Abstract
Aims
SARS-CoV-2, an infectious agent behind the ongoing COVID-19 pandemic, induces high levels of cytokines such as IL-1, IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ etc in infected individuals that play a role in the underlying patho-physiology. Nonetheless, exact association and contribution of every cytokine towards COVID-19 pathology remains poorly understood. Delineation of the roles of cytokines during COVID-19 holds the key to efficient patient management in clinics. This study performed a comprehensive meta-analysis to establish association between induced cytokines and COVID-19 disease severity to help in prognosis and clinical care.
Main methods
Scientific literature was searched to identify 13 cytokines (IL-1β, IL-2, IL-2R, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12, IL-17, TNF-α and IFN-γ) from 18 clinical studies. Standardized mean difference (SMD) for selected 6 cytokines IL-2, IL-4, IL-6, IL-10, TNF-α and IFN-γ between severe and non-severe COVID-19 patient groups were summarized using random effects model. A classifier was built using logistic regression model with cytokines having significant SMD as covariates.
Key findings
Out of the 13 cytokines, IL-6 and IL-10 showed statistically significant SMD across studies synthesized. Classifier with mean values of both IL-6 and IL-10 as covariates performed well with accuracy of ~92% that was significantly higher than accuracy reported in literature with IL-6 and IL-10 as individual covariates.
Significance
Simple panel proposed by us with only two cytokine markers can be used as predictors for fast diagnosis of patients with higher risk of COVID-19 disease deterioration and thus can be managed well for a favourable prognosis.
Keywords: COVID-19, Meta-analysis, Cytokines, Logistic models
COVID-19, Meta-Analysis, Cytokines, Logistic Models.
1. Introduction
COVID-19 pandemic caused by SARS-CoV-2 has emerged as a major threat to mankind affecting 18 + million people resulting in over 2.2 million deaths [1] worldwide. Exuberant inflammation manifested as elevated levels of cytokines, commonly referred as “cytokine storm” (CS) often leads to critical conditions like ARDS (acute respiratory distress syndrome) and death due to multi-organ failure [2].
Innate immune response is the first step of defence mechanism against viral infection. Pattern recognition receptors in host dendritic cells recognize viral genomic DNA or RNA to initiate production of cytokines and chemokines [3] which in turn recruit immune cells like macrophages, neutrophils and T-cells to the site of infection based on their source and target cells [4]. Pro-inflammatory cytokines including, interleukins (IL) such as IL-1, IL-6 and Tumour Necrosis Factor-α (TNF-α) play major role in initial response, whereas anti-inflammatory molecules like IL-10 are produced during sustained infection to keep a check on inflammation and maintain immune homeostasis [5]. Heightened CS-induced acute lung damage leading to fatality [6] is a signature of coronavirus family reported earlier for MERS-CoV and SARS-CoV infections [7, 8].
In COVID-19, elevated levels of both pro-inflammatory and anti-inflammatory cytokines are reported in multiple clinical studies [9, 10, 11]. A recently published extensive meta-analysis summarized elevated levels of IL-2, IL-2R, IL-4, IL-6, IL-8, IL-10, TNF-α and Interferon-γ (IFN-γ) in severe group of patients, whereas no significant increase was found in the levels of IL-1β and IL-17 [12]. A two-arm meta-analysis study synthesized from individual patient data reported statistically significant odds ratio (p < 0.05) to severe disease for only two cytokines - IL-6 and IL-10 [13]. Some meta-analysis efforts reported difference in IL-6 level between severe and non-severe COVID-19 patients in terms of Standardized Mean Difference (SMD) [14], mean difference [15] or ratio of means [16] values that can potentially be used as cut-off to discriminate between severe and non-severe patients. All these meta-analysis summary results were associated with high level of heterogeneity (I2 ~ 98–100%). Another synthesis over three clinical studies determined the elevated ratio IL-6/IFN-γ in severe patients [17] with substantial heterogeneity (I2 = 79%). A recent meta-analysis covering 6242 patients over 24 studies established elevated levels of IL-6 and IL-10 in severe COVID-19 patients with practically no reported heterogeneity for IL-10 [18].
Clinical studies with COVID-19 patient cohorts explored the role of IL-6 alone [19] or IL-6 along with other cytokines including IL-10, IL-2, IL-4, TNF-α and IFN-γ as prognosticator for severe disease [20]. Meta-analysis studies described before [12, 14, 15, 16, 21, 22] concluded elevated levels of cytokines in severe COVID-19 patients but did not attempt to establish their prognostic significance, except the study by Elshazli et al who performed decision tree and Receiver Operating Characteristics (ROC) curve analysis to assess prognostic potential of multiple laboratory parameters including IL-6 [13]. Another study with a cohort of 501 patients [23] attempted to establish a mortality risk model using multiple clinical parameters and IL-6 level. However, despite numerous clinical studies and meta-analyses any reliable prognostic method that can predict the progression of a patient to severe form of disease based on on-admission cytokine levels remains elusive. In this study, we attempt to arrive at a prognostic method through meta-analysis of the levels of commonly used 13 cytokine markers between severe and non-severe patient groups by building a classifier using a logistic regression model.
2. Methods
2.1. Literature search
We performed the meta-analysis following PRISMA guidelines [24]. Literature search was performed in Pubmed, Google Scholar and in preprint archives such as medRxiv, bioRxiv and SSRN library for articles in English published in year 2020 till 31 May 2020. Search terms included COVID-19-specific terms in title of article ("2019-nCov” OR "nCoV-2019″ OR "novel coronavirus" OR "SARS-CoV-2″ OR "COVID-19″ OR COVID19 OR "novel corona virus") along with terms such as “cytokine level” and combinations of common cytokine names and gene symbols. Search strategy was reviewed by all authors and it was decided to retain non-peer reviewed articles in selection in view of the present emerging situation. Identified articles were screened and shortlisted with inclusion criteria as clinical studies with laboratory data for at least two cytokines for severe and non-severe COVID-19 patient groups. Exclusion criteria for shortlisting included review articles, opinions and commentaries, studies that include other pathological conditions or complications associated with COVID-19 and studies without mean or median data of cytokines and their variances for each group.
2.2. Data extraction
Selected articles were reviewed independently by two authors (VK and SKD). Data was extracted from articles using standardized forms and was cross-validated. Cytokine levels reported as median and inter-quartile range (IQR) were converted to mean and standard deviation (SD) using published methods [25]. One study reported non-severe patient data in two clinical groups [26] which were combined using recommendations in Cochrane Handbook [27] section 6.5.2.10. Another study represented data in subgroups of IgG levels and Neutrophil-to-Lymphocyte ratio (NLR) [28], for which we retained data from only the IgG-low subgroups (as that would be closest to early stage of infections), and combined subgroups of high and low NLR values.
2.3. Meta-analysis and meta-regression
To assess the effect of each marker, we used standardized mean difference (SMD) of measured cytokine level (in pg/mL) between severe and non-severe groups including Hedges' correction for positive bias. All calculations were carried out using the metafor library [29] on R statistical software platform. Meta-regression of SMD of a marker was carried out using mixed-effects model with differences in age and sex (measured as percentage of male patients). Publication bias in the studies was analyzed using Funnel Plot and Egger's regression test. Sensitivity of meta-analysis results was assessed by repeating the analysis with leaving out one study at a time to detect any significant change in heterogeneity and summary effect.
2.4. Classifier development
We built a classifier using logistic regression model for categorization of patient groups in to severe and non-severe categories based on mean values of cytokines in the groups. Classifier performance using individual cytokine values or combination of multiple cytokine values are assessed from the Receiver Operating Characteristics (ROC) curve analysis and the area under the curve (AUC).
3. Results
3.1. Study details
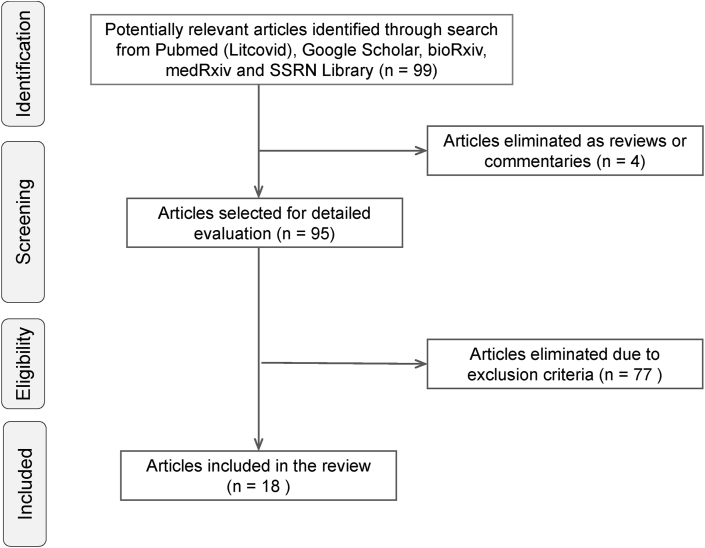
The search yielded a total of 99 “hits” which were screened to select 18 articles containing cytokine levels of severe and non-severe patients reported from clinical studies (Figure 1). All 18 selected studies were conducted in China and included 1,242 non-severe and 915 severe COVID-19 patients (Table 1). Out of the 18 studies, 7 were non-peer reviewed studies [26, 28, 30, 31, 32, 33, 34] published online in preprint archives.
Figure 1.
Schematic diagram depicting workflow for literature search and selection of studies.
Table 1.
Baseline characteristics of included studies for investigation of cytokine levels between severe and non-severe COVID-19 patients.
| Author (year) | Non-severe patients |
Severe Patients |
Clinical Definitions of Severe Patients | Cytokines Measured | ||||
|---|---|---|---|---|---|---|---|---|
| n | Age | Male % | n | Age | Male % | |||
| Chen, 2020 [35] | 10 | 50.3 (10.1) | 70.0 | 11 | 61.2 (7.3) | 90.9 | NHC guideline, pulmonary lesion | IL-6, IL-10, TNF-α |
| Feng, 2020 [30] | 94 | 62.9 (13.7) | 61.7 | 20 | 69.2 (11.1) | 65.0 | NHC guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| He, 2020 [36] | 12 | 42.3 (16.8) | 31.1 | 16 | 62.3 (16.7) | 53.6 | NHC guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Lv, 2020 [37] | 115 | 54.3 (41.5) | 50.4 | 155 | 58.7 (47.4) | 49.7 | Not specified | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Nie, 2020 [26] | 72 | 40.3 (19.3) | 29.1 | 25 | 57.3 (15) | 52.0 | NHC guideline, pulmonary lesion | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Qin, 2020 [38] | 166 | 52.1 (15.4) | 48.2 | 286 | 57 (13.3) | 54.2 | NHC guideline | IL-6, IL-10, TNF-α |
| Shi, 2020 [31] | 22 | 49.7 (12) | 59.7 | 21 | 59 (17.3) | 72.4 | NHC guideline, pulmonary lesion | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Song, 2020 [32] | 31 | 48 (16.5) | 51.6 | 42 | 55.9 (12.2) | 71.4 | NHC guideline, pulmonary lesion | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Wan, 2020 [39] | 102 | 43.1 (13.1) | 53.9 | 21 | 61.3 (15.6) | 52.4 | NHC guideline | IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Wang, 2020 [40] | 55 | 40 (14.1) | 45.0 | 14 | 69.8 (11.4) | 50.0 | NHC guideline with SpO2 < 90% | IL-2, IL-4, IL-6, IL-10, TNF-α |
| Wei, Su, 2020 [41] | 131 | 60.1 (12.4) | 45.0 | 98 | 69.7 (11.6) | 60.0 | NHC guideline, pulmonary lesion | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Xu, 2020 [42] | 80 | 55.7 (17) | 37.5 | 45 | 57.7 (15.7) | 68.9 | NHC guideline | IL-6, IL-10 |
| Zhang, Zhou, 2020 [28] | 44 | 53.5 (20.1) | 47.7 | 21 | 58.3 (13.9) | 71.4 | Not specified | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Zhang, Wang, 2020 [33] | 29 | 44.3 (15.8) | 58.6 | 14 | 61.7 (9.2) | 35.7 | NHC guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Zhang, Yu, 2020 [43] | 93 | 38.2 (12.2) | 34.4 | 18 | 63.3 (24.9) | 77.8 | WHO guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Zhao, 2020 [34] | 53 | 44.8 (15.9) | 43.4 | 18 | 63.9 (17.2) | 38.9 | NHC guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Zheng, 2020 [44] | 22 | 46.9 (14.7) | 40.9 | 74 | 56.8 (13.7) | 66.2 | NHC guideline | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
| Zhu, 2020 [45] | 111 | 50 (15.5) | 65.8 | 16 | 57.5 (11.7) | 56.3 | NHC guideline, pulmonary lesion | IL-2, IL-4, IL-6, IL-10, TNF-α, INF-γ |
NHC guideline: (a) Respiratory distress, RR ≥ 30/min, (b) Oxygen saturation level (SpO2) ≤ 93% at rest and (c) Arterial blood oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤300 mm Hg.
Pulmonary lesion: >50% lesion progression within 24–48h in pulmonary imaging.
WHO guideline: Adolescent or adult with clinical signs of pneumonia (fever, cough, dyspnoea, fast breathing) plus one of the following: respiratory rate >30 breaths/min; severe respiratory distress; or SpO2 < 90% on room air.
Age in mean (SD).
In most studies the severe patient group was designated based on National Health Commission of China guidelines [46] that specify (a) Respiratory distress, RR ≥ 30/min, (b) Oxygen saturation level (SpO2) ≤ 93% at rest and (c) Arterial blood oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤300 mm Hg. For some studies, pulmonary lesion progression >50% within 24–48 h as seen on imaging was used as additional criteria. One study used WHO guideline [47] that categorizes severe patients as adolescent or adult with clinical signs of pneumonia (fever, cough, dyspnoea, fast breathing) plus one of the following: respiratory rate >30 breaths/min; severe respiratory distress; or SpO2 < 90% on room air.
Weighted mean of age of patients in severe group (59.7y, range 55.9–69.8) was higher than the non-severe group (50.2y, range 38.2–62.9). Severe group also had larger percentage of male patients (57.8%, range 35.7–90.9) than non-severe group (48.4%, range 29.1–70.0).
Measurement of total 13 cytokines (IL-1β, IL-2, IL-2R, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12, IL-17, TNF-α and IFN-γ) and various lymphocyte subsets were reported in the selected 18 studies. Time of recording of these parameters is an important information that was disclosed in some of the studies as “on admission” [31, 35, 38, 42, 43, 44], “at early stage of disease” [36] or “on first day after admission before initiation of treatment” [39]. Method of measurement of the cytokines was indicated as flow cytometry-based immunoassay [39, 42, 45, 48], chemiluminescent immunoassay [41] or as multiplexed suspension array immunoassay [28, 34]. However, many articles did not explicitly mention the time of measurement or the specific immunoassay method for measurement of the cytokines. Finally, 13 studies were considered for further analysis which reported measurement of at least six cytokines (IL-2, IL-4, IL-6, IL-10, TNF-α and IFN-γ) aggregating minimum 1000 patients for each cytokine (Supplementary Table S1).
3.2. Meta-analysis of cytokine levels
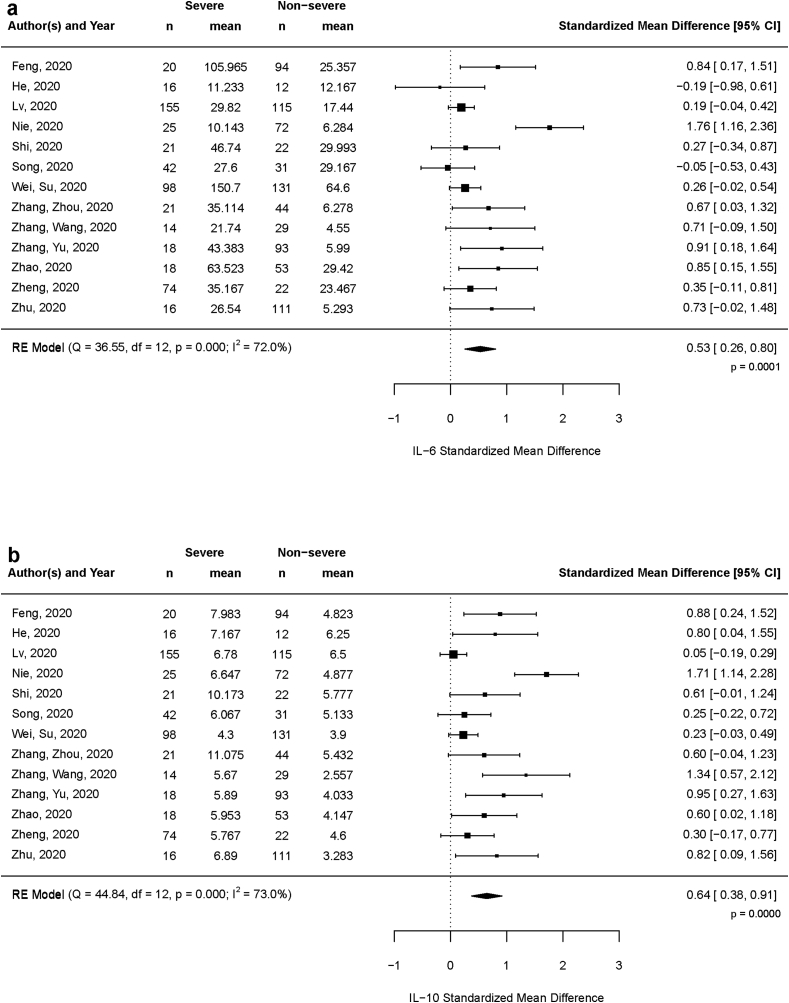
Meta-analysis for SMD value of each marker using a random-effects model showed moderate and statistically significant elevation in severe patients for two cytokines viz. IL-6 (SMD 0.53, 95% CI: 0.26–0.80, p < 0.001) (Figure 2a) and IL-10 (SMD 0.64, 95% CI: 0.38–0.91, p < 0.0001) (Figure 2b). Level of IFN-γ, the type II interferon, showed a weak elevation in severe group (SMD 0.11, 95% CI: -0,01–0.23, p = 0.078) with moderate significance. Summary effect size of other markers IL-2 (p = 0.281), IL-4 (p = 0.305) and TNF-α (p = 0.258) were not significant (Table 2).
Figure 2.
Forest plot showing standardized mean difference of the two cytokines (a) IL-6 and (b) IL-10 between severe and non-severe COVID-19 patients over 13 selected studies.
Table 2.
Meta-analysis result summary of six cytokines IL-2, IL-4, IL-6, IL-10, TNF-α and IFN-γ depicting the observed summary effect (standardized mean difference) and heterogeneity.
| Cytokine | Random Effect Model SMD (95% CI) | Summary Effect p.value | Heterogeneity (I2) |
|---|---|---|---|
| IL-2 | 0.12 (-0.10, 0.35) | 0.281 | 66.9% |
| IL-4 | 0.15 (-0.13, 0.42) | 0.305 | 79.2% |
| IL-6 | 0.53 (0.26, 0.80) | 0.0001 | 72.0% |
| IL-10 | 0.65 (0.38, 0.91) | <0.0001 | 73.0% |
| TNF-α | 0.10 (-0.08, 0.28) | 0.258 | 50.8% |
| IFN-γ | 0.11 (-0.01, 0.23) | 0.078 | 0% |
Bold indicates statistically significant summary effect (p< 0.05)
3.3. Meta-regression with age and gender difference
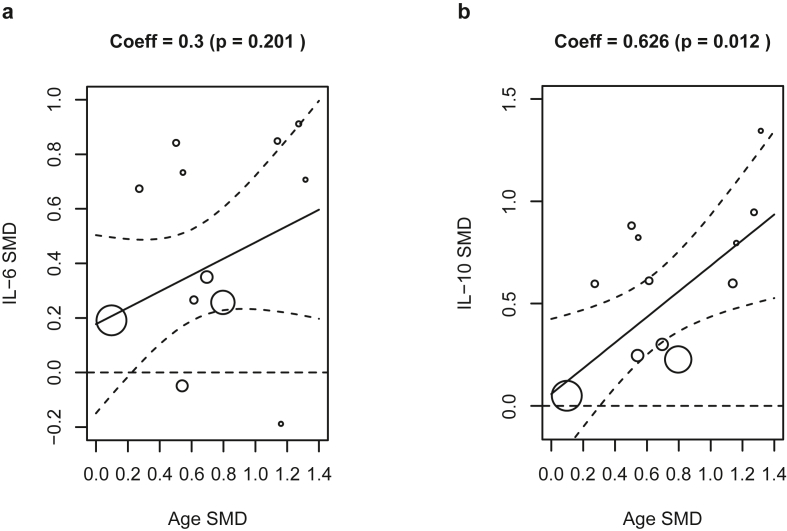
Summary effects of both IL-6 and IL-10 showed substantial heterogeneity (I2 ~ 72–73%) source of which was explored by carrying out meta-regression of IL-6 and IL-10 using (i) SMD of patient age and (ii) difference in male percentage between severe and non-severe groups as moderators in a mixed-effects model. Results revealed IL-10 levels having a dependence on the age difference (coefficient 0.63, p = 0.012), whereas the dependence of IL-6 on age was not significant (p > 0.05, Figure 3). No dependence of IL-6 and IL-10 levels were found on gender expressed as difference of male percentage between the two groups (Supplementary Figure S1). Above results ascribe some of the observed heterogeneity in IL-6 and IL-10 SMD to difference in average age between severe and non-severe group.
Figure 3.
Bubble plots showing meta-regression of the standardized mean difference of (a) IL-6 and (b) IL-10 between severe and non-severe groups with standardized mean difference of age between the two groups.
3.4. Classifier performance
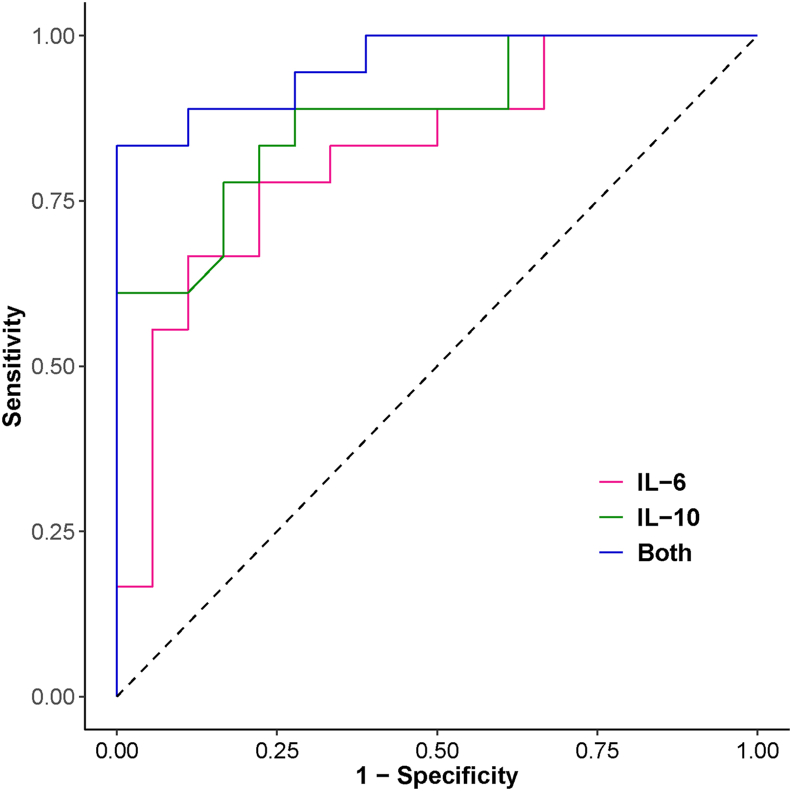
Our results indicate that out of the analysed cytokines, levels of only the pro-inflammatory IL-6 and the immuno-suppressive as well as immuno-stimulatory IL-10 are significantly elevated in the severe group of patients, an observation reported in other studies as well [20,48]. This prompted us to assess the prognostic potential of these markers by developing a logistic regression model using IL-6 and IL-10 mean values as covariates to classify the patient groups as severe and non-severe across all 18 studies. Model using both IL-6 and IL-10 as covariates showed an overall accuracy of 91.7% and area under the corresponding ROC curve as 0.957 (95% CI: 0.898–1), which is higher than the corresponding values using IL-6 (accuracy 77.8%, AUC 0.821) and IL-10 (accuracy 80.6%, AUC 0.878) individually as covariates (Table 3, Figure 4). Model using both IL-6 and IL-10 showed nearly 100% specificity and 83.3% sensitivity for classification in severe and non-severe categories.
Table 3.
Classifier performance summary using different sets of input variables –(i) only IL-6, (ii) only IL-10 and (iii) both IL-6 and IL-10.
| Classifier | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (95% CI) |
|---|---|---|---|---|
| IL-6 | 77.8 | 66.7 | 88.9 | 0.821 (0.680, 0.962) |
| IL-10 | 80.6 | 61.1 | 100 | 0.878 (0.766, 0.990) |
| Both IL-6 and IL-10 | 91.7 | 83.3 | 100 | 0.957 (0.898, 1) |
Figure 4.
ROC curve showing classifier performance using three different sets of covariates – (i) only IL-6 (red line), (ii) only IL-10 (green line) and (iii) both IL-6 and IL-10 (blue line).
3.5. Publication bias and sensitivity
Publication bias towards higher SMD of IL-10 levels reported in smaller-size studies manifested as asymmetry in Funnel plot and Egger's regression test (p < 0.001), whereas the corresponding bias was not significant (p = 0.151) for IL-6 (Supplementary Figure S2). Sensitivity analysis of the studies showed sizeable reduction of heterogeneity in IL-6 (I2 value from 72% to 25%) and moderate reduction for IL-10 (73%–55%) by leaving out one particular study from the analysis (Supplementary Figure S3).
3.6. Limitations
We perceive several limitations of this meta-analysis, including (a) limitation to Chinese ethnicity, which was not by design but due to the fact that most initial studies were reported from this geographic region which reported the first outbreak, (b) selection of non-peer reviewed publications (7 of the 18 studies) as per search strategy, (c) some heterogeneity in clinical criteria for severe disease as shown in Table 1, (d) un-specified time of measurement for the cytokines in many studies and (e) differing and un-specified immunoassay methods of cytokine measurement across studies. However, none of these limitations appear to contribute to a significant bias or heterogeneity as seen in the sensitivity analysis reported above.
A more significant limitation could be due to different treatment options used across studies that may have an effect on disease severity. Out of the 18 studies only two specified the treatment regime as anti-microbial and anti-viral agents along with anti-inflammatory corticosteroid (methylprednisolone) administered to some patients in one study [35] and immunoglobulin treatment in another [42]. However, none of the studies reported usage of targeted anti-inflammatory agents such as tocilizumab. infliximab etc.
4. Discussion
With the sustained spread of COVID-19 in most societies, a suitable prognostic test that can predict progression of patients to severe state of disease with reasonable accuracy is need of the hour for efficient management and care. Early research on COVID-19 indicated the role of pro-inflammatory cytokines such as IL-1, IL-6 and TNF-α released by activated mast cells in respiratory tract submucosa aggravating the inflammatory state and pathogenesis [49,50,51] and the potential of inhibiting some of these cytokines as a possible supportive therapy. Conti and co-workers have extensively studied the respiratory dysfunction caused through induction of IL-1 family of cytokines in pathogenic viral infections and proposed anti-inflammatory cytokines such as IL-37 or IL-38 as potential therapeutics for severe COVID-19 cases [49,52,53]. However, subsequent clinical studies and meta-analyses including the present study have not been able to unequivocally establish a significant elevation in IL-1 levels in the patients.
Elevated levels of IL-6, a pro-inflammatory molecule, is known to down-modulate NK cell activity and are also found to be associated with reduction in granzyme and perforin levels causing impairment of lytic activities [54]. In COVID-19 patients, exacerbation symptoms such as increased body temperature, elevation in inflammation markers like CRP and serum ferritin and progressed chest computed tomography images were associated with increased IL-6 levels which showed a downturn during recovery [55]. This association of IL-6 with pulmonary conditions were reported earlier in patients with pneumonia [56] or severe pneumonitis caused by radiation therapy [57].
IL-10, on the other hand, is an anti-inflammatory cytokine that was found elevated in severe COVID-19 patients [2, 35, 38]. Levels of IL-6, IL-10 and TNF-α were also found to be indicators of T-cell exhaustion in COVID-19 patients [58]. IL-10 is a multifunctional cytokine whose primary function is to limit the inflammatory response. IL-10 is also known to introduce anergy or non-responsiveness of T-cells in anti-tumour cell response [59] as well as in viral infection [60]. Blockade of IL-10 using antibody against IL-10 or its receptor or genetic removal of IL-10 resulted in elimination of infection by virus [60,61,62] or bacterial pathogen [63]. Thus the elevated levels of IL-10 in severe COVID-19 patients was initially ascribed to a negative feedback mechanism through its anti-inflammatory activities [34].
However, in other contexts IL-10 was also observed to have an immuno-stimulatory effect. Administration of recombinant IL-10 caused elevation in levels of LPS-induced IFN-γ release in healthy volunteers [64], and levels of phytohaemagglutinin-induced IFN-γ in moderate Crohn's Disease patients [65]. In view of its pro-inflammatory nature, Lu et al [66] have proposed a two-stage participatory role of IL-10 in COVID-19 disease progression. At the initiation of viral infection, IL-10 acts as a countermeasure to the pro-inflammatory modulators in a negative feedback mechanism. However, with progression of the disease endogenous level of IL-10 increases bringing into action its proinflammatory aspect and making contribution to the raging cytokine storm. However, this remains a hypothetical mechanism that needs to be tested in detailed longitudinal studies.
IL-4 is another cytokine, originally discovered as a B-cell growth factor assisting production of antibodies [67], that was found to suppress the levels of pro-inflammatory cytokines such as IL-1, IL-6 and TNF-α in PBMC isolated from healthy volunteers [68,69,70]. Similar anti-inflammatory nature of IL-4 was also observed in a mouse model of arthritis [71]. Studies with transgenic mice and further reviews have suggested differential effects of IL-4 on different T-cell populations that can affect T-cell infiltration and viral clearance for infections [72,73] However, our meta-analysis failed to establish any significant difference of IL-4 level between severe and non-severe patients, probably indicating a less prominent role of IL-4 in the acute inflammatory phase of COVID-19.
Though the levels of cytokines are increased in severe COVID-19 patients, it's implication in a therapeutic perspective remains unclear. Corticosteroids that can potentially suppress cytokines by inhibiting NF-κB transcription factor have been used on COVID-19 patients. The RECOVERY trial with 2104 patients receiving Dexamethasone compared to 4321 patients with standard-of-care showed that Dexamethasone reduces mortality in severe patients with invasive ventilation or oxygen support, but did not have any effect on patients with mild symptoms [74]. However, a meta-analysis across studies involving infection of SARS-CoV, SARS-CoV-2 and MERS-CoV showed increased mortality risk ratio (RR 2.11, 95% CI: 1.13–3.94) for patients treated with corticosteroids [75]. Theoharides and Conti [76] argued that use of dexamethasone as immunosuppressor may be beneficial in the short-term for severe COVID-19 patients but in the long-term it would negatively affect recovery due to the damaging effect of dexamethasone on protective function of T-cells and on antibody generation ability of B cells.
In an unique study, anti-TNF-α antibody drug, infliximab, was administered to a small group (n = 7) of severe COVID-19 patients without any pre-existing auto-immune condition [77]. Though the patients showed decrease in IL-6 level and increase in lymphocyte count post-administration, the cohort size was not adequate to make any conclusion on the end-point benefits. Few other studies used etanercept [78], adalimumab [79] and infliximab [80] therapy successfully for COVID-19 patients, but with pre-existing auto-immune conditions. On the other hand, there have been several studies targeting IL-6 levels using siltuximab, an IL-6 inhibitor or tocilizumab, an IL-6 receptor inhibitor in severe COVID-19 patients. A retrospective analysis of 88 COVID-19 pneumonia patients who received at list one dose of tocilizumab showed clinical improvement in 44% and 74% patients by day 7 and day 14 respectively [81], though without any control group in the study. However a placebo controlled trial with 243 moderately ill and hospitalized COVID-19 patients who received either a single dose of tocilizumab (n = 161) or placebo (n = 82) did not indicate any efficacy for tocilizumab to prevent intubation or death [82]. A recent review [83] on few such studies did not reveal any conclusive benefits due to lack of appropriate control groups in design or inadequate statistical significance. Further, the timing for anti-IL-6 agent administration remains critical since an early administration may negatively affect the innate response, while the late administration may not yield desired benefits. Some of the ongoing clinical results using anti-IL-6 therapy will shed more light in near future.
Therapeutic strategy of targeting IL-10 has been a nascent topic for infective diseases. Mice models treated with the immunomodulator compound Ammonium trichloro (dioxyethylene-O-O′)tellurate (AS101) that inhibits IL-10 transcription showed an improvement in survival from sepsis-related death [84]. Similarly, lymphocytic choriomeningitis virus (LCMV) infected mice when treated with anti-IL-10 antibody exhibited enhanced T-cell responses resulting in increased IFN-γ production and reduction in viral load [60]. Blockade of IL-10 signalling using an antibody that targets IL-10 receptor also reduced viral titre and persistence in LCMV-infected mice [61,62]. On the other hand, studies with administration of recombinant IL-10 or its pegylated form in cancer patients showed an escalation in CD8+ T-cell response manifested by increase of serum IFN-γ [85,86]. Such promiscuity of IL-10 both as an immune-response suppressor as well as an immuno-stimulator renders it as an unreliable drug target in any disease context.
Despite the unclarity on therapeutic potential of IL-6 and IL-10, their observed levels of elevation in severe COVID-19 patients have prompted clinical researchers to explore use of them as prognosticators. In an earlier study with pneumonia-affected children, IL-6/IL-10 ratio on admission was found to be an indicator of severe disease with sensitivity and specificity of 76.5% and 83.3% respectively [56]. In COVID-19 context, ROC curve analysis on an Italian cohort of 77 patients showed IL-6 as a prognostic factor for combined end-point of progression to severe disease and/or in-hospital mortality with AUC = 0.8 (19). However, performance of prognosis for progression to severe disease was poorer with AUC = 0.75. In the recently reported meta-analysis over a large number of studies, classifier performance was found to be higher for neutrophil count (AUC = 0.831), lymphocyte (AUC = 0.867) and D-dimer (AUC = 0.876) than using IL-6 as covariate (AUC = 0.632) [13]. Another research with 102 COVID-19 patients used logistic regression to evaluate potential of multiple cytokines to diagnose severe and critical patients [20]. Results revealed best prognostic performance for IL-6 (AUC = 0.83), followed by IL-10 (AUC = 0.73) as individual covariates whereas the model with all cytokines showed marginally better performance (AUC 0.86, sensitivity 80.0%, specificity 75.9%). A more recent study with 80 COVID-19 patients established a prognostic score with the slope of IL-6:IL-10 ration over the study period of 7 days predicting decline in condition of patients at an improved classifier performance (AUC >0.95) than using IL-6 level alone for prognosis [87]. All of these results support our finding of both IL-6 and IL-10 as prognosticator for severe COVID-19 disease.
Other than these cytokines, D-Dimer, CRP level and NLR are explored as diagnostic or prognostic markers in COVID-19. Though a couple of studies focused only on their diagnostic potential for COVID-19 positivity [88,89], ROC analysis in a study with 84 patients hospitalized with COVID-19 pneumonia [90] showed modest performance of NLR and CRP to predict a 7-day endpoint as AUC values of 0.69 (CRP), 0.76 (NLR) and 0.080 (both). Another study with 191 confirmed COVID-19 patients [89] established a slightly higher performance with AUC values as 0.84(NLR) and 0.71(CRP) to distinguish severe and non-severe patients. Pooled ROC curve analysis in a couple of meta-analysis studies showed modest classifier performance for CRP and D-Dimer with AUC values as 0.84 and 0.69 [91] and 0.88 and 0.77 [92] respectively to discriminate severe and non-sever patients. Since in all these cases the AUC values are below 0.9, predictive values for NLR, CRP and D-Dimer to diagnose severe patients may remain restricted. A recent proteomics study [93] predicted excellent classification performance (AUC = 0.96) for severe and non-severe COVID-19 patients using 22 serum proteins and 7 metabolites as covariates. However, a proteomics infrastructure is not so common in a clinic, limiting clinical utility of this test.
In this context, results of the meta-analysis and classification obtained in this study assumes importance in multiple aspects. Firstly, synthesis of a large number of studies (more than many of the cited meta-analyses) demonstrates a significant difference in values for only two cytokines – IL-6 and IL-10 with counter-acting mechanisms, reaffirming the dysregulation of immune response in progression to a severe state. Secondly, even at the absence of individual patient data, ROC curve analysis with mean values of IL-6 and IL-10 levels manifests a high level of classifier performance (AUC >0.9) in distinguishing severe from non-severe group patients, which is higher than classifier performance from individual studies [19, 20]. Our results are likely to be more robust since variations from multiple patient cohorts are included in this synthesis. And lastly, it opens the possibility of further research to establish a validated diagnostic test using these two markers for management of COVID-19 patients. Since we are working with mean values of markers in severe or non-severe patient groups and not individual patient-level values, we have refrained from using IL-6/IL-10 ratio as the classifier as was done in the pneumonia study [56]. However, availability of patient data from multiple cohorts of severe and non-severe patients can firmly establish and validate the test with the ratio or any other transform as the basis of classification.
5. Conclusion
Our findings, summarized over multiple studies, indicate a possible dysregulation in immune response against COVID-19, characterized by two cytokines IL-6 and IL-10, shifts the balance between non-severe to severe category of patients, and hence measurement of both markers is necessary to demarcate the boundary. Measurement of serum levels IL-6 and IL-10 are inexpensive and can be performed on admission to clinics or care centres with minimum facilities. Such measurement will be key to identify patients with a greater likelihood of progression to severe disease and thereby to adopt necessary precautionary measures.
Further trials with multiple cohorts of COVID-19 patients or assimilation of patient-level data from existing cohorts will be needed to establish and validate the test. Other than the diagnostic potential, option of possible therapeutic strategy targeting either IL-6 or IL-10 or both is likely to emerge through analysis of such data.
Declarations
Author contribution statement
S. Dhar: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
M. Das: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
K. Vishnupriyan: Performed the experiments.
S. Damodar: Analyzed and interpreted the data.
S. Gujar: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.1 February 2021, 2021. WHO Weekly Operational Update on COVID-19. (Accessed 2 February 2021).
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 4.Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843(11):2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Rojas J.M., Avia M., Martín V., Sevilla N. IL-10: a multifunctional cytokine in viral infections. J Immunol Res. 2017;2017 doi: 10.1155/2017/6104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau S.K.P., Lau C.C.Y., Chan K.H., Li C.P.Y., Chen H., Jin D.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J. Gen. Virol. 2013;94(PART 12):2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 8.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11(June):1–4. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.001. (May):0–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akbari H., Tabrizi R., Lankarani K.B., Aria H., Vakili S., Asadian F. The role of cytokine profile and lymphocyte subsets in the severity of coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Life Sci. 2020;258(July):118167. doi: 10.1016/j.lfs.2020.118167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elshazli R., Toraih E.A., Elgaml A., El-Mowafy M., El-Mesery M., Amin M. Diagnostic and prognostic value of hematological and immunological markers in COVID-19 infection: a meta-analysis of 6320 patients. PloS One. 2020;15(8) doi: 10.1371/journal.pone.0238160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng F., Huang Y., Guo Y., Yin M., Chen X., Xiao L. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int. J. Infect. Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz M., Fatima R., Assaly R. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25948. 0–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coomes E.A., Haghbayan H. Interleukin-6 in COVID-19: a systematic review and meta-analysis. medRxiv. 2020 Apr 3:2020. doi: 10.1002/rmv.2141. 03.30.20048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagunas-Rangel F.A., Chávez-Valencia V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udomsinprasert W., Jittikoon J., Sangroongruangsri S., Chaikledkaew U. Circulating levels of interleukin-6 and interleukin-10, but not tumor necrosis factor-alpha, as potential biomarkers of severity and mortality for COVID-19: systematic review with meta-analysis. J. Clin. Immunol. 2020 doi: 10.1007/s10875-020-00899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni E., Valoriani A., Cei F., Lamanna R., Gelli A.M.G., Ciambotti B. Interleukin-6 as prognosticator in patients with COVID-19. J. Infect. 2020;81:452–482. doi: 10.1016/j.jinf.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han H., Ma Q., Li C., Liu R., Zhao L., Wang W. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microb. Infect. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., Long W., Tu M., Chen S., Huang Y., Wang S. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry B.M., De Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 23.Laguna-Goya R., Utrero-Rico A., Talayero P., Lasa-Lazaro M., Ramirez-Fernandez A., Naranjo L. IL-6–based mortality risk model for hospitalized patients with COVID-19. J. Allergy Clin. Immunol. 2020;146(4):799–807. doi: 10.1016/j.jaci.2020.07.009. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Wan X., Wang W., Liu J., Tong T. Estimating the sample mean and standard deviation from the sample size , median , range and/or interquartile range. BMC Med. Res. Methodol. 2014;14(1):135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie S., Zhao X., Zhao K., Zhang Z., Zhang Z., Zhang Z. Metabolic disturbances and inflammatory dysfunction predict severity of coronavirus disease 2019 (COVID-19): a retrospective study. medRxiv. 2020:2020. 03.24.20042283. [Google Scholar]
- 27.Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ WV (editors), editor. Cochrane Handbook for Systematic Reviews of Interventions Version 6.0. Cochrane
- 28.Zhang B., Zhou X., Zhu C., Feng F., Qiu Y., Feng J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020:2020. doi: 10.3389/fmolb.2020.00157. 03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viechtbauer W. Conducting meta-analyses in R with the metafor. J. Stat. Software. 2010;36(3):1–48. [Google Scholar]
- 30.Feng X., Li P., Ma L., Liang H., Lei J., Li W. Clinical characteristics and short-term outcomes of severe patients with COVID-19 in wuhan, China. medRxiv. 2020:2020. doi: 10.3389/fmed.2020.00491. 04.24.20078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi H., He L., Sun W., Xu J., Wang M., Chen X. Clinical characteristics and prognostic factors of 148 COVID-19 cases in a secondary epidemic area. SSRN Electron J. 2020 [Google Scholar]
- 32.Song C.-Y., Xu J., He J.-Q., Lu Y.-Q. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020;2020 03.05.20031906. [Google Scholar]
- 33.Zhang H., Wang X., Fu Z., Luo M., Zhang Z., Zhang K. Potential factors for prediction of disease severity of COVID-19 patients. medRxiv. 2020:2020. 03.20.20039818. [Google Scholar]
- 34.Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J. Longitudinal profiling of cytokines and chemokines in COVID-19 reveals inhibitory mediators IL-1Ra and IL-10 are associated with disease severity while elevated RANTES is an early predictor of mild disease. JCI Insight. 2020;5(13) doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H. Clinical and immunologic features in severe and moderate forms of Coronavirus Disease 2019. J. Clin. Invest. 2020;27(1095):16. doi: 10.1172/JCI137244. 20023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He R., Lu Z., Zhang L., Fan T., Xiong R., Shen X. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020;127(April):104361. doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv Z., Cheng S., Le J., Huang J., Feng L., Zhang B. Clinical characteristics and co-infections of 354 hospitalized patients with COVID-19 in Wuhan, China: a retrospective cohort study. Microb. Infect. 2020 doi: 10.1016/j.micinf.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID-19) infected patients. Br. J. Haematol. 2020;(April):428–437. doi: 10.1111/bjh.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in wuhan, China. Clin. Infect. Dis. 2020;71(15):769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X., Su J., Yang K., Wei J., Wan H., Cao X. Elevations of serum cancer biomarkers correlate with severity of COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25957. 0–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu B., Fan C yu, Wang A lu, Zou Y long, han Yu Y., He C. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J. Infect. 2020;81(1):e51–60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Yu M., Tong S., Liu L.Y., Tang L.V. Predictive factors for disease progression in hospitalized patients with coronavirus disease 2019 in Wuhan, China. J. Clin. Virol. 2020;127(March):104392. doi: 10.1016/j.jcv.2020.104392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369(March):1–8. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Z., Cai T., Fan L., Lou K., Hua X., Huang Z. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int. J. Infect. Dis. 2020;95:332–339. doi: 10.1016/j.ijid.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Health Commission & State Administration of Traditional Chinese Medicine . National Health Commission of the People’s Republic of China; 2020. Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia. [Google Scholar]
- 47.World Health Organization . World Health Organization; 2020. (2020). Clinical Management of COVID-19: Interim Guidance, 27 May 2020. [Google Scholar]
- 48.Wang F., Hou H., Luo Y., Tang G., Wu S., Huang M. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI insight. 2020 May 21;5(10) doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020 Mar 1;34(2):327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 50.Kritas S.K., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Conti P. Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. J. Biol. Regul. Homeost. Agents. 2020 Jan 1;34(1):9–14. doi: 10.23812/20-Editorial-Kritas. [DOI] [PubMed] [Google Scholar]
- 51.Ronconi G., Teté G., Kritas S.K., Gallenga C.E., Caraffa A., Ross R. COVID-19 induced by SARS-CoV-2 causes Kawasaki-like disease in children: role of pro-inflammatory and anti-inflammatory cytokines. J. Biol. Regul. Homeost. Agents. 2020;34(3):767–773. doi: 10.23812/EDITORIAL-RONCONI-E-59. [DOI] [PubMed] [Google Scholar]
- 52.Conti P., Caraffa A., Gallenga C.E., Ross R., Kritas S.K., Frydas I. IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: inhibitory effect of the IL-1 receptor antagonist (IL-1Ra) J. Biol. Regul. Homeost. Agents. 2020 Sep 1;34(5):1623–1627. doi: 10.23812/20-34-4EDIT-65. [DOI] [PubMed] [Google Scholar]
- 53.Conti P., Gallenga C.E., Tetè G., Caraffa A., Ronconi G., Younes A. How to reduce the likelihood of coronavirus-19 (CoV-19 or SARS-CoV-2) infection and lung inflammation mediated by IL-1. J. Biol. Regul. Homeost. Agents. 2020;34(2):333–338. doi: 10.23812/Editorial-Conti-2. [DOI] [PubMed] [Google Scholar]
- 54.Cifaldi L., Prencipe G., Caiello I., Bracaglia C., Locatelli F., De Benedetti F. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheum. 2015;67(11):3037–3046. doi: 10.1002/art.39295. [DOI] [PubMed] [Google Scholar]
- 55.Liu T., Zhang J., Yang Y., Ma H., Li Z., Zhang J. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol. Med. 2020;12(7) doi: 10.15252/emmm.202012421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Brito R de C.C.M., Lucena-Silva N., Torres L.C., Luna C.F., Correia J de B, da Silva G.A.P. The balance between the serum levels of IL-6 and IL-10 cytokines discriminates mild and severe acute pneumonia. BMC Pulm. Med. 2016;16(1):19–21. doi: 10.1186/s12890-016-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y., Rubin P., Williams J., Hernady E., Smudzin T., Okunieff P. Circulating IL-6 as a predictor of radiation pneumonitis. Int. J. Radiat. Oncol. Biol. Phys. 2001;49(3):641–648. doi: 10.1016/s0360-3016(00)01445-0. [DOI] [PubMed] [Google Scholar]
- 58.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11(May):1–7. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore K.W., Malefyt R.D.W., Robert L., Garra A.O. Interleukin-10 and the interleukin-10 receptor. Mol. Cell Biol. 2001;1(1):683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 60.Maris C.H., Chappell C.P., Jacob J. Interleukin-10 plays an early role in generating virus-specific T cell anergy. BMC Immunol. 2007;8:9–12. doi: 10.1186/1471-2172-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brooks D.G., Trifilo M.J., Edelmann K.H., Teyton L., McGavern D.B., Oldstone M.B.A. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006;12(11):1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ejrnaes M., Filippi C.M., Martinic M.M., Ling E.M., Togher L.M., Crotty S. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J. Exp. Med. 2006;203(11):2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biswas P.S., Pedicord V., Ploss A., Menet E., Leiner I., Pamer E.G. Pathogen-specific CD8 T cell responses are directly inhibited by IL-10. J. Immunol. 2007;179(7):4520–4528. doi: 10.4049/jimmunol.179.7.4520. [DOI] [PubMed] [Google Scholar]
- 64.Lauw F.N., Pajkrt D., Hack C.E., Kurimoto M., van Deventer S.J.H., van der Poll T. Proinflammatory effects of IL-10 during human endotoxemia. J. Immunol. 2000;165(5):2783–2789. doi: 10.4049/jimmunol.165.5.2783. [DOI] [PubMed] [Google Scholar]
- 65.Tilg H., Van Montfrans C., Van den Ende A., Kaser A., Van Deventer S.J.H., Schreiber S. Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon γ. Gut. 2002;50(2):191–195. doi: 10.1136/gut.50.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu L., Zhang H., Dauphars D.J., He Y.W. A potential role of interleukin 10 in COVID-19 pathogenesis. Trends Immunol. 2021;42(1):3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howard M., Farrar J., Hilfiker M., Johnson B., Takatsu K., Hamaoka T. Identification of a T cell-derived B cell growth factor distinct from interleukin 2. J. Exp. Med. 1982;155(3):914–923. doi: 10.1084/jem.155.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Essner R., Rhoades K., McBride W.H., Morton D.L., Economou J.S. IL-4 down-regulates IL-1 and TNF gene expression in human monocytes. J. Immunol. 1989;142(11) [PubMed] [Google Scholar]
- 69.te Velde A., Huijbens R., Heije K., de Vries J., Figdor C. Interleukin-4 (IL-4) inhibits secretion of IL-1 beta, tumor necrosis factor alpha, and IL-6 by human monocytes. Blood. 1990 Oct 1;76(7):1392–1397. [PubMed] [Google Scholar]
- 70.Lee J.D., Swisher S.G., Minehart E.H., McBride W.H., Economou J.S. Interleukin-4 downregulates interleukin-6 production in human peripheral blood mononuclear cells. J. Leukoc. Biol. 1990 May 1;47(5):475–479. doi: 10.1002/jlb.47.5.475. [DOI] [PubMed] [Google Scholar]
- 71.Kawalkowska J.Z., Hemmerle T., Pretto F., Matasci M., Neri D., Williams R.O. Targeted IL-4 therapy synergizes with dexamethasone to induce a state of tolerance by promoting Treg cells and macrophages in mice with arthritis. Eur. J. Immunol. 2016 May 1;46(5):1246–1257. doi: 10.1002/eji.201546221. [DOI] [PubMed] [Google Scholar]
- 72.Silva-Filho J.L., Caruso-Neves C., Pinheiro A.A.S. IL-4: an important cytokine in determining the fate of T cells. Biophys Rev. 2014 Mar 9;6(1):111–118. doi: 10.1007/s12551-013-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bot A., Holz A., Christen U., Wolfe T., Temann A., Flavell R. Local IL-4 expression in the lung reduces pulmonary influenza-virus- specific secondary cytotoxic T cell responses. Virology. 2000 Mar 30;269(1):66–77. doi: 10.1006/viro.2000.0187. [DOI] [PubMed] [Google Scholar]
- 74.Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Z., Liu J., Zhou Y., Zhao X., Zhao Q., Liu J. The effect of corticosteroid treatment on patients with coronavirus infection: a systematic review and meta-analysis. J. Infect. 2020;81:13–20. doi: 10.1016/j.jinf.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Theoharides T.C., Conti P. Dexamethasone for COVID-19? Not so fast. J. Biol. Regul. Homeost. Agents. 2020 Jun 4;34(3):1241–1243. doi: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 77.Stallmach A., Kortgen A., Gonnert F., Coldewey S.M., Reuken P., Bauer M. Infliximab against severe COVID-19-induced cytokine storm syndrome with organ failure - a cautionary case series. Crit. Care. 2020 Jul 17;24(1):444. doi: 10.1186/s13054-020-03158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duret P.M., Sebbag E., Mallick A., Gravier S., Spielmann L., Messer L. Recovery from COVID-19 in a patient with spondyloarthritis treated with TNF-alpha inhibitor etanercept. Ann. Rheum. Dis. 2020 May 22 doi: 10.1136/annrheumdis-2020-217362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tursi A., Angarano G., Monno L., Saracino A., Signorile F., Ricciardi A. COVID-19 infection in Crohn’s disease under treatment with adalimumab. Gut. 2020;69:1364–1365. doi: 10.1136/gutjnl-2020-321240. BMJ Publishing Group. [DOI] [PubMed] [Google Scholar]
- 80.Bezzio C., Manes G., Bini F., Pellegrini L., Saibeni S. Infliximab for severe ulcerative colitis and subsequent SARS-CoV-2 pneumonia: a stone for two birds. Gut. NLM (Medline) 2020 doi: 10.1136/gutjnl-2020-321760. [DOI] [PubMed] [Google Scholar]
- 81.Fernández-Ruiz M., López-Medrano F., Pérez-Jacoiste Asín M.A., Maestro de la Calle G., Bueno H., Caro-Teller J.M. Tocilizumab for the treatment of adult patients with severe COVID-19 pneumonia: a single-center cohort study. J. Med. Virol. 2020 Jul 27 doi: 10.1002/jmv.26308. jmv.26308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S. Efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020 Oct 21 doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020;(April):102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalechman Y., Gafter U., Gal R., Rushkin G., Yan D., Albeck M. Anti-IL-10 therapeutic strategy using the immunomodulator AS101 in protecting mice from sepsis-induced death: dependence on timing of immunomodulating intervention. J. Immunol. 2002;169(1):384–392. doi: 10.4049/jimmunol.169.1.384. [DOI] [PubMed] [Google Scholar]
- 85.Naing A., Papadopoulos K.P., Autio K.A., Ott P.A., Patel M.R., Wong D.J. Safety, antitumor activity, and immune activation of pegylated recombinant human interleukin-10 (AM0010) in patients with advanced solid tumors. J. Clin. Oncol. 2016;34(29):3562–3569. doi: 10.1200/JCO.2016.68.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naing A., Infante J.R., Papadopoulos K.P., Chan I.H., Shen C., Ratti N.P. PEGylated IL-10 (pegilodecakin) induces systemic immune activation, CD8+ T cell invigoration and polyclonal T cell expansion in cancer patients. Canc. Cell. 2018;34(5):775–791. doi: 10.1016/j.ccell.2018.10.007. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McElvaney O.J., Hobbs B.D., Qiao D., McElvaney O.F., Moll M., McEvoy N.L. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. EBioMedicine. 2020:61. doi: 10.1016/j.ebiom.2020.103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nalbant A., Kaya T., Varim C., Yaylaci S., Tamer A., Cinemre H. Can the neutrophil/lymphocyte ratio (NLR) have a role in the diagnosis of coronavirus 2019 disease (COVID-19)? Rev. Assoc. Med. Bras. 2020;66(6):746–751. doi: 10.1590/1806-9282.66.6.746. [DOI] [PubMed] [Google Scholar]
- 89.Yufei Y., Mingli L., Xuejiao L., Xuemei D., Yiming J., Qin Q. Utility of the neutrophil-to-lymphocyte ratio and C-reactive protein level for coronavirus disease 2019 (COVID-19) Scand. J. Clin. Lab. Invest. 2020:1–5. doi: 10.1080/00365513.2020.1803587. 0(0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y., Du X., Chen J., Jin Y., Peng L., Wang H.H.X. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020 Jul 1;81(1):e6–12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang I., Pranata R., Lim M.A., Oehadian A., Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther. Adv. Respir. Dis. 2020;14:1–14. doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soraya G.V., Ulhaq Z.S. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: an updated meta-analysis. Med. Clin. 2020 Aug 28;155(4):143–151. doi: 10.1016/j.medcli.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020 Jul 9;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.