Abstract
Abscess is the highest cause of liver condemnation and is estimated to cost the beef industry US$64 million annually. Fusobacterium necrophorum, commonly found in the bovine rumen, is the primary bacteria associated with liver abscess in cattle. Theoretically, damage to the rumen wall allows F. necrophorum to invade the bloodstream and colonize the liver. The objective of this study was to determine the changes in gene expression in the rumen epithelium and microbial populations adherent to the rumen epithelium and in the rumen contents of beef cattle with liver abscesses compared with those with no liver abscesses. Rumen epithelial tissue and rumen content were collected from 31 steers and heifers with liver abscesses and 30 animals with no liver abscesses. Ribonucleic acid (RNA) sequencing was performed on the rumen epithelium, and a total of 221 genes were identified as differentially expressed in the animals with liver abscesses compared with animals with no abscesses, after removal of genes that were identified as a result of interaction with sex. The nuclear factor kappa-light-chain enhancer of activated B cells signaling and interferon signaling pathways were significantly enriched in the differentially expressed gene (DEG) set. The majority of the genes in these pathways were downregulated in animals with liver abscesses. In addition, RNA translation and protein processing genes were also downregulated, suggesting that protein synthesis may be compromised in animals with liver abscesses. The rumen content bacterial communities were significantly different from the rumen wall epimural bacterial communities. Permutational multivariate analysis of variance (PERMANOVA) analysis did not identify global differences in the microbiome of the rumen contents but did identify differences in the epimural bacterial communities on the rumen wall of animals without and with liver abscesses. In addition, associations between DEG and specific bacterial amplicon sequence variants of epimural bacteria were observed. The DEG and bacterial profile on the rumen papillae identified in this study may serve as a method to monitor animals with existing liver abscesses or to predict those that are more likely to develop liver abscesses.
Keywords: cattle, liver abscess, rumen, transcriptome
Introduction
Liver abscesses in feedlot steers have been associated with high-concentrate grain finishing diets. The prevalence of liver abscesses in feedlot steers ranges between 10% and 20% with a slight rise in the prevalence of liver abscesses in beef cattle in the United States over the last decade (Scarth, 2006; Reinhardt and Hubbert, 2015) and liver condemnation rose almost 10% between 2011 and 2016 with abscess accounting for 58% of all liver condemnation (Harris et al, 2017). While the frequency of liver abscesses is mitigated with tylosin phosphate (i.e., Tylan; Reinhardt and Hubbert, 2015), a macrolide antibiotic, there is pressure to reduce or eliminate the use of prophylactic antibiotics in the United States.
Liver abscesses are the result of bacteria, especially Fusobacterium necrophorum, entering the liver through the portal vein as a result of damage to the rumen wall (Nagaraja and Chengappa, 1998). Injury to the rumen wall may be due to acidosis or physical damage from infiltration by ingested hair or debris (Jensen et al., 1954; Amachawadi and Nagaraja, 2016). Large or multiple small liver abscesses are known to reduce cattle body weight gain and feed intake (Brink et al., 1990). The reduction in hot carcass weight of animals with liver abscesses is estimated to reduce producer profitability by US$38 per animal (Scarth, 2006). Liver abscesses may also affect animal well-being; however, there is currently no effective method available to test whether animals are affected by liver abscesses prior to slaughter.
This study was part of a larger study designed to evaluate whether the essential oil limonene fed as a supplement to cattle would reduce liver abscesses. Essential oils and their components are active against numerous bacterial species, including many Gram-negative pathogens (Dabbah et al., 1970). Limonene is a monocyclic monoterpene found in lemons, orange, and grapefruits (Castillejos et al., 2006) and has been reported to decrease F. necrophorum populations in vitro (Samii, 2016). However, the limonene treatment had no effect on liver abscess frequency, microbial bacterial community populations, dry matter intake, average daily gain, or hot carcass weight of cattle on the study (data not published).
The underlying molecular mechanisms involved in liver abscesses have not been well studied, especially those occurring within the rumen. An understanding of the etiology of liver abscess may aid in the development of tools for the prediction of animals that are more susceptible to liver abscesses. The purpose of this study was to determine whether the expression of various rumen epithelial genes and changes in the microbial populations in the rumen epithelial tissue and content were associated with liver abscesses.
Materials and Methods
Animal care and use
The U.S. Meat Animal Research Center (USMARC) Animal Care and Use Committee reviewed and approved all animal procedures. The procedures for handling cattle complied with the Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching (FASS, 2010).
Animal population
Heifer and steer calves for this study were born in the fall of 2016 and were from the continuous phase of the USMARC Germplasm Evaluation project (Schiermiester et al., 2015). This breeding population includes contributions from the following 18 breeds: Angus, Beefmaster, Brahman, Brangus, Braunvieh, Charolais, Chiangus, Gelbvieh, Hereford, Limousin, Maine Anjou, Red Angus, Salers, Santa Gertrudis, Shorthorn, South Devon, Simmental, and Tarentaise. Calves were part of a population subset developed to monitor disease susceptibility. In addition, these calves were also part of a larger study to evaluate the use of the essential oil limonene to reduce liver abscesses. Animals were fed a diet of 63% corn, 26.5% wet distillers grain with solubles, 8% corn silage, and either 2.5% mineral and vitamin supplement with monensin (control) or an identical mineral and vitamin supplement with limonene, a generally recognized as a safe essential oil. The experiment included six pens of steers (33 to 34 steers within each pen) and three pens of heifers (60 to 62 heifers within each pen) fed a control and six pens of steers and three pens of heifers fed the limonene treatment. All cattle were fed once daily at 0800 hours and on dietary treatments for approximately 180 d.
Rumen tissue sampling
A total of 370 heifers and 405 steers were harvested and livers were scored at the packing plant by the same USMARC scientist who had extensive training scoring the livers. Livers were scored as 0 for no detection of liver abscess, A− for the detection of one small liver abscess or abscesses, A for the detection of multiple small liver abscesses, or A+ for the detection of multiple large lesions. The rumen of animals with liver abscess scores of A+ and control animals with no liver abscesses were tagged for sampling. Rumens were hung and rumen contents were collected using a sterile tube after incision, and the tube was immediately placed into a bed of powdered dry ice. Two pieces of rumen tissue were sampled from the cranial sac of each rumen after contents were emptied. A 150-cm2 piece of rumen tissue was removed and placed into a clean resealable bag and placed on ice for rumen wall bacterial microbiome analyses. A 5-cm2 piece of rumen tissue was placed into a sterile tube and immediately embedded into powdered dry ice for gene expression. Samples remained on dry ice for approximately 4 to 6 h through transport back to the USMARC. Upon arrival, the samples were transferred to an ultracold freezer and stored at −80 °C until further processing.
Ribonucleic acid isolation
Rumen tissue samples were rinsed with sterile water and papillae were clipped from the rumen wall with scissors that were cleaned with RNAZap and RNase/DNase-free water between samples. Total ribonucleic acid (RNA) was isolated from the rumen tissue using the RNeasy Mini Plus kit and QiaShredder columns (Qiagen). Briefly, 800 µL of RLT buffer with β-mercaptoethanol was added to 50 to 100 mg of rumen papillae tissue and homogenized for 40 s using an Omni Prep 6-station homogenizer (Omni International, Kennesaw, GA, USA). The homogenate was centrifuged through a QiaShredder column at full speed for 3 min. The RNeasy Mini Plus kit manufacturer’s protocol was then followed, and the total RNA was eluted in 50 µL of RNase-free water. Total RNA was quantified with a NanoDrop One spectrophotometer (Thermo Scientific, Wilmington, DE). The 260/280 ratios were ≥ 1.8, and the total RNA from each animal was analyzed for quality on a Tapestation 2200 (Agilent, Santa Clara, CA, USA) and produced an average RNA integrity number (RIN) of 8.5 with a range of 6.2 to 9.8.
RNA sequencing
Samples were prepared for RNA sequencing (RNA-Seq) with the Illumina TruSeq Stranded mRNA High Throughput Sample kit and protocol (Illumina Inc., San Diego, CA, USA). The libraries were diluted to 4 nM in Illumina resuspension buffer. The libraries were sequenced as 75 bp paired-end reads using the 150-cycle high output sequencing kits for the Illumina NextSeq.
Processing RNA-Seq data
The quality of the raw paired-end sequence reads in individual fastq files was assessed using FastQC (version 0.11.5; www.bioinformatics.babraham.ac.uk/projects/fastqc), and then reads were trimmed to remove adapter sequences and low-quality bases using the Trimmomatic software (version 0.35) (Bolger et al., 2014). The remaining reads were mapped to the Agricultural Research Service-University of California, Davis (ARS-UCD)1.2 genome assembly (Genbank Accession GCA_002263795.2) using Hisat2 (version 2.1.0) (Kim et al., 2015). The National Center for Biotechnology Information (NCBI) annotation for ARS-UCD1.2 (Release 106) was used to guide the alignment. StringTie (Pertea et al., 2015) was used to determine read counts for each of the 34,624 annotated genes in the ARS-UCD1.2 genome assembly. Genes with low read counts were filtered out of the dataset when there were <15 reads in at least 30 of the samples. This produced a set of 14,288 genes for downstream analyses. Two libraries were removed from the analysis due to low read counts and lower read mapping percentage. The raw sequencing data can be accessed at sequence read archive database with accession number PRJNA555558. The data were analyzed using the DESeq2 package (Love et al., 2014) with the following generalized linear model:
Differentially expressed genes (DEGs) for the three main effects were identified using DESeq2 (PFDR < 0.05). DEG lists were filtered by removing genes that were significant for any of the four interaction terms.
Functional gene annotation and pathway analyses
Gene functions and pathways of overrepresented DEG (PFDR < 0.05) were determined using the protein analysis through evolutionary relationships (PANTHER) classification system (version 13.1) (Mi et al., 2016) and the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (Huang et al., 2009a, 2009b). Enrichment analysis of gene function was performed using PANTHER’s implementation of the binomial test of overrepresentation. The significance of gene ontology (GO) terms was assessed using the default Ensembl Bos taurus GO annotation as background for the enrichment analysis. The default parameters for the Kyoto encyclopedia of genes and genomes pathway and gene function analyses in DAVID were used with B. taurus annotation and official gene symbols to evaluate genes that were overrepresented in the list of DEG (PFDR < 0.05).
Ingenuity pathway analysis (IPA; QIAGEN Redwood City, CA, USA; www.qiagen.com/ingenuity) was used to identify direct and indirect molecular relationships among DEGs (Krämer et al., 2014). Each of the data sets was imported with a Flexible Format using Gene symbol as the identifier. A core analysis was performed on genes in each set, where a P-value for each network is calculated according to the fit of the user’s set of significant genes and the size of the network.
Deoxynucleic acid extraction
Total deoxynucleic acid (DNA) was extracted from total rumen content (rumen fluid and solid particles) and rumen papillae samples (0.25 g) using the Mag-Bind Soil DNA 96 Kit (Omega Bio-tek, Inc., Norwalk, CT, USA) according to the manufacturer’s protocol with the modifications described below. During cell lysis, two bead-beating steps were performed in a TissueLyser (Qiagen Inc., Valencia, CA, USA) for 10 min at 20 Hz, and samples were incubated in a 95 °C water bath for 5 min to ensure cell lysis. Following the removal of polymerase chain reaction (PCR) inhibitors, nucleic acids were precipitated like the procedure described by Yu and Morrison (2004). Briefly, 850 µL of sample supernatant and 260 μL of sodium acetate (10 mM) were mixed in 1.5 mL Eppendorf tubes, vortexed, and incubated on ice for 5 min followed by a centrifugation at 16,000 × g for 15 min at 4 °C. One volume (650 µL) of supernatant was mixed with one volume of isopropanol and incubated on ice for 30 min followed by a centrifugation at 16,000 × g for 15 min at 4 °C. The nucleic acid pellet was washed with ice-cold ethanol (70%) and then dried under vacuum for 3 min. The pellet was resuspended in 450 µL of Tris (10 mM, pH 8).
16S Ribosomal RNA gene amplicon libraries and sequencing
Amplicon libraries of the 16S ribosomal RNA (rRNA) gene (V4 region) were prepared as described by Kozich et al. (2013). Briefly, each 20 μL PCR amplification reaction contained 0.5 μL Terra PCR Direct Polymerase Mix (0.625 Units), 7.5 μL nuclease-free, sterile water, 10 μL 2× Terra PCR Direct Buffer, 1 μL indexed fusion primers (10 μM), and 1 μL DNA (20 to 70 ng DNA). The cycling conditions included an initial denaturation of 98 °C for 3 min, followed by 25 cycles of 98 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s, and a final extension of 68 °C for 4 min. Following amplification, PCR products from each sample were normalized (1 to 2 ng/µL) using the Just-a-Plate 96 PCR Purification and Normalization kit (Charm Biotech, MO, USA) as described by the manufacturer. The normalized libraries were pooled (10 µL/sample) and purified using the Nucleospin Gel and PCR Cleanup kit (Takara Bio USA, Inc., Mountain View, CA, USA) according to the manufacturer’s protocol. Libraries were quality controlled using the BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and quantified using the DeNovix QFX Fluorometer (DeNovix dsDNA Fluorescence Quantification Assay). The libraries were sequenced using the Illumina Miseq System (Illumina, San Diego, CA, USA) using the V2 500 cycles kit according to the manufacturer’s protocol.
16S rRNA gene amplicon sequence data processing
The Illumina 2 × 250 fastq sequencing files (V4) were processed as described by Callahan et al. (2016a) using R (R Core Team, 2018). The forward and reverse reads were trimmed at a constant length based on their quality score (QC > 30), and the reads with more than two expected errors were filtered out from the data (Edgar and Flyvbjerg, 2015). The DADA2 method was used to distinguish the biological variation from PCR or sequencing errors in the filtered reads, and this method infers the sequence variants after removing the substitution errors from the data (Callahan et al., 2016a). The quality-controlled forward and reverse reads were merged and an amplicon sequence variants (ASVs) table with abundance was generated. The chimeras were removed from the data (Callahan et al., 2016b), and taxonomy was assigned down to the species level by using the silva (silva_nr_v132_train_set. fa) database with naive Bayesian classifier method (Wang et al., 2007). The singletons, Archaea, mitochondria, and Cyanobacteria and Rickettsiales taxa were removed from the data. The sequencing depth for each sample was estimated with R package vegan (Oksanen et al., 2017).
Statistical analysis
The bacterial community differences between rumen content and rumen papillae were tested with nested permutational multivariate analysis of variance (PERMANOVA) (Anderson, 2014) with R package BiodiversityR (Kindt and Coe, 2005). For alpha diversity, we calculated the observed species and Simpson’s diversity index (1-D), which takes account of the abundances of the species, and differences in diversity between rumen content and rumen papillae were tested with Friedman’s test (Friedman, 1937). The rumen content and rumen papillae microbiome data were analyzed (PERMANOVA) separately to determine whether the differences in the bacterial community were due to changes in health (healthy vs. liver abscess), sex (steers vs. heifers), or diet (control vs. limonene). Bacterial community differences for main effects (diet, sex, and liver abscess) were determined using the Bray–Curtis distance matrix for principal coordinate analysis (PCoA) and PERMANOVA analysis. Alpha diversity (observed species and Simpson’s diversity index) was calculated from rumen content and rumen papillae data separately, and its differences for main effects (liver abscess, sex, and diet) were tested with the Wilcox test (Gehan, 1965). The P-value < 0.05 was considered significant for all the microbiome analysis. The prevalent ASVs (n = 194) that were present in at least 50% of the rumen papillae samples were used to run a Pearson correlation with DEG genes (n = 221). The log10-transformed relative abundances of prevalent ASVs and DEG genes were used to generate a correlation matrix. The correlations lower than 0.5 and higher than −0.5 were considered moderate while correlations higher than 0.5 and lower than −0.5 were considered high correlations. The data visualization and statistical analysis were performed using the R programming language.
Results
Liver abscess prevalence and RNA-Seq statistics
A total of 370 heifers and 405 steers were evaluated for liver abscess. Liver abscesses were detected in 12.7% of the heifers and 22.5% of the steers (Table 1). The RNA-Seq libraries from the rumen papillae of a total of 61 steers (n = 40) and heifers (n = 21) with liver abscess scores of 0 (no abscesses) or A+ (severe) and with or without limonene treatment were prepared and sequenced. We selected 61 crossbred heifers and steers with and without liver abscesses in order to identify DEGs that are more likely to be robust across sex and population. Fewer heifers were selected because fewer female animals developed liver abscesses. Table 2 presents the number of animals selected by sex, limonene treatment, and liver abscess score. An average of over 42 million 75-bp paired-end reads was generated per animal. Sequence reads were mapped to the B. taurus ARS-UCD1.2 genome assembly with an average 98% read mapping rate.
Table 1.
Liver abscess scores for steers (n = 405) and heifers (n = 370)
| Liver score1 | Steers2 | Heifers |
|---|---|---|
| 0 | 313 (77.3) | 323 (87.2) |
| A | 18 (4.4) | 11 (3) |
| A− | 14 (3.5) | 18 (4.9) |
| A+ | 60 (14.8) | 18 (4.9) |
1Liver abscess scores of 0 = no liver abscess present; A = multiple small liver abscesses; A− = one small liver abscess; A+ = multiple large liver abscesses.
2Number of animals with or without liver abscesses followed by the percentage in parentheses.
Table 2.
Steers and heifers selected for the study by limonene or control treatment and the presence or absence of liver abscess
| Treatment group1 | Steers | Heifers |
|---|---|---|
| LIM(−) ABS (−) | 11 | 6 |
| LIM(−) ABS (+) | 11 | 5 |
| LIM (+) ABS (−) | 8 | 5 |
| LIM (+) ABS (+) | 10 | 5 |
1LIM, limonene. (+) treated with limonene in the feed supplement. (−) no limonene in the supplement. ABS, abscess. (+) animals with liver abscess (score of A+). (−) animals with no detectable liver abscess (score of 0).
DEGs in rumen papillae
A total of 430 genes were identified as differentially expressed in the comparison between animals with liver abscesses and those with no liver abscess (Supplementary Table S1). The analysis for sex (steers compared with heifers) identified the highest number of DEG with a total of 3,396 (PFDR < 0.05; Supplementary Table S2). Only two genes (PRKCA and LOC112441508) were identified in the analysis for limonene vs. control animals (PFDR < 0.05; Supplementary Table S3). After removing genes identified in the analyses of the interaction terms, there were 221 DEGs associated with liver abscesses (Supplementary Table S1). Of these, 96 were upregulated and 125 were downregulated in animals with no liver abscesses.
Gene functions and pathways
Pathways identified by DAVID as overrepresented from the list of DEGs for liver abscess were the transcriptional misregulation in cancer, cyclic adenosine monophosphate (cAMP) signaling, ribosome, cardiac muscle contraction, and nuclear factor kappa B (NFKB) signaling pathways (P < 0.05; Table 3). The PANTHER database search identified the chemokine and cytokine signaling pathway as the pathway with five overrepresented genes (RELA, RELB, RGS4, TYK2, and VWA2). The genes identified in this pathway included: RELA, RELB, TYK2, RGS4, and VWA2. IPA produced the canonical pathways EIF2 signaling, interferon signaling, inducible nitric oxide synthase (iNOS) signaling, hepatic fibrosis/hepatic stellate cell activation, and role of JAK1, JAK2, and TYK in interferon signaling pathways (P < 0.05; Table 4).
Table 3.
Pathway analysis performed with the DAVID v6.8 using a list of DEGs for liver abscesses
| Term | Count | P-value | Genes1 |
|---|---|---|---|
| Transcriptional misregulation in cancer | 6 | 0.02 | IGF1R, RELA, TSPAN7, ETV6, ETV5, HIST1H3G |
| cAMP signaling pathway | 6 | 0.03 | HCN2, ADRB2, ATP1B3, RELA, CREB3L2, SLC9A1 |
| Ribosome | 5 | 0.03 | RPS25, RPL22, RPS27L, RPL38, RPL39 |
| Cardiac muscle contraction | 4 | 0.03 | COX7A1, ATP1B3, SLC9A1, UQCRB |
| NFKB signaling pathway | 4 | 0.05 | LY96, RELA, RELB, TRIM25 |
1 IGF1R, insulin-like growth factor 1 receptor; RELA, RELA proto-oncogene, NFKB subunit; TSPAN7, tetraspanin 7; ETV6, ETS variant transcription factor 6; ETV5, ETS variant transcription factor 5; HIST1H3G, histone H3; HCN2, hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2; ADRB2, adrenoceptor beta 2; ATP1B3, ATPase Na+/K+ transporting subunit beta 3; CREB3L2, CAMP responsive element-binding protein 3 like 2; SCL9A1, sodium/hydrogen exchanger NHE1; RPS25, ribosomal protein S25; RPL22, ribosomal protein L22; RPS27L, ribosomal protein S27 like; RPL38, ribosomal protein L38; RPL39, ribosomal protein L39; COX7A1, cytochrome C oxidase subunit 7A1; UQCRB, ubiquinol-cytochrome C reductase-binding protein; LY96, lymphocyte antigen 96; RELB, RELB proto-oncogene, NFKB subunit; TRIM25, tripartite motif containing 25.
Table 4.
Canonical pathways identified by IPA using a list of DEGs for liver abscesses
| Canonical pathway | P-value | No. of genes | Gene names1 |
|---|---|---|---|
| EIF2 signaling | 0.0003 | 8 | EIF3A, IGF1R, RPL22, RPL38, RPL39, RPL36AL, RPS25, RPS27L |
| Interferon signaling | 0.003 | 3 | IFI6, RELA, TYK2 |
| iNOS signaling | 0.005 | 3 | LY96, RELA, TYK2 |
| Hepatic fibrosis/ hepatic stellate cell activation | 0.01 | 5 | COL7A1, IGF2, IGF1R, LY96, RELA |
| Role of JAK1, JAK2 and TYK2 in interferon signaling | 0.01 | 2 | RELA, TYK2 |
1 EIF3A, eukaryotic translation initiation factor 3 subunit A; IGF1R, insulin-like growth factor 1 receptor; RPL22, ribosomal protein L22; RPL38, ribosomal protein L38; RPL39, ribosomal protein L39; RPL36AL, ribosomal protein L36a Like; RPS25, ribosomal protein S25; RPS27L, ribosomal protein S27 like; IFI6, interferon alpha inducible protein 6; RELA, RELA proto-oncogene, NFKB subunit; TYK2, tyrosine kinase 2; COX7A1, cytochrome C oxidase subunit 7A1; IGF2, insulin-like growth factor 2; LY96, lymphocyte antigen 96.
Differences between rumen content and rumen papillae microbiota
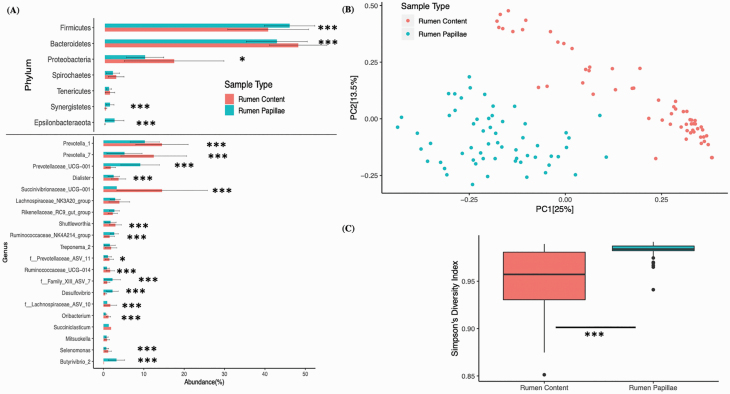
The processing of 3,655,108 reads from all the samples resulted in 4,477 unique ASVs, and 89% and 50% of the identified ASVs were assigned taxonomy at phylum and genus level, respectively. The nested PERMANOVA analysis of the Bray–Curtis distance matrix showed that the bacterial community composition associated with rumen content was different (P < 0.001) from rumen papillae. At the phyla level, significant differences were identified in five of the phylogenetic groups. The rumen content had a higher abundance of Bacteroidetes and Proteobacteria, whereas rumen papillae had a higher abundance of Firmicutes, Synergistetes, and Epsilonbacteraeota (Campylobacterota). At the genus level, there were significant differences in 15 genera. The rumen content had a higher abundance of 10 of the genera, including Prevotella_1, Prevotella_7, and Succinivibrionaceae_UCG-001, whereas rumen papillae had a higher abundance of 5 genera, including Prevotellaceae_UCG-001, Desulfovibrio, and Butyrivibrio_2 (Figure 1A). The PCoA analysis plot (Figure 1B) indicated clustering based on sample location (rumen content vs. rumen epithelium) and the first two principal coordinate components (PC1 and PC2) explained 25% and 13.5% of the total variation, respectively. Alpha diversity analysis showed that rumen papillae samples to have a higher number of associated bacterial species (P = 0.05) and high diversity index (P < 0.001) as compared with rumen content (Figure 1C and Table 5).
Figure 1.
The phylum and genus level differences in relative abundance between rumen papillae and rumen content. (A) Bar plot with error bars is showing the phylum and genus level differences in relative abundance between rumen papillae and rumen content. The y-axis is representing the phyla and genera with at least 1% relative abundance. The Friedman’s test was used to see the differences in relative abundances of different phyla and genera between rumen content and rumen papillae samples (*P < 0.05; ***P < 0.001). (B) The PCoA plot is showing the bacterial community differences between rumen papillae and rumen content samples. Bray–Curtis distance matrix was used to run the PCoA and PERMANOVA analysis. Each dot represents the bacterial community of individual sample and the distance between two dots shows the difference in the bacterial community between two samples. (C) This plot is showing the Simpson’s diversity index (y-axis) in rumen content and rumen papillae samples (x-axis). The Friedman’s test was used to test the difference in alpha diversity between sample sources.
Table 5.
Alpha diversity of rumen content and rumen papillae
| Observed bacterial species | P-value1 | Simpson’s Diversity Index (1-D) | P-value1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample source | Rumen content (n = 59) | 274 ± 63 | 0.0508 | 0.95 ± 0.03 | <0.001 | ||||
| Rumen papillae (n = 59) | 303 ± 64 | 0.98 ± 0.01 | |||||||
| Rumen content | P-value2 | Rumen papillae | P-value2 | Rumen content | P-value2 | Rumen papillae | P-value2 | ||
| Liver abscesses | Healthy (n = 30) | 282 ± 60 | >0.05 | 308 ± 71 | >0.05 | 0.96 ± 0.03 | >0.05 | 0.98 ± 0.01 | >0.05 |
| Liver abscessed (n = 29) | 265 ± 66 | 298 ± 56 | 0.95 ± 0.04 | 0.98 ± 0.01 | |||||
| Sex | Steers (n = 39) | 272 ± 63 | >0.05 | 301 ± 66 | >0.05 | 0.96 ± 0.03 | >0.05 | 0.98 ± 0.01 | >0.05 |
| Heifers (n = 20) | 280 ± 65 | 308 ± 60 | 0.94 ± 0.04 | 0.98 ± 0.01 | |||||
| Diet | Limonene (28) | 274 ± 73 | >0.05 | 310 ± 68 | >0.05 | 0.95 ± 0.04 | >0.05 | 0.98 ± 0.01 | >0.05 |
| No limonene (31) | 274 ± 53 | 297 ± 60 | 0.96 ± 0.03 | 0.98 ± 0.01 | |||||
1Alpha diversity differences between different sample sources (rumen content vs. rumen papillae) were tested with Friedman’s test.
2Alpha diversity differences between different variables (healthy vs. liver abscess, Steer vs. Heifers, and limonene vs. no limonene) within rumen content or rumen papillae samples were tested with Wilcox test.
Bacterial community differences between healthy and liver-abscessed animals
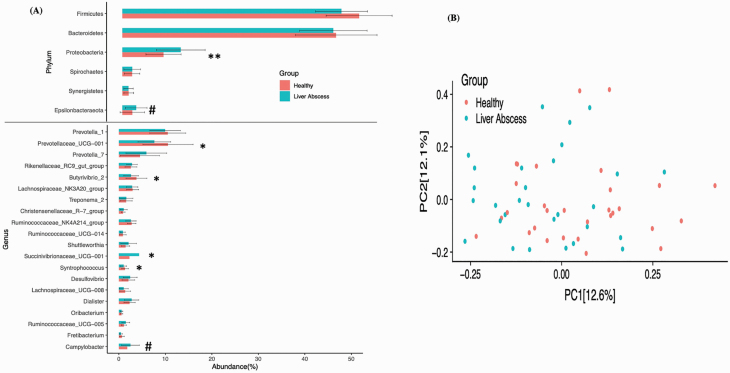
The rumen papillae samples were further analyzed for phylum and genus level differences between animals without and with liver abscesses. At the phyla level, liver-abscessed animals had a higher (P < 0.05) abundance of Proteobacteria and tendency (P = 0.053) for a higher abundance of Epsilonbacteraeota (Campylobacterota) as compared with healthy animals (Figure 2A). At the genus level, there were significant differences in four genera. The epimural microbiota of liver-abscessed animals had a higher abundance of Succinivibrionaceae_UCG-001 and, as a note, a strong tendency for higher Campylobacter (P = 0.053), whereas healthy animals had a significantly higher abundance of Prevotellaceae_UCG-001, Butyrivibrio_2, and Syntrophococcus (P < 0.05).
Figure 2.
Phylum and genus differences in the rumen papillae of cattle with and without liver abscesses. (A) The bar plot with error bars is showing the phylum and genus level difference in relative abundance between healthy and liver-abscessed animals. The phyla and genera with at least 1% abundances are shown on y-axis and their abundances on x-axis. The difference in relative abundance of different phyla and genera between healthy and liver-abscessed animals was tested with Wilcox text (#P < 0.1; *P < 0.05; **P < 0.01). (B) The Bray–Curtis distance matrix was used to run PCoA and PERMANOVA analysis. The first two components (PC1 and PC2) of PCoA were plotted to see the bacterial community differences between healthy and liver-abscessed animals.
For rumen papillae data, the PERMANOVA analysis showed that limonene supplementation did not affect the epimural bacterial communities, but a difference in epimural bacterial communities was observed between heifers and steers (P = 0.048). The Bray–Curtis distance matrix was used to generate the PCoA plot (Figure 2B), which did not show any clustering between animals with and without liver abscesses, but PERMANOVA analysis showed that healthy and liver-abscessed animals had different (P = 0.02) epimural microbiota. For rumen content data, PERMANOVA analysis showed that diet, sex, and health of the animal did not affect (P > 0.05) the bacterial communities in rumen content.
Interaction between rumen papillae-associated microbiome and rumen epithelial expressed genes
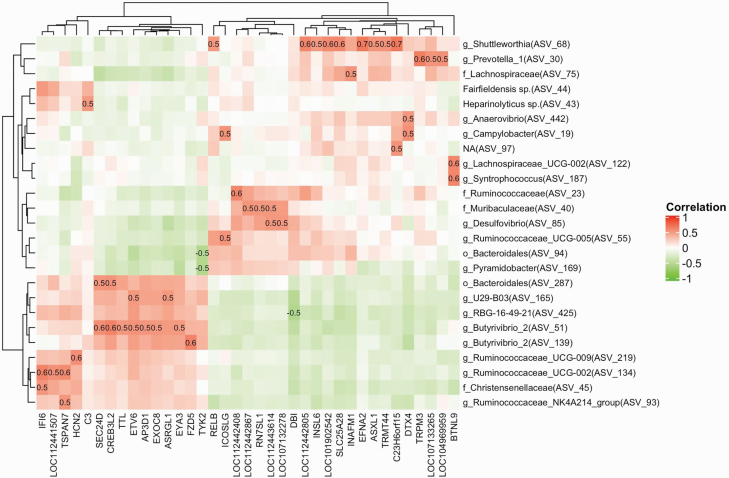
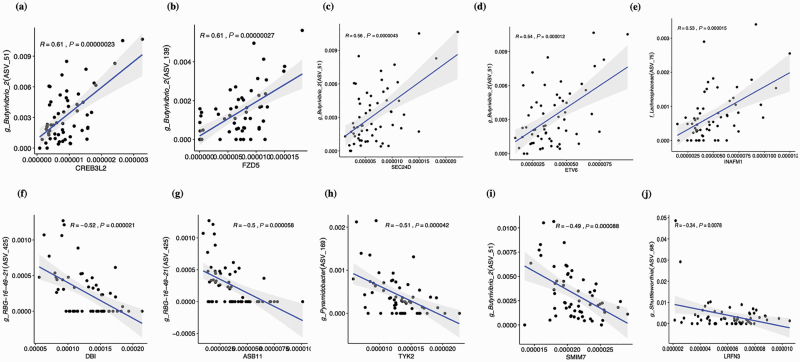
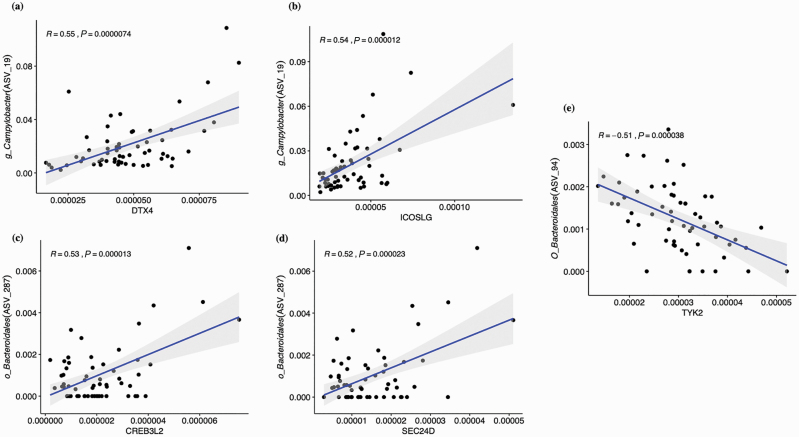
A wide range of correlations (−0.52 to 0.67) was observed. There were 165 ASVs that exhibited at least a moderate correlation (−0.3 > R > 0.3; 1,433 positive and 640 negative) to at least one of 221 DEG genes, and 34 of these ASVs exhibited moderate correlations to a minimum of 20 genes and were plotted in a Heatmap (Supplemental Figure 1). The ASV51 (Butryivibrio_2) exhibited the highest number of associations overall with 96 moderate correlations with DEG genes (76 positive and 20 negative), and a significant number of associations were observed with other ASVs, including in order, ASV2 (Dialister; n = 82), ASV425 (RBG-16-49-21; n = 77), ASV165 (U29-B03; n = 68), and ASV213 (Treponema_2; n = 65). Collectively, five ASVs that were taxonomically identified as Butyrivibrio_2 exhibited 271 moderate correlations. At a higher degree, there were 25 ASVs that exhibited at least a high correlation (Supplementary Table S5, −0.5 > R > 0.5; 48 positive and 3 negative) to at least one of 37 DEG genes as shown in a Heatmap (Figure 3), and 10 of these ASVs exhibited a high correlation to more than one of 12 genes. The ASV68 alone was positively correlated with expression differences of nine DEGs, including EFNA2 and RELB, and this ASV identified as the genus Shuttleworthia. The most significant five positive and five negative correlations between prevalent ASVs and DEG genes are shown in Figure 4, and correlations between the ASVs that identify with taxonomical groups of bacteria associated with bovine liver abscesses and DEG genes are shown in Figure 5.
Figure 3.
Heatmap of the highest correlated (−0.5 > R > 0.5) ASVs and DEG genes present in cattle rumen papillae. The prevalent ASVs (n = 194; present in at least 50% of the rumen papillae samples) were used to run a Pearson correlation with DEG genes (n = 221). The log10-transformed relative abundances of prevalent ASVs and DEG genes were used to generate correlation matrix. The x-axis is representing the ASVs whereas the DEG genes are on y-axis.
Figure 4.
Correlations between the core ASVs and DEG. (A)–(E) illustrate positive and (F)–(J) are negative correlations between core ASVs and DEG genes. The log10-transformed relative abundances were used to calculate the Pearson correlation. The x-axis is the DEG gene and y-axis is the ASV with taxonomy.
Figure 5.
Correlations between the core ASVs and DEG of bacterial ASVs potentially associated with liver abscesses (A–E). The log10-transformed relative abundances were used to calculate the Pearson correlation. The x-axis is the DEG gene and y-axis is the ASV with taxonomy.
Discussion
Because the etiology of liver abscess is thought to stem from damage to the rumen epithelial tissue, we chose to evaluate the transcriptome and microbiome of rumen papillae from heifers and steers with and without liver abscesses. While we do not know when these animals started developing liver abscesses, it is likely that they began to form during dietary adaptation or during the initial phase of feedlot finishing rations. However, the evaluation for liver abscesses and the collection of tissue for this study were performed at the time of slaughter. Currently, there is no test or assay to determine when an animal develops liver abscesses; thus, performing this study at harvest seemed a reasonable time point. To our knowledge, this is the first study to evaluate the expression of genes in the rumen, and the rumen epimural bacterial communities for differences associated with liver abscesses detected at slaughter.
The animals used in this study were part of a larger study to evaluate whether limonene was an effective supplement to reduce liver abscesses. The limonene treatment did not reduce the incidence of liver abscesses or have any effect on performance characteristics (data not published). The transcriptome data from this study support these findings with only two genes differentially expressed between limonene-treated and untreated animals.
There were approximately 3,400 genes identified as differentially expressed by sex in the rumen papillae in this study. Many of these genes were members of canonical pathways involved in growth and energy production. There are known differences in the finishing production of heifers and steers (Williams et al., 1993) that may offer insight into the pathways identified. Growth curves for heifers and steers on a feedlot diet are different (Sorensen, 1972), and mature body weight has long been associated with sex (Emmans, 1997; Zinn et al., 2008). Growth and body weight gain are complex, polygenic traits, and with additional variation due to sex, it stands to reason that genes involved in cellular energy production, transcription, and protein synthesis would be differentially expressed between steers and heifers.
Animals with liver abscesses exhibited differences in the rumen expression of inflammatory response genes. The wgenes LY96, RELA, RELB, and TRIM25 were identified by DAVID as overrepresented in the NFKB pathway and the genes IFI6, RELA, and TYK2 were associated with the interferon signaling pathway by IPA. Four of these six genes (RELA, TRIM25, IFI6, and TYK2) were downregulated in the cattle with liver abscesses; two (RELB and LY96) were upregulated. The lower expression of cytokines in the NFKB pathway does not appear to be a common mechanism for disease or tissue damage, so this may be a rather novel finding, as NFKB is a common host response to microbial pathogens. However, some pathogens have mechanisms to inhibit host inflammatory and immune responses by inhibiting the NFKB pathway during various stages of their lifecycle (Rahman and McFadden, 2011). For example, Bordetella pertussis produces an adhesin that induces the early activation of the NFKB pathway but longer exposure to the adhesin inhibits NFKB activation (Abramson et al., 2008; Rahman and McFadden, 2011).
Several ribosomal genes were identified as overexpressed in the ribosome pathway by DAVID (RPL22, RPL38, RPL39, RPS25, and RPS27L) and the EIF2 signaling pathway by IPA (RPL22, RPL38, RPL39, RPL36AL, RPS25, RPS27L, EIF3A, and IGF1R). All of the ribosomal genes were downregulated in the rumen tissue of animals with liver abscesses compared with those without, while EIF3A and IGF1R were upregulated. The reduced expression of ribosomal genes may suggest that protein synthesis is compromised in the rumen tissue of animals with liver abscesses. While this is the first report of the transcriptome of the rumen in animals with and without liver abscesses, there is precedence for the involvement of EIF2 phosphorylation in the liver of mice fed a high fructose diet, which causes nonalcoholic fatty liver disease. Mice that were deficient in EIF2α phosphorylation showed adverse effects on antioxidant capacity, inflammation, and cell viability (Choi et al., 2017).
While we believe that it may be challenging to visualize small lesions that could allow bacteria to move from the rumen to the liver, we did visually inspect the rumen samples and rumen epithelium at the time of collection, and the tissues were similar in appearance for animals without and with liver abscesses. Not only it is possible that a chronic abscessed liver might alter some cytokine signals in distant tissues like the rumen, but it is also a possibility that the lower level of cytokine genes expressed in the rumen of some animals predisposes calves to the infiltration of F. necrophorum, which ultimately causes liver abscesses.
Several bacteria have been associated with liver abscesses in cattle, including F. necrophorum and Trueperella pyogenes (Amachawadi and Nagaraja, 2016). The rumens of cattle are a reservoir for F. necrophorum, and we evaluated the potential relationships between the rumen bacterial communities and liver abscesses. More importantly, the rumen epithelium is a barrier to F. necrophorum, and the epimural bacteria that populate the papillae not only could be important to rumen function but might also impact the ability of F. necrophorum to infiltrate from the rumen into the blood that goes to the liver. From a community diversity standpoint, the bacterial community structure of the rumen content was distinctly different from the epimural populations found on the rumen papillae in this study and that of Reyes et al. (2019). Although differences in populations between the rumen content and epimural bacteria were expected, there is little available literature reporting these differences (Dinsdale et al., 1980; Reyes et al., 2019). The predominance of the phylum Bacteroidetes in the rumen observed in this current research is well recognized (Myer et al., 2017; Reyes et al., 2019). In contrast, the epimural populations were predominated by Firmicutes which, although studies are fewer, agrees with previous research (Chen et al., 2011; Petri et al., 2013).
The relationships between dietary supplementation of limonene or gender on the community structure of the rumen content bacteria were minimal and not significant relative to diversity analyses or PERMANOVA analysis. Furthermore, the relationship between liver abscess status and bacterial community population structure of the rumen content was not significant. Considering that the rumen content collected for the current study was done at harvest, it would appear that acute population shifts attributed to diet shifts or acidosis may be transient and not reflected in the long-term community structure.
A PERMANOVA analysis of the epimural bacterial communities collected from the rumen papillae did indicate differences in the animals without and with liver abscesses, and significant differences at the phyla level were observed. Animals with liver abscesses had a higher abundance of Proteobacteria, which is a major phylum of phylogenetically related but phenotypically diverse bacteria that includes a variety of opportunistic pathogens (Rizzatti et al., 2017). Considering that this phylum was observed to be more abundant in the rumen content than on the papillae in general, the observation that its higher abundance in the epimural population for the liver-abscessed animal suggests a dysbiosis of the rumen epimural bacteria in the diseased animal. The phylum Epsilonbacteraeota, also recognized as Campylobacterota (Waite et al., 2017), exhibited a strong tendency for greater abundance in the liver-abscessed epimural bacterial populations, and this phylum includes the genera Campylobacter, Helicobacter, and Arcobacter, all pathogenic species found in food animals that invade epithelial tissue (Chan et al., 1992; Wooldridge and Ketley, 1997; Levican et al., 2013).
At the genus level, the epimural population differed significantly for only a few bacterial groups between the animals with and without liver abscesses. The genus Butyrivibrio_2 was found almost exclusively in the epimural population relative to the rumen content, and this same genus was of greater abundance on the papillae of the healthy animals. Furthermore, the genus Butyrivibrio_2 had five ASVs that were moderately correlated with transcription of 221 genes, over 13% of the moderate correlations of bacteria and DEGs, and at higher correlations included two ASVs that had significant associations with the transcription of 8 rumen tissue genes. Feeding a concentrate diet has previously been shown to increase the epimural Butyrivibrio levels in the rumen of goats and increased expression of Toll-like receptors in the rumen epithelium using quantitative polymerase chain reaction (Liu et al., 2015). In the current study, the epimural abundance of ASV51, a member of Buturivibrio_2 genus, was highly and positively correlated with the rumen epithelial expression of CREB3L2 and ETV6, two transcriptional regulator genes that were observed to be differentially expressed in the animals based on the presence or absence of liver abscesses. The genus Shuttleworthia is a bacterial group associated with periodontal disease (Downes et al., 2002). This bacterial group was significantly lower in the epimural population and, within the epimural population, was found to be differentially abundant in the animals with liver abscesses; however, this bacterial group stood out in the analyses as it was highly and positively correlated with nine DEGs, including EFNA2, a gene associated with epithelial development, and RELB, a gene associated with immune function.
Several disease-associated bacteria were observed in the current study. The genus Campylobacter has been previously associated with rumen epimural populations, particularly in animals fed low-grain diets (Wetzels et al., 2016; Petri et al., 2018), and its observation in the current study was not surprising. The microbial composition of liver abscesses in cattle has been described previously and Campylobacter was the 15th most abundant genus reported (Weinroth et al., 2017). Campylobacter was the only predominant taxa group observed in both the current research with the epimural bacteria and this previous research with liver abscesses. Previous research did not observe changes in Campylobacter when animals were subjected to subacute rumen acidosis (Wetzels et al., 2016), so the observed strong tendency for Campylobacter to be associated with the epimural population of cattle that exhibited liver abscesses is novel and needs further study. Interestingly, Campylobacter is recognized as intestinal invasive bacterial species (Wooldridge and Ketley, 1997), and there were significant associations between Campylobacter abundance and rumen epithelial gene expression of DTX4 and ICOSLG, two genes that play a role in immune function. The genus Bacteroides was identified as the most abundant taxa in liver abscesses (Weinroth et al., 2017). While no differences were observed of this bacterial group in the epimural bacteria of healthy and liver-abscessed animals, an ASV that taxonomically classified as the species Bacteroides heparinolyticus was associated with rumen epithelial expression of the C3 gene, a gene associated with antimicrobial and inflammation activities. In addition, two ASVs that taxonomically clustered into an order Bacteroidales group were significantly associated with differences in several DEGs, but this bacterial group was specifically associated with significant reductions in the expression of TYK2, a gene that plays a role in immune function and differentially expressed in animals with liver abscesses. The phylum Synergistetes was significantly associated with the epimural population, and this phylum is often associated with soft tissue disease (Vartoukian et al., 2007). However, neither the phyla nor genera members were observed to be differentially abundant in the rumen epimural samples of the cattle with or without liver abscesses. The genus Pyramidobacter is a member of the phylum Synergistetes, and this genus was associated with significant reductions in the expression of TYK2, a gene associated with cytokine gene expression.
There are currently no methods to predict whether an animal is more susceptible to liver abscesses or determine whether the animal currently has liver abscesses prior to viewing the liver at harvest. A method to determine susceptibility or identify animals with liver abscess prior to harvest could be a useful tool for producers to monitor these animals more closely or to manage them differently. This study identified expression differences between inflammatory response genes in the rumen tissue of animals with liver abscesses compared with control animals upon slaughter. To better understand the molecular mechanisms of liver abscesses prior to or during development, samples from live animals must be evaluated. However, the collection of rumen tissue on large numbers of live animals is time-consuming and invasive. There is evidence that animals with liver abscesses can be distinguished from healthy animals by collecting blood. A recent study by Macdonald et al. (2017) showed that there were differences in plasma and blood parameters in cattle with and without liver abscesses. Because inflammatory signals are a means for tissues to communicate, it is possible that the differences we have detected in inflammatory response genes or proteins in the rumen may also be reflected in the circulating blood or may cause changes in the levels of other genes or proteins that could be detected in the blood, serum, or plasma of animals with liver abscesses. This study was a first step in evaluating the mechanisms in the rumen papillae that may be contributing to or were the result of damage to the rumen wall and the development of liver abscesses in cattle on a finishing diet. This study produced several functional candidate genes and epimural bacterial differences to pursue to gain more insight into the molecular mechanisms underlying liver abscess development in cattle.
Supplementary Data
Supplementary data are available at Journal of Animal Science online.
Supplementary Figure S1: Heatmap of the moderately correlated (−0.3 > R > 0.3) ASVs and DEG genes present in cattle rumen papillae. The prevalent ASVs (n = 194; present in at least 50% of the rumen papillae samples) were used to run a Pearson correlation with DEG genes (n = 221). The log10-transformed relative abundances of prevalent ASVs and DEG genes were used to generate correlation matrix. The x-axis is representing the ASVs, whereas the DEG genes are on y-axis.
Supplementary Table S1. DEGs associated with the main effect of sex. The expression is presented as the log2-fold change. Green cells are genes that were upregulated in steers compared with heifers, and red represents genes that were downregulated in steers
Supplementary Table S2. Genes differentially expressed in the rumen papillae of steers and heifers treated with the essential oil limonene vs. control animals (no limonene). The expression is presented as the log2-fold change. Green cells are genes that were upregulated in animals treated with limonene vs. control animals, and red represents genes that were downregulated. Genes with gray cells indicate those that were excluded because they were also significant for sex
Supplementary Table S3. Genes differentially expressed in the rumen papillae of steers and heifers with and without liver abscesses. The expression is presented as the log2-fold change. Green cells are genes that were upregulated in animals with liver abscess compared with those with no liver abscesses, and red represents genes that were downregulated. Genes with gray cells indicate those that were excluded because they were also significant for sex or limonene
Supplementary Table S4. List of rumen epimural bacterial ASVs that met criteria for abundance and distribution and used to establish the correlations with genes differentially expressed in the rumen papillae of steers and heifers with and without liver abscesses
Supplementary Table S5. Correlations between most abundant rumen epimural bacterial ASVs and genes differentially expressed in the rumen papillae of steers and heifers with and without liver abscesses
Acknowledgments
We would like to gratefully acknowledge the assistance of McKenzie Beals with rumen sample collection, Linda Flathman for laboratory expertise, and Donna Griess with manuscript preparation. Mention of a trade name, proprietary product, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. The USDA is an equal opportunity provider and employer. This work was partially supported by Animal Nutrition, Growth and Lactation grant (2018-67015-27496), Effective Mitigation Strategies for Antimicrobial Resistance grant (2018-68003-27545), and Multi-state research project accession no. 1000579 from the USDA National Institute of Food and Agriculture awarded to S.C.F.
Glossary
Abbreviations
- DAVID
database for annotation visualization, and integrated discovery
- DEG
differentially expressed genes
- DNA
deoxynucleic acid
- GO
gene ontology
- IPA
Ingenuity pathway analysis
- NFKB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PANTHER
protein analysis through evolutionary relationships
- PCoA
principal coordinate analysis
- PCR
polymerase chain reaction
- PERMANOVA
permutational multivariate analysis of variance
- RNA
ribonucleic acid
- RNA-Seq
ribonucleic acid sequencing
- USMARC
U.S. Meat Animal Research Center
Conflict of interest statement
S.C.F., an author of this publication, has disclosed a significant financial interest in NuGUT LLC. In accordance with its Conflict of Interest policy, the University of Nebraska-Lincoln’s Conflict of Interest in Research Committee has determined that this must be disclosed. The rest of the authors have nothing to disclose. The other authors do not have a conflict of interest.
Literature Cited
- Abramson T., Kedem H., and Relman D. A.. . 2008. Modulation of the NF-kappaB pathway by Bordetella pertussis filamentous hemagglutinin. PLoS One 3:e3825. doi: 10.1371/journal.pone.0003825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amachawadi R. G., and Nagaraja T. G.. . 2016. Liver abscesses in cattle: a review of incidence in Holsteins and of bacteriology and vaccine approaches to control in feedlot cattle. J. Anim. Sci. 94:1620–1632. doi: 10.2527/jas.2015-0261 [DOI] [PubMed] [Google Scholar]
- Anderson M. J. 2014. Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. doi: 10.1002/9781118445112.stat07841 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B.. . 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink D. R., Lowry S. R., Stock R. A., and Parrott J. C.. . 1990. Severity of liver abscesses and efficiency of feed utilization of feedlot cattle. J. Anim. Sci. 68:1201–1207. doi: 10.2527/1990.6851201 [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., and Holmes S. P.. . 2016a. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B. J., Sankaran K., Fukuyama J. A., McMurdie P. J., and Holmes S. P.. . 2016b. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 5:1492. doi: 10.12688/f1000research.8986.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejos L., Calsamiglia S., and Ferret A.. . 2006. Effect of essential oil active compounds on rumen microbial fermentation and nutrient flow in in vitro systems. J. Dairy Sci. 89:2649–2658. doi: 10.3168/jds.S0022-0302(06)72341-4 [DOI] [PubMed] [Google Scholar]
- Chan W. Y., Hui P. K., Leung K. M., and Thomas T. M.. . 1992. Modes of Helicobacter colonization and gastric epithelial damage. Histopathology 21:521–528. doi: 10.1111/j.1365-2559.1992.tb00439.x [DOI] [PubMed] [Google Scholar]
- Chen Y., Penner P. B., Li M., Oba M., and Guan L. L.. . 2011. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77:5770–5781. doi: 10.1128/AEM.00375-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. G., Han J., Kim J. H., Kim M. J., Park J. W., Song B., Cha H. J., Choi H. S., Chung H. T., Lee I. K., . et al. 2017. eIF2α phosphorylation is required to prevent hepatocyte death and liver fibrosis in mice challenged with a high fructose diet. Nutr. Metab. (Lond). 14:48. doi: 10.1186/s12986-017-0202-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbah R., Edwards V. M., and Moats W. A.. . 1970. Antimicrobial action of some citrus fruit oils on selected food-borne bacteria. Appl. Microbiol. 19:27–31. PMCID: PMC376603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale D., Cheng K. J., Wallace R. J., and Goodlad R. A.. . 1980. Digestion of epithelial tissue of the rumen wall by adherent bacteria in infused and conventionally fed sheep. Appl. Environ. Microbiol. 39:1059–1066. doi: 10.1128/AEM.39.5.1059-1066.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes J., Munson M. A., Radford D. R., Spratt D. A., and Wade W. G.. . 2002. Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 52:1469–1475. doi: 10.1099/00207713-52-5-1469 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., and Flyvbjerg H.. . 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482. doi: 10.1093/bioinformatics/btv401 [DOI] [PubMed] [Google Scholar]
- Emmans G. C. 1997. A method to predict the food intake of domestic animals from birth to maturity as a function of time. J. Theor. Biol. 186:189–200. doi: 10.1006/jtbi.1996.0357 [DOI] [Google Scholar]
- FASS 2010. Guide for care and use of agricultural animals in agricultural research and teaching. Savoy (IL):Federation of Animal Science Society. [Google Scholar]
- Friedman M. 1937. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J. Amer. Statist. Assoc. 32:675–701. doi: 10.1080/01621459.1937.10503522 [DOI] [Google Scholar]
- Gehan E. A. 1965. A generalized Wilcoxon test for comparing arbitrarily singly-censored samples. Biometrika 52:203–223. doi: 10.1093/biomet/52.1-2.203 [DOI] [PubMed] [Google Scholar]
- Harris M. K., Eastwood L. C., Boykin C. A., Arnold A. N., Hale D. S., Kerth C. R., Griffin D. B., Savell J. W., Belk K. E., Woerner D. R., et al. 2017. National Beef Quality Audit-2016: Transportation, mobility, and harvest-floor assessments of targeted characteristics that affect quality and value of cattle, carcasses, and by-products. Transl. Anim. Sci. 1:229–238. doi: 10.2527/tas2017.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. A. W., Sherman B. T., and Lempicki R. A.. . 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. A. W., Sherman B. T., and Lempicki R. A.. . 2009b. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R., Deane H. M., Cooper L. J., Miller V. A., and Graham W. R.. . 1954. The rumenitis-liver abscess complex in beef cattle. Am. J. Vet. Res. 15: 202–216. PMID: 13148469. [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L.. . 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt R., and Coe. R.. 2005. Tree diversity analysis. A manual and software for common statistical methods for ecological and biodiversity studies. Nairobi (Kenya):World Agroforestry Centre (ICRAF). [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., and Schloss P. D.. . 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. doi: 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J. Jr, and Tugendreich S.. . 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30:523–530. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican A., Alkeskas A., Günter C., Forsythe S. J., and Figueras M. J.. . 2013. Adherence to and invasion of human intestinal cells by Arcobacter species and their virulence genotypes. Appl. Environ. Microbiol. 79:4951–4957. doi: 10.1128/AEM.01073-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Bian G. R., Zhu W. Y., and Mao S. Y.. . 2015. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 6:167. doi: 10.3389/fmicb.2015.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S.. . 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald A. G. C., Bourgon S. L., Palme R., Miller S. P., and Montanholi Y. R.. . 2017. Evaluation of blood metabolites reflects presence or absence of liver abscesses in beef cattle. Vet. Rec. Open 4:e000170. doi: 10.1136/vetreco-2016-000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Poudel S., Muruganujan A., Casagrande J. T., and Thomas P. D.. . 2016. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44(D1):D336–D342. doi: 10.1093/nar/gkv1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P. R., Freetly H. C., Wells J. E., Smith T. P. L., and Kuehn L. A.. . 2017. Analysis of the gut bacterial communities in beef cattle and their association with feed intake, growth, and efficiency. J. Anim. Sci. 95:3215–3224. doi: 10.2527/jas.2016.1059 [DOI] [PubMed] [Google Scholar]
- Nagaraja T. G., and Chengappa M. M.. . 1998. Liver abscesses in feedlot cattle: a review. J. Anim. Sci. 76:287–298. doi: 10.2527/1998.761287x [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., . et al. 2017. vegan: Community Ecology Package. R package version 2.4–5. Available from https://CRAN.Rproject.org/package=vegan. Accessed August 2019.
- Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., and Salzberg S. L.. . 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33:290–295. doi: 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R. M., Kleefisch M. T., Metzler-Zebeli B. U., Zebeli Q., and Klevenhusen. F.. 2018. Changes in the rumen epithelial microbiota of cattle and host gene expression in response to alterations in dietary carbohydrate composition. Appl. Environ. Microbiol. 84:e00384–18. doi: 10.1128/AEM.00384-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R. M., Schwaiger T., Penner G. B., Beauchemin K. A., Forster R. J., McKinnon J. J., and McAllister T. A.. . 2013. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microbiol. 79:3744–55. doi: 10.1128/AEM.03983-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2018. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; Available from https://www.R-project.org. Accessed August 2019. [Google Scholar]
- Rahman M. M., and McFadden. G.. 2011. Modulation of NF-kB signaling by microbial pathogens. Nat. Rev. Microbiol. 9:291–306. doi: 10.1038/nrmicro2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt C. D., and Hubbert M. E.. . 2015. Control of liver abscesses in feedlot cattle: a review. Prof. Anim. Sci. 31:101–108. doi: 10.15232/pas.2014-01364 [DOI] [Google Scholar]
- Reyes A., Weinroth M., Wolfe C., Delmore R., Engle T., Morley P., and Belk K.. . 2019. PSXIV-23 characterization of microbial communities associated with the rumen lining, digesta and rumen fluid from beef cattle consuming a high energy diet using 16S rRNA gene amplicon sequencing. J. Anim. Sci. 97 (Supplement 3):446. doi: 10.1093/jas/skz258.878 [DOI] [Google Scholar]
- Rizzatti G., Lopetuso L. R., Gibiino G., Binda C., and Gasbarrini. A.. 2017. Proteobacteria: a common factor in human diseases. Biomed. Res. Int. 2017:9351507. doi: 10.1155/2017/9351507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samii S. S., Wallace N., Nagaraja T. G., Engstrom M. A., Miesner M. D., Armendariz C. K., Titgemeyer E. C.. 2016. Effects of limonene on ruminal Fusobacterium necrophorum concentrations, fermentation, and lysine degradation in cattle. J. Anim. Sci. 94:3420–3430. doi: 10.2527/jas.2016-0455 [DOI] [PubMed] [Google Scholar]
- Scarth L. L. 2006. The Merck veterinary manual online (8th edition)”. Ref. Rev. 20(2):40. doi: 10.1108/09504120610647492 [DOI] [Google Scholar]
- Schiermiester L. N., Thallman R. M., Kuehn L. A., Kachman S. D., and Spangler M. L.. . 2015. Estimation of breed-specific heterosis effects for birth, weaning, and yearling weight in cattle. J. Anim. Sci. 93:46–52. doi: 10.2527/jas.2014-8493 [DOI] [PubMed] [Google Scholar]
- Sorensen D. 1972. An economic analysis of feeding steers versus heifers [master’s thesis]. Logan (UT): Utah State University. [Google Scholar]
- Vartoukian S. R., Palmer R. M., and Wade W. G.. . 2007. The division “Synergistes”. Anaerobe. 13:99–106. doi: 10.1016/j.anaerobe.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., Sievert S. M., Simon J., Campbell B. J., Hanson T. E.. et al. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (Phyl. nov.). Front. Microbiol. 8:682. doi: 10.3389/fmicb.2017.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., and Cole J. R.. . 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73(16):5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinroth M. D., Carlson C. R., Martin J. N., Metcalf J. L., Morley P. S., and Belk K. E.. . 2017. Rapid Communication: 16S ribosomal ribonucleic acid characterization of liver abscesses in feedlot cattle from three states in the United States. J. Anim. Sci. 95:4520–4525. doi: 10.2527/jas2017.1743 [DOI] [PubMed] [Google Scholar]
- Wetzels S. U., Mann E., Metzler-Zebeli B. U., Pourazad P., Qumar M., Klevenhusen F., Pinior B., Wagner M., Zebeli Q., and Schmitz-Esser S.. . 2016. Epimural indicator phylotypes of transiently-induced subacute ruminal acidosis in dairy cattle. Front. Microbiol. 7:274. doi: 10.3389/fmicb.2016.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Langemeier M. R., Mintert J., and Schroeder T. C.. . 1993. Profitability differences between steers and heifers. Manhattan (KS): Kansas State University Cooperative Extension Service. MF-1075. [Google Scholar]
- Wooldridge K. G., and Ketley J. M.. . 1997. Campylobacter-host cell interactions. Trends Microbiol. 5:96–102. doi: 10.1016/S0966-842X(97)01004-4 [DOI] [PubMed] [Google Scholar]
- Yu Z., and Morrison M.. . 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812. doi: 10.2144/04365ST04 [DOI] [PubMed] [Google Scholar]
- Zinn R. A., Barreras A., Owens F. N., and Plascencia A.. . 2008. Performance by feedlot steers and heifers: daily gain, mature body weight, dry matter intake, and dietary energetics. J. Anim. Sci. 86:2680–2689. doi: 10.2527/jas.2007-0561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.