Abstract
Facilitating clinical translation of quantitative imaging techniques has been suggested as means of improving interobserver agreement and diagnostic accuracy of multiparametric magnetic resonance imaging (mpMRI) of the prostate. One such technique, magnetic resonance fingerprinting (MRF), has significant competitive advantages over conventional mapping techniques in terms of its multi-site reproducibility, short scanning time and inherent robustness to motion. It has also been shown to improve the detection of clinically significant prostate cancer when added to standard mpMRI sequences, however, the existing studies have all been conducted on 3.0 T MRI systems, limiting the technique’s use on 1.5 T MRI scanners that are still more widely used for prostate imaging across the globe. The aim of this proof-of-concept study was, therefore, to evaluate the cross-system reproducibility of prostate MRF T1 in healthy volunteers (HVs) using 1.5 and 3.0 T MRI systems. The initial validation of MRF T1 against gold standard inversion recovery fast spin echo (IR-FSE) T1 in the ISMRM/NIST MRI system revealed a strong linear correlation between phantom-derived MRF and IR-FSE T1 values was observed at both field strengths (R2 = 0.998 at 1.5T and R2 = 0.993 at 3T; p = < 0.0001 for both). In young HVs, inter-scanner CVs demonstrated marginal differences across all tissues with the highest difference of 3% observed in fat (2% at 1.5T vs 5% at 3T). At both field strengths, MRF T1 could confidently differentiate prostate peripheral zone from transition zone, which highlights the high quantitative potential of the technique given the known difficulty of tissue differentiation in this age group. The high cross-system reproducibility of MRF T1 relaxometry of the healthy prostate observed in this preliminary study, therefore, supports the technique’s prospective clinical validation as part of larger trials employing 1.5 T MRI systems, which are still widely used clinically for routine mpMRI of the prostate.
Introduction
Multiparametric magnetic resonance imaging (mpMRI) has recently been adopted by major European and American prostate cancer (PCa) guidelines as the key diagnostic tool for detection, local staging and active surveillance of the disease [1, 2]. Despite significant advantages over the traditional systematic biopsy pathway, such as high negative predictive value for clinically significant PCa, mpMRI has limitations including a known learning curve and considerable interobserver variation for image assessment, even amongst experience readers [3–8]. Making prostate MRI more objective by facilitating clinical translation of quantitative imaging techniques is viewed by many experts as key to solving the aforementioned challenges [9, 10]. This move is also supported by the Prostate Imaging Reporting and Data System (PI-RADS) steering committee with a view to complement the current recommendations that are based exclusively on qualitative visual assessment of images [11, 12].
MR fingerprinting is a rapidly developing quantitative imaging technique enabling fast and simultaneous generation of tissue T1 and T2 property maps that are inherently spatially registered and robust to motion [13, 14]. The key competitive advantages of MRF over standard mapping techniques, which also make it attractive for PCa imaging, include its higher multi-site reproducibility and significantly shorter acquisition time, which is important given the current move towards developing abbreviated prostate imaging protocols in order to reduce scanning times whilst maintaining quality [14–19].
Several pilot studies investigating the added value of MRF to prostate mpMRI have demonstrated significantly improved detection of clinically significant peripheral zone (PZ) and transition zone (TZ) tumors when MRF was used in combination with apparent diffusion coefficient (ADC) mapping [20–23]. However, wider clinical validation of MRF as part of an mpMRI protocol is limited by the fact studies have only validated performance on 3T MRI systems, whist the majority of prostate mpMRI scans even in developed countries are still performed on 1.5T scanners [24–27]. The most recent version of the PI-RADS guidelines (v2.1) suggest that both 1.5T and 3T allow for reliable diagnostic exams in the context of adequate hardware and software, and when optimal acquisition parameters are ensured. [10]
Whilst MRF at both 1.5T and 3T has been used in cardiac and brain imaging, there are surprisingly few studies dedicated to cross-system validation of MRF in different body areas other than the brain [28–30]. It is known that longitudinal T1 relaxation times are more sensitive to an increase of the magnetic field strength compared to T2 relaxation and quantitative magnetisation transfer parameters [31]. In the prostate, which generally has longer T1 values compared to other pelvic and abdominal organs, field-dependent variation of T1 can reach up to 24%, which may affect the cross-system reproducibility of MRF [15, 32].
Therefore, the objective of this proof-of-concept study was to evaluate the cross-system reproducibility of MRF-based T1 mapping of the healthy prostate with data acquired from both phantoms and healthy volunteers using 1.5T and 3T MRI systems.
Methods
Phantom study
To evaluate the accuracy of the introduced T1 and T2 measurements, MRF and standard relaxation mapping data were obtained from the ISMRM/NIST phantom. [27] Phantom data were obtained on 1.5T MR450 and 3T MR750w scanners (both GE Healthcare, Waukesha, WI, USA) using a 32-channel receiver coil. Regions-of-interest (ROIs) were created from the spheres in either the T1 or T2 layer of the phantom. MR Fingerprinting was performed as further described in section 2.4.
Conventional T1 and T2 mapping was performed with inversion recovery fast spin echo (IR-FSE) and multiple spin echo (MSE) imaging, respectively. The field-of-view (FOV) = 260x260 mm2, matrix = 256x256 and slice thickness = 3 mm matched in all techniques. For 2D IR-FSE T1 mapping, data were acquired with inversion times (TIs) of 50, 100, 200, 400, 800, 1600, and 2400 ms. The IR-FSE sequence used a repetition time (TR) = 8000 ms and echo time (TE) = 13 ms at both 1.5 T and 3.0 T. T1 data for all non-MRF techniques were fit using non-linear fitting of the signal equations in Python. Multiple spin echo (MSE) data for T2 estimations were acquired with TEs = 8.1, 16.3, 24.4, 32.6, 40.7, 58.9, 57.0, 65.2 ms. MSE data were fit with a log linear least squares algorithm. The acquisition matched the FOV, matrix, and slices as the MRF acquisition.
Healthy volunteers study
All elements of this prospective study were carried out in accordance with the Declaration of Helsinki and were approved by the institutional ethics board (NRES Committee East of England, UK), with written informed consent obtained from all participants. The volunteers were aged at least 18 years were included in this study, with the exclusion criteria being previous diagnosis or treatment for prostate cancer and clinical contraindication to MRI.
Anatomical imaging
Healthy volunteers underwent prostate MRI on the same 1.5T MR450 and 3T MR750 scanners (both GE Healthcare, Waukesha, WI, USA) used in the phantom study. As per clinical mpMRI acquisition, the protocol included Axial T1 and multiplanar high-resolution T2-weighted 2D fast recovery FSE (FOV = 18x18 cm; voxel size 0.35x0.35 mm2; slice thickness = 3–4 mm; gap = 0–0.5 mm). Diffusion-weighted imaging (DWI) was performed using a spin-echo echo-planar imaging pulse sequence (FOV 28cm; slice thickness 3mm; gap 0mm; b-values: b-150, b-750, and b-1,400 s/mm2) and an additional small FOV (24 cm) b-2,000s/mm2 DWI sequence; apparent diffusion coefficient (ADC) maps were calculated automatically.
MR fingerprinting protocol
MRF at both 1.5 T and 3.0 T was acquired with an inversion-prepared 2D steady-state-free-precession (SSFP) MRF sequence [13]. The acquisition consisted of 979 undersampled interleaved spirals with 656 points per spiral, and with sequential spirals rotated by the golden angle. The maximum gradient strength per spiral was 28 mT/m and the maximum slew rate was 108 T/m/s. The imaging parameters were: field-of-view (FOV) = 260x260 mm2, matrix = 256x256, slices = 3 for ISMRM/NIST imaging and 12–14 for in vivo imaging, in vivo slice thickness = 6 mm and 3 mm at 1.5T and 3T, respectively, spacing = 5mm and 6mm at 1.5T and 3T (39 mm for both for ISMRM/NIST imaging), respectively, sampling bandwidth = ±250 kHz, slice dephasing = 8π, echo time (TE) = 2.5 ms, repetition time (TR) = 10 ms, acquisition time = 9.79 seconds/slice. The flip angle lists matched those in Jiang et al. [33]. Axial images were acquired to match the standard acquisition planes for other prostate image assessments.
Image analysis
MRF T1 values for the whole prostate, peripheral zone (PZ), transition zone (TZ), internal obturator muscle and adipose tissue in the ischioanal fossa were calculated from ROIs drawn by a single fellowship-trained uro-radiologist using the open-source segmentation software ITK-SNAP [34]. ROIs were drawn using anatomical T2WI as reference and subsequently transposed on to the MRF T1 with their size and location being matched to the appropriate FOV parameters and anatomical position of the outlined structures using in-house software developed within Python using the PyQtGraph and PyDicom libraries [35].
Statistics
In the phantom study, simple linear regression was used to evaluate the relationship between T1 and T2 values obtained using IR-FSE, MSE and MRF mapping techniques with their agreement assessed using Bland-Altman analysis. In the healthy volunteers study, the D’Agostino and Pearson test was applied to assess the distribution of imaging values with their intergroup comparison performed using either paired t-test or Wilcoxon matched-pairs signed rank test as appropriate. Both in the phantom and healthy volunteers studies, the variation of imaging values as a means of their cross-system reproducibility was evaluated using a coefficient of variation (CV). All plots and figures were created in Prism 8 (GraphPad Software, San Diego, CA).
Results
Phantom study
Summary mean inter-scanner T1 and T2 values obtained from the corresponding ISMRM/NIST phantom layers using MRF, inversion recovery fast spin echo (IR-FSE) and multiple spin echo (MSE) imaging are presented in Table 1. Paired t-test showed no inter-scanner differences between IR-FSE and MRF-based T1 phantom values (p = 0.795 and p = 0.102, respectively). As a measure of reproducibility, coefficients of variations (CVs) were calculated and are also presented in Table 1, showing inter-scanner differences of 0.17% and 2.57% for IR-FSE and MRF T1, respectively. Inter-scanner T2 values obtained using MRF and MSE imaging were, however, significantly different in both cases (p = 0.0002 and p = 0.0006, respectively) with differences in their CVs reaching 0.10% and 7.66%, respectively.
Table 1. Summary ISMRM/NIST phantom T1 and T2 relaxation times and coefficients of variation obtained using inversion recovery fast spin echo (IR-FSE), multiple spin echo (MSE) and magnetic resonance fingerprinting (MRF) at 1.5T and 3T systems.
| Mean ± SD | Coefficient of variation (%) | ||||
|---|---|---|---|---|---|
| Sequence | 1.5T | 3T | P | 1.5T | 3T |
| IR-FSE T1 | 616.1 ± 400.9 | 615.8 ± 401.8 | 0.7951 | 65.08 | 65.25 |
| MRF T1 | 559.3 ± 402.2 | 544.1 ± 405.3 | 0.1019 | 71.91 | 74.48 |
| MSE T2 | 137.8 ± 112.2 | 92.8 ± 82.7 | 0.0002 | 116.80 | 116.90 |
| MRF T2 | 471.2 ± 550.2 | 95.6 ± 111.8 | 0.0006 | 81.46 | 89.12 |
SD = standard deviation.
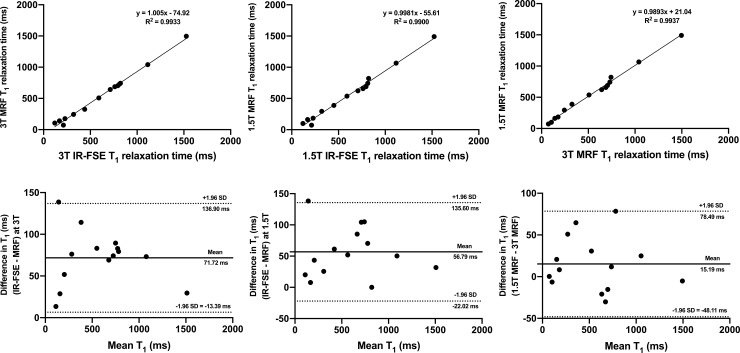
Fig 1A and 1B show the mean ISMRM/NIST phantom MRF T1 values plotted against T1 values obtained using inversion recovery fast spin echo (IR-FSE) imaging at 1.5T and 3T, respectively. The analysis showed a strong linear correlation between MRF and IR-FSE at both field strengths (R2 = 0.998 at 1.5T and R2 = 0.993 at 3T; p = < 0.0001 for both) with slopes of the linear fits and y-intercepts presented in the corresponding figures. The Bland-Altman analysis revealed good agreement between the two techniques at both 1.5T and 3T with the corresponding mean bias and 95% limits of agreement (LOA) presented in Fig 1D and 1E, respectively. When plotted against each other, MRF T1 values obtained at 1.5T and 3T also demonstrated a strong linear correlation (R2 = 0.994; p < 0.0001) and acceptable agreement (bias 15.19 ms, 95% LOA -48.11 ms to 78.49 ms) with corresponding plots presented in Fig 1C and 1F, respectively.
Fig 1.
Linear regression plots (a-c) and Bland-Altman plots (d-f) comparing MRF T1 ISMRM/NIST phantom values with those obtained using IR-FSE at 3T (a, d) and 1.5T (b, e). An inter-scanner comparison of MRF T1 values is also presented (c, f). On Fig (d-f), dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, IR-FSE = inversion recovery fast spin echo, SD = standard deviation.
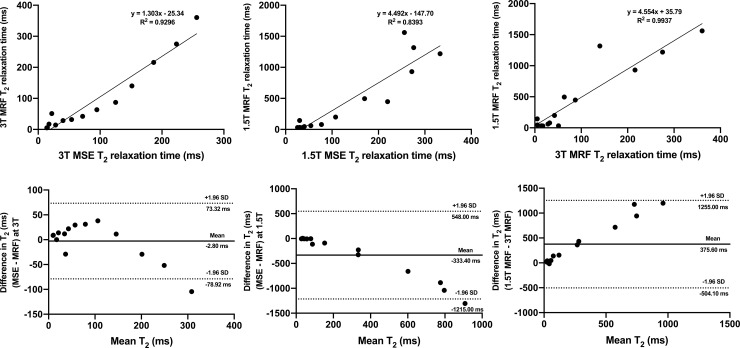
The outcomes of a similar analysis comparing the MRF and multiple spin echo (MSE) T2 values obtained at 1.5T and 3T are presented in Fig 2. It is of note that T2 values obtained in this study showed consistently poorer reproducibility as demonstrated by both linear regression and Bland-Altman analysis, prompting us to focus the following presentation and analysis of the in vivo results on T1 relaxometry.
Fig 2.
Linear regression plots (a-c) and Bland-Altman plots (d-f) comparing MRF T2 ISMRM/NIST phantom values with those obtained using MSE at 3T (a, d) and 1.5T (b, e). An inter-scanner comparison of MRF T2 values is also presented (c, f). On Fig (d-f), dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, MSE = multiple spin echo, SD = standard deviation.
Healthy volunteers study
The study included 10 healthy male volunteers (mean age 32.3 years, range 24–41) who underwent MRF of the prostate at 1.5T and 3T within the same imaging session.
Summary mean inter-scanner MRF T1 values and coefficients of variation (CVs) obtained from the whole prostate, peripheral zone (PZ), transition zone (TZ), internal obturator muscle and fat in the ischioanal fossa are presented in Table 2. As expected, 3T MRF T1 values were significantly longer in all tissues compared to 1.5T MRF (P < 0.0001 for all). Inter-scanner CVs demonstrated only marginal differences in all tissues with the highest difference of 3% observed in fat (2% at 1.5T vs 5% at 3T). T1 values obtained from the PZ showed the highest overall variation with CVs at 1.5T and 3T being 10.9% and 10.3%, respectively. The outcomes of a similar analysis conducted for in vivo MRF T2 relaxometry is presented in S1 Table.
Table 2. Summary MRF T1 relaxation times and coefficients of variation obtained at 1.5T and 3T from the whole prostate, peripheral zone, transition zone, internal obturator muscle and fat from the ischioanal fossa.
| Mean ± SD | Coefficient of variation (%) | ||||
|---|---|---|---|---|---|
| Tissue | 1.5T | 3T | P | 1.5T | 3T |
| Prostate | 1054.0 ± 80.1 | 1769.0 ± 112.7 | < 0.0001 | 7.60 | 6.37 |
| Peripheral zone | 1202.0 ± 131.3 | 2041.0 ± 209.4 | < 0.0001 | 10.93 | 10.26 |
| Transition zone | 968.8 ± 60.7 | 1552.0 ± 105.2 | < 0.0001 | 6.26 | 6.78 |
| Muscle | 725.6 ± 23.8 | 1446.0 ± 73.95 | < 0.0001 | 3.29 | 5.12 |
| Fat | 192.1 ± 3.8 | 345.1 ± 17.1 | < 0.0001 | 1.99 | 4.96 |
SD = standard deviation.
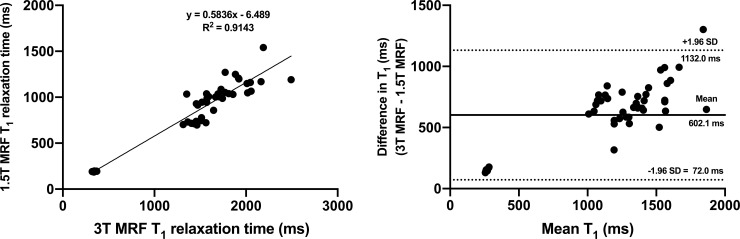
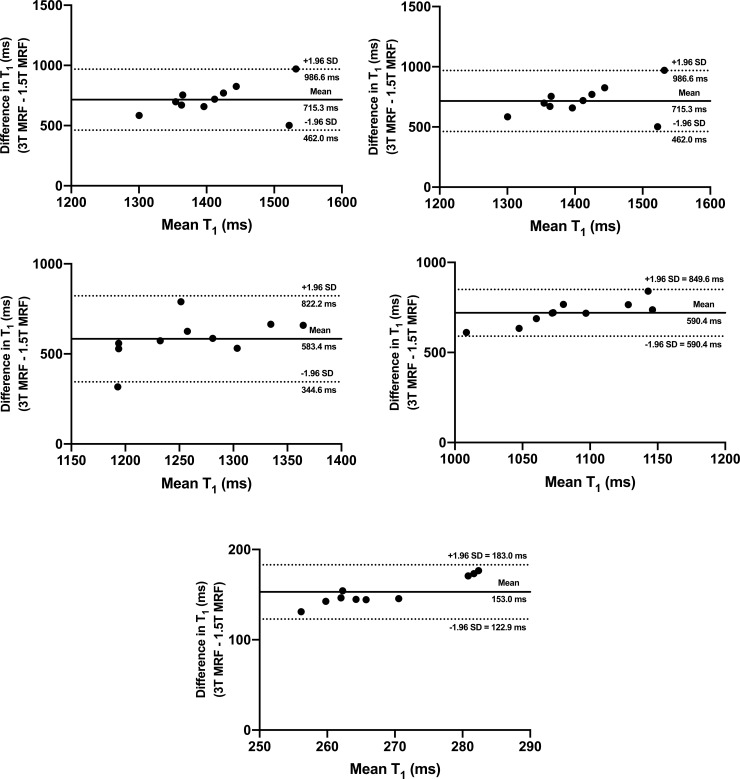
Albeit weaker than in the phantom experiment, a strong linear correlation was noted between the combined in vivo 1.5T and 3T MRF T1 values obtained from all tissues included in the analysis (R2 = 0.914; p < 0.0001); Fig 3A. Fig 3B illustrates the inter-scanner agreement between MRF T1 relaxation times (bias 602.1 ms, 95% LOA 72.0 ms to 1132.0 ms) with only a single value being outside the 95% LOA. The outcomes of the Bland-Altman analysis for each individual tissue are presented in Fig 4, with acceptable agreement noted for all tissues included in the analysis. S2 and S3 Figs depict the outcomes of a similar analysis for MRF T2 relaxometry.
Fig 3.
Linear regression (a) and Bland-Altman (b) plots comparing in vivo MRF T1 values obtained from all tissues combined at 1.5T and 3T systems. On Fig (b), dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
Fig 4.
Bland-Altman plots comparing in vivo MRF T1 values obtained from the whole prostate (a), peripheral zone (b), transition zone (c), internal obturator muscle (d) and fat in the ischioanal fossa (e) at 1.5T and 3T systems. Dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
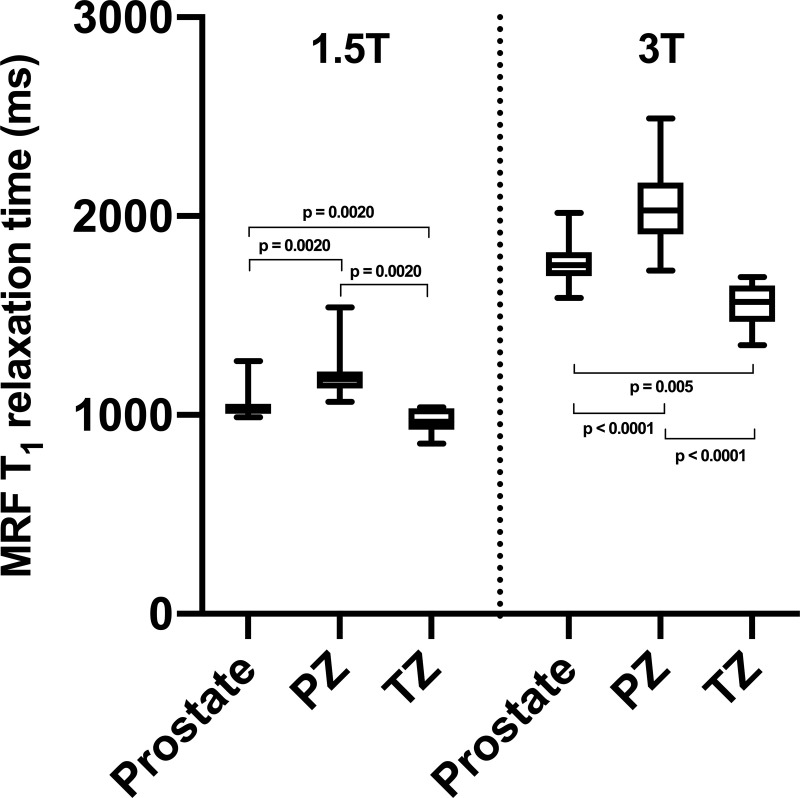
At both 1.5T and 3T, MRF T1 relaxometry enabled confident differentiation between the whole prostate, PZ and TZ (Fig 5). It is of note that the difference between MRF T1 relaxation times derived from PZ and TZ was slightly more prominent at 3T (24%) compared to 1.5T (19%). S3 Fig demonstrates the outcomes of a similar comparison for MRF T2 relaxometry.
Fig 5.
Box-and-whiskers plots representing MRF T1 relaxation times obtained from the whole prostate, peripheral zone (PZ) and transition zone (TZ) at 1.5T and 3T (left and right sides of the graph, respectively). Top and bottom of boxes represent 25th and 75th percentiles of data, respectively; line in boxes represents the median value and bars represent minimum and maximum values. MRF = magnetic resonance fingerprinting.
Discussion
This prospective proof-of-concept study investigated the cross-system reproducibility of SSFP MRF T1 of the prostate in healthy volunteers who were scanned using 1.5T and 3T systems with the same sequence parameters and reconstruction methods. The study was preceded by a phantom experiment that confirmed high reliability of MRF T1 mapping used when validated against IR-FSE imaging. The healthy volunteers study revealed acceptable inter-scanner agreement and excellent reproducibility of MRF T1 relaxometry in all tissues. At 3T, the magnitude of difference between MRF T1 obtained from the peripheral and transition zones of the prostate was slightly higher than at 1.5T, being, however, within the limits of statistical significance at both field strengths. These results, therefore, support further validation of prostate MRF at 1.5T as part of a standard-of-care mpMRI protocol.
To our knowledge, this is the first study to report MRF of the prostate performed at 1.5T, which is still commonly used for routine prostate imaging [26]. The observed high cross-system reproducibility of prostate MRF T1 is similar to that reported in the brain by Buonincontri et al., suggesting the overall robustness of MRF to field-dependent variation of T1 [30]. Our 3T MRF T1 values obtained from the healthy PZ and TZ are similar to those obtained in a patient population by other authors (2247.0 ms ± 450 ms for PZ [22] and 1800 ms ± 150 ms for TZ [20]) with subtle differences being expected due to the known age-related changes in histological and imaging features of the prostate [36, 37]. The observed ability of MRF to differentiate PZ and TZ in healthy volunteers at both 1.5T and 3T highlights MRF’s sensitivity to the underlying tissue organisation, which may be even more prominent in the older patients, in whom even visual differentiation of the prostatic zones is much less challenging [36, 38].
These preliminary results pave the way for future studies investigating the diagnostic performance of prostate MRF at 1.5T. It is known that prostate mpMRI at 1.5T is able to produce T2-weighted images of comparable diagnostic quality, however, the key argument in favour of 3T imaging is its ability to achieve higher signal-to-noise and contrast-to-noise ratios for diffusion-weighted imaging (DWI), which is the key sequence for PZ assessment, enabling DWI at higher b-values [39–42]. If 1.5T MRF retains its added value to ADC mapping previously demonstrated at 3T, it can potentially compensate for the inherent inferiority of DWI at 1.5T [20–22]. Furthermore, MRF’s inherent robustness to motion may help further establish its role as an auxiliary sequence to DWI, which is often degraded by motion artefact or rectal loading [43–46]. Finally, the added value of quantitative MRI can be harnessed clinically in the active surveillance setting where time-series change of tumour-derived relaxometry can serve as a more objective means of tracking the disease progression [47].
Our study has some limitations. The healthy volunteers study population was a small cohort of healthy volunteers of a younger age than typical patients with PCa. Although this may have introduced age-related heterogeneity, the reported high cross-system reproducibility of MRF is further complemented by its ability to differentiate normal PZ and TZ, which is considered less straightforward in younger adults [48]. Furthermore, the inclusion of older adults would limit our ability to sample healthy prostatic tissue due to high prevalence of benign conditions such as benign prostatic hyperplasia and prostatitis, or even undiagnosed PCa in otherwise healthy older men [49, 50]. To achieve comparable image quality, MRF at 1.5T was acquired with higher slice thickness and gap than at 3T, however, this mirrors standard clinical practice [51]. Having shown broadly similar results, MRF T2 mapping was unreliable in this study, and stronger consideration will be given in future studies to methods increasing its signal-to-noise ratio and improving the pattern matching algorithms.
In conclusion, the excellent reproducibility and high cross-system agreement of MRF T1 relaxometry of the healthy prostate observed in this preliminary study support the technique’s prospective clinical validation on 1.5T MRI systems. The reported findings could be used to justify further investigations for incorporating MRF into clinical prostate MRI protocols, larger population studies, and multi-site/multi-vendor investigations. If this method can be translated into clinical practice and improve both the speed and diagnostic value of imaging, it would add significant value given the current unprecedented demand on imaging services.
Supporting information
SD = standard deviation.
(DOCX)
Linear regression (a) and Bland-Altman (b) plots comparing in vivo MRF T2 values obtained from all tissues combined at 1.5T and 3T systems. On Fig (b), dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
(TIF)
Bland-Altman plots comparing in vivo MRF T2 values obtained from the whole prostate (a), peripheral zone (b), transition zone (c), internal obturator muscle (d) and fat in the ischioanal fossa (e) at 1.5T and 3T systems. Dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
(TIF)
Top and bottom of boxes represent 25th and 75th percentiles of data, respectively; line in boxes represents the median value and bars represent minimum and maximum values. MRF = magnetic resonance fingerprinting.
(TIF)
Data Availability
The data underlying this study are available on Mendeley (DOI: 10.17632/pgfrksc9z5.1).
Funding Statement
The authors acknowledge support from National Institute of Health Research Cambridge Biomedical Research Centre, Cancer Research UK (Cambridge Imaging Centre grant number C197/A16465), the Engineering and Physical Sciences Research Council Imaging Centre in Cambridge and Manchester, and the Cambridge Experimental Cancer Medicine Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.EAU Guidelines: Prostate Cancer | Uroweb. [cited 2 Jun 2020]. Available: https://uroweb.org/guideline/prostate-cancer/
- 2.Standard Operating Procedure for Multiparametric Magnetic Resonance Imaging in the Diagnosis, Staging and Management of Prostate Cancer—American Urological Association. [cited 2 Jun 2020]. Available: https://www.auanet.org/guidelines/mri-of-the-prostate-sop
- 3.Gaziev G, Wadhwa K, Barrett T, Koo BC, Gallagher FA, Serrao E, et al. Defining the learning curve for multiparametric magnetic resonance imaging (MRI) of the prostate using MRI-transrectal ultrasonography (TRUS) fusion-guided transperineal prostate biopsies as a validation tool. BJU Int. 2016;117: 80–86. 10.1111/bju.12892 [DOI] [PubMed] [Google Scholar]
- 4.Le JD, Tan N, Shkolyar E, Lu DY, Kwan L, Marks LS, et al. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: Correlation with whole-mount histopathology. Eur Urol. 2015;67: 569–576. 10.1016/j.eururo.2014.08.079 [DOI] [PubMed] [Google Scholar]
- 5.Greer MD, Shih JH, Lay N, Barrett T, Bittencourt L, Borofsky S, et al. Interreader variability of prostate imaging reporting and data system version 2 in detecting and assessing prostate cancer lesions at prostate MRI. Am J Roentgenol. 2019;212: 1197–1205. 10.2214/AJR.18.20536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenkrantz AB, Ginocchio LA, Cornfeld D, Froemming AT, Gupta RT, Turkbey B, et al. Interobserver reproducibility of the PI-RADS version 2 lexicon: A multicenter study of six experienced prostate radiologists. Radiology. 2016;280: 793–804. 10.1148/radiol.2016152542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Rooij M, Israël B, Tummers M, Ahmed HU, Barrett T, Giganti F, et al. ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: quality requirements for image acquisition, interpretation and radiologists’ training. Eur Radiol. 2020. [cited 25 Jun 2020]. 10.1007/s00330-020-06929-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, Padhani AR, Patel A, Ahmed HU, Allen C, Bardgett H, et al. Certification in reporting multiparametric magnetic resonance imaging of the prostate: recommendations of a UK consensus meeting. BJU Int. 2020 [cited 6 Jan 2021]. 10.1111/bju.15285 [DOI] [PubMed] [Google Scholar]
- 9.Schieda N, Lim CS, Zabihollahy F, Abreu‐Gomez J, Krishna S, Woo S, et al. Quantitative Prostate MRI. J Magn Reson Imaging. 2020; jmri.27191 10.1002/jmri.27191 [DOI] [PubMed] [Google Scholar]
- 10.Panda A, Gulani V. Quantitative Imaging of Prostate: Scope and Future Directions Reading MRI of the Prostate. Springer International Publishing; 2020. pp. 97–108. 10.1007/978-3-319-99357-7_10 [DOI] [Google Scholar]
- 11.Barrett T, Rajesh A, Rosenkrantz AB, Choyke PL, Turkbey B. PI-RADS version 2.1: one small step for prostate MRI Clinical Radiology. W.B. Saunders Ltd; 2019. pp. 841–852. 10.1016/j.crad.2019.05.019 [DOI] [PubMed] [Google Scholar]
- 12.Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2 European Urology. Elsevier B.V.; 2019. pp. 340–351. 10.1016/j.eururo.2019.02.033 [DOI] [PubMed] [Google Scholar]
- 13.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, et al. Magnetic resonance fingerprinting. Nature. 2013;495: 187–192. 10.1038/nature11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGivney DF, Boyacıoğlu R, Jiang Y, Poorman ME, Seiberlich N, Gulani V, et al. Magnetic resonance fingerprinting review part 2: Technique and directions. J Magn Reson Imaging. 2020;51: 993–1007. 10.1002/jmri.26877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bipin Mehta B, Coppo S, Frances McGivney D, Ian Hamilton J, Chen Y, Jiang Y, et al. Magnetic resonance fingerprinting: a technical review. Magn Reson Med. 2019;81: 25–46. 10.1002/mrm.27403 [DOI] [PubMed] [Google Scholar]
- 16.Poorman ME, Martin MN, Ma D, McGivney DF, Gulani V, Griswold MA, et al. Magnetic resonance fingerprinting Part 1: Potential uses, current challenges, and recommendations Journal of Magnetic Resonance Imaging. John Wiley and Sons Inc.; 2020. pp. 675–692. 10.1002/jmri.26836 [DOI] [PubMed] [Google Scholar]
- 17.Sushentsev N, Caglic I, Sala E, Shaida N, Slough RA, Carmo B, et al. The effect of capped biparametric magnetic resonance imaging slots on weekly prostate cancer imaging workload. Br J Radiol. 2020;93: 20190929 10.1259/bjr.20190929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Leest M, Israël B, Cornel EB, Zámecnik P, Schoots IG, van der Lelij H, et al. High Diagnostic Performance of Short Magnetic Resonance Imaging Protocols for Prostate Cancer Detection in Biopsy-naïve Men: The Next Step in Magnetic Resonance Imaging Accessibility. Eur Urol. 2019;76: 574–581. 10.1016/j.eururo.2019.05.029 [DOI] [PubMed] [Google Scholar]
- 19.Zawaideh JP, Sala E, Shaida N, Koo B, Warren AY, Carmisciano L, et al. Diagnostic accuracy of biparametric versus multiparametric prostate MRI: assessment of contrast benefit in clinical practice. Eur Radiol. 2020;30 10.1007/s00330-020-06782-0 [DOI] [PubMed] [Google Scholar]
- 20.Panda A, Obmann VC, Lo WC, Margevicius S, Jiang Y, Schluchter M, et al. MR fingerprinting and ADC mapping for characterization of lesions in the transition zone of the prostate gland. Radiology. 2019;292: 685–694. 10.1148/radiol.2019181705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu AC, Badve C, Ponsky LE, Pahwa S, Dastmalchian S, Rogers M, et al. Development of a Combined MR Fingerprinting and Diffusion Examination for Prostate Cancer. Radiology. 2017;283: 729–738. 10.1148/radiol.2017161599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panda A, O’connor G, Lo WC, Jiang Y, Margevicius S, Schluchter M, et al. Targeted Biopsy Validation of Peripheral Zone Prostate Cancer Characterization With Magnetic Resonance Fingerprinting and Diffusion Mapping. Invest Radiol. 2019;54: 485–493. 10.1097/RLI.0000000000000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sushentsev N, Kaggie JD, Buonincontri G, Schulte RF, Graves MJ, Gnanapragasam VJ, et al. The effect of gadolinium-based contrast agent administration on magnetic resonance fingerprinting-based T1 relaxometry in patients with prostate cancer. Sci Rep. 2020;10 10.1038/s41598-019-56089-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegde J V., Mulkern R V., Panych LP, Fennessy FM, Fedorov A, Maier SE, et al. Multiparametric MRI of prostate cancer: An update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer Journal of Magnetic Resonance Imaging. NIH Public Access; 2013. pp. 1035–1054. 10.1002/jmri.23860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnetic resonance imaging (MRI) equipment, operations and planning in the NHS Report from the Clinical Imaging Board. 2017. Available: www.ipem.ac.ukwww.sor.org [Google Scholar]
- 26.Davies C, Castle JT, Stalbow K, Haslam PJ. Prostate mpMRI in the UK: the state of the nation. Clin Radiol. 2019;74: 894.e11–894.e18. 10.1016/j.crad.2019.09.129 [DOI] [PubMed] [Google Scholar]
- 27.Brizmohun Appayya M, Adshead J, Ahmed HU, Allen C, Bainbridge A, Barrett T, et al. National implementation of multi-parametric magnetic resonance imaging for prostate cancer detection–recommendations from a UK consensus meeting BJU International. Blackwell Publishing Ltd; 2018. pp. 13–25. 10.1111/bju.14361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton JI, Jiang Y, Chen Y, Ma D, Lo WC, Griswold M, et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med. 2017;77: 1446–1458. 10.1002/mrm.26216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz G, Jaubert O, Botnar RM, Prieto C. Cardiac Magnetic Resonance Fingerprinting: Technical Developments and Initial Clinical Validation Current Cardiology Reports. Current Medicine Group LLC 1; 2019. pp. 1–10. 10.1007/s11886-019-1086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buonincontri G, Biagi L, Retico A, Cecchi P, Cosottini M, Gallagher FA, et al. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0 T. Neuroimage. 2019;195: 362–372. 10.1016/j.neuroimage.2019.03.047 [DOI] [PubMed] [Google Scholar]
- 31.Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, et al. T 1, T 2 Relaxation and Magnetization Transfer in Tissue at 3T. 2005. [cited 2 Jun 2020]. 10.1002/mrm.20605 [DOI] [PubMed] [Google Scholar]
- 32.De Bazelaire CMJ, Duhamel GD, Rofsky NM, Alsop DC. MR Imaging Relaxation Times of Abdominal and Pelvic Tissues Measured in Vivo at 3.0 T: Preliminary Results. Radiology. 2004;230: 652–659. 10.1148/radiol.2303021331 [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74: 1621–1631. 10.1002/mrm.25559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31: 1116–1128. 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- 35.Mason D. SU-E-T-33: Pydicom: An Open Source DICOM Library. Med Phys. 2011;38: 3493–3493. 10.1118/1.3611983 [DOI] [Google Scholar]
- 36.De Visschere PJL, Vral A, Perletti G, Pattyn E, Praet M, Magri V, et al. Multiparametric magnetic resonance imaging characteristics of normal, benign and malignant conditions in the prostate. Eur Radiol. 2017;27: 2095–2109. 10.1007/s00330-016-4479-z [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Tian WZ, Hu CH, Niu TL, Wang XL, Chen XY. Age-related changes of normal prostate: Evaluation by MR diffusion tensor imaging. Int J Clin Exp Med. 2015;8: 11220–11224. Available: www.ijcem.com/ [PMC free article] [PubMed] [Google Scholar]
- 38.Allen KS, Kressel HY, Arger PH, Pollack HM. Age-related changes of the prostate: Evaluation by MR imaging. Am J Roentgenol. 1989;152: 77–81. 10.2214/ajr.152.1.77 [DOI] [PubMed] [Google Scholar]
- 39.Van Nieuwenhove S, Saussez TP, Thiry S, Trefois P, Annet L, Michoux N, et al. Prospective comparison of a fast 1.5-T biparametric with the 3.0-T multiparametric ESUR magnetic resonance imaging protocol as a triage test for men at risk of prostate cancer. BJU Int. 2019;123: 411–420. 10.1111/bju.14538 [DOI] [PubMed] [Google Scholar]
- 40.Shah ZK, Elias SN, Abaza R, Zynger DL, DeRenne LA, Knopp M V., et al. Performance Comparison of 1.5-T Endorectal Coil MRI with 3.0-T Nonendorectal Coil MRI in Patients with Prostate Cancer. Acad Radiol. 2015;22: 467–474. 10.1016/j.acra.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullrich T, Quentin M, Oelers C, Dietzel F, Sawicki LM, Arsov C, et al. Magnetic resonance imaging of the prostate at 1.5 versus 3.0 T: A prospective comparison study of image quality. Eur J Radiol. 2017;90: 192–197. 10.1016/j.ejrad.2017.02.044 [DOI] [PubMed] [Google Scholar]
- 42.Engels RRM, Israël B, Padhani AR, Barentsz JO. Multiparametric Magnetic Resonance Imaging for the Detection of Clinically Significant Prostate Cancer: What Urologists Need to Know. Part 1: Acquisition. Eur Urol. 2020;77: 457–468. 10.1016/j.eururo.2019.09.021 [DOI] [PubMed] [Google Scholar]
- 43.Caglic I, Hansen NL, Slough RA, Patterson AJ, Barrett T. Evaluating the effect of rectal distension on prostate multiparametric MRI image quality. Eur J Radiol. 2017;90: 174–180. 10.1016/j.ejrad.2017.02.029 [DOI] [PubMed] [Google Scholar]
- 44.Slough RA, Caglic I, Hansen NL, Patterson AJ, Barrett T. Effect of hyoscine butylbromide on prostate multiparametric {MRI} anatomical and functional image quality. Clin Radiol. 2018;73: 216.e9—216.e14. 10.1016/j.crad.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 45.Hsieh JJL, Svalbe I. Magnetic resonance fingerprinting: from evolution to clinical applications. J Med Radiat Sci. 2020; jmrs.413 10.1002/jmrs.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett T, Lawrence EM, Priest AN, Warren AY, Gnanapragasam VJ, Gallagher FA, et al. Repeatability of diffusion-weighted MRI of the prostate using whole lesion ADC values, skew and histogram analysis. Eur J Radiol. 2019;110: 22–29. 10.1016/j.ejrad.2018.11.014 [DOI] [PubMed] [Google Scholar]
- 47.Caglic I, Sushentsev N, Gnanapragasam V, Sala E, Shaida N, Koo B, et al. MRI-derived PRECISE scores for predicting pathologically-confirmed radiological progression in prostate cancer patients on active surveillance. Eur Radiol 2020. 2020; 1–10. 10.1007/s00330-020-07336-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bura V, Caglic I, Snoj Z, Sushentsev N, Berghe AS, Priest AN, et al. MRI features of the normal prostatic peripheral zone: the relationship between age and signal heterogeneity on T2WI, DWI, and DCE sequences. Eur Radiol. 2021; 1–10. 10.1007/s00330-020-07108-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitzing YX, Prando A, Varol C, Karczmar GS, Maclean F, Oto A. Benign conditions that mimic prostate carcinoma: MR imaging features with histopathologic correlation. Radiographics. 2016;36: 162–175. 10.1148/rg.2016150030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell KJL, Del Mar C, Wright G, Dickinson J, Glasziou P. Prevalence of incidental prostate cancer: A systematic review of autopsy studies. Int J Cancer. 2015;137: 1749–1757. 10.1002/ijc.29538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burn PR, Freeman SJ, Andreou A, Burns-Cox N, Persad R, Barrett T. A multicentre assessment of prostate MRI quality and compliance with UK and international standards. Clin Radiol. 2019;74: 894.e19–894.e25. 10.1016/j.crad.2019.03.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SD = standard deviation.
(DOCX)
Linear regression (a) and Bland-Altman (b) plots comparing in vivo MRF T2 values obtained from all tissues combined at 1.5T and 3T systems. On Fig (b), dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
(TIF)
Bland-Altman plots comparing in vivo MRF T2 values obtained from the whole prostate (a), peripheral zone (b), transition zone (c), internal obturator muscle (d) and fat in the ischioanal fossa (e) at 1.5T and 3T systems. Dotted lines represent upper and lower 95% limits of agreement and bold lines represent the mean biases with appropriate captions included. MRF = magnetic resonance fingerprinting, SD = standard deviation.
(TIF)
Top and bottom of boxes represent 25th and 75th percentiles of data, respectively; line in boxes represents the median value and bars represent minimum and maximum values. MRF = magnetic resonance fingerprinting.
(TIF)
Data Availability Statement
The data underlying this study are available on Mendeley (DOI: 10.17632/pgfrksc9z5.1).