosopis juliflora imperils grasslands and the endangered Grevy’s Zebra in Northeastern Ethiopia. Rangeland Ecology and Management 68:402–407. [Google Scholar]
- Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA. 2013. diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution 4:782–788. [Google Scholar]
- Kilawe CJ, Mbwambo JR, Kajembe GC, Mwakalukwa EE, Amri AM, Mushi GV, Athumani AM, Eckert S, Eschen R. 2017. Mrashia: Prosopis invading pastures and agricultural lands in Tanzania. Insights Report. Morogoro: Woody Weeds. [Google Scholar]
- Lachmuth S, Durka W, Schurr FM. 2010. The making of a rapid plant invader: genetic diversity and differentiation in the native and invaded range of Senecio inaequidens. Molecular Ecology 19:3952–3967. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J. 2007. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences of the United States of America 104:3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Brown GK, Byrne M, Ndlovu J, Richardson DM, Thompson GD, Wilson JRU. 2011. Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) in South Africa. Diversity and Distributions 17:861–871. [Google Scholar]
- Le Roux J, Wieczorek AM. 2009. Molecular systematics and population genetics of biological invasions: towards a better understanding of invasive species management. Annals of Applied Biology 154:1–17. [Google Scholar]
- Le Roux JJ, Wieczorek AM, Tran CT, Vorsino AE. 2010. Disentangling the dynamics of invasive fireweed (Senecio madagascariensis Poir. species complex) in the Hawaiian Islands. Biological Invasions 12:1–16. [Google Scholar]
- Little PD 2019. When “Green” equals thorny and mean: the politics and costs of an environmental experiment in East Africa. African Studies Review 62:132–163. [Google Scholar]
- Mazibuko DM 2012. Phylogenetic relationships of Prosopis in South Africa: an assessment of the extent of hybridization, and the role of genome size and seed size in the invasion dynamics. Master of Science Thesis, Stellenbosch University, South Africa. [Google Scholar]
- Mbaabu PR, Ng W-T, Schaffner U, Gichaba M, Olago D, Choge S, Oriaso S, Eckert S. 2019. Spatial evolution of Prosopis invasion and its effects on LULC and livelihoods in Baringo, Kenya. Remote Sensing 11:1217. [Google Scholar]
- Meirmans PG 2020. genodive version 3.0: easy-to-use software for the analysis of genetic data of diploids and polyploids. Molecular Ecology Resources 20:1126–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW. 2011. Assessing population structure: F(ST) and related measures. Molecular Ecology Resources 11:5–18. [DOI] [PubMed] [Google Scholar]
- Moncada X, Plaza D, Stoll A, Payacan C, Seelenfreund D, Martínez E, Bertin A, Squeo FA. 2019. Genetic diversity and structure of the vulnerable species Prosopis chilensis (Molina) Stuntz in the Coquimbo Region, Chile. Gayana Botánica 76:91–104. [Google Scholar]
- Muturi GM 2012. Ecological impacts of Prosopis invasion in riverine forest of Kenya. PhD Thesis, Wageningen University, The Netherlands. [Google Scholar]
- Naranjo A, Poggio L, Zeiger SE. 1984. Phenol chromatography, morphology and cytogenetics in three species and natural hybrids of Prosopis (Leguminosae - Mimosoideae). Plant Systematics and Evolution 144:257–276. [Google Scholar]
- Nybom H 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13:1143–1155. [DOI] [PubMed] [Google Scholar]
- Otsamo A, Maua JO. 1993. Observations on pod production of planted Prosopis juliflora in Bura, Tana River District, Kenya. East African Agricultural and Forestry Journal 58:111–114. [Google Scholar]
- Palacios RA, Bravo LD. 1981. Hibridación natural en Prosopis (Leguminosae) en la región Chaqueña Argentina. Evidencias morfológicas y cromatográficas. Darviniana 23:3–35. [Google Scholar]
- Palacios RA, Burghardt AD, Frías-Hernández JT, Olalde-Portugal V, Grados N, Alban L, Martínez-de la Vega O. 2012. Comparative study (AFLP and morphology) of three species of Prosopis of the Section Algarobia: P. juliflora, P. pallida, and P. limensis. Evidence for resolution of the “P. pallida-P. juliflora complex.” Plant Systematics and Evolution 298:165–171. [Google Scholar]
- Paradis E 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. [DOI] [PubMed] [Google Scholar]
- Pasiecznik NM, Tewari JC, Harsh LN, Cruz G, Tewari JC, Cadoret K, Maldonado LJ. 2001. The Prosopis juliflora - Prosopis pallida complex: a monograph. Coventry, UK: HDRA. [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics 52:273–288. [Google Scholar]
- Poynton RJ 2009. Tree planting in Southern Africa. Volume 3: other genera. Department of Agriculture, Forestry and Fisheries, Pretoria. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyšek P, Hulme PE, Meyerson LA, Smith GF, Boatwright JS, Crouch NR, Figueiredo E, Foxcroft LC, Jarošík V, Richardson DM, Suda J, Wilson JRU. Hitting the right target: taxonomic challenges for, and of, plant invasions. AoB PLANTS 5:plt042; doi: 10.1093/aobpla/plt042. [DOI] [Google Scholar]
- Ramírez L, de la Vega A, Razkin N, Luna V, Harris PJC. 1999. Analysis of the relationships between species of the genus Prosopis revealed by the use of molecular markers. Agronomie 19:31–43. [Google Scholar]
- Ramírez-Martínez A 2015. Análisis biogeográfico de la distribución conocida y potencial de cuatro taxa del género Prosopis L. (Leguminosae-Mimosoideae). Master Thesis, Universidad Autónoma Metropolitana-Iztapalapa, Mexico. [Google Scholar]
- Saidman BO, Bessega C, Ferreyra LI, Julio N, Vilardi JC. 2000. The use of genetic markers to assess populations structure and relationships among species of the genus Prosopis (Leguminosae). Boletín de la Sociedad Argentina de Botánica 35:315–324. [Google Scholar]
- Saidman BO, Vilardi J. 1987. Analysis of the genetic similarities among seven species of Prosopis (Leguminosae:Mimosoideae). Theoretical and Applied Genetics 75:109–116. [Google Scholar]
- Schierenbeck KA, Ellstrand ÆNC. 2009. Hybridization and the evolution of invasiveness in plants and other organisms. Biological Invasions 11:1093–1105. [Google Scholar]
- Seebens H, Bacher S, Blackburn TM, Capinha C, Dawson W, Dullinger S, Genovesi P, Hulme PE, van Kleunen M, Kühn I, Jeschke JM, Lenzner B, Liebhold AM, Pattison Z, Pergl J, Pyšek P, Winter M, Essl F. 2020. Projecting the continental accumulation of alien species through to 2050. Global Change Biology 00:1–13. [DOI] [PubMed] [Google Scholar]
- Shackleton RT, Le Maitre DC, Pasiecznik NM, Richardson DM. 2014. Prosopis: a global assessment of the biogeography, benefits, impacts and management of one of the world’s worst woody invasive plant taxa. AoB PLANTS 6:plu027; doi: 10.1093/aobpla/plu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 94:275–288. [DOI] [PubMed] [Google Scholar]
- Sherry M, Smith S, Patel A, Harris P, Hand P, Trenchard L, Henderson J. 2011. RAPD and microsatellite transferability studies in selected species of Prosopis (section Algarobia) with emphasis on Prosopis juliflora and P. pallida. Journal of Genetics 90:251–264. [DOI] [PubMed] [Google Scholar]
- Shiferaw H, Schaffner U, Bewket W, Alamirew T, Zeleke G, Teketay D, Eckert S. 2019. Modelling the current fractional cover of an invasive alien plant and drivers of its invasion in a dryland ecosystem. Scientific Reports 9:1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Keller SR. 2007. Historical range expansion determines the phylogenetic diversity introduced during contemporary species invasion. Evolution 61:334–345. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesová M, Pysek P. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany 109:19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GD, Bellstedt DU, Byrne M, Millar MA, Richardson DM, Wilson JR, Le Roux JJ. 2012. Cultivation shapes genetic novelty in a globally important invader. Molecular Ecology 21:3187–3199. [DOI] [PubMed] [Google Scholar]
- Trenchard LIZJ, Harris PJC, Smith SJ, Pasiecznik NM. 2008. A review of ploidy in the genus Prosopis (Leguminosae). Botanical Journal of the Linnean Society 156:425–438. [Google Scholar]
- van Klinken RD 2012. Prosopis spp. — mesquite In: Julien M, McFadyen R, Cullen J, eds. Biological control of weeds in Australia. Melbourne, Australia: CSIRO Publishing, 477–485. [Google Scholar]
- van Klinken RD, Campbell SD. 2001. Australian weeds series: Prosopis species. Plant Protection Quarterly 16:2–20. [Google Scholar]
- van Klinken RD, Graham J, Flack LK. 2006. Population ecology of hybrid mesquite (Prosopis species) in Western Australia: how does it differ from native range invasions and what are the implications for impacts and management? Biological Invasions 8:727–741. [Google Scholar]
- van Wilgen BW, Dyer C, Hoffmann JH, Ivey P, Le Maitre DC, Moore JL, Richardson DM, Rouget M, Wannenburgh A, Wilson JRU. 2011. National-scale strategic approaches for managing introduced plants: insights from Australian acacias in South Africa. Diversity and Distributions 17:1060–1075. [Google Scholar]
- Vega MV, Hernández P. 2005. Molecular evidence for natural interspecific hybridization in Prosopis. Agroforestry Systems 64:197–202. [Google Scholar]
- Verga A 2005. Recursos genéticos, mejoramiento y conservación de especies del género Prosopis. In: Norverto CA, ed. Mejores árboles para más forestadores: el programa de producción de material de propagación mejorado y el mejoramiento genético en el Proyecto Forestal de Desarrollo. Buenos Aires, Argentina: SAGPyA, 205–221. [Google Scholar]
- Verga A, López Lauenstein D, López C, Navall M, Joseau M, Gómez C, Royo O, Degano W, Marcó M. 2009. Caracterización morfológica de los algarrobos (Prosopis sp.) en las regiones fitogeográficas Chaqueña y Espinal norte de Argentina. Quebracho 17:31–40. [Google Scholar]
- Vilardi JC, Saidman BO, Palacios RA. 1988. Muestreo según variabilidad In: Prosopis en Argentina. Documento preliminar elaborado para el I taller internacional sobre recurso genético y conservación de germoplasma en Prosopis. Facultad de Ciencias Agropecuarias, UNC-FAO, PIRB, Córdoba, Argentina: FAO (Food and Agriculture Organization of the United Nations), 119–124. [Google Scholar]
- Wakie TT, Evangelista PH, Jarnevich CS, Laituri M. 2014. Mapping current and potential distribution of non-native Prosopis juliflora in the Afar region of Ethiopia. PLoS ONE 9:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS 1996. Genetic data analysis II. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Zalapa JE, Brunet J, Guries RP. 2010. The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evolutionary Applications 3:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann HG 1991. Biological control of mesquite, Prosopis spp. (Fabaceae), in South Africa. Agriculture, Ecosystems & Environment 37:175–186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotype data generated in the study are available on the DRYAD online repository (doi:10.5061/dryad.zgmsbcc97).