Abstract
COVID-19 pandemic has started in December 2019 in China and quickly extended to become a worldwide health and economic emergency issue. It is caused by the novel coronavirus; SARS-CoV-2. COVID-19 patients’ clinical presentations vary from asymptomatic infection or flu like symptoms to serious pneumonia which could be associated with multiple organ failure possibly leading to death. It is understood that the immune response to SARS-CoV-2 includes all elements of the immune system which could altogether succeed in viral elimination and complete cure. Meanwhile, this immune response may also lead to disease progression and could be responsible for the patient’s death. Many trials have been done recently to create therapies and vaccines against human coronavirus infections such as MERS or SARS, however, till now, there is some controversy about the effectiveness and safety of antiviral drugs and vaccines which have been developed to treat and prevent this disease and its management depends mainly on supportive care. The spike glycoprotein or protein S of SARS-CoV-2 is the main promoter that induces development of neutralizing antibodies; hence, many attempts of vaccines and antiviral drugs development have been designed to be directed specifically against this protein. While some of these attempts have been proved to be efficient in in vitro settings, only few of them have been proceeded to randomized animal trials and human studies which makes COVID-19 prevention an ongoing challenge.
This review describes the natural immune response scenario during COVID-19 and the vaccines development trials to create efficient vaccines thus helping to build more effective approaches for prophylaxis and management.
Keywords: Novel coronavirus, COVID-19, SARS COV-2, Immune response, Cytokine storm, Vaccines
1. Introduction
COVID-19 was announced as a pandemic on March 11, 2020 with records of more than 86 million confirmed cases and 1.874.732 reported deaths all over the world by January 6, 2021 [1]. The origin of this viral infection was in Wuhan city, Hubei province, China, where a series of cases were first discovered in December 2019 [2].
The etiology was immediately determined as beta coronavirus with high sequence homology to bat coronaviruses (CoVs) which uses the angiotensin-converting enzyme 2 (ACE2) receptor as the main cell entry process [3]. Its human-to-human transmission was confirmed following possible zoonotic spillover. SARS-CoV-2 is also linked to SARS (severe acute respiratory syndrome) which was previously named as SARS-CoV-1 and Middle Eastern Respiratory Syndrome (MERS) Corona viruses, which resulted in zoonotic and local outbreaks in 2003 and 2012, respectively. COVID-19 patients present clinically with wide range of symptoms varying from no or mild symptoms like influenza clinical picture to more severe forms of pyrexia, cough, dyspnea, sometimes followed by respiratory failure and multi system failure then death [4]. Whereas SARS-CoV-2 is less deadly than SARS or MERS; its lethality rate is estimated to be 2.7% versus 9.6% for SARS and 35% for MERS [5], however its global extension has led to immense uncertainty and devastating effects in many countries due to its high infectivity rate requiring specialized medical care in intensive care units (ICU) [6] and revealing the unseen vulnerabilities of health systems and the importance of global health cooperation.
The most seriously affected population is the old age group, especially those suffering from chronic diseases as well as the immunocompromised patients. Additionally, there are some regional differences in COVID-19 infection patterns whose causes are not clearly understood [7].
Although a fast and coordinated immune response exerts the first line of defense in COVID-19, exaggerated production of inflammatory cytokines during the innate response could result in tissue injury either at the site of infection or systemically. Moreover, a dysregulated cell mediated, and humoral response may worsen the condition. It was reported that significant changes occur in both the innate and adaptive immune response while encountering SARS-CoV-2 leading to enormous release of cytokines or the “cytokine storm” which represents the ongoing hysterical activation of the immune system [8].
There is no fully effective therapy till now particularly for the less immunocompetent patients which makes evading complications a real challenge. Most of the suggested therapies for COVID-19 are derived from those used previously in treatment of related viruses such as SARS and MERS or other viruses as Zika or Ebola. Examples of treatments that showed some success till now are remdesivir (adenosine analogue), lopinavir/ritonavir (protease inhibitors) alone or combined with interferon-β, chloroquine, hydroxychloroquine, and plasma therapy [9].
Since there is limited control of the pandemic even by physical distancing and good hygiene measurements and there is minimal understanding of the cytokine storm nature and the changes that occur in the signaling pathways stimulated by SARS-CoV-2, thus, better identification of the immune response scenario in COVID-19 patients especially at the molecular level could help finding the molecular targets either for therapy or vaccines.
The aim of this review is to focus on the main aspects of both the innate and adaptive immune responses as well as the effective vaccines strategies for SARS-CoV-2.
The primary objectives of this review are to determine the immune response responsible for fighting SARS-CoV-2 specifically and discuss its vaccines development strategies especially those showing promising results.
2. Virology
The virus was identified by Wu et al. [10] and named it WH-Human 1, and by Zhou et al., simultaneously, who named it 2019-nCoV [11]. Later on, the virus name was changed to “SARS-CoV-2” by the Coronavirus Study Group (CSG) of International Committee on Taxonomy of Viruses (ICTV), since it was found to be the sister virus of severe acute respiratory syndrome coronavirus (SARS) [12]. Based on this, the WHO officially announced the virus name as SARS-CoV-2 and the infection as COVID-19 by February 11, 2020 [13].
Coronaviruses belong to the family Coronaviridae (subfamily Coronavirinae) and are capable of infecting various wild animals as well as humans where they can cause diseases ranging from mild flu to severe respiratory infections and sometimes fatal complications [12]. Among the 6 Corona viruses (CoVs) which are pathogenic to humans, 4 of them have led to mild respiratory infections, however, the other 2 viruses; SARS and MERS were responsible for epidemics of severe respiratory infections in 2003 and 2012 respectively [14]. SARS and MERS have a lower infectivity rate but a higher lethality than SARS-CoV-2 which showed tremendously higher infectivity with apparently lower lethality rate [15].
SARS-CoV-2 is the 7th identified Corona virus and the 3rd zoonotic virus of CoVs that has been transmitted from animals to humans after SARS and MERS [16], [17]. In fact, the Chinese horseshoe bats have been proposed to serve as the primary source for SARS-CoV-2. It was reported that SARS-CoV-2 genome has shown around 80% similarity with SARS [18] and nearly all its encoded proteins are homologous with those of SARS [8].
Corona viruses are enveloped single stranded positive sense RNA viruses; their RNA is approximately 30 Kbp. They have a spheroidal shape, their diameter is 80–120 nm and their envelope holds the structural proteins; spike (S), membrane (M), and envelope (E), and they include the nucleocapsid (N) inside the virion which covers the RNA [19].
S is a glycoprotein that projects from the viral membrane, giving it the crown shape and hence the corona virus name, it also helps in the attachment of the virus to different surfaces leading to its high stability and infectivity. Glycoprotein S is composed of 2 subunits: S1 which contains the receptor binding domain (RBD) that has epitopes which could be recognized by T and B lymphocytes and induce the production of neutralizing antibodies, and S2 which induces the fusion of the virion with the host cell membrane [15].
The surface angiotensin-converting enzyme 2 (ACE2), which is expressed on type-I and -II alveolar cells of the lungs is the main receptor that allows the entrance of SARS-CoV-2 into human host cells. Noteworthy, the expression of ACE2 receptors is not only restricted to the lung cells but their expression on small intestine enterocytes, kidney proximal tubules cells, endothelial cells of arteries and veins, and the arterial smooth muscle was also demonstrated which may explain the extrapulmonary spread of SARS-CoV-2 [20]. Furthermore, homology modeling showed great degree of structural resemblance between the receptor-binding domains of SARS and SARS-CoV-2 [21]. However, the degree of affinity of the 2 viruses to their receptors may vary which may be the cause of the higher infectivity and virulence of SARS-CoV-2 in comparison to SARS [22].
Binding between RBD of the virus and its receptor initiates conformational changes of S protein which causes cleavage of S1 and S2 and this is considered as a fundamental step that enables S2 to induce the fusion of the virus envelope with the cell membrane followed by the internalization of the viral RNA into the cytoplasm of host cells by endocytosis [19], [23]. Next, the viral RNA acts as a template for the translation of the polyproteins pp1a and pp1b which are then cleaved into 5– 16 non-structural proteins (nsp2-nsp9), which in turn induce reorganization of the membranes to form the vesicles where viral replication and transcription complexes are formed then assembly of the virions starts and the mature virions are released from the cells by the secretory pathway to infect neighboring cells [19].
In addition, it was found that entry of the virus is helped by the cellular host type 2 transmembrane serine protease (TMPRSS2) which is expressed on many cell types and has a role in priming the S protein leading to its cleavage at the S1/S2 site to enable fusion of the viral envelope with the host cell membrane. Thus, TMPRSS2 could be a possible biological target in therapies and vaccines strategies and the entry of SARS-CoV-2 into the host cells could be prevented by TMPRSS2 inhibitor (camostat mesylate). In fact, TMPRS2 inhibitor (camostat mesylate) is already in use in Japan for treatment of pancreatitis which proposes its possible benefit in treating COVID-19 cases [22].
3. Pathological features of COVID-19
Pathological features of COVID-19 involve different organs such as lungs, GIT, liver, kidneys, heart, blood, brain, etc. [24].
The respiratory system is primarily affected by SARS-CoV-2; severely affected patients commonly develop acute respiratory distress and pneumonia. Macroscopically, increased pulmonary weight due to large quantities of viscid secretions was noticed postmortem in addition to congestion, consolidation, and pleurisy. Microscopically, variable pathologic features were found such as diffuse alveolar damage, lymphocytic infiltration, giant cells formation and fibrosis [25]. Moreover, there were microangiopathy, intra-capillary clot formation, thickened capillary walls and congestion in autopsy samples [26].
The heart seems also to be affected during this infection; cardiomegaly especially right ventricular hypertrophy was the major observation. In addition, increased infiltration of the myocardium with macrophages and lymphocytes causing myocarditis, was noticed and may be caused by the high levels of proinflammatory cytokines secretion such as IL-6 and TNF-α. Also, myocardial microvascular thrombi formation and right ventricular strain injury were detected [27]. Blood vessels showed signs of inflammation with lymphocytic infiltration in addition to endothelial cell death in postmortem samples of COVID-19 patients [28].
SARS-CoV-2 appears to cause certain pathologic findings in the GIT; many patients have complained of diarrhea and the virus was found in their stools. Lymphocytic infiltration was noticed in the epithelium of the esophagus as well as the gastric, duodenal, and rectal lamina propria which contained also some viral nucleocapsid proteins [29]. In addition, increased levels of liver enzymes were reported in COVID-19 patients. Macroscopically, hepatomegaly with darkened tissue biopsies were observed while on the microscopic side, hepatic cell degeneration with focal necrosis, thrombosis, portal fibrosis, lymphocytic infiltration and collaterals formation were noticed [30].
Kidneys could be affected in some patients; acute proximal tubular injury with loss of brush border and vacuole degeneration, necrosis and pigmented casts formation were among the major pathological features in COVID-19 autopsy samples. Furthermore, there were other changes such as capillary microthrombi formation, swollen endothelial cells and formation of protein exudate which may explain the acute renal failure and proteinuria reported in some patients [31].
Regarding the skin, redness with vesicles or pustules formation and allergy were the most clinical presentations among severe COVID-19 patients. On the microscopic level, there is lymphocytic infiltration, signs of inflammation, vascular microthrombi formation in the dermis and Langerhans cells formation in the epidermis [31].
Postmortem brain specimens showed some degree of necrosis and infarction in addition to acute disseminated encephalomyelitis like appearance [32]. The pathological changes that occur in other organs such as spleen, lymph nodes, thyroid gland and pancreas are less clarified and larger studies are needed to demonstrate them, however, one common finding that seems to affect most of the organs is microthrombi formation which favors the explanation of SARS-CoV-2 induced DIC in many severe cases of COVID-19 [31].
4. Immune response to SARS- CoV-2
The main aim of both innate and adaptive immune responses against viral infections, in general, is to block the viral infection and eliminate the infected cells. Type I interferons secreted during innate immunity and neutralizing antibodies produced through adaptive immune system could prevent the viral infection. However, if the cells got already infected, then it is the role of the natural killer (NK) cells as a part of the innate immune system and the cytotoxic CD8 T cells of the adaptive immune system to eliminate them [33].
In specific, SARS-CoV-2 infection involves the activation of both innate and adaptive immune systems to clear the infection [34]. However, the exaggerated proinflammatory cytokines production has been proposed to cause pathologic condition in lungs characterized by occurrence of respiratory distress which could be followed by pulmonary failure with or without multi organ failure [10].
In the same context, marked increase in proinflammatory cytokines and chemokines such as, IL-1β, IL-2, IL-6, IL-8, IL-17, and TNF-α was also noticed in COVID-19 cases [35]. Moreover, elevated levels of neutrophil lymphocyte ratio (NLR), proinflammatory cytokines and chemokines in COVID-19 patients were correlated with more severe form of the disease and worse prognosis hypothesizing the relation between the inflammatory responses in patients and the immunopathologic nature of the disease [36].
Another study has shown that SARS-CoV-2 patients with severe complications had increased concentrations of inflammatory markers such as high-sensitivity C-reactive protein (Hs-CRP) [37] which is a member of the pentraxin family and an important acute phase protein that plays a fundamental role in both innate and adaptive immune responses [38], [39].
5. Innate immunity to SARS-CoV-2
The main process by which the innate immune system fights any viral infection is production of type I interferons by the infected cells particularly the plasmacytoid dendritic cells and cytotoxic killing of the viral infected cells by the NK cells [33].
Production of interferons is initiated by recognizing the viral nucleic acid by the pattern recognition receptors (PRRs) such as endosomal Toll like receptors (TLRs) and retinoic acid-inducible gene (RIG) like receptors. After ligand-receptor binding, PRRs stimulate certain adaptor proteins which, in turn leads to the activation of critical down-stream transcription factors, including the interferon regulatory factor (IRF) transcription factor which initiates interferon gene transcription leading to production of interferons. Type I interferons acts by suppressing the viral replication in infected and neighboring cells. Moreover, PRRs activate other signaling pathways such as NF-κB, and AP-1, leading to secretion of many other cytokines and chemokines [40].
The produced chemokines serve in recruiting cells of innate immunity as neutrophils, monocytes, NK cells and dendritic cells which in turn help in producing more chemokines like MIG, IP-10, and MCP-1 which play important role in attracting lymphocytes and initiating the adaptive immune response [41].
The early immune response initiated by SARS-CoV-2 in comparison to other coronaviruses' infections was monitored by several studies. For example, a recent in vitro study reported that both SARS and SARS-CoV-2 have equal ability to infect type I and type II pneumocytes and alveolar macrophages with better capability of intracellular replication for SARS-CoV-2 over SARS. However, SARS-CoV-2 was not able to initiate the production of type I, II and III interferons, in addition it had less effective production of other cytokines when compared to SARS immune response at the same early stage of infection. Only 5 cytokines (IL-6, MCP1, CXCL1, CXCL5, and CXCL10/IP10) were expressed by SARS-CoV-2 infection while all the 11 cytokines measured in this study were produced through SARS immune response [42].
Moreover, it was shown that SARS-CoV-2 is associated with less interferons production and increased proinflammatory cytokines release including IL-1B, IL-6, TNF-α, and IL-1 receptor antagonist in a study that investigated the immune response of SARS-CoV-2 in relation to other respiratory viruses such as SARS, MERS, respiratory syncytial virus, human parainfluenza virus type 3, and influenza A virus. The study involved respiratory cell lines infection, in vivo infection of ferrets as well as lung samples obtained from dead COVID-19 cases [43].
These findings hypothesize that SARS-CoV-2 has an altered immune response compared to the other coronaviruses in term of more capability of intracellular replication in lungs, increased activation of innate immunity associated with higher levels of proinflammatory cytokines as well as high evasion mechanisms abilities that allow it to skip the production of interferons and their antiviral response [15].
On the other side, NK cells play a potential role in fighting viral infection early in the disease before development of adaptive immune response. As an evasion mechanism, the virus infected cells decrease the expression of MHC class I on their surfaces so the immune system could not catch them especially CD8 cytotoxic cells, however, this leads to activation of the NK cells which are inhibited naturally through recognition of MHC class I on the target cells enabling NK cells to exert their cytotoxic actions against the infected cells and hence eradication of the viral infection [33].
6. Signaling pathways activated by SARS-CoV-2
Knowing the intracellular molecules involved in activation of host immune response may help targeting them in designing therapeutic and vaccines strategies. This may have better effect than targeting the viral peptides themselves which are liable for viral mutations and evasion mechanisms [44]
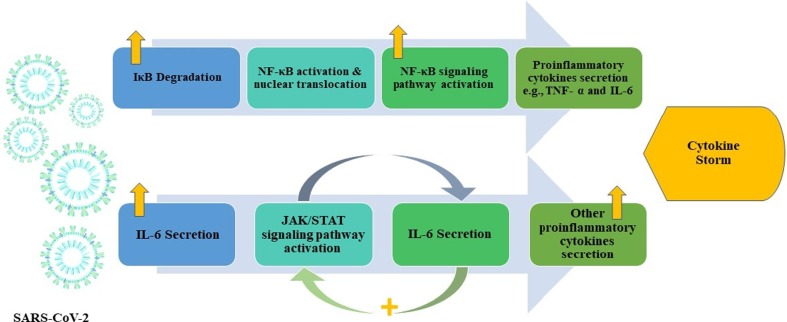
Since there are many similarities between SARS-CoV-2 and SARS in structure and mode of infection, it is postulated that they share innate immunity methods of signaling pathways activation. Following the attachment of SARS-CoV-2 S protein to ACE2 receptors expressed on the host cells, the viral RNA is recognized through TLR 3, 7 and 8 and the cytosolic RNA receptors; RIG-I [45].This recognition especially through TLR3, 7 and 8 initiates signaling pathways activation in monocytes such as IRF3 (IFN regulatory factor-3), nuclear factor κB (NF-κB), JAK (Janus kinase)/STAT (signal transducer and activator of transcription) leading to interferon type I and other cytokines production which in turn leads to differentiation of T cells towards CD4 T helper cells (Fig. 1 .) [46].
Fig. 1.
Supposed signaling pathways initiated by SARS-CoV-2. Illustration representing main signaling pathways hypothesized to be triggered by SARS-CoV-2 leading to cytokine storm in complicated cases. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; IκB, inhibitor of nuclear factor kappa B; NF-κB, nuclear factor kappa B; TNF- α; tumor necrosis factor alpha, IL-6, interleukin 6; JAK/STAT, Janus kinase/ signal transducer and activator of transcription [8].
6.1. The NF-κB/TNFα signaling pathway
Nuclear factor Kappa B (NF-κB) is an important transcription factor which plays a significant role in regulating innate and adaptive immune responses. Various pathogens stimulate nuclear translocation of NF-κB which exists normally in the cytoplasm of cells [47]. It, then, initiates the expression of genes whose products are necessary for the inflammatory response such as cytokines and chemokines [48]. Although this activation is essential for an optimum immune response, it has been suggested to lead to exaggerated inflammatory response leading to lung injury and respiratory distress caused by respiratory viruses such as SARS [49]. Furthermore, NF-κB activation has been implicated in enormous production of IL-6 and TNF-α cytokines in murine macrophages after exposure to recombinant SARS S protein. This was postulated to be due SARS S protein related degradation of IκBα, a normal inhibitor of NF-κB, leading to augmented activation of NF-κB signaling pathway [50].
Another study confirms these findings; they found that certain therapies which can suppress NF-κB signaling pathway such as caffeic acid phenethyl ester and parthenolide led to decreased inflammatory process by inhibiting the expression of genes encoding inflammatory cytokines and chemokines as TNF-α, CXCL2, and MCP-1 in the lungs of mice having SARS infection. This in turn helped in preventing disease progression in those mice and decreased their mortality rates after the infection [49]. Similarly, it was reported that infection with SARS in macaques has led to increased translocation of NF-κB secondary to its activation especially in old ones more than the younger macaques leading to a strong inflammatory viral response [51].
All these data suggest that suppression of NF-κB could be an efficient way to escape the undesirable inflammatory process caused by SARS, however, designing therapies that aim to target this molecule specifically may be problematic and could instead affect the normal innate immunity process leading to exacerbation of infections. In addition, many viruses have the ability to successfully block this signaling pathway leading to in-effectivity of such therapy and highlights the hope of targeting its inflammatory products instead such as TNF-α [52]. Furthermore, Anti-TNF-α biological treatments such as infliximab and adalimumab have been tried in treating a variety of autoimmune diseases such as rheumatoid arthritis and psoriasis with a reasonable success rate which favors targeting this cytokine specifically among the other inflammatory cytokines which are involved in the cytokine storm [53]. In fact, patients with active rheumatoid arthritis who were treated with such therapies had also diminished levels of other inflammatory cytokines including IL-6 and IL-1 leading to fast amelioration of their inflammatory conditions. Indeed, when an anti-TNF-α is administrated in patients with active rheumatoid arthritis, it has been demonstrated to induce a rapid decrease of a broad spectrum of cytokines (e.g., IL-6 and IL-1), as well as of others acute-phase related proteins and vascular permeability factor [54]
Based on these data and on the high similarity between SARS-CoV-2 and SARS, monoclonal antibodies against TNF-α were hypothesized to suppress the cytokine storm occurring in COVID-19 patients and decrease its possible consequences. A clinical trial study using Adalimumab as TNF-α inhibitor has been started in Chinese COVID-19 patients to investigate its efficacy and safety as well [8].
Moreover, it was found that TNF-α converting enzyme (TACE) mediated shedding of ACE2 which is required for virus internalization into the host cells is enhanced by SARS S protein. This means that monoclonal antibodies against TNF-α could exert their therapeutic effects through double hypotheses: prevention of viral entry and weakening of the inflammatory process and cytokine storm [55].
6.2. The IL-6/JAK/STAT signaling pathway
IL-6 was found to be elevated significantly following SARS-CoV-2 infection where it was postulated to participate strongly in the cytokine storm in infected patients [56]. IL-6 interacts with its receptors expressed on the immune cells such as glycoprotein 130 (gp 130) receptor and membrane bound IL-6 receptor as well as soluble receptor for gp130 leading to activation of the JAK/STAT signaling pathway [57]. There is bidirectional relationship between both IL-6 and JAK/STAT signaling pathway meaning that they could stimulate each other as activation of JAK/STAT signaling leads to more IL-6 secretion and vice-versa [58].
Many cell types are known to produce IL-6 such as macrophages, endothelial and smooth muscle cells and once produced, it stimulates the production of other cytokines especially MCP-1 which induces atherogenesis [59], increased expression of adhesion molecules [60] and proliferation of vascular smooth muscle cells [61]. This may consequently explain the cardiovascular complications occurring in COVID-19 patients where high levels of IL-6 are detected [62].
The production of IL-6 is seen to be caused by angiotensin II, which is locally generated by inflamed vessels to bind to its receptor; angiotensin II receptor type 1 (AT1 receptor) leading to activation of JAK/STAT signaling. The augmented production of angiotensin II promotes more IL-6 secretion in AT1/JAK/STAT-dependent way, thus entering in a vicious circle of inflammatory response [63].
Remarkably, it was found that SARS S protein has an important role in decreasing the expression of ACE2 with a subsequent increase of angiotensin II [64]. Similarly, it was postulated that SARS-CoV-2 could exert same actions and decrease ACE2 receptors expression leading to increased accumulation of angiotensin II and hence increased IL-6 secretion leading to cardiovascular complications and pulmonary damage [8]. In addition, this inflammatory pathway has been implicated in the activation of NF-κB and ADAM pathways. Particularly, ADAM17 causes ACE2 cleavage thus inactivating it, and increasing angiotensin II, accordingly, leading to hypertension and other cardiovascular pathologies [65].
It was demonstrated that the metalloprotease ADAM17 transforms the membrane form of IL-6 receptor α (IL-6Rα) to its soluble form (sIL-6Rα) with the subsequent activation of STAT3 which in turn activates NF-κB signaling. Therefore, SARS-CoV-2 could lead to activation of both NF-κB and STAT3 pathways promoting the enhanced production of IL-6 which is in turn implicated in more activation of NF-κB by STAT3 leading to a hyper-inflammatory response and may proceed to development of autoimmune diseases [66]. It was proposed that the amplified production of IL-6 induces the secretion of many other inflammatory cytokines and chemokines leading to migration of lymphocytes and leucocytes to the site of inflammation and maintaining the IL-6 mediated inflammatory response at its highest level [67].
6.3. Sphingosine-1-phosphate receptor 1 pathway
Sphingosine-1-phosphate (S1P) 1 is a key signaling pathway that plays a role in regulating the inflammatory process; it involves lymphoid cells recruitment, vascular permeability and production of cytokines and chemokines. It exerts its functions after binding to its receptors; five G-protein-coupled receptors (S1PRs1–5) which exist in different types of cells [68].
The activation of S1P1 receptor which is usually bound to a G inhibitory protein and widely expressed on many types of cells leads to stimulation of Ras/ERK signaling pathway [69]. Interestingly, the S1P1 receptor signaling was found to limit the immunopathological injury caused by both innate and adaptive responses, hence suppressing the cytokine storm formation following Influenza viral infection mainly through diminished production of IFN-α, CCL2, IL-6, TNF-α, and IFN-γ which helped in decreasing the mortality rates in the infected mice [44]. Similarly, in another study done later, it was found that stimulation of S1P1 has led to blockage of cytokines secretion and inhibited migration of inflammatory cells to the lungs of H1N1 influenza infected mice. It was demonstrated that these actions were exerted through reduction of cytokine storm independently of TLR3 and TLR7 signaling pathways but rather through targeting MyD88 (myeloid differentiation primary response gene 88)/TRIF (TIR-domain-containing adapter-inducing IFN-β) signaling which are main players in the NF-κB pathway [70].
7. Cytokine storm
It is well observed clinically that one of the main causes of deterioration of patients’ clinical conditions is the significant increase of proinflammatory cytokines levels in their sera suggesting the importance of this immune defense mechanism in the pathological processes of this disease [35]. This huge increase in cytokines levels or what is known clinically with cytokine storm, could lead to serious pathologic findings including increased vascular permeability, increased blood viscosity, deteriorated pulmonary functions which could be associated with multiple organ failure and death [71].
A study which compared COVID-19 patients who were admitted to ICU units with clinically stable patients and normal controls found that there was noticeable increase in serum levels of certain cytokines including IL-1β, IL-1 receptor antagonist, IL-7, IL-8, IL-9, IL-10, basic FGF, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF, TNF-α, and VEGF in ICU and stable patients in relation to the third group. In specific, IL-2, IL-7, IL-10, G-CSF, IP-10, MCP-1, MIP-1α, and TNF-α were even higher in those admitted to ICU compared with the clinically stable patients highlighting the role of the cytokine storm in worsening the patients’ clinical conditions [72]. Another study showed similar findings suggesting the relation between the increase in proinflammatory cytokines levels and the occurrence of serious lung pathology [73].
The cytokine storm has been proposed to be in part due to abnormal GIT microbiota features which have normally important role in maintaining a balanced host immune response. In a study which involved 366 study subjects, it was found that there is a significant correlation between microbiota disturbance and uncontrolled production of inflammatory cytokines suggesting a relation between COVID-19 cytokine storm and microbiota patterns. Examples of these microbiota are Bacteroides genus, Streptococcus genus and Clostridiales which had negative correlation with the measured proinflammatory cytokines while Ruminococcus genus, Blautia genus and Lactobacillus genus had positive correlation [74].
8. Adaptive immunity to SARS-CoV-2
Adaptive immune response against viruses in general, is directed mainly by humoral immunity which secretes neutralizing antibodies to block the binding of viruses to their receptors on the target host cells and hence prevent their internalization inside these cells. The other arm of the adaptive immunity is cell mediated T cell response that is concerned with cytotoxic killing of virus infected cells which leads to eradication of the infection. The antibodies produced by T-dependent germinal center interactions are high affinity antibodies making them more efficient in neutralizing the viruses in their extracellular stage; before their entry into the host cells, during virus budding to infect neighboring cells or after death of the infected cells. These antibodies neutralize the viral infection by attaching to the viral envelope or capsid antigens and then prevent the virus binding and internalization to the host cells, consequently, the cycle of viral infection and spread to the near cells is broken down. Among the neutralizing antibodies, IgA immunoglobulins are the most effective for blocking the viral infection in respiratory and intestinal systems. Furthermore, these antibodies may also opsonize the viruses and initiates their phagocytosis. In addition, complement system may help the humoral immunity by opsonizing the viruses-antibodies complexes and promoting their phagocytosis as well as formation of membrane attack complexes (MAC) in the envelope of certain viruses leading to their lysis directly [33].
Once the virus succeeds in entering the cell, then, it is not accessible anymore to the neutralizing antibodies and they have no effect on the viral infection at this stage. Then it is the role of cell mediated immunity to interfere, mainly the CD8 T cells which recognize the virus antigens expressed on the groove of MHC class I of the infected cells. This leads to activation of CD 8 cytotoxic T lymphocytes (CTLs), usually with the help of T helper lymphocytes cytokines, and the costimulatory molecules expressed on the surface of infected cells in order to reach the full activation and clonal expansion process. The effector CTLs exert their cytotoxic actions against the viral infected cells leading to their killing and elimination of the infection. They may also eradicate the viral infecting cells by other ways such as activation of nucleases inside the target cells leading to degradation of the viral genome [33].
It is reported that SARS-CoV-2 affect largely total lymphocytic count especially CD3, CD4 and CD8 cells which were found to be much lower in patients with severe clinical conditions and mortalities in relation to cured ones [37]. In fact, lymphocytopenia associated with change in the total leucocytic count is seen constantly in COVID-19 patients suggesting the disease severity and prognosis [18]. Moreover, in severe infections, flowcytometric analysis showed a significant decrease in all components of total leucocytic count including peripheral B lymphocytes, CD4 T helper lymphocytes, CD8 cytotoxic lymphocytes, NK cells, monocytes, eosinophils and basophils [75].
Another study that was done in Wuhan on 452 SARS-CoV-2 patients showed decrease in total count of T lymphocytes including both helper T cells, regulatory T cells (T reg) and memory T cells, however naïve T cells count was reported to be elevated [36]. Both naïve T cells and memory T cells are necessary to keep the immune response well-coordinated and effective as Naïve T cells are responsible of fighting new infections while memory T cells induce antigen specific adaptive immunity. If there is an increase in naïve T cells in relation to regulatory T cells, then this favors an exaggerated production of pro-inflammatory cytokines leading to a hyperinflammatory response whereas if there is a decrease in the memory T cells count, this in turn could lead to reinfection with SARS-CoV-2 as these cells are in charge of defensing the body against the same infection in case of re-exposure [18]. One of the explanations of decreased lymphocytic count in these patients is the direct infection of lymphocytes by SARS-CoV-2 leading to their lysis [76].
Regarding humoral immune system, Wen et al. showed marked decrease in naïve B lymphocytes count with significant increase of plasma cells count in the peripheral blood mononuclear cells of COVID-19 patients during their recovery period. Furthermore, some modifications in B cell receptors were noticed [77]. As B cells play a significant role in controlling viral infections, monitoring the seroconversion patterns in COVID-19 patients is crucial for assessing their conditions clinically. It was reported that 96.8% of patients had seroconversion of IgG or IgM within 20 days from the onset of symptoms followed by a plateau phase for 6 days later. All the studied subjects had virus specific IgG antibodies while only 94.1% of them achieved virus specific IgM antibodies around 20–22 days after the start of symptoms [78].
9. Immunologic effects on SARS-CoV-2 vaccine strategies
Identifying the SARS-CoV-2 specific immune response is necessary to understand the basics of therapies, passive and active immunization that should be developed against the virus. In fact, the virus induced short-term immune response could have strong inferences in the immunogenicity and effectiveness of the proposed vaccines. Moreover, knowing the stability and duration of the immune response induced by the virus and its epitopes that are recognized by the host B and T lymphocytes might help in developing new vaccine strategies [79].
It was reported that CoV N protein could induce the activation and differentiation of effector specific cytotoxic T cells. Furthermore, the induced neutralizing antibody titers significantly correlated with the count of these specific cytotoxic T lymphocytes. Same was noticed for the viral M protein which induced the production of neutralizing antibodies which could highlight the role of M protein as a promising target for efficient protein-based vaccines development. A recent research has found that activation of CD4 and CD8 T lymphocytes was mainly induced by the S, M, and N proteins and partly by nsp3, nsp4, and ORF8 [80]. This information indicates that adding more viral epitopes derived from the viral antigens other than the S protein such as M, N, ORF3a or nsp6 could help in improving the efficacy of the candidate vaccine designed specifically against SARS-CoV-2 infection [81].
Vaccine optimization depends mainly on augmenting the vaccine immunogenicity while lowering its non-specific protein domains that might cause undesirable effects or toxicity. The first step of SARS-CoV-2 vaccine optimization is the proper design of the specific antigen. This explains how recognition of B and T lymphocyte immunogenic epitopes is crucial for vaccine development. These immunogenic epitopes may be determined through comparison with the most promising epitopes of SARS giving the best results of immunogenicity based on the sequence homology between the 2 viruses [81]. In fact, many T cell and B cell epitopes were recognized as identical between SARS and SARS-CoV-2, most of them were derived from the S or N antigens [82]. In addition, 25 immunogenic epitopes from SARS-CoV-2 proteins; 4 derived from M antigen, 8 from N antigen and 13 from S antigen were recognized through immunoinformatics and found to be safe and effective in inducing strong immunogenicity with low incidence of side effects [83]. Researchers found some other epitopes that could be beneficial in designing an efficient vaccine able to protect against this viral infection. sixty five peptides that are not similar to self-antigens were expected to activate an adaptive immune response against SARS-CoV-2 and hypothesized to be utilized in nucleic acid vaccines [84].
10. Vaccines overview
Recently, there have been several attempts to create vaccines against human Corona virus infections, however, all were limited due to their wide diversity of sequences [85]. Several vaccines and immunotherapies have been tried during the latest viral epidemics such as Zika, Ebola and the previous CoVs family infections. Most of these trials have been investigated firmly to assess their applicability and usefulness in preventing the current pandemic [86].
Most of the current CoV vaccines attempts are targeting the S protein of the virus as it is the principal promotor of antibodies development and T-cell responses making it the perfect candidate in vaccines development strategies. Examples of these vaccines are the ones involving the full length S protein or other appropriate parts of it or S1 receptor binding domain or virus like particles (VLP), viral vectors or DNA [85], [87], [88], [89].
The most efficient vaccine should enhance the production of blocking antibodies that target the S1 subunit receptor binding domain to block the binding of the S1 protein of the virus to its receptor, in addition to blocking the viral RNA uncoating. Chen et al., has demonstrated that the C-terminal domain of the S1 subunit of porcine Deltacoronavirus contains the immunodominant region that evolves the strongest blocking effect [90]. Furthermore, because of RBD capability to trigger the formation of neutralizing antibodies, both recombinant peptides containing RBD and recombinant vectors encoding RBD may be promising for the production of successful SARS vaccines [88].
Kim et al has shown that nasal administration of recombinant adenovirus-based vaccines that express MERS Spike protein into mice, enhances the production of IgG and secretory IgA antibodies as well as inducing the activation of T lymphocytes and development of memory cells which reside mainly in lungs giving those mice life-long immune responses [91].
Moreover, a study which compared the effect of rabies virus as a viral vector against Gram-positive enhancer matrix (GEM) as a bacterial vector; both expressing MERS Spike protein; they found that the viral vector vaccine gave the mice markedly stronger cell mediated immune response and faster humoral immunity [92]. Additionally, knowing that there are many similarities between SARS and MERS, the applicability of designing one vaccine working against all CoV family viruses was investigated and they found that there could be a possible cross-reactivity among CoVs [93].
Also, since SARS-CoV-2 and SARS exhibit antigenic similarity, vaccines developed against SARS could be cross-reactive against SARS-CoV-2 [94]. However, when the sequences of the full-length S protein of SARS-CoV-2 were compared with those of SARS, they found that the most variable regions exist in the S1 subunit of spike protein which is normally the main target of most developed vaccines [95]. This may assume the difficulty of designing one common efficient vaccine for both viruses [96].
The nucleocapsid (N) protein and the possible B cell epitopes of MERS E protein have been recommended as feasible targets that could initiate cell mediated and humoral immune interactions [97]. In addition, reverse genetic approaches were applied in live-attenuated vaccines to deactivate the exonuclease effects of non-structural protein 14 (nsp14) or to eliminate the envelope protein in SARS [85]. Avian infectious bronchitis virus (IBV) is a chicken Corona virus and it was suggested by Bijlenga that strain H of avian live virus IBV vaccine might be helpful in protecting against SARS [98]. Since this vaccine is depending mainly on the production of neutralizing antibodies, so it could be suggested as another effective choice for protection against SARS-CoV-2 after assessing its efficacy in monkeys [99].
11. Passive immunization
Observing that infected individuals with SARS infection have efficient neutralizing antibodies after their recovery, it was postulated that the use of monoclonal antibodies may be efficient in controlling CoV infections after the exposure to the virus [87].
A clinical trial study involving the administration of a group of monoclonal antibodies directed against six specific epitopes in MERS S protein which are responsible for receptor binding and membrane fusion has started [100]. Similarly, targeting many S protein epitopes may be a promising strategy to augment the humoral immune response against CoVs infections. The cross-reactivity of monoclonal antibodies directed against SARS RBD is significantly based on the similarity among RBDs of CoVs [101].
These cross-reacting SARS RBD-specific antibodies can be tested for effectiveness in SARS-CoV-2 patients. Hence, comparative studies are needed to compare receptor binding domains of SARS-CoV-2 with those of SARS in order to determine the appropriate antibodies to be investigated clinically [102].
The development of totally human antibodies as human single-chain antibodies or humanized-nanobodies could be possible through technology, these antibodies (transbodies) could traverse the host cell membrane to interrupt the viral replication process through binding to one or more of the viral proteins. Examples of these transbodies are those directed against influenza, hepatitis C virus(HCV), Ebola, and Dengue viruses [103].Therefore, it could be a good option in treating COVID-19 patients with transbodies directed against the SARS-CoV-2 intracellular peptides as papain-like proteases, cysteine-like protease or any other non-structural proteins to inhibit the viral replication machinery safely [104].
12. Animal models used in vaccine development
Unfortunately, adequate animal models for assessing Corona viruses’ vaccines are not available easily rendering the vaccine production cycle difficult. Succeeding in developing an appropriate animal model which could imitate the clinical condition in humans would be an effective tool to explore the pathophysiology of the disease and help to evaluate the suitable vaccines and treatments with the minor possible harmful effects. In fact, different types of animals have been used to assess SARS, MERS, and SARS-CoV-2 infections. Examples of these animals are mice, guinea pigs, hamsters, rabbits and rhesus macaques [105].
Several attempts were done to develop suitable animal models for SARS, however, the specificity of the virus to ACE2 was a significant obstacle. Then, Yang et al., succeeded in developing an appropriate transgenic mouse model through the introduction of hACE2 gene into the mouse genome [106]. As for MERS, the initial animal model used for development of its vaccine was rhesus macaques. The animal models showed clinical symptoms similar to those occurring in humans such as pyrexia, cough and decreased appetite [107]. In addition, other animal models were used for MERS including the common marmoset where the clinical condition progressed to fatal pneumonia. Fortunately, antibodies production as well as activated cell mediated immune response could be identified in these animals after exposure to MERS vaccine [108]. Furthermore, golden Syrian hamster was used as animal model to evaluate the vaccine development process against several strains of SARS to assess the virus pathophysiology as well as the vaccine effectiveness and safety [109].
Incapability of MERS to replicate in the lungs of mice, hamsters or ferrets made these animals inapplicable animal models for it. These animals are naturally susceptible to SARS but resist MERS infection. However, Zhou et al., have been succeeded in modifying them genetically to be able for MERS infection and replication [110]. Such efforts for genetic modification of these small animals including mice and rabbits to enable them to be susceptible to such viral infections, though time consuming, but are favored as these models are more cost-effective and easier to be manipulated when compared with larger animals [111]. More studies are required to identify the most appropriate animal models for the novel SARS-CoV-2. This will involve determining the degree of specificity of the virus to its receptors on host cells as well as investigating its pathophysiology and specific immune response.
13. Strategies of developing specific SARS-CoV-2 vaccine
Some mRNA vaccines have been developed for SARS-CoV-2 (Table 1 .), they encode stable form of S protein before its fusion to the host cell membrane and depend mainly on a recent genetic technique which does not involve viral culture in the laboratory but in the human body instead. This method involves mRNA fragments that encode the viral proteins which are then injected into the body, subsequently, the viral proteins are presented by the antigen presenting cells to be recognized by T and B lymphocytes to start the activation of the host antiviral immune response [7]. Currently, there are many candidate mRNA vaccines that have been approved by WHO [112].
Table 1.
Summary of SARS-CoV-2 vaccines that reached phase 3 of human clinical trials approved by WHO, January 5, 2021.1
| SARS-COV-2 vaccine manufacturer | Vaccine platform | Type of candidate vaccine | Number of doses | Timing of doses | Route of administration | |
|---|---|---|---|---|---|---|
| 1 | University of Oxford/ AstraZeneca | Non-replicating viral vector | ChAdOx1-S - (AZD1222) (Covishield) | 1–2 | 0,28 days | Intramuscular injection |
| 2 | Sinovac Research and Development Co., Ltd | Inactivated virus | SARS-CoV-2 vaccine (inactivated) | 2 | 0,14 days | Intramuscular injection |
| 3 | Wuhan institute of biological products/China National Biotec Group Co/ Sinopharm | Inactivated virus | Inactivated SARS-CoV-2 vaccine (Vero cell) | 2 | 0,21 days | Intramuscular injection |
| 4 | Sinopharm/China National Biotec Group Co/ Beijing Institute of Biological Products | Inactivated virus | Inactivated SARS-CoV-2 vaccine (Vero cell) | 2 | 0,21 days | Intramuscular injection |
| 5 | Moderna/NIAID | RNA vaccine | mRNA −1273 | 2 | 0,28 days | Intramuscular injection |
| 6 | CanSino biological Inc./Beijing institute of biotechnology | Non-replicating viral vector | Recombinant novel coronavirus vaccine (Adenovirus type 5 vector) | 1 | Day 0 | Intramuscular injection |
| 7 | Gamaleya Research Institute; Health Ministry of the Russian Federation | Non-replicating viral vector | Gam-COVID-Vac Adeno-based (rAd26-S + rAd5-S) | 2 | 0,21 days | Intramuscular injection |
| 8 | Janssen Pharmaceutical | Non-replicating viral vector | Ad26.COV2.S | 1–2 | Day 0 or 0,56 days | Intramuscular injection |
| 9 | Novavax | Protein subunit | SARS-CoV-2 rS/Matrix M1-Adjuvant (Full length recombinant SARS CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M) | 2 | 0,21 days | Intramuscular injection |
| 10 | BioNTech + Fosun Pharma; Jiangsu Provincial Center for Disease Prevention and Control + Pfizer | RNA vaccine | BNT162 (3 LNP-mRNAS) | 2 | 0,28 days | Intramuscular injection |
| 11 | Anhui Zhifei Longcom Biopharmaceutical + Institute of Microbiology, Chinese Academy of Sciences | Protein subunit | Recombinant SARS-CoV-2 vaccine (CHO Cell) | 2–3 | 0,28 days or 0, 28, 56 days | Intramuscular injection |
| 12 | CureVac AG | RNA vaccine | CVnCoV Vaccine | 2 | 0,28 days | Intramuscular injection |
| 13 | Institute of Medical Biology + Chinese Academy of Medical Sciences | Inactivated virus | SARS-CoV-2 vaccine (vero cells) | 2 | 0,28 days | Intramuscular injection |
| 14 | Research Institute for Biological Safety Problems, Rep of Kazakhstan | Inactivated virus | QazCovid-in® - COVID-19 inactivated vaccine | 2 | 0,28 days | Intramuscular injection |
| 15 | Cadila Healthcare Ltd. | DNA vaccine | nCov vaccine | 3 | 0, 28, 56 days | Intradermal |
WHO. Draft landscape of COVID-19 candidate vaccines 2020 [Available from: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
DNA vaccines reflect the new evolutionary path of vaccine production. Genetic manipulation is used to generate the vaccinations produced through recombinant DNA processing. Basically, the DNA which encodes the target protein is inserted into the appropriate cell line or microorganism by a plasmid or viral vector where the DNA is then converted to a protein. Afterwards, the product is extracted by purification technique [113]. One of the advantages of DNA vaccines is that it is easy to manufacture such plasmids in big amounts in addition to the long-life immunity they could provide [113]. INO-4800 vaccine is a DNA based vaccine whose preclinical investigations have been done in animal models to test its results and efficacy and fortunately, has shown good primary outcomes, it was then proceeded to enter human clinical trial, phase I where it involved 40 healthy individuals who are receiving, each, 2 doses of the vaccine with 4 weeks interval in between, then their antibody responses were assessed [114]. After showing promising results in this phase, it proceeded to phase II/III clinical trials [112].
Similarly, nineteen SARS-CoV-2 protein subunit candidate vaccines are approved by WHO. Examples include capsid-like particle AdaptVac, Drosophila S2 insect cell expression system VLPs and peptide antigens formulated in lipid nanoparticle formulation [112]. Protein Subunit vaccines include some epitopes of the virus usually produced through recombinant DNA techniques or viral culture [115]. One advantage of this type of vaccines is the relatively fewer number of antigens, hence lower chance of potential side effects. However, this low number of antigenic epitopes could elicit weaker immune response but this disadvantage is treated by conjugating such epitopes with adjuvant proteins to bypass this weakness [116].
Additionally, many replicating and non-replicating viral vector candidate vaccines are being tested for efficacy and safety. Replicating viral vectors such as measles, influenza vector expressing RBD and horsepox vector expressing S protein as well as non-replicating viral vectors such as adenovirus type 5 vector are proceeding to clinical trials with favorable initial results in the preclinical phase [112]. AZD1222 is a non-replicating viral vector vaccine under clinical trial (formerly known as ChAdOx1 nCoV-19) that is using adenovirus vector and targeting SARS-COV-2 S protein resulting in generation of humoral immune response with production of immunoglobulins against the virus and acceptable safety profile [117].
Another advanced strategy is the production of vaccines that are formed of virus like particles (VLPs) such as parts of the viral surface proteins. This is a complex procedure that involves processing these particles into more immunogenic proteins that could initiate the host antibody and cell mediated immunity [118]. VLP vaccine expressing viral RBD as well as plant derived VLP are being tested pre-clinically and approved by WHO [112].
Inactivated viruses’ vaccines work through including the whole virus which has been deactivated either chemically or physically. These vaccines are more stable compared to other types of vaccines as they are usually maintained in powder form, but unfortunately less effective and more expensive due to its complicated production machinery [119].
Currently, the number of vaccines developed against SARS-CoV-2 which are in the clinical phase is 63 in addition to 172 other vaccines in the pre-clinical stage. Many of the 63 clinical phases’ vaccines have already reached phase III of clinical investigations and they are totally diverse due to the critical necessity for fast vaccine development process to fight the emerging serious infection (table 1). They include 6 inactivated virus vaccines given as 2 intramuscular (IM) doses with 14, 21 or 28 days interval [[112], [120]]. The one developed by Sinovac containing aluminum hydroxide has demonstrated good results in the clinical trials showing acceptable safety and efficacy profile in volunteers [81]. The study included 144 vaccinators in phase I and 600 vaccinators in phase II. The study subjects were grouped into 3 groups: those receiving a dose of 3 ug (1st group), those receiving a dose of 6 ug group (2nd group) and placebo group (3rd group). In the 0,14 days dosage timing pattern, side effects occurred at a percentage of 29% in the 1st group, 38% in the 2nd group and 8% in the 3rd group while in the 0, 28 dosage timing pattern, the frequency of side effects was 13% in the 1st group, 17% in the 2nd group and 13% in the 3rd group. Neutralizing immunoglobulins were found in 46% of the 1st group, 50% of the 2nd group and in none of the 3rd group on day 14 after the 0, 14 days vaccination doses while they were found in 83% of the 1st group, 79% of the 2nd group and 4% (one person) in the 3rd group in the 0, 28 days vaccination pattern. Regarding phase II study, it showed occurrence of side effects in 33% of the 1st group, 35% of the 2nd group, and 22% of the 3rd group in the 0, 14 days vaccination pattern while they appeared in 19% of the 1st group, 19% of the 2nd group, and 18% of the 3rd group in the 0, 28 days vaccinations. Neutralizing antibodies were detected in 92% of the 1st group, 98% of the 2nd group, and 3% of the 3rd group in the 0, 14 days vaccinations while in the 0, 28 days vaccinations, they were found in 97% of the 1st group, 100% of the 2nd group, and 0% of the 3rd group. These data recommended that the 3 ug dose of this vaccine should be suggested for the efficacy evaluation in phase III clinical trial [120].
The WHO list also includes non-replicating viral vector vaccines such as AZD1222 manufactured by AstraZeneca and university of Oxford which revealed promising safety, tolerance and immunogenicity in phase I/II clinical trials with few adverse reactions such as pain, tenderness and fever which could be relieved by simple antipyretics and analgesics, the Adenovirus type 5 vector vaccine developed by CanSino Biological Inc./Beijing Institute of Biotechnology which showed a safe and efficient immune response at a dose of 5 × 10 viral particles per mL in the previous clinical trials as well as Gam-COVID-Vac adeno-based vaccine (rAd26-S + rAd5-S) developed by Gamaleya Research Institute and Health Ministry of the Russian Federation and the Ad26.COV2. S vaccine encoding a perfusion stabilized S protein developed by Janssen Pharmaceutical which showed, in its preclinical study, that a single shot of the vaccine was able to induce strong neutralizing antibody response and protect the rhesus macaques [81].
Two protein subunit vaccines have been also joining this phase III list; the SARS-CoV-2 rS/Matrix M1-Adjuvant (Full length recombinant SARS CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M) developed by Novavax and the recombinant SARS-CoV-2 vaccine (CHO Cell) by Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology/Chinese Academy of Sciences [[109], [112]].
RNA based vaccines such as the mRNA −1273 vaccine developed by Moderna/National Institute of Allergy and Infectious Diseases (NIAID), BNT162 vaccine (3 LNP-mRNAs) by BioNTech/Fosun Pharma/Jiangsu Provincial Center for Disease Prevention and Control/Pfizer and the CVnCoV Vaccine by CureVac AG have been also investigated in phase III clinical trials in addition to one DNA vaccine; the INO-4800 + electroporation vaccine by Inovio Pharmaceuticals/International Vaccine Institute [112]. For the BNT162 vaccine, phase I/II clinical trials have demonstrated an efficient neutralizing antibody response after the 2nd shot of the vaccine though most of the volunteers have shown mild to moderate reaction symptoms following the vaccine administration [25]. The antibody titers were evaluated at days: 0,7 and 21 after the 1st shot, at days 7 and 14 after the 2nd shot. For all the 3 different doses of the vaccine at day 21 after the first shot, geometric mean concentrations of specific IgG were between 534 and 1778 U ml−1 which are higher than those found in COVID-19 patients following 14 days of a positive confirmatory SARS-CoV-2 PCR test. At day 7 after the 2nd shot (dosage of 10-μg and 30-μg), the geometric mean concentrations of specific IgG had elevated to 4813 and to 27,872 U ml−1, respectively and maintained their high levels at the last point of evaluation after the 2nd shot while for the dosage of 100 μg, the Ab concentrations did not rise more at day 21 after the first shot [121]. Regarding the mRNA −1273 Moderna vaccine, no severe side effects were noticed among the studied subjects and it elicited neutralizing antibody response in addition to promising CD4 T cell responses involving Th1 lymphocytes [122]. Neutralizing antibody titers increased quickly following the 1st dose of the vaccine; for the dose of 25 μg, the geometric main titer was 323,945 among the vaccinators of age group (56–70 years old) and 1,128,391 among those of 71 years old and older while for those who got the 100 μg dose, the titer was 1,183,066 and 3,638,522, respectively for both age groups. High serum neutralizing antibodies persisted following the 2nd dose of the vaccine in all vaccinators and similar results were reported for the 18–55 age group participants who showed higher neutralizing Ab titers than those obtained from convalescent sera [123].
14. Summary and future perspectives
The novel corona virus, SARS-CoV-2, is one of the most challenging crises that faced the whole world since the 1918 Influenza outbreak unveiling the hidden weaknesses of the health care systems globally and urging the need for efficient and safe vaccines and therapies development. Luckily, vaccine research has advanced dramatically over the last period, with the creation of multiple types of RNA and DNA vaccines, approved viral vector vaccines, recombinant peptides in addition to cell culture based immunotherapies [124]. Based on the high similarity between SARS-CoV-2 and SARS, the identification of S protein, present in both viruses and responsible for virus binding to the host cell membrane and its internalization into the cell has been facilitated and proposed as the main target for many vaccines under trials. This will help in quick production of neutralizing antibodies that bind to the S protein and hence prevent the infection process. Nevertheless, although many S protein targeting vaccines have been developed already aiming to protect against SARS and MERS, most of them did not give the desirable results when tested in animal models or caused severe harmful effects as lung injury or multiorgan failure [[125], [126]]. In addition, it is not well-known whether the infection with this virus involves lifelong immunity nor the possibility of re-infections which urge researchers all over the world to continue their research on this topic to explore the exact pathophysiology of the virus and to develop a highly effective vaccine and treatment.
However, the specific immune signaling pathways stimulated by SARS-CoV-2 are still to be well identified as this may ensure a great hope in revealing the reliable and significant biological markers to be targeted in vaccines and immunotherapies to block the cytokine storm occurring in most of the severe cases. Moreover, since it is known that the innate immunity stimulates many signaling pathways, it would be more promising to target various signaling pathways simultaneously during treatment which should include a combination of specific cytokine blockers, corticosteroids as well as S1PR1 agonists instead of administering only one drug as this may enhance the chance of recovery especially in advanced cases. Therefore, more studies are needed to evaluate if this combination of drugs could protect the patients against the progression of lung damage and multi organ failure compared with single target drug regime plans.
Finally, all the above-mentioned issues illustrate some of the main holes in our understanding of COVID-19 physiological implications, fundamental immune signaling mechanisms, and effective vaccines development strategies that prospective studies desperately need to tackle.
15. Funding
No sources of funding were used for this work.
16. Data sharing statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Asia Cn. Novel Coronavirus Map 2020 [updated 12/08/2020. Available from: https://infographics.channelnewsasia.com/covid-19/map.html.
- 2.WHO. WHO Coronavirus Disease (COVID-19) Dashboard 2020. Available from: https://covid19.who.int/.
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauci A.S., Lane H.C., Redfield R.R. Covid-19 – navigating the uncharted. N. Engl. J. Med. 2020;382(13):1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calina D., Docea A.O., Petrakis D., Egorov A.M., Ishmukhametov A.A., Gabibov A.G., et al. Towards effective COVID19 vaccines: Updates, perspectives and challenges (Review) Int. J. Mol. Med. 2020;46(1):3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michele Catanzaro F.F., Racchi Marco, Corsini Emanuela, Govoni Stefano, Lanni Cristina. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduction and Targeted. Therapy. 2020;5(84):1–10. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Wang Y.M., Xu J.Y., Cao B. Potential antiviral therapeutics for 2019 Novel Coronavirus. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43(3):170–172. doi: 10.3760/cma.j.issn.1001-0939.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. Author Correction: a new coronavirus associated with human respiratory disease in China. Nature. 2020;580(7803):E7. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. Severe acute respiratory syndrome-related coronavirus: the species and its viruses–a statement of the coronavirus study group. BioRxiv. 2020 [Google Scholar]
- 13.WHO. Naming the Coronavirus Disease (COVID-19 and the Virus That Causes it 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/ naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causesit.
- 14.Corman V.M., Lienau J., Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist (Berl). 2019;60(11):1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J.S. Mackenzie, D.W. Smith, COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don't. Microbiol Aust. 2020:MA20013. [DOI] [PMC free article] [PubMed]
- 17.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L., Liu K., Liu H.G. Cause analysis and treatment strategies of recurrence' with novel coronavirus pneumonia (covid-19) patients after discharge from hospital. Chin. J. Tuberc. Respir. Dis. 2020;43(4):281–284. doi: 10.3760/cma.j.cn112147-20200229-00219. [DOI] [PubMed] [Google Scholar]
- 19.Fung T.S., Liu D.X. Human Coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 20.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the Novel Coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabary M., Khanmohammadi S., Araghi F., Dadkhahfar S., Tavangar S.M. Pathologic features of COVID-19: a concise review. Pathol. Res. Pract. 2020;216(9):153097. doi: 10.1016/j.prp.2020.153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Pan H., Su H., Huang B., Yang M., Luo D., Weng M., Ma L., Nie X. Pathological changes of the spleen in ten patients with new coronavirus infection by minimally invasive autopsies. Zhonghua Bing li xue za zhi. 2020;49 doi: 10.3760/cma.j.cn112151-20200401-00278. E014–E. [DOI] [PubMed] [Google Scholar]
- 26.Li G., Summa B., Hu B., Wenk C., Akmatbekov A., Harbert J.L., Vander Heide R.S., Brown J.Q. Multiscale three-dimensional pathology findings of COVID-19 diseased lung using high-resolution cleared tissue microscopy. bioRxiv. 2020 [Google Scholar]
- 27.Basso C., Leone O., Rizzo S., De Gaspari M., van der Wal A.C., Aubry M.C., et al. Pathological features of COVID-19-associated myocardial injury: a multicentre cardiovascular pathology study. Eur. Heart J. 2020;41(39):3827–3835. doi: 10.1093/eurheartj/ehaa664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao F., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. Sonzogni GP, M. Seghezzi, M.G. Alessio, A. Gianatti, L. Licini, P. Zerbi, L. Carsana, R. Rossi, E. Lauri, Liver and COVID 19 Infection: A Very Preliminary Lesson Learnt from Histological Post-Mortem Findings in 48 Patients, 2020.
- 31.Trik M., Bouzidi L., Makni S., Kallel R., Boudawara T. Pathological features of the novel human coronavirus disease (COVID-19): a review. JI M Sfax. 2020;35:1–7. [Google Scholar]
- 32.Reichard R.R., Kashani K.B., Boire N.A., Constantopoulos E., Guo Y., Lucchinetti C.F. Neuropathology of COVID-19: a spectrum of vascular and acute disseminated encephalomyelitis (ADEM)-like pathology. Acta Neuropathol. 2020;140(1):1–6. doi: 10.1007/s00401-020-02166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abul K., Abbas A.H.L., Pillai Shiv. Elsevier saunders; 2015. Cellular and Molecular Immunology. [Google Scholar]
- 34.Control ECfDPa. Immune responses and immunity to SARS-CoV-2 2020.Available from: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/immune-responses.
- 35.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Chuan, Ziwei Hu, Zhang Shuoqi, Sheng Yang Yu, Tao Cuihong Xie, Ma Ke, Shang Ke, Wang Wei, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020:1–7. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Dongze, Hong Liu Yu, Jia Fanghui Li, Wang Wei, Jiang Wu, Zhi Wan Yu, Cao Rui Zeng. Immune dysfunction leads to mortality and organ injury in patients with COVID-19 in China: insights from ERS-COVID-19 study. Signal Transduct. Target Ther. 2020;5(62) doi: 10.1038/s41392-020-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omran A., Maaroof A., Saleh M.H., Abdelwahab A. Salivary C-reactive protein, mean platelet volume and neutrophil lymphocyte ratio as diagnostic markers for neonatal sepsis. J. Pediatr. (Rio J). 2018;94(1):82–87. doi: 10.1016/j.jped.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Omran A., Ali M., Saleh M.H., Zekry O. Salivary C-reactive protein and mean platelet volume in diagnosis of late-onset neonatal pneumonia. Clin Respir J. 2018;12(4):1644–1650. doi: 10.1111/crj.12723. [DOI] [PubMed] [Google Scholar]
- 40.Hur S., Double-Stranded R.N.A. Sensors and modulators in innate immunity. Annu. Rev. Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlman S.D. Immunopathogenesis of coronavirus infections. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–45 e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teijaro J.R., Walsh K.B., Cahalan S., Fremgen D.M., Roberts E., Scott F., et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146(6):980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Marcken M., Dhaliwal K., Danielsen A.C., Gautron A.S., Dominguez-Villar M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci Signal. 2019;12(605) doi: 10.1126/scisignal.aaw1347. [DOI] [PubMed] [Google Scholar]
- 46.Olejnik J., Hume A.J., Muhlberger E. Toll-like receptor 4 in acute viral infection: too much of a good thing. PLoS Pathog. 2018;14(12):e1007390. doi: 10.1371/journal.ppat.1007390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., Verma I.M. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 48.Hayden M.S., West A.P., Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25(51):6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 49.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., et al. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88(2):913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., et al. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;128(1–2):1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smits S.L., de Lang A., van den Brand J.M., Leijten L.M., van I.W.F., Eijkemans M.J., et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6(2) doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hiscott J., Nguyen T.L., Arguello M., Nakhaei P., Paz S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene. 2006;25(51):6844–6867. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva L.C., Ortigosa L.C., Benard G. Anti-TNF-alpha agents in the treatment of immune-mediated inflammatory diseases: mechanisms of action and pitfalls. Immunotherapy. 2010;2(6):817–833. doi: 10.2217/imt.10.67. [DOI] [PubMed] [Google Scholar]
- 54.Feldmann M., Maini R.N. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu. Rev. Immunol. 2001;19:163–196. doi: 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 55.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., et al. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U.S.A. 2008;105(22):7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puja Mehta D.F.M., Brown Michael, Sanchez Emilie, Tattersall Rachel S., Manson Jessica J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5):105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee C., Lim H.K., Sakong J., Lee Y.S., Kim J.R., Baek S.H. Janus kinase-signal transducer and activator of transcription mediates phosphatidic acid-induced interleukin (IL)-1beta and IL-6 production. Mol. Pharmacol. 2006;69(3):1041–1047. doi: 10.1124/mol.105.018481. [DOI] [PubMed] [Google Scholar]
- 59.Biswas P., Delfanti F., Bernasconi S., Mengozzi M., Cota M., Polentarutti N., et al. Interleukin-6 induces monocyte chemotactic protein-1 in peripheral blood mononuclear cells and in the U937 cell line. Blood. 1998;91(1):258–265. [PubMed] [Google Scholar]
- 60.McLoughlin R.M., Hurst S.M., Nowell M.A., Harris D.A., Horiuchi S., Morgan L.W., et al. Differential regulation of neutrophil-activating chemokines by IL-6 and its soluble receptor isoforms. J. Immunol. 2004;172(9):5676–5683. doi: 10.4049/jimmunol.172.9.5676. [DOI] [PubMed] [Google Scholar]
- 61.Xiang S., Dong N.G., Liu J.P., Wang Y., Shi J.W., Wei Z.J., et al. Inhibitory effects of suppressor of cytokine signaling 3 on inflammatory cytokine expression and migration and proliferation of IL-6/IFN-gamma-induced vascular smooth muscle cells. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2013;33(5):615–622. doi: 10.1007/s11596-013-1168-x. [DOI] [PubMed] [Google Scholar]
- 62.Qu D., Liu J., Lau C.W., Huang Y. IL-6 in diabetes and cardiovascular complications. Br. J. Pharmacol. 2014;171(15):3595–3603. doi: 10.1111/bph.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schieffer B., Luchtefeld M., Braun S., Hilfiker A., Hilfiker-Kleiner D., Drexler H. Role of NAD(P)H oxidase in angiotensin II-induced JAK/STAT signaling and cytokine induction. Circ. Res. 2000;87(12):1195–1201. doi: 10.1161/01.res.87.12.1195. [DOI] [PubMed] [Google Scholar]
- 64.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84(2):1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eguchi S., Kawai T., Scalia R., Rizzo V. Understanding angiotensin II type 1 receptor signaling in vascular pathophysiology. Hypertension. 2018;71(5):804–810. doi: 10.1161/HYPERTENSIONAHA.118.10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami M., Kamimura D., Hirano T. Pleiotropy and specificity: insights from the interleukin 6 family of cytokines. Immunity. 2019;50(4):812–831. doi: 10.1016/j.immuni.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 67.Hirano T., Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52(5):731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiegel S., Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011;11(6):403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryan A.M., Del Poeta M. Sphingosine-1-phosphate receptors and innate immunity. Cell. Microbiol. 2018;20(5):e12836. doi: 10.1111/cmi.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teijaro J.R., Walsh K.B., Rice S., Rosen H., Oldstone M.B. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc. Natl. Acad. Sci. U.S .. 2014;111(10):3799–3804. doi: 10.1073/pnas.1400593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhe Xu, Wang Yijin, Zhang Jiyuan, Huang Lei, Zhang Chao, Liu Shuhong, Zhao Peng, Liu Hongxia, Zhu Li, Tai Yanhong, Bai Changqing, Gao Tingting, Song Jinwen, Xia Peng, Dong Jinghui, Zhao Jingmin, Wang Fu-Sheng. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Chaolin, Li Xingwang, Ren Lili, Zhao Jianping, Yi Hu, Zhang Li, Fan Guohui, Jiuyang Xu, Xiaoying Gu, Cheng Zhenshun, Ting Yu, Xia Jiaan, Wei Yuan, Wu Wenjuan. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Yingxia, Huang Fengming, Yang Yang, Wang Fuxiang, Yuan Jing, Zhang Zheng, Qin Yuhao, Li Xiaoyun, Zhao Dandan. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020;7(6):1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gou Wanglong, Yue Liang, Chen Geng-dong, Cai Xue, Shuai Menglei, Fengzhe Xu, Yi Xiao, Chen Hao, Zhu Yi Judy, Xiao Mian-li, Jiang Zengliang, Miao Zelei, Xiao Congmei, Shen Bo, Xiaomai Wu. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. MedRxiv. 2020 [Google Scholar]
- 75.Shi Yaling, Chen Xing, Liu Yanxia, Huang Jide, Jingyi Ou, Deng Xilong. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. medRxiv. 2020 doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan Li, Zhang Duanyang, Ding Jinya, Huang Qianchuan, Tang Yi-Quan, Wang Qiongshu, Miao Hongming. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target Ther. 2020;5(1):33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen Wen, Tang Hao, Le Wenqing, Zhang Xiaopeng, Zheng Yingfeng, Liu XiuXing, Xie Lihui, Li Jianmin, Ye Jinguo, Cui Xiuliang, Miao Yushan, Wang Depeng, Dong Jiantao, Xiao Chuan-Le, Chen Wei, Wang Hongyang. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. medRxiv. 2020 doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quan-Xin Long B.-Z.L., Deng Hai-Jun, Gui-Cheng Wu, Deng Kun, Yao-Kai Chen Pu, Liao Jing-Fu Qiu, Lin Yong, Cai Xue-Fei, Wang De-Qiang, Yuan Hu, Ren Ji-Hua, Tang Ni, Yin-Yin Xu, Li-Hua Yu, Mo Zhan, Gong Fang, Zhang Xiao-Li, Tian Wen-Guang, Li Hu, Xi Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 79.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther. 2020;5(1):237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee S., Tworowski D., Detroja R., Mukherjee S.B., Frenkel-Morgenstern M. Immunoinformatics and structural analysis for identification of immunodominant epitopes in SARS-CoV-2 as potential vaccine targets. Vaccines (Basel) 2020;8(2) doi: 10.3390/vaccines8020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yarmarkovich M., Warrington J.M., Farrel A., Maris J.M. Identification of SARS-CoV-2 vaccine epitopes predicted to induce long-term population-scale immunity. Cell Rep Med. 2020;1(3):100036. doi: 10.1016/j.xcrm.2020.100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang S., He Y., Liu S. SARS vaccine development. Emerg. Infect. Dis. 2005;11(7):1016–1020. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Widjaja I., Wang C., van Haperen R., Gutierrez-Alvarez J., van Dieren B., Okba N.M.A., et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes. Infect. 2019;8(1):516–530. doi: 10.1080/22221751.2019.1597644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen R., Fu J., Hu J., Li C., Zhao Y., Qu H., et al. Identification of the immunodominant neutralizing regions in the spike glycoprotein of porcine deltacoronavirus. Virus Res. 2020;276:197834. doi: 10.1016/j.virusres.2019.197834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim M.H., Kim H.J., Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length Spike protein of Middle East respiratory syndrome coronavirus. PLoS ONE. 2019;14(7):e0220196. doi: 10.1371/journal.pone.0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li E., Yan F., Huang P., Chi H., Xu S., Li G., et al. Characterization of the immune response of MERS-CoV vaccine candidates derived from two different vectors in mice. Viruses. 2020;12(1) doi: 10.3390/v12010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., et al. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect. 2020;9(1):275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22(2):74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie Q., He X., Yang F., Liu X., Li Y., Liu Y., et al. Analysis of the genome sequence and prediction of B-cell epitopes of the envelope protein of middle east respiratory syndrome-Coronavirus. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018;15(4):1344–1350. doi: 10.1109/TCBB.2017.2702588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bijlenga G. Proposal for vaccination against SARS coronavirus using avian infectious bronchitis virus strain H from The Netherlands. J. Infect. 2005;51(3):263–265. doi: 10.1016/j.jinf.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92(5):479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goo J., Jeong Y., Park Y.S., Yang E., Jung D.I., Rho S., et al. Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Res. 2020;278:197863. doi: 10.1016/j.virusres.2020.197863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng L.P., Ge X.Y., Peng C., Tai W., Jiang S., Du L., et al. Cross-neutralization of SARS coronavirus-specific antibodies against bat SARS-like coronaviruses. Sci. China Life Sci. 2017;60(12):1399–1402. doi: 10.1007/s11427-017-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen J. New coronavirus threat galvanizes scientists. Science. 2020;367(6477):492–493. doi: 10.1126/science.367.6477.492. [DOI] [PubMed] [Google Scholar]
- 103.Seesuay W., Jittavisutthikul S., Sae-Lim N., Sookrung N., Sakolvaree Y., Chaicumpa W. Human transbodies that interfere with the functions of Ebola virus VP35 protein in genome replication and transcription and innate immune antagonism. Emerg. Microbes. Infect. 2018;7(1):41. doi: 10.1038/s41426-018-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 2020;16(6):1232–1238. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gretebeck L.M., Subbarao K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57(5):450–459. [PubMed] [Google Scholar]
- 107.Munster V.J., de Wit E., Feldmann H. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 2013;368(16):1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X., et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10(8):e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roberts A., Lamirande E.W., Vogel L., Jackson J.P., Paddock C.D., Guarner J., et al. Animal models and vaccines for SARS-CoV infection. Virus Res. 2008;133(1):20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccines. 2018;17(8):677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.WHO. Draft landscape of COVID-19 candidate vaccines 2020. Available from: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 113.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines. 2016;15(3):313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ferraro B., Morrow M.P., Hutnick N.A., Shin T.H., Lucke C.E., Weiner D.B. Clinical applications of DNA vaccines: current progress. Clin. Infect. Dis. 2011;53(3):296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arena CT. Inovio commences Phase I trial of DNA vaccine for Covid-19 2020 [Available from: https://www.clinicaltrialsarena.com/news/inovio-SARS-COV-2-vaccine-trial/urisimplehttps://www.clinicaltrialsarena.com/news/inovio-SARS-COV-2-vaccine-trial/.
- 116.Zhang N., Tang J., Lu L., Jiang S., Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee N.H., Lee J.A., Park S.Y., Song C.S., Choi I.S., Lee J.B. A review of vaccine development and research for industry animals in Korea. Clin. Exp. Vaccine Res. 2012;1(1):18–34. doi: 10.7774/cevr.2012.1.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arena CT. University of Oxford starts enrolment for Covid-19 vaccine trial 2020. Available from: https://www.clinicaltrialsarena.com/news/oxford-university-covid-19-vaccine-trial/urisimplehttps://www.clinicaltrialsarena.com/news/oxford-university-covid-19-vaccine-trial/.
- 119.Sarkar B.I.S., Zohora U.S., Ullah M.A. Virus like particles – a recent advancement in vaccine development. Korean J. Microbiol. 2019;55:327–343. [Google Scholar]
- 120.WHO, WHO, WHO2020.
- 121.Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 123.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]