Abstract
Pedigree information was traditionally used to assess inbreeding. The availability of high-density marker panels provides an alternative to assess inbreeding, particularly in the presence of incomplete and error-prone pedigrees. Assessment of autozygosity across chromosomal segments using runs of homozygosity (ROH) has emerged as a valuable tool to estimate inbreeding due to its general flexibility and ability to quantify the chromosomal contribution to genome-wide inbreeding. Unfortunately, the identification of ROH segments is sensitive to the parameters used during the search process. These parameters are heuristically set, leading to significant variation in the results. The minimum length required to identify an ROH segment has major effects on the estimation of inbreeding and inbreeding depression, yet it is arbitrarily set. To overcome this limitation, a search algorithm to approximate mutation enrichment was developed to determine the minimum length of ROH segments. It consists of finding genome segments with significant effect differences in trait means between animals with high and low burdens of autozygous intervals with a specific length. The minimum length could be determined heuristically as the smallest interval at which a significant signal is detected. The proposed method was tested in an inbred Hereford cattle population genotyped for 30,220 SNPs. Phenotypes recorded for six traits were used for the approximation of mutation loads. The estimated minimum length was around 1 Mb for yearling weight (YW) and average daily gain (ADG) and 4 Mb for birth weight and weaning weight. These trait-specific thresholds estimated using the proposed method could be attributed to a trait-dependent effect of homozygosity. The detection of significant inbreeding effects was well aligned with the estimated thresholds, especially for YW and ADG. Although highly deleterious alleles are expected to be more frequent in recent inbreeding (long ROH), short ROH segments (<5 Mb) could contain a large number of less deleterious mutations with substantial joint effects on some traits (YW and ADG). Our results highlight the importance of accurate estimation of the ROH-based inbreeding and the necessity to consider a trait-specific minimum length threshold for the identification of ROH segments in inbreeding depression analyses. These thresholds could be determined using the proposed method provided the availability of phenotypic information.
Keywords: inbreeding, inbreeding depression, mutation load, runs of homozygosity
Introduction
Inbreeding leads to an increase in autozygosity throughout the genome largely in the form of runs of homozygosity (ROH) resulting in an increased risk of homozygosity for deleterious alleles (Ku et al., 2011). These ROH would contribute to inbreeding depression if they contain recessive deleterious alleles. Due to their general flexibility and ability to quantify chromosomal and segments contribution to genome-wide inbreeding, analysis of ROH has become a tool to estimate inbreeding and its impact on traits and a valuable alternative to the traditional pedigree-based approach, particularly in the presence of incomplete and error-prone pedigrees (Cassell et al., 2003; Zhang et al., 2015a; Curik et al., 2017; Sumreddee et al., 2019). Additionally, the length of an ROH can be used to track the ontogeny of an autozygous segment. Short ROH segments are likely to have risen from remote ancestors compared with long ROH segments. Thus, the former will reflect the older origin of inbreeding as opposed to the latter that will reflect the recent origin of inbreeding (Keller et al., 2011).
Several methods to identify ROH segments from stretches of homozygous markers have been proposed. Rule-based approaches, such as those implemented in the popular software PLINK (Purcell et al., 2007), are commonly used for ROH analysis in humans and animal applications due to their simplicity (Ceballos et al., 2018; Meyermans et al., 2020). Despite their popularity, the identification of ROH segments is often based on a set of heuristically predefined parameters leading to significant variations in the estimation of genomic inbreeding (Howrigan et al., 2011; Ku et al., 2011; Yengo et al., 2017). In fact, the minimum length to declare a segment as an ROH has major effects on the estimation of inbreeding and its effects (Pryce et al., 2014; Bérénos et al., 2016; Ferenčaković et al., 2017), yet different parameters settings were used within and across species (Keller et al., 2012; Kim et al., 2015; Martikainen et al., 2018; Addo et al., 2019; Szmatoła et al., 2019). The complexity resides in trying to balance the ability to identify short ROH segments representing ancient inbreeding and lowering the probability of false detection of nonidentical by descent (IBD) regions (Purfield et al., 2012). This complex problem could be reasonably addressed if actual or functionally predicted deleterious variants are available. In fact, Keller et al. (2011) showed that inbreeding estimated from ROH is highly correlated with homozygous mutation loads using simulated data. Furthermore, several studies have shown a significant enrichment of deleterious alleles (known and functionally predicted mutations) in ROH regions (Szpiech et al., 2013; Zhang et al., 2015b; Pemberton and Szpiech, 2018; Sams and Boyko, 2019). Thus, knowledge of deleterious mutation enrichment has been used to determine the minimum length threshold for identifying ROH regions. Using known mutations associated with rare diseases in dogs, Sams and Boyko (2019) were able to assess the minimum length of an ROH segment based on identifying enriched mutations for varying size ROH tracts.
Unfortunately, the actual mutation loads contributing to inbreeding depression are seldom known in practice, especially for complex traits. Thus, inferring mutational load from available phenotypic information could be useful in assessing the minimum length threshold to declare an autozygous segment as an ROH. Furthermore, understanding the genesis of ROH segments, the purging of deleterious mutations, and the effects of the latter would require the ability to discriminate between ancient and more recent inbreeding. However, thresholds to discriminate between short and long ROH segments are largely unknown. Thus, the objectives of this study are to: 1) develop an approach to determine the minimum length of an autozygous segment to be declared as an ROH and 2) assess the implications of the minimum length threshold on measuring inbreeding depression in a purebred Hereford beef cattle population.
Materials and Methods
As this study was carried out using previously compiled data and no live animal experiments were performed, therefore, Animal Care and Use Committee approval was not required.
Animals, phenotypes, and genotypes
Data used in this study were collected on line 1 Hereford cattle herd maintained in isolation, at USDA-ARS, Fort Keogh Livestock and Range Research Laboratory, Miles City, MT (Knapp et al., 1951; MacNeil, 2009; Leesburg et al., 2014). Briefly, the line 1 Hereford herd was founded in 1934 based on two paternal half-sib males and 50 unrelated females and was maintained under a careful mating scheme to minimize inbreeding (MacNeil et al., 1992; MacNeil, 2009). Management specific to this herd has been previously described (MacNeil, 2009; MacNeil and Vermeire, 2012; MacNeil et al., 2017). Historically, the objective of selection in line 1 was focused on increasing growth rate from weaning to 1 yr of age (MacNeil et al., 1992). Since 2011, the selection was primarily focused on improving calving ease (direct and maternal) while maintaining weaning weight (WW) and yearling weight (YW) at least to herd averages.
Pedigree, genotypes, and phenotypes used in this study were previously described by Sumreddee et al. (2019). The pedigree consisted of 10,186 animals (639 sires and 3,315 dams). The average pedigree-based inbreeding coefficient (Fped) of genotyped animals (n = 785) was 29.2% (SD = 5.3%) with an average of 17.15 equivalent complete generations (Sumreddee et al., 2019). Further characterization of the genetic architecture of the population can be found in Huang et al. (2012).
Genomic information consisted of single-nucleotide polymorphism (SNP) genotypes. Data quality control and marker genotype imputation from low (3k, 9k, 20k, and 27k) to medium-density SNP panels (50k) can be found in Sumreddee et al. (2019). After imputation and quality control, the marker data consisted of 785 animals genotyped for 30,810 SNPs. For ROH identification, only autosomal SNPs (30,220) were used with an average distance between consecutive markers of 83.51 kb (SD = 5.83 kb). Only animals born between 1990 and 2016, with complete marker, phenotypic, and parental information, were used in ROH-based analyses. Phenotypic data consisted of birth weight (BW), WW, YW, average daily gain (ADG) between weaning and yearling, age at first calving (AFC), and heifer pregnancy status (HPS) recorded as a binary trait (0 = nonpregnant, 8.7% or 1 = pregnant, 91.3%). A full description of the data could be found in Supplementary Table S1.
Algorithm for estimating the minimum length of an ROH segment
Accumulation of the effects of deleterious mutations across the genome is the predominant cause of inbreeding depression (Charlesworth and Willis, 2009). There is ample evidence of significant enrichment of deleterious alleles in ROH segments (Szpiech et al., 2013; Zhang et al., 2015b; Pemberton and Szpiech, 2018; Sams and Boyko, 2019). Thus, this enrichment could be exploited in the identification of ROH segments. Unfortunately, only a limited number of mutations are known for complex traits. To remedy this situation, a search algorithm to approximate mutation enrichment across the genome was proposed and used to estimate the minimum length of an ROH segment. The proposed method’s main hypothesis is that autozygous segments carrying unknown (hidden) mutations will have a significant effect on traits.
ROH segments were identified using a sliding window approach as implemented in PLINK version 1.9 (Purcell et al., 2007; Chang et al., 2015). Parameter settings for the detection of ROH for this population were previously described by Sumreddee et al. (2019). The --homozyg function was used to perform ROH analysis. A maximum of two heterozygous SNPs were allowed within the sliding window to minimize the impact of potential genotyping errors. The maximum gap between two SNPs in a homozygous segment was set to 500 kb. A minimum SNP density of 1 SNP per 500 kb inside an ROH was used to improve the ROH genome coverage for the current density SNP panel. Additionally, due to a limited density of marker panel used in the current study, very few ROH segments smaller than 0.1 Mb were detected (results not shown). Thus, only autozygous segments larger than 0.1 Mb were used in the search for the minimum threshold for ROH identification. In order to capture short segments (≥0.1 Mb), the minimum number of markers was set to 5 consecutive homozygous SNPs per ROH.
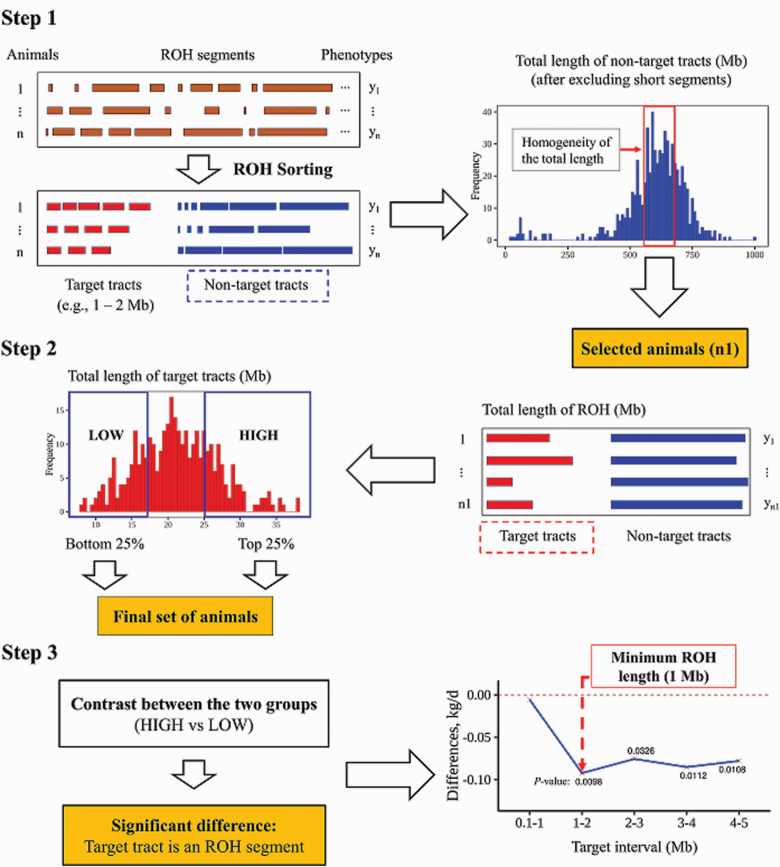
Briefly, the three-step algorithm consists of finding the shortest autozygous segment with a significant effect on the trait. The minimum length threshold was set equal to the lower bound of such interval. The proposed algorithm was implemented separately for each trait. Therefore, the minimum length threshold could be different across traits. A graphical representation of the proposed three-step algorithm is shown in Figure 1.
Figure 1.
A flow chart of the three-step search algorithm to approximate mutation loads within an autozygous segment to estimate the minimum length of an ROH. Step 1: Identification of ROH segments based on the predefined target tract (e.g., 1 to 2 Mb) and selection of animals with similar nontarget tract length. Step 2: Clustering the selected animals into HIGH and LOW groups based on the total length of their target tracts. Step 3: Estimation of the difference in the effect of two groups on the trait of interest and its associated significance level.
Step 1
All autozygous segments were identified for each genotyped animal. A target tract (i.e., 0.1 to 1 Mb) is specified by the user. Autozygous segments shorter than the lower bound of the target tract were removed, and the remaining segments were clustered into “target” group (segments with length that falls within the length of the target tract) and “nontarget” group (segments with length that falls outside the length of the target track). For each animal, the total length of the nontarget segments (segments that fall within the nontarget group) was calculated. Animals with a total length of nontarget segments outside the interquartile range of the distribution (difference between the third and first quartiles) were removed. The remaining animals constituted the “selected set” with size n1. Only these animals were used in the following two steps of the proposed algorithm. This was done to remove, or at least attenuate, the potential effect of nontarget autozygous segments.
Step 2
Animals in the selected set identified in step 1 were sorted based on the total length of their target segments (Figure 1). Animals in the top and bottom quartiles of the distribution of total target segment length were clustered into “HIGH” and “LOW” groups, respectively, and they formed the final set of animals that were used in the search of the minimum length threshold. Note that for female fertility traits, all selected animals from step 1 were retained, and the assignment of animals into HIGH or LOW groups was based on the median value of the total length of target tracts due to a limited number of animals with phenotypes. The number of animals in each group can be found in Supplementary Table S2.
Step 3
Using the final set of animals identified in step 2, the contrast between the effects of the HIGH and LOW groups was estimated for each predefined target tract (Figure 1). For each trait, a linear model was specified, which included the group effects (HIGH and LOW) and the systematic effects of sex and age (for WW and YW) and months of birth (for AFC and HPS). In addition, a regression on the total length of nontarget tracts was included to correct for variation between animals. The estimated effects of HIGH and LOW groups were contrasted and tested for significance (odds ratio between HIGH and LOW groups for HPS).
The minimum length threshold was determined as the lower bound of the shortest target tract with a significant contrast between the two group means (HIGH to LOW). All statistical analyses regarding the estimation of minimum length thresholds were performed in R (R Core Team, 2018).
Inbreeding depression analyses
To evaluate the impact of the minimum length threshold used to identify ROH segment in the estimates of inbreeding depression, the proportion of the autosomal genome that is covered by ROH segments for an individual i () was calculated following the method proposed by McQuillan et al. (2008) as:
| (1) |
where LROH and LAUTO are the total length of all ROH segments and the autosomal genome (2,512,189 kb), respectively.
Inbreeding depression was assessed by regressing the trait phenotypes (BW, WW, YW, and ADG) on inbreeding coefficients using a mixed model:
| (2) |
where y is a vector of phenotypes for the trait of interest; β is the vector of fixed effects of sex, birth year, age (as covariate for WW and YW), and the regression coefficient on genomic inbreeding; and u is a vector of random additive effects. m and p are the vectors of random maternal and maternal permanent environmental effects (only for BW and WW), and e is the vector of random residuals. X, Z, W, and S are known incidence matrices with the appropriate dimensions.
Single-trait analyses were carried out using the BLUPF90 family programs (Misztal et al., 2002). The effect of inbreeding on a given trait was assessed based on the significance of its associated regression coefficient using the t-statistic unit ( > 2).
Results and Discussion
Approximation of minimum length thresholds for ROH
The ability of the proposed algorithm to cluster animals into HIGH and LOW groups was assessed by testing the differences in key parameters between the two groups as illustrated in Supplementary Figure S1. For ADG and across predefined target tract sizes, the two groups of animals in final data sets differed significantly (P < 0.00001) in their number and the total length of target segments (Supplementary Figure S1A and B). Similar results were observed for the other growth (BW, WW, and YW) and female fertility (AFC and HPS) traits (results not shown). For growth traits, there was no significant difference (P > 0.05) in the total length of nontarget tracts for most of the tract sizes as indicated in Supplementary Figure S1C (only results for ADG are shown). The only significant difference was observed for the 0.1 to 1.0 Mb interval (P = 0.026). However, the differences in the total length of nontarget tracts between the HIGH and LOW groups were significant across all intervals for the fertility traits (result not shown). Similar results were observed in terms of the coefficients of variation between the two groups (Supplementary Table S2). Admittedly, for fertility traits (AFC and HPS), variations in their total length of nontarget segments were different between the two groups. This is due to the fact that all animals were kept before the selection step regardless of their variation in nontarget tracts in order to ensure a sufficient number of animals for estimating the effects. However, such observed heterogeneity was accounted for by adding a regression on the total lengths of nontarget tracts in the statistic model for all traits.
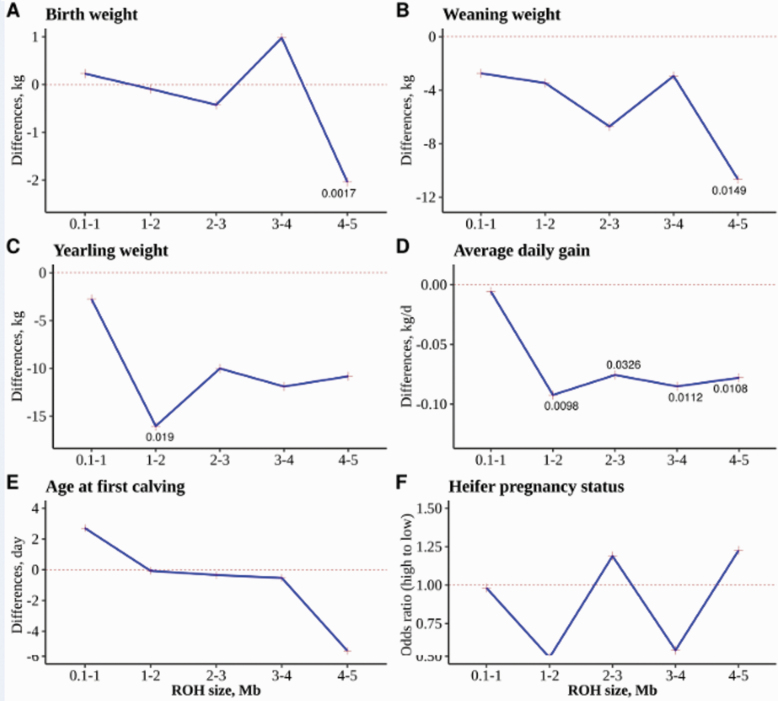
The difference between the HIGH and LOW group effects at varying thresholds for the minimum length of ROH segments is shown in Figure 2. As defined earlier, the HIGH group included animals with a significantly higher fraction of their genome covered with autozygous segments within the specified target tract (e.g., 0.1 to 1.0 Mb) (Supplementary Figure S1A). Under the biological assumption that ROH regions are likely to be enriched with deleterious mutations, HIGH and LOW groups are expected to have different effects on traits. Thus, the lower bound of the shortest predefined target tract at which a significant difference between the LOW and HIGH group effects was observed was declared as the minimum threshold to identify ROH regions.
Figure 2.
Differences in LS means (A to E) or the odds ratio for pregnancy (F) between HIGH and LOW groups for different sizes of target tracts and associated significant P values (<0.05).
For YW and ADG, the 1 to 2 Mb was the shortest segment at which significant difference was observed between the HIGH and LOW groups with P value of 0.02 and 0.009, respectively (Figure 2C and D and Supplementary Table S3). This result indicates that 1 Mb is approximately the minimum threshold to identify ROH for YW and ADG. Higher thresholds were observed for BW (4 Mb) and WW (4 Mb) (Figure 2A and B and Supplementary Table S3). Across the different intervals, the HIGH group had smaller least square (LS) means compared with the LOW group for the growth traits. This seems to indicate that animals with a higher portion of their genome covered by ROH segments carry more deleterious than beneficial mutations for growth traits, which resulted in the reduction of performance. For instance, within the 1 to 2 Mb segments, the LS means for the HIGH group were 16 kg and 93 g/d lower than the LOW group for YW and ADG, respectively (Supplementary Table S3). For WW, the difference in LS means between HIGH and LOW groups was 10.7 kg (P < 0.05) within the 4 to 5 Mb intervals. For BW, a similar trend was observed; although within the 3 to 4 Mb interval, the HIGH group had heavier weights compared with the LOW group (P = 0.11). This could be due to the incomplete recording of BW and the subjective assessment of the trait. For fertility traits, there was little difference between the two groups for AFC, except for the 4 to 5 interval where animals in the HIGH group were 5 d younger (P = 0.052) at their first calving compared with the LOW group (Figure 2E and Supplementary Table S4). No significant odds ratio between HIGH and LOW groups was observed for HPS (P > 0.05; Figure 2F and Supplementary Table S4). For these two fertility traits, extended intervals (up to 10 Mb) were further investigated. However, none of these analyses showed significant differences between groups (results not shown).
The significant differences between the HIGH and LOW groups for short homozygous target segments (i.e., 1 to 2, 2 to 3, 3 to 4, and 4 to 5 Mb long) indicate that these short autozygous tracts are likely to harbor more unfavorable than favorable alleles for the traits analyzed. This result is in agreement with previous findings showing that short ROH regions, of comparable sizes to the current study, are enriched with deleterious variants in several species, including humans (Szpiech et al., 2013), domestic dogs (Sams and Boyko, 2019), and cattle (Zhang et al., 2015b). Furthermore, it highlights their importance in the calculation of inbreeding and the estimation of inbreeding depression. Using cattle data, Zhang et al. (2015b) revealed a significantly higher enrichment of deleterious variants in short and medium compared with long (>3 Mb) ROH regions, which could be in part due to hitchhiking effects. Significant inbreeding effects were found to vary across the different ROH length segments (1 to 2, 2 to 4, 4 to 8, 8 to 16, and >16 Mb) and traits (yield, fertility, and udder health) in dairy cattle, indicating that both short and long ROH contributing to inbreeding depression (Doekes et al., 2019). These results seem to indirectly imply that the specific minimum length thresholds of ROH could be important in assessing inbreeding depression. Although the sensitivity and specificity tend to be lower for the detection of short ROH segments (e.g., <1 Mb), particularly using SNP chip data (Purfield et al., 2012; Zhang et al., 2015a), they should not be overlooked given their relevance for the study of inbreeding depression. Favorable effects of certain autozygous segments observed for BW and AFC may be indicative of target sites of positive selection or regions of favorable mutations. The inconsistent results for the fertility traits may be attributed to the selective culling of inbred females with reproductive problems. Consequently, highly inbred females that remained in the herd had limited impact of inbreeding on their reproductive performance.
A recent study using Holstein cattle data reported significant enrichment of ROH regions in low compared with high-fertility (sire conception rate) bulls. In fact, these enriched regions are found to harbor genes directly implicated in sperm biology and male fertility (Nani and Peñagaricano, 2020). These results strongly support the hypothesis used to develop the proposed method.
Genotype counting, an ROH analysis approach that is used by popular public software such as PLINK, could benefit from our approach as a supplementary tool for determining the minimum ROH length criterion. Lack of consensus criteria for ROH analyses could have resulted in biased results and limited comparison across studies. Recently, a general guideline for ROH analysis using PLINK for medium-density SNP data has just been developed (Meyermans et al., 2020); however, a criterion about a minimum length threshold for ROH remains unsolved. Since our approach allows the detection of homozygous segments affecting a trait, the decision to exclude non-influential short segments from the ROH analysis can be made more accurately. The proposed approach allows for the removal of short segments that are likely not to influence the trait, avoiding an overestimation of ROH-based inbreeding and a biased estimate of its effects. This is especially important when the ROH detection software is not sensitive enough to discriminate IBD ROH segments from linkage disequilibrium (LD) patterns (Ferenčaković et al., 2013a, 2013b). This issue is particularly frequent when the marker density is similar to currently used SNP panels (Zhang et al., 2015a).
Additionally, incorporating the approximated mutation load using the proposed method into the genotype-counting approaches could be an alternative to the more computationally expensive model-based methods, such as the hidden Markov model (HMM) framework (Browning and Browning, 2010) and likelihood-based approach (Broman and Weber, 1999; Pemberton et al., 2012; Kardos et al., 2017). The latter requires the knowledge of the population-allele frequencies, which might not be accurately obtained, especially when the sample size is small. Although model-based approaches (e.g., HMM) do not rely on the prior definition of arbitrary thresholds for indentifying ROH segments (Druet and Gautier, 2017), the identified autozygous segments may not have effects on phenotypes. Calculation of genomic inbreeding that incorporates ROH segments with potentially no effects (particularly short segments) has shown to reduce power to detect inbreeding depression (Pryce et al., 2014). Consequently, such biased estimates of inbreeding effects could have serious herd management consequences.
The effects of homozygosity are trait-dependent (Ku et al., 2011), which will lead to different minimum length of ROH segments across traits using our approach. Using the proposed method, we are not redefining inbreeding depression, although the estimated inbreeding will not necessarily reflect the overall level of autozygosity in the genome.
Patterns of ROH
The patterns and distribution of ROH segments in the autosomes of line 1 Hereford animals were investigated using the minimum length threshold of 1 Mb (ROH1.0) as approximated using the proposed algorithm (obtained from YW and ADG) and two shorter length thresholds (ROH0.1 and ROH0.5). The latter were used for comparison purposes given their ability to discover ancient haplotypes. As expected, the number and total length of ROH per individual decreased with the increase of the minimum length threshold (Table 1). On average, the number (length) of ROH segments was 116 (623.2 Mb), 112 (621.8 Mb), and 105 (616.7 Mb) per animal for ROH0.1, ROH0.5, and ROH1.0, respectively. ROH segments of 4 to 8 Mb long were the most abundant throughout the genome, while longer ROH segments (≥12 Mb) were less common. Using the different minimum length threshold, 12% to 13%, 23% to 25%, 37% to 41%, 11% to 13%, and 7% to 8% of the ROH were 1 to 2, 2 to 4, 4 to 8, 8 to 12, and greater than 12 Mb long, respectively (Figure 3). On average, an animal carried 4, 7, 14, 27, 43, 13, and 8 of ROH segments with length of 0.1 to 0.5, 0.5 to 1, 1 to 2, 2 to 4, 4 to 8, 8 to 12, and greater than 12 Mb long, respectively (Supplementary Table S5). Almost all animals (>96%) had at least five ROH segments for each length class between 1 and 12 Mb. However, only 36% of animals had more than 5 ROH segments for 0.1 to 0.5 Mb segment (Supplementary Table S5). The majority of identical homozygous segments of 4 to 12 Mb long are likely inherited from common ancestors around 6 to 12 generations ago (assuming 1 Morgan equals 100 Mb), while long ROH tracts (>12 Mb) are potentially arisen from closely related parents with common ancestors originated around at most 4 generations ago (Ceballos et al., 2018). Moreover, the sizeable portion of short homozygous segments (<4 Mb) shown in Figure 3 is consistent with the history of line 1 Hereford population that was founded by a small number of animals and was closed since its founding around 85 years ago (Knapp et al., 1951; MacNeil, 2009). Furthermore, the small effective population size (around 56.2) and the high level of LD in the population resulting from extreme bottlenecks since its founding (Huang et al., 2012) are well in agreement with our results. Admittedly, the SNP panel used in this study could not efficiently capture a short ROH segment (e.g., <1 Mb; Supplementary Table S5), which potentially originated from remotely ancestral relatedness even before the pedigree records began. Therefore, the inference about short ROH segments and their effects cannot be made unless higher SNP density or sequence data are available.
Table 1.
Summary description of ROH segments identified using minimum length thresholds of 0.1 (ROH0.1), 0.5 (ROH0.5), and 1.0 Mb (ROH1.0)
| ROH detection | Parameter1 | Mean | SD | Min. | Max. |
|---|---|---|---|---|---|
| ROH0.1 | ROH_n | 115.7 | 24.1 | 9 | 176 |
| ROH_L | 5.39 | 4.52 | 0.11 | 64.86 | |
| ROH_TL | 623.19 | 138.06 | 27.02 | 1,045.75 | |
| ROH_nsnp | 70.4 | 62.4 | 5 | 929 | |
| ROH0.5 | ROH_n | 111.8 | 23.3 | 9 | 164 |
| ROH_L | 5.56 | 4.50 | 0.50 | 64.86 | |
| ROH_TL | 621.79 | 137.81 | 27.02 | 1,042.63 | |
| ROH_nsnp | 72.6 | 62.3 | 5 | 929 | |
| ROH1.0 | ROH_n | 105.2 | 21.9 | 7 | 152 |
| ROH_L | 5.86 | 4.47 | 1.00 | 64.86 | |
| ROH_TL | 616.74 | 136.83 | 25.91 | 1,034.06 | |
| ROH_nsnp | 76.7 | 62.1 | 5 | 929 |
1ROH_n, number of ROH per animal; ROH_L, individual ROH segment length, Mb; ROH_TL, total ROH length per animal, Mb; ROH_nsnp, number of SNPs per ROH.
Figure 3.
Distribution of ROH segments using different minimum length thresholds of 0.1 (ROH0.1), 0.5 (ROH0.5), and 1.0 Mb (ROH1.0).
Compared with other cattle populations (Ferenčaković et al., 2013a; Gurgul et al., 2016; Szmatoła et al., 2016; Peripolli et al., 2018), a larger extent of long ROH segments (>8 Mb) observed in the line 1 population are not surprising given that the population was maintained as a closed population since its formation, in which inbreeding is likely unavoidable (MacNeil et al., 1992; MacNeil, 2009). These observed recent haplotypes resulting from recent parental relatedness could have a crucial contribution to the overall level of autozygosity in the population.
Estimation of inbreeding depression
Table 2 presents the estimates of inbreeding and inbreeding depression for growth traits using different minimum length thresholds (from 0.5 to 8 Mb) to identify ROH segments. As expected, genome-wide inbreeding decreases with the increase in the minimum threshold with an average rate of 0.0176 per a 1-Mb increase in minimum threshold. For example, the means (SD) of ROH-based inbreeding coefficient (FROH) were 0.243 (0.054), 0.228 (0.051), 0.205 (0.048), and 0.111 (0.031) with the minimum length thresholds of 0.5, 2, 4, and 8 Mb, respectively. Estimation of inbreeding effects was restricted to sets of ROH identified using a minimum length of at least 0.5 Mb because additional shorter ROH segments (i.e., ≥0.1 to 0.5 Mb) were found to be relatively less abundant with the marker density panels used in the current study (Table 1).
Table 2.
Estimates of inbreeding depression for growth traits when FROH was calculated using different minimum length thresholds
| Regression coefficient (SE)3 | ||||||
|---|---|---|---|---|---|---|
| Minimum Length1 |
n 2 | FROH (SD) | BW | WW | YW | ADG |
| 0.5 | 785 | 0.243 (0.054) | −0.043 (0.043) | −0.184 (0.227) | −0.888 (0.364) | −4.97 (1.30) |
| 1.0 | 785 | 0.229 (0.051) | −0.041 (0.045) | −0.187 (0.239) | −0.887 (0.383) | −5.03 (1.37) |
| 2.0 | 785 | 0.228 (0.051) | −0.041 (0.046) | −0.187 (0.239) | −0.891 (0.383) | −5.05 (1.37) |
| 3.0 | 785 | 0.223 (0.050) | −0.040 (0.046) | −0.189 (0.242) | −0.912 (0.388) | −5.18 (1.39) |
| 4.0 | 785 | 0.205 (0.048) | −0.045 (0.048) | −0.170 (0.250) | −0.847 (0.403) | −5.11 (1.44) |
| 5.0 | 783 | 0.179 (0.044) | −0.066 (0.051) | −0.153 (0.267) | −0.906 (0.432) | −5.70 (1.56) |
| 6.0 | 780 | 0.152 (0.039) | −0.083 (0.054) | −0.216 (0.285) | −0.908 (0.466) | −5.31 (1.72) |
| 7.0 | 776 | 0.129 (0.035) | −0.074 (0.058) | −0.186 (0.306) | −0.903 (0.499) | −5.36 (1.89) |
| 8.0 | 770 | 0.111 (0.031) | −0.097 (0.062) | −0.154 (0.328) | −0.833 (0.541) | −4.62 (2.10) |
1The minimum length threshold (in Mb) used to identify ROH segments.
2Number of genotyped animals.
3Significant effects ( > 2) are in bold font. Values set in italics were the estimates associated with the approximate minimum length of ROH obtained using the search algorithm proposed in the present study.
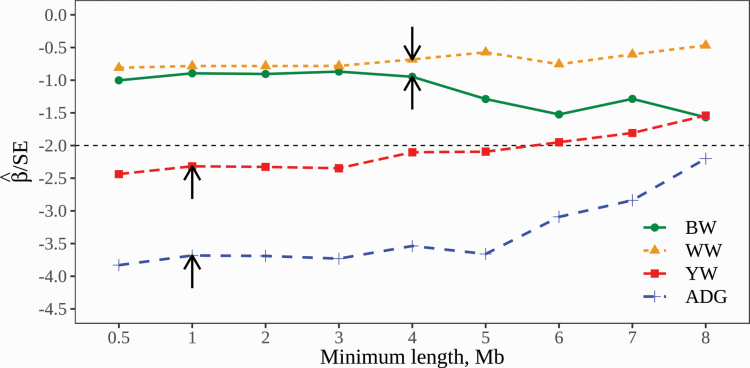
Across all the thresholds for the four growth traits, the negative estimates of the regression coefficient indicate a detrimental impact of inbreeding (Table 2). A 1% increase in FROH was associated with a decrease of 0.04 to 0.1 kg for BW, 0.15 to 0.22 kg for WW, 0.83 to 0.91 kg for YW, and 4.6 to 5.7 g/d for ADG; however, only YW and ADG showed significant inbreeding depression (t-statistic < −2). Inbreeding depression was not significant when the minimum threshold was greater than 5 Mb for YW. However, it persisted up to a minimum threshold of 8 Mb for ADG. This detrimental effect increases with the increase of the minimum threshold to reach a maximum at around 3 (YW) to 5 Mb (ADG). This is expected as longer ROH segments reflect more recent common ancestors with highly deleterious alleles that the population did not manage to purge given the short amount of time since their rise. When the minimum threshold is large (≥6 Mb), the identified ROH segments will account only for less than two-thirds of the genome-wide inbreeding (62.6%). Thus, in spite of accounting for highly deleterious recent mutations, a considerable number of mutations with potentially substantial collective effect on traits (e.g., YW) will not be considered as they fall in shorter segments (<6 Mb; Figure 3).
Significant effects of inbreeding depend on the trait and minimum threshold length. In fact, inbreeding has a significant negative effect on ADG and YW across all threshold values and when the threshold is smaller or equal to 5 Mb, respectively. However, it has no significant effect on BW and WW (Table 2). The trend and strength of unfavorable effects are generally in agreement with previous research using similar data set and a fixed minimum threshold at 1 Mb (Sumreddee et al., 2019). Using trait-specific thresholds (1 Mb for YW and ADG) estimated by the proposed method clearly indicates its ability to identify the minimum ROH segment length that allows the detection of the significant effect of inbreeding depression (Table 2 and Figure 4). Although inbreeding depression was not significant for BW and WW, the minimum length was possible to identify for both traits (4 Mb). In spite of the fact that our proposed method uses the effects on the trait to identify the minimum length, it does not require the presence of a significant inbreeding depression as the latter depends on the effects of all deleterious alleles across the genomes (Charlesworth and Willis, 2009) and not only those harbored in the target ROH segments. The minimum length of ROH segments has been explicitly observed to affect the estimation of inbreeding depression in previous studies. Similar results were observed in a study of inbreeding depression for fitness-related traits (lamb body size) in sheep when ROH segments were identified using either a 5 or 10 Mb minimum length threshold (Bérénos et al., 2016). Although the estimates of inbreeding depression slightly differed for the two thresholds, Bérénos et al. (2016) reported that inbreeding depression was less pronounced (larger SEs) when a longer minimum length threshold (10 Mb) was used. Ferenčaković et al. (2017) also reported a lower ability to detect significant inbreeding effects on semen quality traits in cattle when ROH segments of 4 Mb or longer were used compared with shorter ROH segments (>2 Mb). Inconsistent results were observed when higher thresholds (>60 SNPs or the equivalence of >3.5 Mb) for the identification of ROH segments were used leading to a greater reduction in milk yield in dairy cattle breeds even after correction for the general homozygosity (Pryce et al., 2014). Collectively, these results support the hypothesis that the effect of homozygosity regions could be trait and population specific. Shorter ROH regions arising from more ancient common ancestors are expected to have less or no detrimental impacts on traits compared with longer segments (arising from more recent common ancestors) as selection eliminates deleterious alleles following inbreeding over time. Pryce et al. (2014) and Saura et al. (2015) concluded that purging is a likely cause to explain the higher inbreeding depression effects of long ROH segments (more recent inbreeding) compared with their short counterparts in farm animals. Although in general there is a well-supported relationship between purging and the age of inbreeding, the efficiency of purging of deleterious mutations from shorter ROH segments (reflecting older inbreeding) is likely to be influenced by factors such as selection pressure (Hedrick, 1994; Mc Parland et al., 2009) and rate of inbreeding (Holt et al., 2005) within the population, leading to variability in the effects of segments. Furthermore, purging does not affect all short ROH segments equally. Short autozygous regions may still have significant deleterious effects despite their distant origin due to some specific management and mating practices (e.g., allowing inbreeding only from distant common ancestors after several generations of inbreeding avoidance). Although imperfect, there is a well-supported relationship between purging and the age of inbreeding that we tried to exploit in this study. In addition, the accumulation of a large number of short homozygous segments (reflecting ancient inbreeding) with potentially small effects could substantially contribute to the expression of inbreeding depression as observed in the current study. This could partially explain the inconsistency of the findings from different studies regarding the effects of the minimum length threshold for ROH on inbreeding depression.
Figure 4.
Estimates of inbreeding depression across different minimum length thresholds used to identify ROH for growth traits. Arrows indicate the approximate location of the minimum length thresholds obtained using the proposed search algorithm. The horizontal dashed line indicates the threshold ( < −2) to declare significance.
High thresholds (>4 Mb) were recommended for the identification of ROH segments in the presence of medium-density SNP panels due to the difficulties in precisely identifying shorter segments (Ferenčaković et al., 2013b). Similarly, the 50k SNP panel is found to be suitable for detecting ROH segments longer than 5 Mb as concluded by Purfield et al. (2012). This general recommendation seems to be partially supported by our results for BW and WW (the estimated minimum length threshold > 4 Mb). Substantially lower thresholds (>1 Mb) were accurately estimated for YW and ADG despite the limited size of the marker panel and phenotypic data. Discarding these short autozygous segments that are likely to contain deleterious alleles not only results in an underestimation of the autozygosity originated from more distant ancestors but could also lead to substantial bias in the estimation of inbreeding depression. The proposed method provides a simple but useful approach to approximate the minimum length threshold to identify ROH segments for complex traits in the absence of detailed genetic/genomic information. The data used in this study were generated in a small and well-controlled population that is not representative of common commercial livestock populations. Our primary goal was to test the ability of the proposed method to estimate the minimum threshold to identify ROH segments and the data set was largely adequate for that purpose. Thresholds are trait specific and likely to be data set specific. Further evaluation with other datasets is needed and it is currently in progress.
Conclusions
In this study, a search algorithm was developed to identify the minimum length of autozygous segments to be declared as ROH. The algorithm approximates the mutation enrichment at the individual level using available phenotypic information. This is of valuable practical importance as the direct assessment of mutation enrichment using genetic/genomic information is seldom available for the majority of livestock populations and complex traits. The proposed approach is heuristic in nature and the estimated thresholds are trait specific. Although ROH are signatures of the genome evolution and are not specific to each trait, their impact on traits could be different. Including short ROH segments that carry no mutations relevant to a specific trait will likely “bias” the estimation of inbreeding depression. The assessment of our proposed method showed its potential benefits in terms of the estimates of the genome-wide level of autozygosity as well as the calculation of inbreeding depression. The history of line 1 Hereford cattle population was reasonably revealed by the patterns of ROH length distribution, where the intermediate-size ROH segments were predominant followed by a significant portion of short autozygous segments, reflecting immediate population bottleneck and breeding management of the herd. In addition, the burden of relatively long ROH segments (≥8 Mb) reflects recent inbreeding that could occur due to the mating of closely related individuals. This pattern is different from what has been observed in humans or even other cattle populations where short autozygous segments are often more abundant.
Supplementary Material
Acknowledgments
The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual’s income is derived from any public assistance program. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA’s TARGET Center at +1 (202) 720-2600 (voice and TDD). To file a complaint of discrimination, write to USDA, Director, Office of Civil Rights, 1400 Independence Avenue, S.W., Washington, D.C. 20250-9410, or call +1 (800) 795-3272 (voice) or +1 (202) 720-6382 (TDD). USDA is an equal opportunity provider and employer.
Glossary
Abbreviations
- ADG
average daily gain between weaning and yearling
- AFC
age at first calving
- BW
birth weight
- FROH
ROH-based inbreeding coefficient
- HMM
hidden Markov model
- IBD
identical by descent
- ROH
runs of homozygosity
- SNP
single-nucleotide polymorphism
- WW
weaning weight
- YW
yearling weight
Conflict of interest statement
The authors declare no conflict of interest.
Literature Cited
- Addo S., Klingel S., Hinrichs D., and Thaller G.. . 2019. Runs of Homozygosity and NetView analyses provide new insight into the genome-wide diversity and admixture of three German cattle breeds. PLoS One. 14:e0225847. doi: 10.1371/journal.pone.0225847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérénos C., Ellis P. A., Pilkington J. G., and Pemberton J. M.. . 2016. Genomic analysis reveals depression due to both individual and maternal inbreeding in a free-living mammal population. Mol. Ecol. 25:3152–3168. doi: 10.1111/mec.13681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., and Weber J. L.. . 1999. Long homozygous chromosomal segments in reference families from the centre d’Etude du polymorphisme humain. Am. J. Hum. Genet. 65:1493–1500. doi: 10.1086/302661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., and Browning B. L.. . 2010. High-resolution detection of identity by descent in unrelated individuals. Am. J. Hum. Genet. 86:526–539. doi: 10.1016/j.ajhg.2010.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassell B. G., Adamec V., and Pearson R. E.. . 2003. Effect of incomplete pedigrees on estimates of inbreeding and inbreeding depression for days to first service and summit milk yield in Holsteins and Jerseys. J. Dairy Sci. 86:2967–2976. doi: 10.3168/jds.S0022-0302(03)73894-6 [DOI] [PubMed] [Google Scholar]
- Ceballos F. C., Joshi P. K., Clark D. W., Ramsay M., and Wilson J. F.. . 2018. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 19:220–234. doi: 10.1038/nrg.2017.109 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., and Lee J. J.. . 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., and Willis J. H.. . 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10:783–796. doi: 10.1038/nrg2664 [DOI] [PubMed] [Google Scholar]
- Curik I., Ferenčaković M., and Sölkner J.. . 2017. Genomic dissection of inbreeding depression: a gate to new opportunities. R. Bras. Zootec. 46:773–782. doi: 10.1590/s1806-92902017000900010 [DOI] [Google Scholar]
- Doekes H. P., Veerkamp R. F., Bijma P., de Jong G., Hiemstra S. J., and Windig J. J.. . 2019. Inbreeding depression due to recent and ancient inbreeding in Dutch Holstein–Friesian dairy cattle. Genet. Sel. Evol. 51:54. doi: 10.1186/s12711-019-0497-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druet T., and Gautier M.. . 2017. A model-based approach to characterize individual inbreeding at both global and local genomic scales. Mol. Ecol. 26:5820–5841. doi: 10.1111/mec.14324 [DOI] [PubMed] [Google Scholar]
- Ferenčaković M., Hamzić E., Gredler B., Solberg T. R., Klemetsdal G., Curik I., and Sölkner J.. . 2013a. Estimates of autozygosity derived from runs of homozygosity: empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 130: 286–293. doi: 10.1111/jbg.12012 [DOI] [PubMed] [Google Scholar]
- Ferenčaković M., Sölkner J., and Curik I.. . 2013b. Estimating autozygosity from high-throughput information: effects of SNP density and genotyping errors. Genet. Sel. Evol. 45:42. doi: 10.1186/1297-9686-45-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenčaković M., Sölkner J., Kapš M., and Curik I.. . 2017. Genome-wide mapping and estimation of inbreeding depression of semen quality traits in a cattle population. J. Dairy Sci. 100:4721–4730. doi: 10.3168/jds.2016-12164 [DOI] [PubMed] [Google Scholar]
- Gurgul A., Szmatoła T., Topolski P., Jasielczuk I., Żukowski K., and Bugno-Poniewierska M.. . 2016. The use of runs of homozygosity for estimation of recent inbreeding in Holstein cattle. J. Appl. Genet. 57:527–530. doi: 10.1007/s13353-016-0337-6 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W. 1994. Purging inbreeding depression and the probability of extinction: full-sib mating. Heredity (Edinb). 73(Pt 4):363–372. doi: 10.1038/hdy.1994.183 [DOI] [PubMed] [Google Scholar]
- Holt M., Meuwissen T., and Vangen O.. . 2005. The effect of fast created inbreeding on litter size and body weights in mice. Genet. Sel. Evol. 37:523–537. doi: 10.1186/1297-9686-37-6-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howrigan D. P., Simonson M. A., and Keller M. C.. . 2011. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics 12:460. doi: 10.1186/1471-2164-12-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Maltecca C., Macneil M. D., Alexander L. J., Snelling W. M., and Cassady J. P.. . 2012. Using 50 k single nucleotide polymorphisms to elucidate genomic architecture of line 1 Hereford cattle. Front. Genet. 3:285. doi: 10.3389/fgene.2012.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M., Qvarnström A., and Ellegren H.. . 2017. Inferring individual inbreeding and demographic history from segments of identity by descent in Ficedula flycatcher genome sequences. Genetics 205:1319–1334. doi: 10.1534/genetics.116.198861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. C., Simonson M. A., Ripke S., Neale B. M., Gejman P. V., Howrigan D. P., Lee S. H., Lencz T., Levinson D. F., and Sullivan P. F.; Schizophrenia Psychiatric Genome-Wide Association Study Consortium 2012. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 8:e1002656. doi: 10.1371/journal.pgen.1002656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. C., Visscher P. M., and Goddard M. E.. . 2011. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics 189:237–249. doi: 10.1534/genetics.111.130922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. S., Sonstegard T. S., Van Tassell C. P., Wiggans G., and Rothschild M. F.. . 2015. The relationship between runs of homozygosity and inbreeding in Jersey cattle under selection. PLoS One. 10:e0129967. doi: 10.1371/journal.pone.0129967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp B., Church R. C., and Flower A. E.. . 1951. Genetic history of the line 1 Hereford cattle at the United States range livestock experiment station. Montana Agri. Exp. Sta. Bull. 479:3–27. [Google Scholar]
- Ku C. S., Naidoo N., Teo S. M., and Pawitan Y.. . 2011. Regions of homozygosity and their impact on complex diseases and traits. Hum. Genet. 129:1–15. doi: 10.1007/s00439-010-0920-6 [DOI] [PubMed] [Google Scholar]
- Leesburg V. L. R., MacNeil M. D., and Neser F. W. C.. . 2014. Influence of miles city line 1 on the United States Hereford population1,2,3. J. Anim. Sci. 92: 2387–2394. doi: 10.2527/jas.2013-6890 [DOI] [PubMed] [Google Scholar]
- MacNeil M. D. 2009. Invited Review: Research contributions from seventy-five years of breeding line 1 Hereford cattle at miles city, Montana1,2. J. Anim. Sci. 87:2489–2501. doi: 10.2527/jas.2009-1909 [DOI] [PubMed] [Google Scholar]
- MacNeil M. D., Cardoso F. F., and Hay E.. . 2017. Genotype by environment interaction effects in genetic evaluation of preweaning gain for line 1 Hereford cattle from Miles City, Montana. J. Anim. Sci. 95:3833–3838. doi: 10.2527/jas2017.1829 [DOI] [PubMed] [Google Scholar]
- MacNeil M. D., Urick J. J., Newman S., and Knapp B. W.. . 1992. Selection for postweaning growth in inbred Hereford cattle: the Fort Keogh, Montana line 1 example. J. Anim. Sci. 70:723–733. doi: 10.2527/1992.703723x [DOI] [PubMed] [Google Scholar]
- MacNeil M., and Vermeire L.. . 2012. Effect of weather patterns on preweaning growth of beef calves in the Northern Great Plains. Agric. Sci. 3:929. doi: 10.4236/as.2012.37113 [DOI] [Google Scholar]
- Martikainen K., Sironen A., and Uimari P.. . 2018. Estimation of intrachromosomal inbreeding depression on female fertility using runs of homozygosity in Finnish Ayrshire cattle. J. Dairy Sci. 101:11097–11107. doi: 10.3168/jds.2018-14805 [DOI] [PubMed] [Google Scholar]
- Mc Parland S., Kearney F., and Berry D. P.. . 2009. Purging of inbreeding depression within the Irish Holstein-Friesian population. Genet. Sel. Evol. 41:16. doi: 10.1186/1297-9686-41-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A. L., Abdel-Rahman R., Franklin C. S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A., . et al. 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83:359–372. doi: 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans R., Gorssen W., Buys N., and Janssens S.. . 2020. How to study runs of homozygosity using PLINK? A guide for analyzing medium density SNP data in livestock and pet species. BMC Genomics 21:94. doi: 10.1186/s12864-020-6463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Strabel T., Auvray B., Druet T., and Lee D. H.. . 2002. BLUPF90 and related programs (BGF90). In: Proceedings of the 7th World Congress on Genetics Applied to Livestock Production; August 19–23, 2002. Montpellier (France); Organising Committee; p. 28. [Google Scholar]
- Nani J. P., and Peñagaricano F.. . 2020. Whole-genome homozygosity mapping reveals candidate regions affecting bull fertility in US Holstein cattle. BMC Genomics 21:338. doi: 10.1186/s12864-020-6758-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton T. J., Absher D., Feldman M. W., Myers R. M., Rosenberg N. A., and Li J. Z.. . 2012. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91:275–292. doi: 10.1016/j.ajhg.2012.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton T. J., and Szpiech Z. A.. . 2018. Relationship between deleterious variation, genomic autozygosity, and disease risk: insights from the 1000 genomes project. Am. J. Hum. Genet. 102:658–675. doi: 10.1016/j.ajhg.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peripolli E., Stafuzza N. B., Munari D. P., Lima A. L. F., Irgang R., Machado M. A., Panetto J. C. D. C., Ventura R. V., Baldi F., and da Silva M. V. G. B.. . 2018. Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics 19:34. doi: 10.1186/s12864-017-4365-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J. E., Haile-Mariam M., Goddard M. E., and Hayes B. J.. . 2014. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 46:71. doi: 10.1186/s12711-014-0071-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., . et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D. C., Berry D. P., McParland S., and Bradley D. G.. . 2012. Runs of homozygosity and population history in cattle. BMC Genet. 13:70. doi: 10.1186/1471-2156-13-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from https://www.R-project.org/. [Google Scholar]
- Sams A. J., and Boyko A. R.. . 2019. Fine-scale resolution of runs of homozygosity reveal patterns of inbreeding and substantial overlap with recessive disease genotypes in domestic dogs. G3 (Bethesda). 9:117–123. doi: 10.1534/g3.118.200836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura M., Fernández A., Varona L., Fernández A. I., de Cara M. Á., Barragán C., and Villanueva B.. . 2015. Detecting inbreeding depression for reproductive traits in Iberian pigs using genome-wide data. Genet. Sel. Evol. 47:1. doi: 10.1186/s12711-014-0081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumreddee P., Toghiani S., Hay E. H., Roberts A., Agrrey S. E., and Rekaya R.. . 2019. Inbreeding depression in line 1 Hereford cattle population using pedigree and genomic information. J. Anim. Sci. 97:1–18. doi: 10.1093/jas/sky385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmatoła T., Gurgul A., Jasielczuk I., Ząbek T., Ropka-Molik K., Litwińczuk Z., and Bugno-Poniewierska M.. . 2019. A comprehensive analysis of runs of homozygosity of eleven cattle breeds representing different production types. Animals (Basel) 9:1024. doi: 10.3390/ani9121024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmatoła T., Gurgul A., Ropka-Molik K., Jasielczuk I., Ząbek T., and Bugno-Poniewierska M.. . 2016. Characteristics of runs of homozygosity in selected cattle breeds maintained in Poland. Livest. Sci. 188:72–80. doi: 10.1016/j.livsci.2016.04.006 [DOI] [Google Scholar]
- Szpiech Z. A., Xu J., Pemberton T. J., Peng W., Zöllner S., Rosenberg N. A., and Li J. Z.. . 2013. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 93:90–102. doi: 10.1016/j.ajhg.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L., Zhu Z., Wray N. R., Weir B. S., Yang J., Robinson M. R., and Visscher P. M.. . 2017. Detection and quantification of inbreeding depression for complex traits from SNP data. Proc. Natl. Acad. Sci. U. S. A. 114:8602–8607. doi: 10.1073/pnas.1621096114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Calus M. P., Guldbrandtsen B., Lund M. S., and Sahana G.. . 2015a. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet. 16:88. doi: 10.1186/s12863-015-0227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Guldbrandtsen B., Bosse M., Lund M. S., and Sahana G.. . 2015b. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics 16:542. doi: 10.1186/s12864-015-1715-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.