Microbiota control of maternal behavior regulates early postnatal growth of offspring.
Abstract
Maternal behavior is necessary for optimal development and growth of offspring. The intestinal microbiota has emerged as a critical regulator of growth and development in the early postnatal period life. Here, we describe the identification of an intestinal Escherichia coli strain that is pathogenic to the maternal-offspring system during the early postnatal stage of life and results in growth stunting of the offspring. However, rather than having a direct pathogenic effect on the infant, we found that this particular E. coli strain was pathogenic to the dams by interfering with the maturation of maternal behavior. This resulted in malnourishment of the pups and impaired insulin-like growth factor 1 (IGF-1) signaling, leading to the consequential stunted growth. Our work provides a new understanding of how the microbiota regulates postnatal growth and an additional variable that must be considered when studying the regulation of maternal behavior.
INTRODUCTION
The early postnatal period of life is a critical stage for growth and development. During this phase, there is rapid maturation of all physiologies including immune, metabolic, endocrine, and neural processes, the proper function of which is necessary for healthy growth and development. Maturation of these pathways is dependent on internal factors including genetic, physiological, and epigenetic mechanisms as well as external environmental cues (1–3). Disruption of any of these intrinsic or extrinsic factors can lead to states of nutritional challenge, where the infant is undernourished, leading to the adverse development of these processes, resulting in stunted growth or wasting (1, 3). The mammalian microbiota—the trillions of microbes that reside on the body surfaces exposed to the environment—has emerged as a critical regulator of postnatal growth and development, influencing both wasting and growth-stunting pathogenesis (4–6). In a mouse model of infection-induced muscle wasting, a commensal Escherichia coli strain was found to be protective against muscle wasting via manipulation of an innate immune-endocrine signaling axis that enables the skeletal muscle to maintain a healthy mass during the infection (4). In children, malnutrition was associated with an immature fecal microbiota profile [Subramanian et al. (7)], and colonization of germ-free (GF) mice with the fecal microbiota of undernourished children was sufficient to cause growth stunting in the gnotobiotic mice (6). Conversely, gavage of gnotobiotic mice with malnourished microbiota with microbiota constituents harvested from healthy donors was sufficient to promote weight and lean mass gain (8). The mechanisms by which the intestinal microbiota will influence early postnatal development and growth are likely complex and multifactorial; however, one common theme that has emerged from these initial studies is that the intestinal microbiota is an important regulator of the growth hormone (GH; somatotropin)/insulin-like growth factor 1 (IGF-1) axis, which is important for promoting healthy growth of the host (9–11).
While the focus has been to understand how dysbiotic microbiotas, and the direct interactions between the microbiota and the infant, can cause impaired growth and development, maternal factors that are important for healthy postnatal early development and growth will also be regulated by the microbiota. Here, we describe a mechanism by which the microbiota regulates growth and development during the early postnatal stage of life. Our analysis of different E. coli strains in gnotobiotic mice demonstrates that there is significant variability in the ability of E. coli strains to regulate linear growth and body weight in a murine postnatal model. We identified one E. coli strain, E. coli O16:H48 MG1655, that was pathogenic to infant mice, causing stunted linear growth and body weight gain. Consistent with previous reports, we found this stunting to be dependent on impaired IGF-1 signaling in the pups. However, rather than this being due to direct interactions between the microbe and the pup, we found that the defects in endocrine signaling and consequential stunting to be due to impaired maternal factors. Specifically, we found that dams monocolonized with E. coli O16:H48 MG1655 exhibited poor maternal behavior, resulting in malnourishment of their offspring, which likely contributed to the attenuated IGF-1 signaling in the offspring. Cross-fostering of E. coli O16:H48 MG1655 pups onto dams that exhibited optimal maternal behavior was sufficient to prevent the defective IGF-1 and growth stunting phenotypes. Together, our findings support a model by which the intestinal microbiota is necessary for the proper maturation of maternal behaviors after birth and that disruption of these behaviors leads to malnourishment of offspring that results in growth stunting.
RESULTS
Identification of E. coli strains that differentially affect postnatal growth
Escherichia species are one of the first colonizers of the infant gut after birth (12). The pan-genome of E. coli comprises ~16,000 genes, which represent the core E. coli genome (13). Thus, 90% of the pan-genome among E. coli strains are considered to be accessory. We hypothesized that, despite taxonomic similarity, this genomic variation can lead to phenotypic differences in various aspects of postnatal health including growth and development. To test this, we examined how different E. coli strains affect growth in gnotobiotic mice. We gavaged adult female GF Swiss Webster mice with one of four different human- or mouse-derived E. coli strains to establish monocolonized gnotobiotic mice (Fig. 1, A and B). We then bred these mice to male Swiss Webster mice that were monocolonized with the respective E. coli strain. F1 progenies were then bred, and the resulting F2 progenies were examined for body weight and linear growth at postnatal day (P) 21 and compared with GF and specific pathogen–free (SPF) Swiss Webster control mice of the same age and litter size (Fig. 1, C to F). We used Swiss Webster mice for our experiments because the investigation of the effects of the microbiome on postnatal growth and development requires mice that exhibit normal maternal behavior in the SPF and GF states. SPF mice had greater linear growth compared with GF mice, indicating that the microbiota promotes linear growth during the postnatal phase (Fig. 1, C and D). Similarly, SPF mice exhibited significantly greater body weight compared with GF mice (Fig. 1, E and F), indicating that the microbiota is necessary for normal growth during the postnatal stage as has been previously described (7, 10, 14).
Fig. 1. Identification of E. coli strains that differentially affect postnatal growth.
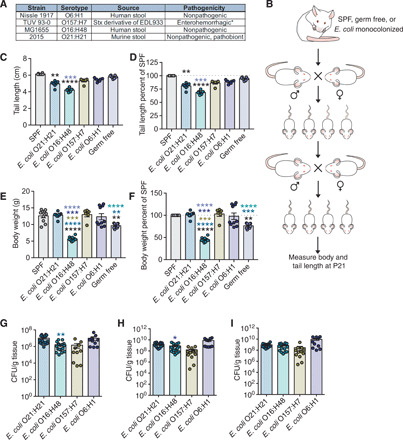
(A) Details of E. coli strains used. Pathogenicity indicates behavior in mice with the exception of E. coli O157:H7. * indicates that E. coli O157:H7 is pathogenic in humans. (B) Experimental scheme. Each group of GF mice was orally gavaged with single strain culture of E. coli [5 × 108 colony-forming units (CFU) per mouse], mated, and maintained in gnotobiotic isolators. The monocolonized 6- to 8-week-old F1 progenies were mated, and the resulting F2 progenies were analyzed for postnatal development. (C) Tail length measurements taken at P21 to assess linear growth of gnotobiotic litters. SPF (n = 6), O21:H21 (n = 7), O16:H48 (n = 7), O157:H7 (n = 6), O6:H1 (n = 6), and GF (n = 7). (D) Tail length measurements shown in (C) as percentage of average SPF length. (E) Body mass of E. coli gnotobiotic litters at P21. SPF (n = 10), O21:H21 (n = 9), O16:H48 (n = 11), O157:H7 (n = 6), O6:H1 (n = 10), and GF (n = 8). (F) Body weight in (E) as percentage of average SPF weight. (G to I) Intestinal colonization levels of bacteria at P21 (n = 10 to 18 per condition). Statistical significance was determined using one-way analysis of variance (ANOVA) with post Tukey test or Kruskal-Wallis test with Dunn’s multiple comparison for pairwise analyses. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Significance stars are colored to indicate which condition significance is referring to. Error bars represent means ± SEM. E. coli O21:H21 described in (4), E. coli O16:H48 described in (15), E. coli O157:H7 described in (61), and E. coli O6:H1 described in (62).
From our analysis, we found that all E. coli strains were capable of stably colonizing the gastrointestinal tract (Fig. 1, G to I). Colonization with any of the tested E. coli strains was not sufficient to promote linear growth of pups that was comparable to that observed in SPF pups (Fig. 1, C and D). Mice monocolonized with E. coli O157:H7 or O6:H1 exhibited comparable linear growth to GF mice (Fig. 1, C and D). Mice monocolonized with E. coli O21:H21 or O16:H48 exhibited reduced linear growth compared with GF pups (Fig. 1, C and D). E. coli O16:H48 had the most marked negative effect on linear growth, with pups exhibiting ~26% reduction in linear growth compared with GF mice and ~32% reduction compared with SPF mice (Fig. 1D). This indicates that monocolonization with any of the E. coli strains tested was not sufficient to promote linear growth observed in SPF mice and that some strains can be pathogenic to linear growth.
We found that the effects the E. coli strains had on postnatal linear growth could be decoupled for their effects on weight gain. Pups monocolonized with E. coli O6:H1, E. coli O21:H21, or E. coli O157:H7 had higher body weight compared with GF mice and comparable weights to SPF mice, indicating that colonization with one of these strains was sufficient to promote normal body weight gain (Fig. 1, E and F). By contrast, colonization with the strain E. coli O16:H48 resulted in a reduced weight phenotype compared with GF mice (Fig. 1, E and F), indicating that colonization with E. coli O16:H48 had a pathogenic effect on both postnatal weight and linear development. E. coli O16:H48 pups exhibited body weights that were ~30% less than GF pups and greater than 60% less than SPF pups of the same age and from similar litter size (Fig. 1F). Together, our results demonstrate that intestinal E. coli strains have differential effects on postnatal growth in the mouse and fall into three categories: (i) strains that have no effect on linear growth and positive effects on weight gain; (ii) strains that have a pathogenic effect on linear growth but are sufficient to promote normal weight gain; and (iii) strains that have pathogenic effects on both linear growth and weight gain.
Postnatal care provided by E. coli O16:H48 colonized dams results in growth stunting
From our analysis, we determined that pups birthed by E. coli O16:H48–monocolonized dams exhibited a growth-stunting phenotype (Fig. 1, C to F). E. coli O16:H48 is a derivative of the human isolate K-12 strain that was isolated from the stool of a patient with diphtheria in the 1920s (15). Since its isolation, it has been widely used in laboratories as a standard, nonpathogenic E. coli strain (16–18). Thus, we were surprised to reveal a pathogenic effect of E. coli O16:H48 for mouse development in this maternal-offspring system. Low birth weight is a risk factor for growth stunting (19). We found that E. coli O16:H48 pups had comparable birth weights to both GF and E. coli O21:H21 pups, indicating that the failure to thrive phenotype observed in E. coli O16:H48 pups is not due to low birth weight (Fig. 2A), We tracked the rate of weight gain of pups for 7 to 8 weeks after birth and found that from P0 to P21 when pups are with their mothers, E. coli O16:H48 pups exhibited a reduced growth rate compared with SPF, E. coli O21:H21, and GF pups (Fig. 2, B to D, and fig. S1). After weaning at P21, the growth rate of E. coli O16:H48 mice increased, with female mice reaching comparable weights as control mice around 5 weeks old and males reaching comparable weights as control mice around 8 weeks old (Fig. 2, B and C, and fig. S1).
Fig. 2. Cross-fostering rescues stunted growth exhibited by E. coli O16:H48 pups.
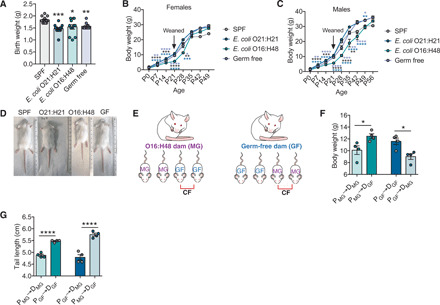
(A) Birth weights of pups (n = 10 to 12 per condition). (B and C) Body weights at birth (P0) to adulthood (P49 to P56) for (B) females (P0, n = 10 to 12; P7, n = 9 to 11; P14, n = 10 to 11; P21, n = 7 to 11; P28, n = 4 to 10; P35, n = 7 to 10; P42, n = 7 to 10; P49, n = 6 to 11) and (C) males (P0, n = 10 to 12; P7, n = 9 to 11; P14, n = 10 to 11; P21, n = 7 to 11; P28, n = 10 to 13; P35, n = 9 to 11; P42, n = 9 to 13; P49, n = 9 to 12; P56, n = 5 to 9). From P0 to P21, the data represent the weights of both male and female littermates and therefore show the same data in (B) and (C) for those time points. Males and females were separated at the time of weaning, and data points beginning at P28 represent the weights of the sex indicated. (D) Representative images of pups at P21 to P24. (E) Experimental scheme of maternal cross-fostering. Immediately after birth, half of the litter is fostered by a different dam, and half of the litter remains with their biological dam. MG (for MG1655) indicates E. coli O16:H48 pups and dams; GF indicates GF pups and dams. (F) Body weight of pups at P21 after cross-fostering (P, pup; D, dam; MG, E. coli O16:H48; n = 4 per condition). (G) Tail length of cross-fostered pups at P21 (P, pup; D, dam; MG, E. coli O16:H48; n = 4 per condition). Statistical significance was determined using one-way ANOVA with post Tukey test or Kruskal-Wallis test with Dunn’s multiple comparison for pairwise analyses. Significance stars are colored to indicate which condition significance is referring to. *P < 0.05 [P = 0.05 for (A)], **P < 0.01, ***P < 0.001, and ****P < 0.0001. Error bars represent means ± SEM. Scatter plot data for each time point for (B) and (C) are shown in fig. S1. Photo credit: Yujung Michelle Lee, Salk Institute.
Growth during the preweaning phase in a maternal-offspring system is dependent on both maternal and offspring factors. Thus, the pathogenic effects of E. coli O16:H48 could be due to direct effects of the microbe on the pups or indirectly by influencing maternal factors that are important for offspring growth. To distinguish between these two possibilities, we performed cross-fostering experiments where we placed E. coli O16:H48 pups with GF dams, and GF pups with E. coli O16:H48 dams and monitored pup growth (Fig. 2E). We found that cross-fostering of E. coli O16:H48 pups by GF dams was sufficient to prevent the growth-stunting phenotype in these pups. E. coli O16:H48 pups fostered by GF dams were comparable in weight and linear length to GF pups cared for by the same GF dams and ~120% the weight and length (tail length) of E. coli O16:H48 pups cared for by E. coli O16:H48 dams (Fig. 2, F and G). GF pups fostered by E. coli O16:H48 dams exhibited a stunted phenotype comparable to E. coli O16:H48 pups cared for by E. coli O16:H48 dams and were 80% of the average body weight and tail length of GF pups with GF dams (Fig. 2, F and G). Thus, the growth-stunting phenotype in E. coli O16:H48 pups was due to indirect effects of E. coli O16:H48 that disrupt maternal factors that are important for offspring growth.
E. coli O16:H48 pups are malnourished and exhibit reduced milk consumption
Growth stunting is caused by a lack of nutrients (20). Low muscle and fat mass are important indicators of malnutrition (21). Consistent with this, body composition analysis revealed that E. coli O16:H48 pups had significantly reduced fat and muscle composition compared with SPF, GF, and E. coli O21:H21 pups (Fig. 3, A and B). This suggests that E. coli O16:H48 negatively affected offspring growth by disrupting maternal derived nutrients. This can be due to changes in the quality or quantity of milk provided by dams to their pups. Sialylated milk oligosaccharides promote microbiota-dependent growth in infants (22). To determine whether milk produced by E. coli O16:H48 dams had altered levels of milk oligosaccharides, we quantified sialyl(α2,3)lactose (3SL) and sialyl(α2,6)lactose (6SL), which are the only milk oligosaccharides in mice (23, 24). We found no differences in 3SL or 6SL levels in milk collected from SPF, GF, and E. coli O16:H48–monocolonized dams (Fig. 3, C and D), suggesting that E. coli O16:H48 does not cause growth stunting in pups by negatively affecting the ability of dams to produce milk composed of the major sialylated milk oligosaccharides.
Fig. 3. E. coli O16:H48 pups are malnourished and exhibit reduced milk consumption.
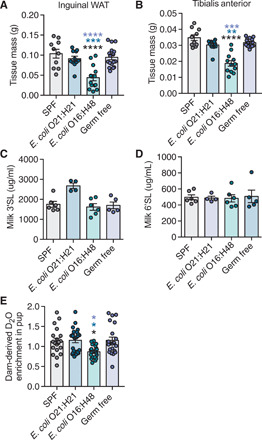
(A) Inguinal white adipose tissue (WAT) and (B) skeletal leg muscle tibialis anterior (TA) masses of P21 pups (n = 10 to 17 mice per condition). (C and D) Levels of (C) 3’SL and (D) 6’SL in milk collected from dams with litters aged P9 and P10 (during peak milk production) (n = 4 to 6 per condition). (E) Percent enrichment of D2O detected in pup sera after 48 hours of nursing on D2O-treated dams (n = 19 to 24 per condition). Statistical significance was determined using one-way ANOVA with post Tukey test. Significance stars are colored to indicate which condition significance is referring to. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Error bars represent means ± SEM.
We next determined whether the growth-stunting phenotype exhibited by E. coli O16:H48 pups was associated with overall reduced feeding. To do this, we used a D2O-based labeling approach to quantify milk consumption of pups. Lactating dams were injected with a bolus of D2O-saline and then maintained on 8% D2O water for stable enrichment of D2O in the body. Pups were allowed to nurse on D2O-enriched dams for 48 hours, after which we quantified the percent enrichment of D2O in the serum of individual pups as a proxy for milk consumption amount over the 48-hour testing period. We found that D2O enrichment in the sera of E. coli O16:H48 pups was significantly reduced, compared with SPF, GF, and gnotobiotic E. coli O21:H21 mice (Fig. 3E), suggesting that in this E. coli O16:H48 dam-offspring system, growth stunting is associated with disturbances in the quantity of milk provided by the dam to offspring.
E. coli O16:H48 dams exhibit impaired maternal behavior
There are physiological and behavioral maternal factors that influence milk availability for offspring. We performed a nipple function and suckling assay to determine whether E. coli O16:H48–monocolonized dams had defects in milk production or nipple function (25). Pups were removed from their mothers to fast, after which they were weighed and returned to their sedated mothers that were turned on their backs with nipples exposed. The pups were placed next to their mothers and allowed to latch on her nipples and suckle. The pups were allowed to feed for 1 hour, after which they were weighed. The change in weight was used as an indirect measurement for the amount of milk consumed. We found that the change in weight exhibited by E. coli O16:H48 pups was comparable to SPF and E. coli O21:H21, all of which were greater than the weight change exhibited by GF pups (Fig. 4A). This suggests that E. coli O16:H48 dams have functional nipples and produce and secrete comparable volumes of milk in a defined feeding period. It also demonstrates, consistent with our cross-fostering assay, that the reduced milk intake exhibited by E. coli O16:H48 pups is not due to defects in the pups ability to latch and suckle.
Fig. 4. Impaired maternal behavior in E. coli O16:H48 dams.
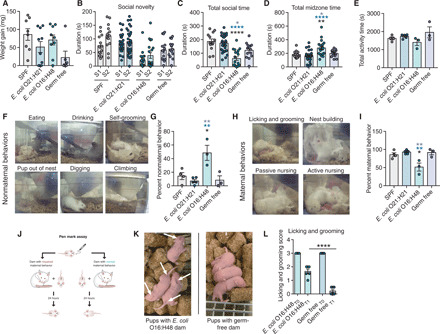
(A) Weight gain of pups after feeding on sedated dams for 1 hour (n = 6 to 10 per condition). (B) Social novelty test (n = 13 to 30 per condition). S1, familiar mouse; S2, novel mouse. (C) Total socialization time calculated from social novelty test in (B) (n = 13 to 30 per condition). (D) Total time in midzone calculated from the social novelty test in (B) (13 to 30 per condition). (E) Dams were recorded for ~30 min, and the total amount of active time was measured (n = 3 to 5 per condition). (F) Representative images of nonmaternal behaviors. (G) Dams were recorded, and the total amount of time performing the nonmaternal behaviors in (G) was measured (n = 3 to 5 per condition). (H) Representative images of maternal behaviors. (I) Dams were recorded, and the total amount of time performing the maternal behaviors in (H) was measured (n = 3 to 5 per condition). (J) Schematic demonstrating the “pen mark” assay for quantification of licking and grooming. (K) Representative images of pups from the pen mark assay. Pups cared for by E. coli O16:H48 dams have visible pen mark 24 hours later, while pups with GF dams have barely visible pen mark. (L) Quantification of the pen mark assay (n = 6 to 7 pups per condition). Statistical significance was determined using one-way ANOVA with post Tukey test or unpaired Student’s t test for pairwise analyses. Significance stars are colored to indicate which condition significance is referring to. *P < 0.05, **P < 0.01, and ****P < 0.0001. Error bars represent means ± SEM. Photo credit: Yujung Michelle Lee, Salk Institute.
We next considered the possibility that E. coli O16:H48–colonized mice had behavioral defects that influenced their ability to provide adequate care and nutrients to their offspring. We first examined the social behavior of E. coli O16:H48–monocolonized adult females and control mice. We performed a three-chamber social novelty test to assess social behavior. Mice prefer to spend more time with another mouse and prefer a novel social subject over a familiar one (social novelty) (26). In this test, the test subject is allowed to freely explore inside a three-chambered box with openings between the chambers (26). One chamber houses a familiar mouse, and another chamber houses a novel mouse. The amount of time the test mouse interacts with the familiar and novel mouse is recorded. We found that while E. coli O16:H48–monocolonized mice preferred to spend time with the novel mouse, the total amount of time spent socializing by E. coli O16:H48 was significantly reduced compared with SPF, E. coli O21:H21–monocolonized, and GF mice (Fig. 4, B and C). Instead, E. coli O16:H48 mice spent significantly more time not socializing and contained within the midzone of the assay apparatus compared with the three control conditions (Fig. 4D). From video recording analysis, we determined that the decreased socialization time and increased midzone time were not due to reduced activity or mobility issues, as E. coli O16:H48–monocolonized dams exhibit comparable activity times as SPF dams (Fig. 4E).
We next considered that E. coli O16:H48 dams had impaired maternal behavior. We video recorded dams with their litters in ~30-min segments of time from P2 to P4 and measured the amount of time the dams spent performing nonmaternal and maternal behaviors (Fig. 4, F to I). For nonmaternal behaviors, we quantified the amount of time the dam spends digging and climbing, eating and drinking, self-grooming, and when pups were found outside of their nest (Fig. 4F) (27). For maternal behaviors, we looked for nest building, licking and grooming, passive nursing, and active nursing (Fig. 4H) (28). We found that E. coli O16:H48 dams exhibited greater time performing nonmaternal behaviors compared with our three control conditions and significantly less time performing maternal behaviors compared with SPF, E. coli O21:H21, and GF dams (Fig. 4, F to I). We next developed the “pen mark” assay that could allow us to better quantify maternal behaviors such as licking and grooming (Fig. 4J). Briefly, pups were marked on two distinct anatomical regions (head and rear) using a VWR marker and placed back in their home cages containing their respective dams. After 24 hours, each pup was scored for the intensity of the marks on each region of the body as a way to measure the dam’s licking and grooming behavior (Fig. 4, K and L). We found that pups that were with E. coli O16:H48 dams exhibited greater pen mark intensity after 24 hours compared with pups that were placed with GF dams, indicating that E. coli O16:H48 dams had reduced licking and grooming behaviors in the 24-hour period compared with GF dams (Fig. 4, K and L). In conclusion, the growth stunting and impaired milk consumption exhibited by E. coli O16:H48 pups are associated with impaired maternal behavior in E. coli O16:H48 dams.
E. coli comparative analyses reveal differences in tryptophan physiology
To begin to understand the mechanism by which E. coli O16:H48 compromises maternal behavior, we first considered the possibility that colonization with E. coli O16:H48 impaired the oxytocin neuroendocrine system in dams. Oxytocin is a critical regulator of maternal behavior in the postnatal period and for social bonding and is regulated by the intestinal microbiota (29). We first measured the amount of circulating oxytocin in dams at P3 and found that E. coli O16:H48 dams exhibited comparable levels of circulating oxytocin as the other conditions examined (fig. S2). Consistent with this, we found that expression of oxytocin in the hypothalamus of E. coli O16:H48 dams was not significantly different from SPF, E. coli O21:H21, or GF mice (fig. S2).
We next generated and characterized genome assemblies of the four E. coli isolates in silico to identify genomic variants that may explain the observed phenotypic differences in maternal behavior (table S1). Our bioinformatic analyses revealed differences at the whole-genome level (Fig. 5A) with four distinct in silico multilocus sequence types (MLSTs) and serotyping profiles (Fig. 5B). There are 3292 shared genes (i.e., core-genome) of a total 7149 predicted genes (i.e., pan-genome), and the core-genome maximum likelihood phylogenetic tree further supports the presence of distinct E. coli strains with bifurcations at each node (Fig. 5B). Subsystem ontology analysis of the strains revealed a predominantly similar profile across the functional categories, of note, however, is the higher proportion of genomic content predicted to be involved in cell signaling and protein metabolism in E. coli O16:H48 compared with the other three strains and the lower proportion of genomic content predicted to be involved in membrane transport in E. coli O16:H48 compared with the other three strains analyzed (fig. S3).
Fig. 5. Comparative analysis of E. coli strains.
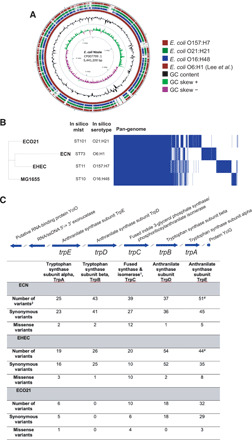
(A) Comparison of E. coli genomes. E. coli Nissle 1917 (accession number CP007799.1) was used as the reference to compare each of the study isolates. Comparative analysis was computed using a BLAST-based approach and illustrates high sequence identity and coverage of E. coli O6:H1 (ECN), the Nissle strain from this study to the reference Nissle genome, and missing genomic regions throughout the genomes of E. coli O16:H48 (MG1655), E. coli O157:H7 (EHEC), and E. coli O21:H21 (ECO21). (B) Phylogenetic comparison of E. coli genomes. A core-genome maximum likelihood phylogenetic tree of the E. coli strains O6:H48 (MG1655), E. coli O6:H1 (ECN), E. coli O157:H7 (EHEC), and E. coli O21:H21 (ECO21). Prepended to the phylogeny are data from in silico mlst, in silico serotyping, and a heatmap displaying pan-genome data of the four strains. There are 3293 core genes represented by shared blocks of blue and 3857 accessory genes. (C) Genomic variants detected in genes associated with tryptophan biosynthesis from strains E. coli O6:H1 (ECN), E. coli O157:H7 (EHEC), and E. coli O21:H21 (ECO21) when E. coli O16:H48 (MG1655) was designated the reference strain. 1Complete annotation is fused indole-3-glycerol phosphate synthase/phosphoribosylanthranilate isomerase. 2Please refer to fig. S4 for a detailed breakdown of genomic variants, including predicted amino acid substitutions. #Additional genomic variant is a splice region.
Serotonin is a monoamine neurotransmitter that regulates maternal behavior (30). Previous work has shown that E. coli Nissle 1917, but not E. coli MG1655, enhances bioavailability of serotonin in the host (31). The essential amino acid tryptophan is the precursor of serotonin, and gut microbes regulate tryptophan availability (32). We therefore directed our analyses toward genes associated with tryptophan metabolism (tnaA) and biosynthesis (trp operon). Analysis of the trp operon, by way of a multiple sequence alignment, revealed a concentrated number of amino acid substitutions in the region annotated as the bifunctional-fused indole-3-glycerol phosphate synthase/phosphoribosylanthranilate isomerase (Fig. 5C and fig. S4). Furthermore, variant analysis identified a suite of missense mutations in genes throughout the trp operon among the four strains, reported in parentheses as E. coli O6:H1 (ECN), E. coli O157:H7 (EHEC), and E. coli O21:H21 (ECO21) when compared with E. coli O16:H48 (MG1655): tryptophan synthase subunit alpha (2, 3, and 1 variants), tryptophan synthase subunit beta (2, 1, and 0 variants), fused indole-3-glycerol phosphate synthase/phosphoribosylanthranilate isomerase (12, 10, and 4 variants), anthranilate synthase subunit TrpD (1, 2, and 0 variants), and anthranilate synthase subunit TrpE (5, 8, and 3 variants) (Fig. 5C, fig. S4, and data S1). Our analyses also included genes annotated as tryptophan transporters AroP (fig. S5), Mtr (fig. S6), and TnaB (fig. S7), and a lyase enzyme, tryptophan decarboxylase (fig. S8). Comparative analysis of the key enzyme associated with tryptophan metabolism, tryptophanase (TnaA), from E. coli O16:H48 (MG1655) and E. coli O6:H1 (ECN), revealed a single amino acid substitution at position 212 from a valine (ECN) to an alanine (MG1655) (fig. S7). Together, our analyses demonstrate genomic differences found in E. coli O16:H48, including differences in precursors of neurotransmitters, that may contribute to the poor maternal behavior found in E. coli O16:H48–colonized dams.
E. coli O16:H48 pups have attenuated IGF-1 signaling
To determine the mechanism by which the poor maternal behavior and malnourishment caused growth stunting, we considered a role for the endocrine system. The GH–liver IGF-1 axis is a critical regulator of postnatal growth and is regulated by the nutritional state of the organism (33, 34). During malnutrition and fasting states, levels of IGF-1 and signaling are reduced (35, 36). We hypothesized that the failure to thrive and malnourished phenotype of E. coli O16:H48 pups were associated with reduced IGF-1 activity. To determine this, we first looked at IGF-1 signaling in the liver and found that there was reduced phosphorylation of the serine/threonine kinase Akt in E. coli O16:H48 pups, indicating that these pups had reduced IGF-1 signaling (Fig. 6A and fig. S9). Reduced IGF-1 signaling in the E. coli O16:H48 pups could be due to reduced production of IGF-1 by the liver, reduced secretion and/or stability of IGF-1 in the circulation, or IGF-1 resistance. To distinguish between these possibilities, we first measured the levels of IGF-1 in the livers. We found that E. coli O16:H48 pups had comparable levels of IGF-1 in their livers (Fig. 6B). Furthermore, the levels of IGF-1 in the white adipose tissue, a second tissue source for IGF-1 that can regulate body size (37), were not significantly different in E. coli O16:H48 pups compared with the three control conditions (Fig. 6C). By contrast, serum levels of IGF-1 were significantly reduced in E. coli O16:H48 pups compared with SPF, E. coli O21:H21, and GF pups, suggesting that the reduced IGF-1 signaling in E. coli O16:H48 pups is because of impaired secretion and/or stability of circulating IGF-1 (Fig. 6D).
Fig. 6. Impaired IGF-1 signaling in E. coli O16:H48 neonates.
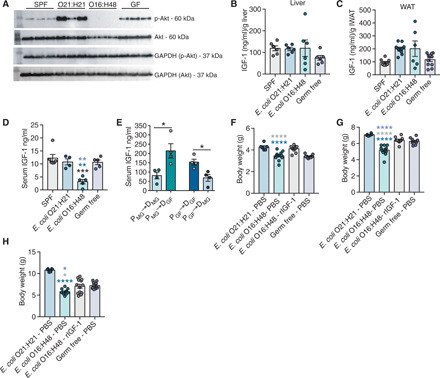
(A) IGF-1 signaling levels in liver at P3 determined by measuring phosphorylated Akt and Akt levels. GAPDH for loading control (n = 3 per condition). Uncropped blots are in fig. S9. (B) IGF-1 protein levels in liver at P3 (n = 6 per condition). (C) Adipose tissue IGF-1 protein levels at P3, n = 6 to 11 per condition. (D) Serum IGF-1 levels at P3 (n = 4 to 6 pups per condition). (E) Serum IGF-1 levels at P21 from cross-fostering experiments (n = 4 per condition). (F to H) Body weight measurements of animals treated with daily intraperitoneal injection of mouse rIGF-1 or PBS vehicle control at (F) P7, (G) P14, and (H) P21; n = 6 to 13 per condition. Statistical significance was determined using one-way ANOVA with post Tukey test or unpaired t test. Significance stars are colored to indicate which condition significance is referring to. *P < 0.05, **P < 0.01, ***P < 0.0005, and ****P < 0.0001. Error bars represent means ± SEM.
Our data thus far indicate that the growth-stunting phenotype exhibited by E. coli O16:H48 is associated with reduced IGF-1 levels and downstream signaling. Our data further show that the failure to thrive phenotype is regulated by maternal factors. To determine whether the effects of E. coli O16:H48 on IGF-1 in pups are due to direct actions of the microbe on the pups or indirectly via interactions between the microbe and the dam, we measured the circulating levels of IGF-1 in pups from our cross-fostering experiment (Fig. 2, E to G). Cross-fostering of E. coli O16:H48 pups by GF dams exhibited significantly higher circulating levels of IGF-1 compared with their littermates that were left to nurse on their E. coli O16:H48 dams (Fig. 6E). GF pups that were fostered by E. coli O16:H48 dams exhibited significantly reduced IGF-1 serum levels compared with their littermates that were left to nurse on their GF dams (Fig. 6E). Thus, monocolonization with E. coli O16:H48 impairs maternal factors (presumably maternal behavior and the ability of the dam to provide nutrients) of the dam that are necessary for promoting circulating IGF-1 levels in their offspring.
To determine whether exogenous supplementation of IGF-1 could rescue the failure to thrive phenotype in E. coli O16:H48 pups, we intraperitoneally injected neonates with recombinant IGF-1 (rIGF-1) daily and compared their body weights with those of phosphate-buffered saline (PBS)–injected control groups. We found that E. coli O16:H48 pups that were injected with exogenous rIGF-1 had increased growth compared with E. coli O16:H48 pups that were injected with PBS (Fig. 6, F to H). The body weight of E. coli O16:H48 pups treated with rIGF-1 was comparable to that of GF pups injected with PBS (Fig. 6, F to H). However, the body weight of rIGF-1–treated pups was significantly reduced compared with E. coli O21:H21 pups injected with PBS (Fig. 6, F to H). Together, our data suggest that administration of exogenous IGF-1 was sufficient to rescue E. coli O16:H48 pups from the stunted growth phenotype, enabling them to reach comparable size as GF mice. However, IGF-1 administration was not sufficient to promote growth comparable to E. coli O21:H21 or SPF in E. coli O16:H48 pups, indicating that there are additional microbial factors that are necessary to promote optimal weight gain and that are lacking in the E. coli O16:H48 maternal-offspring system.
DISCUSSION
We present our approach to investigate the effects of different E. coli strains on postnatal development. Our work supports a model by which E. coli O16:H48 monocolonization impairs the maturation of maternal behaviors in dams after birth. This results in malnourishment of the offspring and negatively affects IGF-1 levels and signaling in the offspring, presumably as a consequence of the malnutrition. Administration of exogenous IGF-1 was sufficient to rescue the pathogenic effects of E. coli O16:H48 but was not sufficient to promote healthy growth similar to SPF mice and like the other E. coli strains tested (Fig. 7). Our study adds to the ever-growing evidence that the microbiota is necessary for optimal growth and development and that it is necessary for normal behavioral processes.
Fig. 7. E. coli O16:H48 impairs maternal behavior resulting in offspring malnutrition and growth stunting.
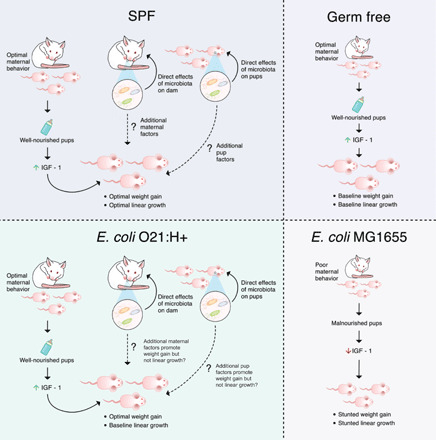
Our work supports a model by which E. coli O16:H48 monocolonization impairs the maturation of maternal behaviors in dams after birth. This results in malnourishment of the offspring and negatively affects IGF-1 levels and signaling in the offspring, presumably as a consequence of the malnutrition. Administration of exogenous IGF-1 was sufficient to rescue the pathogenic effects of E. coli O16:H48 but was not sufficient to promote healthy weight growth similar to SPF mice and like the other E. coli strains tested. By contrast, dams monocolonized with E. coli O21:H21 exhibited maternal behavior comparable to GF and SPF mice. Their offspring exhibited linear growth similar to the offspring of GF dams, indicating that monocolonization with E. coli O21:H21 was not sufficient to promote linear growth observed in offspring of SPF mice. Monocolonization with E. coli O21:H21 was sufficient to promote weight gain in offspring comparable to that of SPF offspring.
Shortly after birth, the infant intestine becomes colonized by maternal and environmentally derived facultative anaerobic bacteria such as E. coli (12). From our analyses, we identified E. coli strains that varied in their ability to promote normal postnatal growth in an infant mouse model. We found that while none of the E. coli strains tested could promote normal linear growth compared with SPF mice, the E. coli strains varied in their ability to promote normal weight gain during the preweaning period. Colonization with E. coli O21:H21, O6:H1, or O157:H7 was sufficient to promote weight gain comparable to that exhibited by SPF pups and greater than GF pups. Our data demonstrate that the effects of different microbiota E. coli strains have on linear growth and weight gain can be decoupled. Impaired linear growth with normal weight gain in humans is associated with metabolic and cognitive disorders later in life (38, 39). Furthermore, disruptions in microbiota homeostasis contribute to metabolic and cognitive disorders (29, 40, 41). Our findings suggest that one way in which infant intestinal dysbiosis may contribute to health problems later in life may be due to the disruption in the relationship between linear growth and weight gain and adds to the ever-growing evidence that the intestinal microbiota early in life has influence on multiple aspects of health at later life stages. In adults, numerous disease settings are associated with intestinal dysbiosis characterized by an overrepresentation of E. coli and other members of the Enterobacteriaceae family, suggesting that these bacteria may cause or exacerbate disease states in adults (42, 43). Our findings that E. coli O21:H21, O6:H1, and O157:H7 promote healthy infant postnatal growth suggest that the effects microbes have on the health of their hosts are complex and can vary over the different life stages of the individual—being beneficial for some life stages but pathogenic at other life stages.
We identified one E. coli strain, E. coli O16:H48, that was pathogenic to infant mice and resulted in a growth-stunted phenotype. In addition to impaired linear growth, E. coli O16:H48 pups exhibited significantly reduced weight compared with not only SPF mice but also GF control mice. We found that the growth stunting of E. coli O16:H48–monocolonized pups was associated with reduced circulating levels of IGF-1 and signaling. Administration of exogenous IGF-1 was sufficient to rescue the pathogenic effects of E. coli O16:H48 and the resulting stunted phenotype, enabling the E. coli O16:H48 pups to reach similar body weight of GF pups. However, E. coli O16:H48 pups treated with rIGF-1 still did not reach comparable size of pups monocolonized with E. coli O21:H21 or SPF pups, suggesting that E. coli O16:H48 lacks the ability to promote normal weight gain like other E. coli strains we used in our study. As E. coli O16:H48 is a derivative of the original human isolate K-12 strain that was initially isolated from a patient with diphtheria and is now a laboratory-adapted strain (16), it is possible that it lost factors that enable it to function similarly to our other E. coli strains and promote healthy weight gain during the preweaning phase of infant mice either indirectly by influencing other factors of the mothers or via direct pathogenic interactions on the pups. For example, breast milk contains maternally derived IgA antibodies that are administered to infants during the early postnatal period and important for offspring health (44–46). Another possibility is that the lipopolysaccharide (LPS) of O16:H48 is phenotypically rough and lacks the O antigen, which is its repeated oligosaccharide unit (47). From our comparative genomic analysis, we found that E. coli O16:H48 genome is missing genes that encode LPS core heptose (I) kinase, LPS glucosyltransferase I, and LPS core heptosyltransferase 3, while the other three strains in our study contain these genes (data S2). Understanding the microbial factors that contribute to E. coli O16:H48 pathogenicity and those that enable the other E. coli strains to promote healthy growth will be of interest for future studies.
Our discovery that an intestinal microbiota species regulates systemic IGF-1 and body size is in agreement with previous findings from our group and others. We previously reported in a mouse model of infection-induced muscle wasting that the intestinal microbiota regulates circulating levels of IGF-1 and body size (4). Subsequent studies in mice and fruit flies showed that intestinal microbiota regulation of the GH (somatotropin)/IGF-1 axis is important for promoting healthy growth of the host (10, 14). In these previous studies, the microbiota was shown to regulate IGF-1 through direct interactions with the host. In the current study, we demonstrated that in maternal-infant systems, the intestinal microbiota can regulate growth and IGF-1 levels in the offspring indirectly, via effects on maternal factors. With our cross-fostering experiments, we demonstrated that pups that received maternal care from E. coli O16:H48 dams had reduced circulating IGF-1 regardless of intestinal microbiota status of the pups. By contrast, fostering of E. coli O16:H48 pups by GF dams was sufficient to rescue the impaired IGF-1 phenotype in E. coli O16:H48 pups. We found that dams monocolonized with E. coli O16:H48 exhibited impaired social and maternal behaviors, and we suggest that these behavioral defects contribute to E. coli O16:H48’s negative effects on infant IGF-1 levels and growth. Maternal care is necessary to provide infants with nutrients and IGF-1 secretion, and stability is dependent on the nutritional status of the host (33, 34, 36). In agreement with this, we found that E. coli O16:H48 pups consume less milk when receiving maternal care from E. coli O16:H48 dams compared with pups that receive maternal care from GF, SPF, or E. coli O21:H21–monocolonized dams. Thus, E. coli O16:H48 impairment of maternal behavior likely prevents dams from providing adequate nutrients that regulate IGF-1 levels in infants. In future work, it will be important to investigate other aspects of maternal care including bonding, warmth, and grooming, which may also contribute to the IGF-1 deficiency in E. coli O16:H48 pups.
An outstanding question from our study is what is the mechanism by which E. coli O16:H48 impairs maternal behavior. Oxytocin is a critical regulator of maternal behavior in the postnatal period and has been shown to be regulated by the intestinal microbiota. In mice, treatment with Lactobacillus reuteri improved deficits in social behavior and was associated with increased hypothalamic expression of oxytocin (29). We, however, did not find a defect in central or peripheral oxytocin levels in E. coli O16:H48 dams. The essential amino acid tryptophan is a precursor for serotonin, a monoamine neurotransmitter that regulates maternal behavior (30). Previous reports have shown that one strain used in our study, E. coli Nissle 1917, enhances bioavailability of serotonin in the host, but E. coli O16:H48 (MG1655) does not (31). From our comparative genomic analyses, we revealed a suite of missense mutations in genes associated with tryptophan biosynthesis. These mutations are concentrated in the trpC gene, which encodes for a bifunctional enzyme and warrants further investigation by way of site-directed mutagenesis work. Whether the maternal behavior deficiencies are due to direct effects of E. coli O16:H48 colonization on brain health or a consequence of developmental defects in E. coli O16:H48 mice independent of the growth stunting remains to be determined; however, our results with the cross-fostering studies suggest it may be the latter. In our cross-fostering studies, the GF dams become colonized with E. coli O16:H48 when these pups are introduced to the dam. Despite becoming colonized with this microbe, ex-GF dams were able to rescue the E. coli O16:H48 pups from the stunted phenotype and IGF-1 deficiencies. These data suggest that short-term colonization of dams during the postnatal period with E. coli O16:H48 is not sufficient to induce the impaired maternal behavior and stunting phenotype in infants.
Although often overlooked, a major risk factor associated with childhood growth failure is a deficit in maternal neurological health, which can interfere with maternal behaviors. For example, several epidemiological studies of mother-infant pairs in developing countries with high prevalence of childhood undernutrition revealed that there is high association between childhood growth failure and poor maternal mental health (48–50). This association most likely exists because maternal depression interferes with maternal interest in offspring and emotional quality of nurturing, which limit the offspring from reaching maximum growth capacity (51). The factors that mediate such relationship between postnatal growth failure and maternal behavior are not defined by one general mechanism, but most likely driven by multiple environmental parameters that contribute to optimal maternal-offspring interactions. Together, our study adds an additional layer of complexity to our understanding of maternal behavior and demonstrates that the maternal microbiota will affect maternal behavior, and this likely regulates offspring development and growth.
MATERIALS AND METHODS
Bacterial strains
To prepare bacterial inocula for oral gavaging, E. coli O21:H21, E. coli O16:H48, E. coli O157: H7, and E. coli O6:H1 were grown in Luria-Bertani (LB) media at 37°C shaking and stored in 25% glycerol/PBS frozen stocks and used directly for gavaging.
Animals/gnotobiotic mouse model
GF Swiss Webster mice were orally gavaged with 5 × 108 colony-forming units (CFU) of E. coli O21:H21, E. coli O16:H48, E. coli O157: H7, or E. coli O6:H1 and maintained in sterile semiflexible isolators under a 12-hour light-dark cycle and given autoclaved standard chow diet and distilled water ad libitum. The status of GF and gnotobiotic conditions was monitored routinely for bacterial, viral, and fungal contaminations. All animal experiments were approved by our Institutional Animal Care and Use Committee.
Animal husbandry
GF and gnotobiotic Swiss Webster mice were kept under sterile conditions in flexible film isolators inflated with HEPA (high-efficiency particulate air)–filtered air, exposed to 12:12-hour light:dark cycles in the Ayres laboratory gnotobiotic facility at the Salk Institute. SaniChip bedding was used in the cages, and mice were fed LabDiet 5K67 for breeders and LabDiet 5010 for holding and experimental cages ad libitum. All bedding, food, and water were autoclaved and transferred into the isolators using an autoclave transfer cylinder. Cages were changed weekly, and water bottles were refilled with sterile water. All breeder cages were checked for new litters on a daily basis, and when a new litter was born, the date of birth and wean date were written on the cage card. Breeding was set up as either pair or harem mating, and litters were weaned 21 days after birth, separated by sex into cages, with five or less mice per cage. Sterility of each autoclave load was monitored using a self-contained biological indicator (Steris S3061), and each isolator was monitored for any microbial contamination by culturing feces in brain heart infusion (BHI), nutrient, and sabouraud-dextrose media anaerobically and aerobically at 37° and 42°C. Fecal DNA was also extracted and amplified by16S rDNA polymerase chain reaction (PCR) as another method of microbiological monitoring of isolators. Sentinel mouse for each isolator was submitted on a quarterly basis for health assessment including serology, bacteriology, parasitology, and necropsy.
Assessment of development
Body weights of each neonate in a given litter were monitored using Pesola spring scale with crocodile clamp inside GF, gnotobiotic, and SPF semiflexible isolators during the entire duration of the experiment. Linear growth of neonates was quantified by measuring the length of tail and nose-to-anus distance. For body composition analyses, inguinal white adipose tissue and tibialis anterior muscle were dissected and weighed on a Mettler Toledo scale, and weights were recorded.
Quantification of bacterial colony-forming units in tissues
Colonization levels of all E. coli were quantified by serially diluting intestinal homogenates and plating on eosin methylene blue agar (EMB)/ampicillin, vancomycin, neomycin and metroidazole (E. coli O21:H21), or EMB (E. coli O16:H48, E. coli O157: H7, and E. coli O6:H1). Plates were incubated at 37°C overnight. Intestinal tissues were homogenized in 1 ml of sterile PBS supplemented with 1% Triton X-100 using a bead beater. Some GF or gnotobiotic intestinal tissues were homogenized in 5 ml of sterile PBS supplemented with 1% Triton X-100 using a Qiagen tissue shredder.
Neonatal rIGF-1 injections
For rIGF-1 injection experiments, GF and gnotobiotic mice (E. coli O16:H48 and E. coli O21:H21) were transferred from gnotobiotic isolators to autoclaved airtight IsoCage Bioexclusion System with positive pressure and HEPA filtration (Tecniplast SpA, Buguggiate, Varese, Italy), which were prepackaged with Sani-Chips bedding (P. J. Murphy) and standard chow before autoclaving. Autoclaved distilled water was provided to the transferred mice. All handling of mice housed in IsoCages was performed under laminar flow hood with sterilization procedures using the disinfectant Clidox-S (1:3:1 formulation). Three-day-old pups were intraperitoneally injected with sterile murine rIGF-1 (R&D Systems) or sterile PBS daily at a dose of 200 μg/kg until weaning age (21-day old) was reached using a 100-μl gastight glass syringe (Hamilton 7656-01) with 34-gauge needle (Hamilton pt. style 4, 12-30o, 0.375-inch length). Daily body weights were measured using an electronic scale for tracking body weight development.
Enzyme-linked immunosorbent assay
Serum and tissue homogenates were prepared for IGF-1 enzyme-linked immunosorbent assay (ELISA). Tissues were frozen in liquid nitrogen and homogenized in lysis buffer (1% Triton X-100 in PBS) using Bead Mill 24 (Thermo Fisher Scientific). Lysates were centrifuged and supernatants were used for ELISAs. IGF-1 was measured using anti-IGF monoclonal antibody clone #126002 and anti-mouse IGF-1 biotinylated antibody (R&D Systems).
Akt Western blot
Tissues were ground in LN2 and protein was extracted using Life Technologies tissue extraction reagent II. Proteins of interest were probed using p-Akt (Ser473) (193H12) rabbit mAB #4058 T, total Akt rabbit Ab #9272S, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Ab (Cell Signaling Technology).
Food intake measurements
Milk intake/suckling behavior/nipple function assessment
Pups at P3, P7, P11, P14, and P18 were fasted for 7 hours by removing them from the house cage containing the lactating dam and were placed on a heat pad. After collecting the fasted whole-body weights of each pup, the pups in each litter were returned to the home cage containing anesthetized lactating dam placed on her side with the body tilted so that the lines of nipples would be exposed to the pups. To ensure that pups do not lose their way to the nipples, a divider/wall was placed rostral to the pectoral nipples and another placed caudal to the inguinal nipples. After observation of the pups to latch to the nipples, the pups were allowed to suckle for milk for 1 hour, and their final body weights were measured. The body weight gain of each pup during 1 hour of suckling was used to estimate the milk intake.
D2O labeling and serum D2O enrichment measurement
Forty-eight hours before termination, dam was intraperitoneally injected with 0.035 ml/g body weight 0.9% NaCl-D2O, and drinking water was replaced with 8% D2O-enriched water. Dam was fasted for 6 hours before serum collection. At the time of euthanasia, serum from individual pups in the litter was also collected. The 2H labeling of water from samples or standards was determined via deuterium acetone exchange. Five microliters of sample or standard was reacted with 4 μl of 10 N NaOH and 4 μl of a 5% (v/v) solution of acetone in acetonitrile for 24 hours. Acetone was extracted by the addition of 600 μl of chloroform and 0.5 g of Na2SO4 followed by vigorous mixing. One hundred microliters of the chloroform was then transferred to a gas chromatography/mass spectrometry (GC/MS) vial. Acetone was measured using an Agilent DB-35MS column [30 m × 0.25 mm inner diameter (i.d.) × 0.25 mm, Agilent J&W Scientific] installed in an Agilent 7890A gas chromatograph interfaced with an Agilent 5975C mass spectrometer (MS) with the following temperature program: 60°C initial, increase by 20°C/min to 100°C, increase by 50°C/min to 220°C, and hold for 1 min. The split ratio was 40:1 with a helium flow of 1 ml/min. Acetone was eluted at approximately 1.5 min. The MS was operated in the electron impact mode (70 eV). The mass ions 58 and 59 were integrated, and the % M1 [m/z (mass/charge ratio) 59] was calculated. Known standards were used to generate a standard curve, and plasma percent enrichment was determined from this. All samples were analyzed in triplicate. To calculate the pup’s normalized percent D2O enrichment, the dam’s percent D2O enrichment and the pup’s weight were taken into account using the following equation
Social interaction test
Mouse was placed in the central chamber of the three-chamber apparatus and habituated for 5 min. Right and left chambers were isolated by using the dividing Plexi glass walls. One control mouse noted as “social 1” was placed inside a wire containment cup and placed in the left/right chamber, and another mouse noted as “social 2” was placed inside a cup placed in the third chamber. To begin testing, the walls between the chambers were removed, and the mouse was allowed to explore each of the three chambers for 10 min. For quantification social novelty, ANY-maze software linked to an overhead camera was used to score the amount of the time the mouse spends interacting with the “social 1” versus “social 2” subject.
Licking and grooming assay
To assess maternal behavior of licking and grooming of pups, pups were marked with a laboratory marker on the top of head and bottom and placed back in their home cage with their respective dams. Photos were taken at To and T1 for records. After 24 hours, the quality of the marks on the pups was assessed on the basis of the following scores: 0, no mark visible; 1, visible mark is extremely faint; 2, visible mark is clear but faint; and 3, visible mark is clear. The average of two separate scores (one for head and another for bottom) was plotted for each pup.
Maternal behavior assessment
Dams with their respective litters were undisturbed and recorded in their home cages during P2 to P4 using a pet camera (Petcube). For each dam, a 30-min video was recorded during morning (9:00 a.m. to 12:00 p.m.) and afternoon (4 p.m. to 7 p.m.) for each testing day. Recorded videos were used to individually score maternal and nonmaternal behaviors of each dam by measuring the duration the dam performs the following behaviors:
Maternal behaviors
Nest building
Licking and grooming
Active nursing
Passive nursing
Nonmaternal behaviors
Eating and drinking
Self-grooming
Digging and climbing
Pups out of nest
Oxytocin analysis
Hypothalamus gene expression analyses
RNA was extracted from snap-frozen hypothalamus using the Qiagen AllPrep DNA/RNA Mini Kit per the manufacturer’s protocol. RNA was treated with DNase, and complementary DNA (cDNA) was synthesized using SuperScript III reverse transcriptase (Invitrogen) and Oligo(dT). We performed real-time quantitative PCR using the QuantStudio 5 Real-Time PCR system (Applied Biosystems) with iTaq Universal SYBR Green Supermix (Bio-Rad) per the manufacturer’s protocol. The number of gene copies was normalized to Rps17. The following were the list of primer sequences: Rps17F, 5′-CGCCATTATCCCCAGCAAG-3′; Rps17R, 5′-TGTCGGGATCCACCTCAATG-3′; OxytocinF, 5′-CGGATCTCAGACTGAGCACC-3′; OxytocinR, 5′-ACTTGCGCATATCCAGGTCC-3′.
Measurement of circulating oxytocin in dams
Blood from dams nursing litters aged between P3 was collected by cardiac puncture, and serum was isolated using a serum separator tube. Oxytocin levels were measured in the serum using the Luminex-based MILLIPLEX MAP Rat/mouse Neuropeptide Magnetic Bead Panel from Millipore (RMNPMAG-83 K-01) per the manufacturer’s protocol.
Whole-genome isolate sequencing
E. coli colonies were purified for genomic DNA (gDNA) extraction by single colony selections after streaking onto Luria Agar incubated overnight at 37°C. gDNA was extracted from a liquid culture (Luria Bertani broth) using the Qiagen DNeasy Blood and Tissue Kit. Extracted gDNA was sent to The Sequencing Center (Fort Collins, CO) for whole-genome sequencing on the Illumina MiSeq using Nextera XT paired-end 150-bp libraries and protocols (Illumina Inc., San Diego, CA, USA).
Bioinformatic analyses
Illumina paired-end sequence reads were assembled using Shovill v1.1.0 (https://github.com/tseemann/shovill), a genome assembler based on SPades (52), with standard parameters (table S1). Contigs were annotated with Prokka v1.14.6 (53), and the MLST and serotype profiles were determined in silico using BLAST (Basic Local Alignment Search Tool)–based tools (https://github.com/tseemann/mlst and https://github.com/tseemann/abricate). The pan-genome of each study isolate was determined using Roary v3.13.0 (54) and visualized using Phandango (55). MUSCLE v3.8.1551 (56) was used to generate multiple sequence alignments of key coding sequences at the amino acid level and visualized using iTOL (57). Genomic variants between E. coli strains were identified using Snippy v4.6.0 (https://github.com/tseemann/snippy). Artemis (release 18.1.0) facilitated the extraction of amino acid sequences from genes of interest that are associated with tryptophan metabolism and biosynthesis for downstream analyses, including plotting of coding region (CDS) schematics with genoPlotR (58). Last, Illumina paired-end sequence reads for each E. coli isolate were joined and filtered for quality (≥ Phred scores of 25), and analyzed using MG-RAST v4.0.3 (59) to characterize subsystem categories. ggplot2 v3.3.2 (60) was used to create the pie charts for visualizing subsystems data.
Statistical analysis
Data are represented as means ±SEM. Statistical tests were performed using GraphPad Prism version 6.0. For pairwise analyses, a t test was performed. For comparisons of three or more samples, we performed a one-way analysis of variance (ANOVA) with a post Tukey test or Kruskall-Wallis test with a post Dunn’s test. We performed D’Agostino and Pearson and Shapiro-Wilk normality tests, and Brown-Forsythe and Bartlett’s variance tests to determine whether we should use a one-way ANOVA or Kruskall-Wallis test.
Acknowledgments
We thank G. Chen for technical support for the sequencing of the E. coli genomes;,D. Michel for assistance with the GF and gnotobiotic husbandry, as well as technical assistance with experiments done with E. coli O6:H1 and E. coli O157:H7 gnotobiotic mice; R. Hampton, S. Hedrick, V. Nizet, M. Raffatellu, and members of the Ayres laboratory for helpful discussions and guidance. Funding: This work was supported by a Crohn’s and Colitis Foundation Senior Research Award and NIH R01 AI114929, both awarded to J.S.A. Funding from the Lynne and Mason Rosenthal/Leo S. Guthman Foundation was awarded to Y.M.L. Author contributions: J.S.A. conceived the idea, provided technical guidance, analyzed the data, made the figures, and wrote the paper. Y.M.L. performed experiments, wrote the Material and Methods section, plotted the data, and made the figures. A.M. performed the genome analyses. M.W., J.M.G., and C.M.M. developed the D2O milk consumption assay, performed the experiments, and analyzed the data. A.J.F. and L.B. developed the milk oligosaccharide assay, performed the experiments, and analyzed the data. A.M. performed the genomic analyses. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequence data from this study are available through the European Nucleotide Archive (BioProject PRJEB441461). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabe6563/DC1
REFERENCES AND NOTES
- 1.Hindmarsh P. C., Geary M. P. P., Rodeck C. H., Kingdom J. C. P., Cole T. J., Factors predicting ante- and postnatal growth. Pediatr. Res. 63, 99–102 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Knight B., Shields B. M., Turner M., Powell R. J., Yajnik C. S., Hattersley A. T., Evidence of genetic regulation of fetal longitudinal growth. Early Hum. Dev. 81, 823–831 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Vilcins D., Sly P. D., Jagals P., Environmental risk factors associated with child stunting: A systematic review of the literature. Ann. Glob. Health 84, 551–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schieber A. M. P., Lee Y. M., Chang M. W., Leblanc M., Collins B., Downes M., Evans R. M., Ayres J. S., Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 350, 558–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanton L. V., Barratt M. J., Charbonneau M. R., Ahmed T., Gordon J. I., Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352, 1533 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Smith M. I., Yatsunenko T., Manary M. J., Trehan I., Mkakosya R., Cheng J., Kau A. L., Rich S. S., Concannon P., Mychaleckyj J. C., Liu J., Houpt E., Li J. V., Holmes E., Nicholson J., Knights D., Ursell L. K., Knight R., Gordon J. I., Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M. A., Benezra A., DeStefano J., Meier M. F., Muegge B. D., Barratt M. J., VanArendonk L. G., Zhang Q., Province M. A., Petri W. A. Jr., Ahmed T., Gordon J. I., Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanton L. V., Charbonneau M. R., Salih T., Barratt M. J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M. J., Trehan I., Jorgensen J. M., Fan Y. M., Henrissat B., Leyn S. A., Rodionov D. A., Osterman A. L., Maleta K. M., Newgard C. B., Ashorn P., Dewey K. G., Gordon J. I., Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J., Herzog J. W., Tsang K., Brennan C. A., Bower M. A., Garrett W. S., Sartor B. R., Aliprantis A. O., Charles J. F., Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. U.S.A. 113, E7554–E7563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M. E., Balmand S., Hudcovic T., Heddi A., Rieusset J., Kozakova H., Vidal H., Leulier F., Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854–857 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Suzawa M., Muhammad N. M., Joseph B. S., Bland M. L., The toll signaling pathway targets the insulin-like peptide Dilp6 to inhibit growth in Drosophila. Cell Rep. 28, 1439–1446.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Houghteling P. D., Walker W. A., Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 60, 294–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukjancenko O., Wassenaar T. M., Ussery D. W., Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60, 708–720 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storelli G., Defaye A., Erkosar B., Hols P., Royet J., Leulier F., Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Anderson E. S., Viability of, and transfer of a plasmid from, E. coli K12 in human intestine. Nature 255, 502–504 (1975). [DOI] [PubMed] [Google Scholar]
- 16.Blattner F. R., Plunkett G. III, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y., The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B. L., Mori H., Horiuchi T., Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2, 2006.0007 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith H. W., Survival of orally administered E. coli K 12 in alimentary tract of man. Nature 255, 500–502 (1975). [DOI] [PubMed] [Google Scholar]
- 19.Kelleher K. J., Casey P. H., Bradley R. H., Pope S. K., Whiteside L., Barrett K. W., Swanson M. E., Kirby R. S., Risk factors and outcomes for failure to thrive in low birth weight preterm infants. Pediatrics 91, 941–948 (1993). [PubMed] [Google Scholar]
- 20.de Onis M., Branca F., Childhood stunting: A global perspective. Matern. Child Nutr. 12 Suppl. 1, 12–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer M., JeVenn A., Hipskind P., Evaluation of muscle and fat loss as diagnostic criteria for malnutrition. Nutr. Clin. Pract. 30, 239–248 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Charbonneau M. R., O’Donnell D., Blanton L. V., Totten S. M., Davis J. C. C., Barratt M. J., Cheng J., Guruge J., Talcott M., Bain J. R., Muehlbauer M. J., Ilkayeva O., Wu C., Struckmeyer T., Barile D., Mangani C., Jorgensen J., Fan Y.-m., Maleta K., Dewey K. G., Ashorn P., Newgard C. B., Lebrilla C., Mills D. A., Gordon J. I., Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164, 859–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuhrer A., Sprenger N., Kurakevich E., Borsig L., Chassard C., Hennet T., Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J. Exp. Med. 207, 2843–2854 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss G. A., Hennet T., The role of milk sialyllactose in intestinal bacterial colonization. Adv. Nutr. 3, 483S–488S (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al Aïn S., Belin L., Schaal B., Patris B., How does a newly born mouse get to the nipple? Odor substrates eliciting first nipple grasping and sucking responses. Dev. Psychobiol. 55, 888–901 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Yang M., Silverman J. L., Crawley J. N., Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci. Chapter 8, Unit 8.26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capone F., Bonsignore L. T., Cirulli F., Methods in the analysis of maternal behavior in the rodent. Curr. Protoc. Toxicol. Chapter 13, Unit13.19 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Pedersen C. A., Vadlamudi S., Boccia M. L., Moy S. S., Variations in maternal behavior in C57BL/6J mice: Behavioral comparisons between adult offspring of high and low pup-licking mothers. Front. Psych. 2, 42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buffington S. A., di Prisco G. V., Auchtung T. A., Ajami N. J., Petrosino J. F., Costa-Mattioli M., Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angoa-Perez M., Kuhn D. M., Neuronal serotonin in the regulation of maternal behavior in rodents. Neurotransmitter (Houst) 2, e615 (2015). [PMC free article] [PubMed] [Google Scholar]
- 31.Nzakizwanayo J., Dedi C., Standen G., Macfarlane W. M., Patel B. A., Jones B. V., Escherichia coli Nissle 1917 enhances bioavailability of serotonin in gut tissues through modulation of synthesis and clearance. Sci. Rep. 5, 17324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Höglund E., Øverli Ø., Winberg S., Tryptophan metabolic pathways and brain serotonergic activity: A comparative review. Front. Endocrinol. (Lausanne) 10, 158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hintz R. L., Suskind R., Amatayakul K., Thanangkul O., Olson R., Plasma somatomedin and growth hormone values in children with protein-calorie malnutrition. J. Pediatr. 92, 153–156 (1978). [DOI] [PubMed] [Google Scholar]
- 34.Stratikopoulos E., Szabolcs M., Dragatsis I., Klinakis A., Efstratiadis A., The hormonal action of IGF1 in postnatal mouse growth. Proc. Natl. Acad. Sci. U.S.A. 105, 19378–19383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore T., Beltran L., Carbajal S., Strom S., Traag J., Hursting S. D., DiGiovanni J., Dietary energy balance modulates signaling through the Akt/mammalian target of rapamycin pathways in multiple epithelial tissues. Cancer Prev. Res. (Phila.) 1, 65–76 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Hawkes C. P., Grimberg A., Insulin-like growth factor-I is a marker for the nutritional state. Pediatr. Endocrinol. Rev. 13, 499–511 (2015). [PMC free article] [PubMed] [Google Scholar]
- 37.Klöting N., Koch L., Wunderlich T., Kern M., Ruschke K., Krone W., Bruning J. C., Blüher M., Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes 57, 2074–2082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Lucia Rolfe E., Araújo de França G. V., Vianna C. A., Gigante D. P., Miranda J. J., Yudkin J. S., Horta B. L., Ong K. K., Associations of stunting in early childhood with cardiometabolic risk factors in adulthood. PLOS ONE 13, e0192196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie W., Jensen S. K. G., Wade M., Kumar S., Westerlund A., Kakon S. H., Haque R., Petri W. A., Nelson C. A., Growth faltering is associated with altered brain functional connectivity and cognitive outcomes in urban Bangladeshi children exposed to early adversity. BMC Med. 17, 199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S., Kim H., Yim Y. S., Ha S., Atarashi K., Tan T. G., Longman R. S., Honda K., Littman D. R., Choi G. B., Huh J. R., Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley R. E., Backhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I., Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., Orsi R. H., Wiedmann M., McDonough P., Kim S. G., Berg D., Schukken Y., Scherl E., Simpson K. W., Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 1, 403–418 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Shin N. R., Whon T. W., Bae J. W., Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Gopalakrishna K. P., Macadangdang B. R., Rogers M. B., Tometich J. T., Firek B. A., Baker R., Ji J., Burr A. H. P., Ma C., Good M., Morowitz M. J., Hand T. W., Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 25, 1110–1115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koch M. A., Reiner G. L., Lugo K. A., Kreuk L. S. M., Stanbery A. G., Ansaldo E., Seher T. D., Ludington W. B., Barton G. M., Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindner C., Thomsen I., Wahl B., Ugur M., Sethi M. K., Friedrichsen M., Smoczek A., Ott S., Baumann U., Suerbaum S., Schreiber S., Bleich A., Gaboriau-Routhiau V., Cerf-Bensussan N., Hazanov H., Mehr R., Boysen P., Rosenstiel P., Pabst O., Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 16, 880–888 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Browning D. F., Wells T. J., França F. L. S., Morris F. C., Sevastsyanovich Y. R., Bryant J. A., Johnson M. D., Lund P. A., Cunningham A. F., Hobman J. L., May R. C., Webber M. A., Henderson I. R., Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol. Microbiol. 87, 939–950 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Rahman A., Lovel H., Bunn J., Iqbal Z., Harrington R., Mothers’ mental health and infant growth: A case-control study from Rawalpindi, Pakistan. Child Care Health Dev. 30, 21–27 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Surkan P. J., Kennedy C. E., Hurley K. M., Black M. M., Maternal depression and early childhood growth in developing countries: Systematic review and meta-analysis. Bull. World Health Organ. 89, 608–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross J., Hanlon C., Medhin G., Alem A., Tesfaye F., Worku B., Dewey M., Patel V., Prince M., Perinatal mental distress and infant morbidity in Ethiopia: A cohort study. Arch. Dis. Child. Fetal Neonatal Ed. 96, F59–F64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girma S., Fikadu T., Abdisa E., Maternal common mental disorder as predictors of stunting among children aged 6-59 months in western Ethiopia: A case-control study. Int. J. Pediatr. 2019, 4716482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., Fookes M., Falush D., Keane J. A., Parkhill J., Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hadfield J., Croucher N. J., Goater R. J., Abudahab K., Aanensen D. M., Harris S. R., Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guy L., Kultima J. R., Andersson S. G., genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R. A., The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York ISBN 978-3-319-24277-4, 2016); 10.1007/978-3-319-24277-4. [DOI]
- 61.Ferdous M., Zhou K., Mellmann A., Morabito S., Croughs P. D., de Boer R. F., Kooistra-Smid A. M., Rossen J. W., Friedrich A. W., Is shiga toxin-negative Escherichia coli O157:H7 Enteropathogenic or enterohemorrhagic Escherichia coli? Comprehensive molecular analysis using whole-genome sequencing. J. Clin. Microbiol. 53, 3530–3538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reister M., Hoffmeier K., Krezdorn N., Rotter B., Liang C., Rund S., Dandekar T., Sonnenborn U., Oelschlaeger T. A., Complete genome sequence of the gram-negative probiotic Escherichia coli strain Nissle 1917. J. Biotechnol. 187, 106–107 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabe6563/DC1


