Summary
In both animals and plants, development involves anatomical modifications. In the root of Arabidopsis thaliana, maturation of the ground tissue (GT)—a tissue comprising all cells between epidermal and vascular ones—is a paradigmatic example of these modifications, as it generates an additional tissue layer, the middle cortex (MC).1, 2, 3, 4 In early post-embryonic phases, the Arabidopsis root GT is composed of one layer of endodermis and one of cortex. A second cortex layer, the MC, is generated by asymmetric cell divisions in about 80% of Arabidopsis primary roots, in a time window spanning from 7 to 14 days post-germination (dpg). The cell cycle regulator CYCLIN D6;1 (CYCD6;1) plays a central role in this process, as its accumulation in the endodermis triggers the formation of MC.5 The phytohormone gibberellin (GA) is a key regulator of the timing of MC formation, as alterations in its signaling and homeostasis result in precocious endodermal asymmetric cell divisions.3,6,7 However, little is known on how GAs are regulated during GT maturation. Here, we show that the HOMEODOMAIN LEUCINE ZIPPER III (HD-ZIPIII) transcription factor PHABULOSA (PHB) is a master regulator of MC formation, controlling the accumulation of CYCD6;1 in the endodermis in a cell non-autonomous manner. We show that PHB activates the GA catabolic gene GIBBERELLIN 2 OXIDASE 2 (GA2ox2) in the vascular tissue, thus regulating the stability of the DELLA protein GIBBERELLIN INSENSITIVE (GAI)—a GA signaling repressor—in the root and, hence, CYCD6;1 expression in the endodermis.
Keywords: homeodomain leucin zipper III, cyclin D6;1, CYCD6;1, middle cortex, gibberellin, ground tissue, root development, microRNA
Graphical Abstract
Highlights
-
•
PHB regulates cell non-autonomously the timing of MC formation
-
•
A time-dependent rise of PHB expression controls the CYCD6;1 switch in the GT
-
•
PHB regulates GAI stability modulating GA levels
-
•
PHB regulates root GA levels activating GA2ox2 expression in the vasculature
Bertolotti, Unterholzner et al. show that a time-dependent threshold of the transcription factor PHB governs the timing of middle cortex formation in the Arabidopsis root. PHB promotes CYCD6;1 expression in the endodermis cell non-autonomously by reducing gibberellin (GA) levels in the vascular tissue and, hence, stabilizing the GAs repressor GAI.
Results and Discussion
PHB and PHV Control MC Formation via the Regulation of the CYCLIN D6;1 (CYCD6;1) Expression Domain
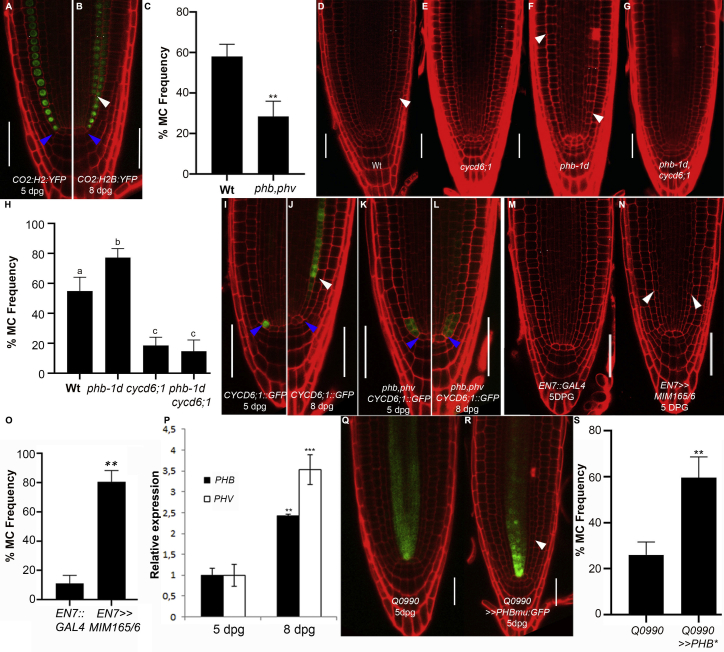
In Arabidopsis, expression of PHABULOSA (PHB) and of its redundant homologous PHAVOLUTA (PHV) is restricted to the vascular tissue due to the repressive activity of microRNA165 (miRNA165) and 166 in the ground tissue (GT).8, 9, 10 We have recently shown that miR165- and 166-resistant mutants of PHB and PHV (phb-1d and phv-1d, respectively), where PHB and PHV are present also in the GT, have supernumerary cortex formation already during early phase of root development, suggesting that these transcription factors regulate GT patterning.11 Because Arabidopsis plants acquire an additional cortical layer in late post-embryonic root development (Figures 1A and 1B), we assessed whether PHB and PHV control middle cortex (MC) formation analyzing MC development in phb,phv loss-of-function plants (phb-13, phv-11).12 Under our conditions, at 8 days post-germination (dpg), about 55% of wild-type (WT) plants start to develop MC, whereas only 25% of the phb,phv roots show a second cortical layer (Figure 1C),13 suggesting that PHB and PHV may control MC development.
Figure 1.
PHB Regulates MC Formation Cell Non-autonomously
(A and B) Confocal images of CO2::H2B:YFP at 5 (A) and 8 (B) dpg.
(C) Histogram depicting the percentage of plants showing MC formation in WT, phb-13, and phv-11 mutants at 8 dpg.
(D–G) Confocal images of 8 dpg old root meristems of WT (D), cycd6;1-1 (E), phb-1d (F), and phb-1d, cycd6;1-1 (G).
(H) Histogram reporting the percentage of MC formation in WT, phb-1d, cycd6;1-1, and phb-1d, cycd6;1-1. p < 0.005; ANOVA.
(I–L) Confocal images of CYCD6;1::GFP:GUS and of phb-13, phv-11, CYCD6;1::GFP:GUS at 5 (I and K) and 8 dpg (J and L).
(M and N) Confocal images of root meristems of EN7::GAL4 (M) and EN7>>MIM165/6 (N) at 5 dpg.
(O) Histogram depicting the percentage of MC formation in EN7::GAL4 and EN7>>MIM165/6 at 5 dpg.
(P) Relative expression of PHB and PHV in WT plants at 5 and 8 dpg. N = 3.
(Q and R) Confocal image of Q0990 and Q0990>>PHBmu:GFP root meristems at 5 dpg.
(S) Histogram depicting the percentage of MC formation in Q0990 and Q0990>>PHBmu:GFP at 5 dpg.
Scale bars, 50 μm; white arrowheads, MC; blue arrowheads, CEI. Student’s t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005 ). n = 20, N = 3. Different letters show statistical significance. Error bars: SD. See also Figure S1.
The CYCD6;1 gene is necessary for MC formation.5 We have recently shown that phb-1d roots have higher expression of CYCD6;1 in the GT.11 To assess whether PHB controls the number of cortical layers via CYCD6;1, we generated a phb-1d,cycd6;1-1 double mutant and analyzed GT development at 8 dpg. Only about 15% of phb-1d,cycd6;1-1 and cycd6;1-1 roots showed an additional cortical layer as compared to 75% of phb-1d (Figures 1D–1H), thus suggesting that PHB requires CYCD6;1 activity to promote MC formation.
CYCD6;1 shows a maximum of expression in the cortex/endodermis initial (CEI) and in its daughter cell (CEID) from embryogenesis up to 5 dpg, although subsequently, it is predominantly expressed in the endodermis. To assess whether PHB and PHV control this time-dependent variation in CYCD6;1 expression, we analyzed CYCD6;1 in WT and phb, phv plants harboring the CYCD6;1 promoter fused to the GREEN FLUORESCENT and GLUCURONIDASE genes (CYCD6;1::GFP:GUS). At 5 dpg, in WT roots, GFP signal is detectable in the CEI, CEID, endodermis, and cortex, although at 8 dpg, it is mostly present in the endodermis and in newly formed MC (Figures 1I and 1J). At the contrary, in phb,phv roots, the GFP signal is detectable in the CEI, CEID, and endodermis both at 5 dpg and at 8 dpg (Figures 1K and 1L).
Altogether, these data suggest that PHB and PHV regulate MC formation controlling the timing of CYCD6;1 expression. As PHB and PHV are both sufficient to promote cortex formation,11 we focused our studies on PHB.
miR165a, 166a, and 166b act from the endodermis to control PHB and PHV expression in the GT and in the vasculature.2,11 qRT-PCR on WT roots and GFP fluorescent signal of the transcriptional reporters of MIR165A and 166a (MIR165A::GFP and MIR166A::GFP) revealed that pre-miR165a and pre-mir166a decrease between 5 and 8 dpg (SD1). To understand whether this decrease results in precocious MC formation, we knocked down miR165 and 166 in the endodermis, expressing MIMICRY165/6 (MIM165/6)14 under the control of the ENDODERMIS7 (EN7) promoter, driving expression specifically in CEI, CEID, and endodermis.11,13 EN7>>MIM165/6 plants show early MC formation (Figures 1M–1O; SD1), suggesting that decreased levels of miR165/6 govern MC timing formation. PHB/PHV/miR165/166 module controls vasculature patterning other than MC formation, nonetheless neither phb,phv8 nor EN7>>MIM165/6 (SD1) show vascular defects, suggesting that the two events might be independent.
As miR165 and 166 levels decrease at 8 dpg, we thought that PHB expression might expand in the GT. Analysis of plants carrying a translational GFP reporter fusion (PHB-GFP) revealed that no PHB expression could be detected in the GT at 8 dpg (SD1), indicating that PHB might act cell non-autonomously from the vascular tissue to promote MC formation.
PHB is expressed in the vasculature already during embryogenesis but promotes MC formation only at 7 to 8 dpg.3 Because phb-1d mutants have higher levels of PHB mRNA than the WT and show precocious MC formation,15 we hypothesized that, in WT plants, PHB might increase between 5 and 8 dpg: indeed, qRT-PCR indicates that PHB level increases between 5 and 8 dpg (Figure 1P). To understand whether increased PHB expression in the vasculature is responsible for MC formation, we overexpressed a miRNA-insensitive version of PHB fused to the GFP (PHBmu:GFP) specifically in this domain from early stages of root development utilizing the GAL4/upstream activating sequence (UAS) transactivation system (Q0990,UAS::PHBmu:GFP).9 Interestingly, Q0990>>PHBmu:GFP roots show MC formation already at 5 dpg (Figures 1Q–1S), supporting the notion that increased PHB levels in the vasculature promote MC formation cell non-autonomously.
Because CYCD6;1 expression in the endodermis is a necessary requirement for MC formation, we hypothesized that PHB might promote CYCD6;1 expression in the endodermis cell non-autonomously from the vasculature. Thus, we generated Q0990>>PHBmu:GFP, CYCD6;1::GFP:GUS plants. Q0990>>PHBmu:GFP plants show CYCD6;1 expression in the endodermis already at 5 dpg, suggesting that an increase of PHB in the vasculature is sufficient to control the switch of CYCD6;1 expression from the CEI/D to the endodermis (SD1).
These results suggest that increased PHB expression in the vasculature is sufficient to control the timing of MC formation regulating the switch of CYCD6;1 expression from the CEI/D to the endodermis.
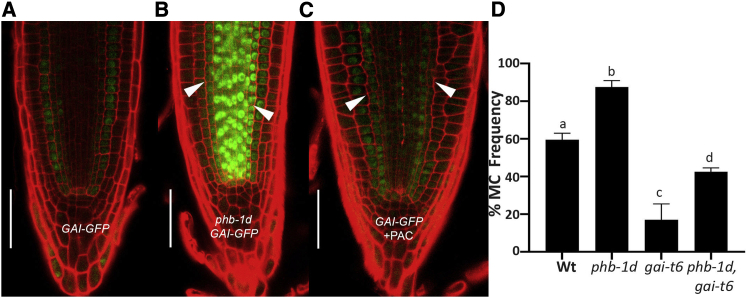
PHB Regulates GAI Stability
Gibberellins (GAs) are key regulators of MC timing formation; high level of GA activity, achieved through the degradation of the DELLA proteins GIBBERELLIC ACID INSENSITIVE (GAI) and REPRESSOR OF GAI (RGA), represses MC formation.3,16 To assess whether PHB regulates GAI and RGA, we analyzed the translational fusions GAI-GFP and RGA-GFP in phb-1d roots. This revealed that GAI is expressed at higher level and with an expanded domain in phb-1d compared to WT roots: in phb-1d roots, the signal is enhanced and present in both the GT and the vasculature, although in WT roots, GAI-GFP fluorescence was detectable only in the GT (Figures 2A and 2B). In contrast, RGA expression pattern in phb-1d remains unchanged (SD2).
Figure 2.
PHB Promotes GAI Stabilization
(A–C) Root meristems of GAI-GFP (A); phb-1d, GAI-GFP (B); and PAC-treated (50 μM; 24 h) GAI-GFP (C) at 5 dpg. Scale bars, 50 μm; white arrowheads, MC.
(D) Histogram reporting the percentage of MC formation in WT; phb-1d, gai-t6; and phb-1d, gai-t6. Student’s t test. Different letters show statistical significance.
n = 20, N = 3. Error bars: SD; p < 0.005; ANOVA. See also Figure S2.
GA activity is fine-tuned by a negative-feedback loop with DELLA proteins, such as GAI: high GA levels promote GAI degradation via the proteasome pathway, enabling the expression of GA-dependent genes; conversely, GAI represses the response to GA, inhibiting the activity of GA-dependent transcription factors.17, 18, 19 To understand whether PHB controls GAI transcription, we measured GAI mRNA level in phb-1d via qRT-PCR; GAI mRNA level does not vary in this background (SD2), suggesting that PHB controls GAI abundance at the protein level. Consistently with this, GAI-GFP plants treated with the GA biosynthesis inhibitor paclobutrazol (PAC) showed the GFP signal is present in both the root GT and the vasculature (Figures 2A–2C), similarly to phb-1d roots. To establish whether PHB regulates MC formation through the control of GA levels, we treated phb,phv mutants for 48 h with PAC. We observed that PAC treatment was sufficient to promote MC formation in phb,phv roots at 5 dpg (SD2), suggesting that the decreased MC formation in phb,phv is due to high GA levels. These results indicate that PHB promotes GAI protein stability via the control of GA levels.
To assess whether GAI is necessary to regulate MC development, we analyzed MC formation in the loss-of-function mutants gai-t6, gai-2, and gai-3: at 8 dpg, only about 20% of the roots from the three gai mutants show formation of MC (Figure 2C; SD3), suggesting that GAI is required for the correct development of the MC.
The control of GA homeostasis is required to regulate the timing of CYCD6;1 expression in the endodermis.16,20 Indeed, PAC treatment on CYCD6;1::GFP:GUS plants is sufficient to promote early expression of this gene in the endodermis at 5 dpg, causing a precocious MC formation (SD3).3,21 Similarly to PAC-treated plants, we observed that the gai-1 gain-of-function mutant—where GAI is insensitive to the GA-dependent degradation22—forms MC earlier and accumulates CYCD6;1::GFP:GUS signal in the endodermis already at 5 dpg (SD3), suggesting that the increase of GAI stability is sufficient to promote CYCD6;1 in the endodermis and, in turn, MC formation.
We then tested whether GAI activity mediates PHB-dependent regulation of MC formation. To this end, we generated gai-t6,phb-1d plants; loss of GAI partially rescues the phenotype of phb-1d mutants (Figure 2D), suggesting that PHB requires GAI activity to promote MC formation.
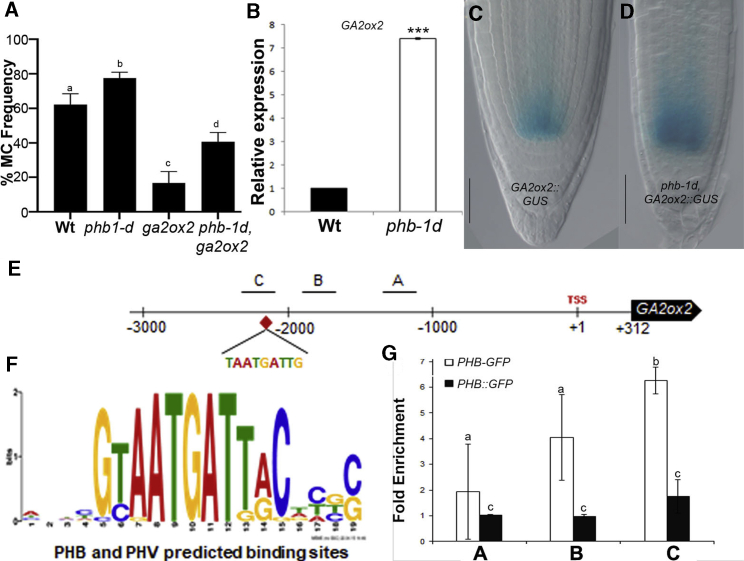
PHB Regulates GA Homeostasis via GA2ox2
GAI stability depends on GA levels, which in turn depend on the rate of GA catabolism and synthesis.23 GA synthesis is controlled by the GA3ox and Ga20ox enzymes, but neither of these genes is expressed in the root meristem.24,25 GA degradation depends on the activity of the GIBBERELLIN 2 OXIDASE (GA2ox) dioxygenases,26,27 among which GA2ox2 is expressed, as PHB, mostly in the vasculature (SD4).28 Thus, we hypothesized that PHB might promote GAI stability via the control of GA2ox2 expression.
We first found that GA2ox2 is required for MC development, as only 20% of ga2ox2-1 loss-of-function mutants29 show MC formation at 8 dpg (Figure 3A). Interestingly, in ga2ox2-1, a strong reduction of GA levels, due to 48-h PAC treatment, results in MC formation at 5 dpg (SD2). This suggests that ga2ox2-1 root phenotype is due to increased GA levels.
Figure 3.
PHB Directly Regulates GA2ox2 Expression
(A) Histogram depicting the percentage of MC formation in WT; phb-1d, ga2ox2-1; and phb-1d, ga2ox2-1 plants at 8 dpg. p < 0.005; ANOVA. Different letters show statistical significance.
(B) GA2ox2 relative expression in WT and phb-1d plants.
(C and D) Root meristems of GA2ox2::GUS (C) and phb-1d, GA2ox2::GUS (D) plants at 5 dpg. Scale bars, 50 μm.
(E and F) GA2ox2 promoter illustration. TSS indicates the transcriptional start site (+1) and the fragments used as probes for the ChIP experiment are marked with A (−1,186/−1,214 bp), B (−1,755/−1,909 bp), and C (−2,123/−2,271 bp). The red rhombus indicates the putative binding site of PHB and PHV, TAATGATTG (PlantPAN2.0) illustrated in (F).
(G) ChIP experiment using root meristems of PHB-GFP at 8 dpg. Fold enrichment of PHB-GFP on the indicated fragments A, B, and C was determined by qRT-PCR and calculated as ratio of anti-GFP IP to control beads immunoprecipitation (IP) of each independent replicate. UBQ10 was used for normalization.
(A and B) n = 20, N = 3; Student’s t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005). Error bars: SD. See also Figure S3.
To assess whether PHB promotes GA2ox2 expression, we analyzed GA2ox2 mRNA level in phb-1d via qRT-PCR and found higher levels of GA2ox2 compared to the WT (Figure 3B). Moreover, analysis of the transcriptional reporter GA2ox2::GUS showed that the GA2ox2 expression domain is wider in phb-1d than in the WT (Figures 3C and 3D).
To evaluate whether GA2ox2 is a PHB direct target, we first performed an in silico analysis30 that revealed a canonical HOMEODOMAIN LEUCINE ZIPPER III (HD-ZIPIII) recognition site in the GA2ox2 promoter (Figures 3E and 3F). Therefore, we performed a chromatin immunoprecipitation (ChIP) assay from 8-dpg-old PHB-GFP and PHB::GFP roots: ChIP-qPCR revealed that the fragment including the putative HD-ZIPIII site was enriched in the GFP-IP chromatin of PHB-GFP, but not in PHB::GFP, indicating that PHB-GFP binds directly to the GA2ox2 promoter (Figure 3G).
We then investigated whether PHB requires GA2ox2 activity to control MC formation, analyzing GT development in phb-1d,ga2ox2-1 double mutants. At 8 dpg, 75% of phb-1d roots, as opposed to 35% of phb-1d,ga2ox2-1 plants, show an additional cortical layer (Figure 3A), indicating that PHB requires GA2ox2 to promote MC formation.
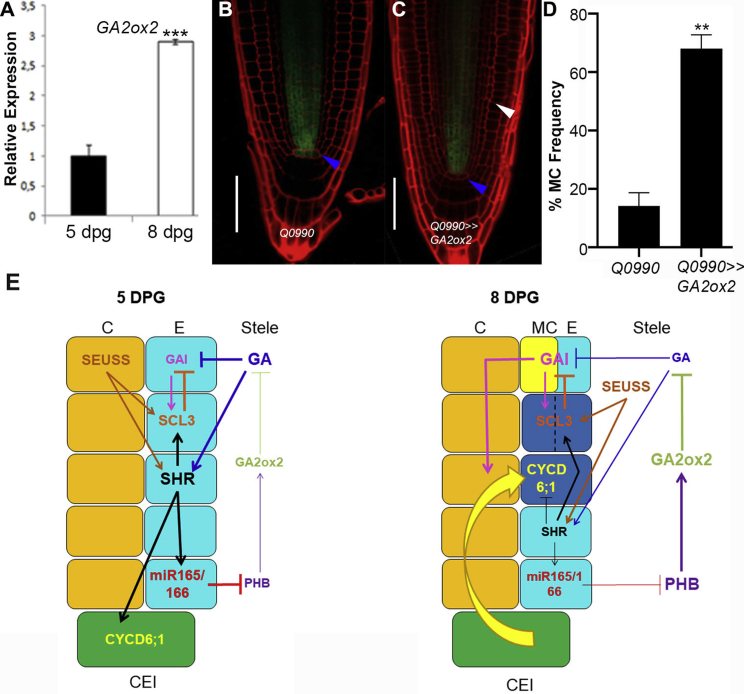
As PHB mRNA increases between 5 and 8 dpg, we wondered whether also GA2ox2 mRNA might increase in this time frame in a PHB- and PHV-dependent fashion: qRT-PCR showed that indeed it does, although the expression pattern does not change (Figure 4A; SD4). This suggests that, similarly to PHB, the increase in GA2ox2 expression in the vasculature may be sufficient to promote MC formation. To verify this possibility, we increased GA2ox2 levels in the vasculature during early stages of root development, generating Q0990,UAS::GA2ox2 plants (Q0990>>GA2ox2): roots of these plants show MC formation already at 5 dpg (Figures 4B–4D), confirming that an increased GA2ox2 expression in the vasculature is sufficient to promote MC formation.
Figure 4.
GA2ox2 Regulates MC Formation Cell Non-autonomously
(A) Relative expression of GA2ox2 in 5 and 8 dpg old WT plants. N = 3.
(B and C) Confocal images of meristems of Q0990 (B) and Q0990>>GA2ox2 plants at 5 dpg (C). Scale bars, 50 μm; white arrowhead indicates MC formative asymmetric division; blue arrowheads indicate the CEI.
(D) Histogram depicting the percentage of MC formation in Q0990 and Q0990>>Ga2ox2 at 5 dpg. n = 20, N = 3.
(A and D) n = 3; Student’s t test (∗∗p < 0.01; ∗∗∗p < 0.005). Error bars: SD.
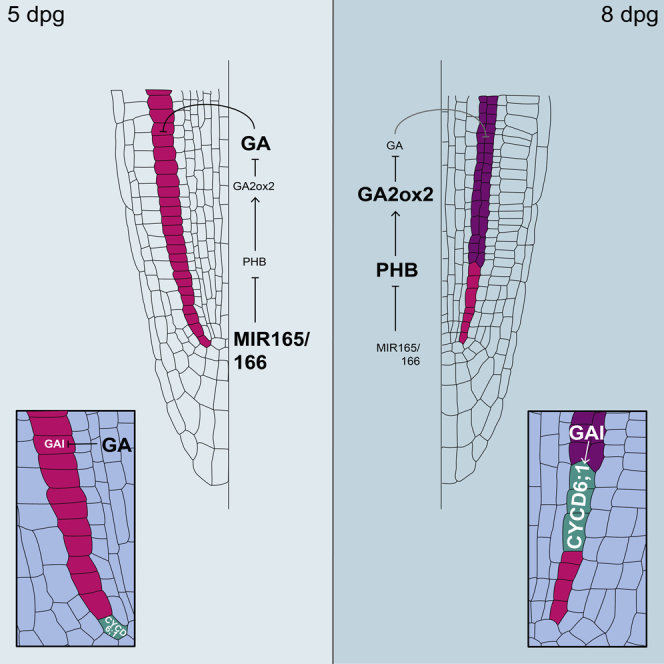
(E) Model: PHB levels increase between 5 and 8 dpg, resulting in increased GA2ox2 expression. Increased GA2ox2 levels promote the degradation of GAs in the vasculature, stabilizing GAI protein. GAI directs the accumulation of CYCD6;1 in the endodermis, promoting MC formation. Decrease levels of GAs after 5 dpg dampens SHR levels that regulate miR165 and 166 and SCL3 that in turn attenuate PHB expression and GAI activity, respectively. Orange, cortex (C); cyan, endodermis (E); yellow, middle cortex (MC); green, cortex/endodermis initial (CEI); blue, periclinally dividing cells (dashed line). Yellow arrow indicates the CYCD6;1 switch.
See also Figure S4.
Our data indicate that increase of PHB expression regulates the timing of MC formation by controlling GA homeostasis in the vasculature; PHB promotes GA2ox2 expression in this tissue, regulating GAs catabolism in the root and, hence, the timing of CYCD6;1 expression in the endodermis cell non-autonomously, and the GA2ox2-dependent decrease of GA level stabilizes GAI, thus promoting CYCD6;1 accumulation in the endodermis and, consequently, MC formation (Figure 4E). PHV might work similarly to PHB, as PHV mRNA increases between 5 and 8 dpg (Figure 1P).
Our data suggest that there might be a threshold of PHB/PHV levels resulting in a precise temporal regulation of GA levels to promote MC formation. SHORTROOT (SHR) and SCARECROW (SCR) promote miR165 and miR166 expression in the GT.8,9,31 GAs promote miR165 and miR166 expression and SHR accumulation in the endodermis, and GA activity decreases after 5 dpg.3,16,32,33 We propose that the decrease in miR165 and 166 levels after 5 dpg may depend on the reduction in GA activity, and hence SHR accumulation, causing an increase of PHB/PHV expression (Figure 4E). Consistently, GA treatments decrease PHB mRNA in WT roots (SD4).
GA activity in the root meristem depends on the coordinated action of SEUSS (SEU), SHR, SCR, and SCARECROWLIKE3 (SCL3).3,16,34,35 SEU induces SHR, SCR, and SCL3, and this latter is also a direct target of the SHR/SCR complex. SCL3 promotes GA signaling, dampening activity of DELLA proteins; GAs repress SCL3 expression, generating a negative feedback loop that fine-tunes GA activity in roots (Figure 4E).3,16,34 The PHB-dependent GA homeostasis control might be coordinated with the SEU/SHR/SCR/SCL3 pathway. Consistently, the scl3,phb,phv triple mutant resembles phb,phv mutant at 8 dpg, suggesting that PHB and PHV are epistatic to SCL3 (SD4). PHB and PHV might control SCL3 either via the GA-dependent regulation of SHR level in the endodermis or through GAI-dependent regulation of SCL3.21,36,37 Future studies will unravel how those two pathways integrate to mediate proper MC development.
The phb-1d,ga2ox2-1 double mutant shows a partially restored root WT phenotype, whereas phb-1d,cycd6;1-1 root resembles cycd6;1-1 ones. This suggests that PHB might act not only via the cell non-autonomous regulation of GA levels to promote CYCD6;1 expression in the endodermis9,11 but also cell-autonomously via some other yet unidentified mechanisms. The presence of two different mechanisms might be at the base of the interspecific variability in GT patterning; plants like Arabidopsis, where PHB expression is confined in the vasculature, acquire post-embryonically an additional cortical layer, whereas other species, such as Cardamine hirsuta, where PHB is expressed in the GT, show multiple cortical layers since embryogenesis.11,38 A PHB-dependent cell non-autonomous mechanism might be sufficient for species whose roots acquire MC only in late stages of development, whereas in roots of species having multiple cortical layers since embryogenesis, this mechanism could be combined with a cell-autonomous one.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli DH5α | N/A | N/A |
| Agrobacterium tumefaciens GV3101 | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Murashige & Skoog Medium | Duchefa | Cat# M0221 |
| MES hydrate | Duchefa | Cat# M1503 |
| Plant-agar | Duchefa | Cat# P1001 |
| Sucrose | Duchefa | Cat# S0809 |
| Kanamycin | Sigma-Aldrich | Cat# K1377 |
| Rifampicin | Duchefa | Cat# R0146 |
| Tetracycline | Duchefa | Cat# T0150 |
| Gentamicin | Duchefa | Cat# G0124 |
| Streptomycin | Duchefa | Cat# S0148 |
| Spectinomycin | Duchefa | Cat# S0188 |
| Phosphinothricin | Sigma-Aldrich | Cat# 77182-82-2 |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530S |
| trans-Zeatin | Sigma-Aldrich | Cat# Z0876 |
| Dexamethasone | Sigma-Aldrich | Cat# D4902 |
| Hpy188III | NEB | Cat# R0622S |
| Complete protease inhibitor cocktail | Roche | Cat# 11697498001 |
| x-GlcA | Duchefa | Cat# X1405.1000 |
| Dimethyl-sulfoxide | Sigma-Aldrich | Cat# 67-68-5 |
| Ethanol | Sigma-Aldrich | Cat# 64-17-5 |
| Na2HPO4 | Duchefa | Cat# 10028-24-7 |
| NaH2PO4 | Carlo Erba | Cat# 7558-80-7 |
| K3 Fe(CN)6 | Sigma-Aldrich | Cat# 13746-66-2 |
| K4Fe(CN)6 | Sigma-Aldrich | Cat# 14459-95-1 |
| Chloral Hydrate | Acros Organics | Cat# 302-17-0 |
| Glycerol | Sigma-Aldrich | Cat# 56-81-5 |
| Paclobutrazol | Duchefa | Cat# P0922.0500 |
| GA4+7 | Duchefa-Biochemie | Cat# G0938 |
| Glycine | BIORAD | Cat# 1610718 |
| Formaldehyde solution 37% | Sigma-Aldrich | Cat# 252549 |
| Miracloth | Merck-Millipore | Cat# 475855 |
| Hydrocloric acid 37% | Fisher scientific | Cat# 1298971 |
| Magnesium chloride | Carlo Erba | Cat# 459337 |
| DL-1,4-Dithiothreitol | Acros Organics | Cat# 327190100 |
| Triton X-100 | Acros Organics | Cat# 215680010 |
| Ethylenediaminetetraacetic acid disodium salt | Carlo Erba | Cat# 405497 |
| Sodium lauryl sulfate | Carlo Erba | Cat# P7600517 |
| Sodium chloride | Duchefa-Biochemie | Cat# S0520.5000 |
| GFP Trap_A beads | Chromotek | Cat# 141205001A |
| GFP-Trap Magnetic Agarose beads | Chromotek | Cat# 90312001MA |
| Tween 20 | Acros Organics | Cat# 233362500 |
| Proteinase K | Invitrogen | Cat# 1657252 |
| Tris Ultrapure | Duchefa-Biochemie | Cat# 010894.04 |
| Sodium acetate | Carlo Erba | Cat# 478137 |
| Basic Fuchsine | BioPlus | Cat#2177006-1 |
| Xylitol | Sigma-Aldrich | Cat# 87-99-0 |
| Sodium deoxycholate | Sigma-Aldrich | Cat# 302-95-4 |
| Urea | Acros Organics | Cat# 57-13-6 |
| Propidium Iodide | Sigma-Aldrich | Cat# MKBV0241V |
| Critical Commercial Assays | ||
| SensiFAST SYBR | Bioline | Cat# BIO-92005 |
| NucleoSpin RNA Plus | Macherey-Nagel | Cat# 740984 |
| qPCRBIO SyGreen Mix | PCR Biosystems | Cat# PB20.11-05 |
| Gel/PCR DNA Fragments Extraction Kit | Geneaid | Cat# DF100 |
| NucleoSpin Plasmid | Macherey-Nagel | Cat# 740588 |
| Gateway BP Clonase II | Thermo-Fisher | Cat# 11789 |
| Gateway LR Clonase II | Thermo-Fisher | Cat# 11791 |
| Superscript VILO cDNA Synthesis Kit | Thermo-Fisher | Cat# 11754 |
| Rneasy Micro Kit | QIAGEN | Cat# 74004 |
| MinElute Reaction Cleanup Kit (50) | QIAGEN | Cat# 28204b |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis: Col-0 | NASC | N/A |
| Arabidopsis: phb-13,phv-11 ER+ | This paper | N/A |
| Arabidopsis: cycd6-1 | NASC | SALK_021738 |
| Arabidopsis: CYCD6;1::GFP:GUS | 5 | N/A |
| Arabidopsis: UAS::PHBmu | 9 | N/A |
| Arabidopsis: Q0990 | NASC | N9217 |
| Arabidopsis: gai-t6 | 39 | N/A |
| Arabidopsis: gai-1ER+,CYCD6;1::GFP:GUS | This paper | N/A |
| Arabidoopsis: CO2::His2B:YFP | 13 | N/A |
| Arabidopsis: gai-2 | NASC | SAIL_587_C02 |
| Arabidopsis: gai-3 | NASC | SALK_208684 |
| Arabidopis: GAI-GFP | 40 | N/A |
| Arabidopsis: GA2ox2::GUS | 41 | N/A |
| Arabidopsis: UAS:GA2ox2 | This paper | N/A |
| Arabidopsis: UAS::MIM165/6 | This paper | N/A |
| Arabidopsis: EN7::GAL4 | This paper | N/A |
| Arabidopsis: phb-1d | 12 | N/A |
| Arabidopsis: ga2ox2-1 | NASC | SALK_051749 |
| Arabidopsis: RGA-GFP | 32 | N/A |
| Arabidopsis: phb-1d,RGA-GFP | This paper | N/A |
| Arabidopsis: phb-1d,GAI-GFP | This paper | N/A |
| Arabidopsis: PHB::GFP | 12 | N/A |
| Arabidopsis: PHB-GFP | 12 | N/A |
| Arabidopsis: scl3-1 | NASC | SALK_002516 |
| Arabidopsis: scl3-1,phb13,phv11 | This paper | N/A |
| Arabidopsis: shr-2 | 42 | CS2972 |
| Arabidopsis: MIR165A::GFP | 9 | N/A |
| Arabidopsis: MIR166A::GFP | 9 | N/A |
| Oligonucleotides | ||
| See STAR Methods section and Tables S1–S3 | N/A | N/A |
| Recombinant DNA | ||
| pB7m43GW | 43 | N/A |
| P4P1-UAS | 43 | N/A |
| P221-GA2OX2 | This paper | N/A |
| P2P3-NOS | 44 | N/A |
| pDONR221-MIM165/6 | This paper | N/A |
| pDONORP4P1-pEN7 | This paper | N/A |
| pDONOR221-GAL4 | 43 | N/A |
| Software and Algorithms | ||
| Excel | Microsoft | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
| GraphPad | https://www.graphpad.com/scientific-software/prism/ | N/A |
| PlantPAN2.0 | http://plantpan2.itps.ncku.edu.tw/ | N/A |
| Other | ||
| Zen 2010 | Zeiss | N/A |
| Fitotron SGC 120 Growth chamber | Weiss Technik, UK | N/A |
| Zeiss Axio Imager A2 | Zeiss | N/A |
| 7500 Fast Real-Time PCR system | Applied Biosystems | N/A |
| Branson Digital Sonifier 450 | Fisher Scientific | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raffaele Dello Ioio (raffaele.delloioio@uniroma1.it).
Materials Availability
Unique materials used in this study will be freely available.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Arabidopsis thaliana background lines Columbia-0 (Col-0) were used for experimentation, with mutants and transgenic lines in these backgrounds as detailed in the Key Resources Table.
Method Details
Plant Material and Growth Conditions
The Arabidopsis thaliana ecotypes Columbia-0 (Col-0) was used as controls as the gai-t6,45 gai-2, gai-3, phb-13, phv-11 ER+,12 phb-1d,12 cycd6;1-15, ga2ox2-1 and scl3-1 are in this background. gai-1 ER+, CYCD6;1::GFP:GUS was obtained by F3 populations of gai-1 ER+, CYCD6;1::GFP:GUS crosses. ER+ was selected by phenotype. scl3-1, gai-2, gai-3 and ga2ox2-1 mutants were obtained from the NASC collection (SALK_002516, SAIL_587_C02, SALK_208684 and SALK_051749 respectively). Homozygous mutants from the Salk T-DNA were identified by PCR as described (http://signal.salk.edu/tdnaprimers.2.html). gai-1 and gai-t6 mutants were genotyped as described in Dill and Sung.45 phb-13, phv-11 ER+ were obtained by crossing phb-13, phv-1146 with wild-type (Wt) Col-0 background. ER+ was selected by phenotype. Enhancer trap line Q0990 was obtained from the NASC. CO2::H2B:YFP, CYCD6;1::GFP:GUS, RGA-GFP and GAI-GFP transgenic plants have been described previously.5,13,40 phb-1d,RGA-GFP, phb1-d,GAI-GFP, phb1-d,GA2ox2::GUS, and phb-13,phv-11,ER+,CYCD6;1::GFP:GUS were obtained by crossing. UAS::PHBmu:GFP plants were obtained transforming the UAS::PHBmu:GFP plasmid9 in Wt Col-0 background via floral dip.47 For growth conditions, Arabidopsis seeds were surface sterilized, and seedlings were grown on 1/2 Murashige and Skoog (MS) medium containing 0.8% agar at 22°C in long-day conditions (16-h-light/8-h-dark cycle) as previously described.48
MC analysis and confocal imaging
For root MC analysis, root meristems of 5 and 8 days post germination (dpg) plants were analyzed utilizing a differential Interference Contrast (DIC) with Nomarski technology microscopy (Zeiss Axio Imager A2). Plants were mounted in a chloral hydrate solution (8:3:1 mixture of chloral hydrate:water:glycerol). Confocal images were obtained using a confocal laser scanning microscope (Zeiss LSM 780). For confocal laser scanning analysis, the cell wall was stained with 10 μM propidium iodide (Sigma-Aldrich). For vascular analysis Basic fuchsin (BioPlus) was combined with Clearsee as described in Ursache et al.49 For each experiments, a minimum of 20 roots for three biological replicates were analyzed.
Data reported in histograms represent the average of the three biological replicates. MC formation frequency is calculated as percentage of plants presenting periclinal divisions in the endodermis. Statistics has been calculated utilizing GraphPad Prism Version (https://www.graphpad.com/scientific-software/prism/).
Generation and Characterization of Transgenic Plants
Standard molecular biology techniques and the Gateway system (Invitrogen) were used for the cloning procedures. For the UAS::GA2ox2 transgenic plant, the genomic sequence of GA2ox2 (2450 bp) was amplified from genomic DNA of Arabidopsis Columbia-0 ecotype using specific primers (GA2ox2 FW 5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTGGATGGTGGTTTTGCCACAGC −3′, GA2ox2 REV 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTGTCATACAAGGGTTTTATGATTGAG −3′) and cloned in a pDONOR221 (pDONOR221-gGA2ox2). A LR reaction was then conducted by using the pDONORP4P1-UAS, pDONOR221-gGA2ox2 and a pDONORP2P3-NOS vector. The LR products were sub-cloned in the Gateway pBm43GW destination vector. Plasmids were transformed into Q0990 plants by floral dipping.44
For EN7>>MIM165/6 transgenic plant, UAS::MIM165/6 transcriptional fusion was obtained as follow: the sequence of MIM165/166 was amplified from vector generated by Todesco et al.48 using specific primers (MIM165FW 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGG CCGCAAAACACCACAAAAACA-3′, MIM165REV 5′-GGGGACCACTTTGTACAAGAAAGC TGGGTGAACTAGTGGATCCCCCATCACCAC-3′) and cloned in pDONR221 Gateway vector by BP recombination (Invitrogen). Subsequently pDONRP4P1-UAS, pDONR221-MIM165/6 and pDONR P2P3-NOS were recombined into a pB7m34GW destination vector via LR reaction (Invitrogen). To generate EN7::GAL4 construct, pDONORP4P1-pEN7 and pDONOR221-GAL4 were recombined with pDONOR P2P3-NOS into a pB7m34GW destination vector via LR reaction (Invitrogen). Plasmids were transformed into Col-0 plants by floral dipping.47 Then, EN7::GAL4 plants were crossed with UAS::MIM165/6 plants.
Drug treatments
3 dpg seedlings were transferred with tweezers onto solid 1/2 MS medium plates containing PAC (PACLOBUTRAZOL) (Duchefa) at a final concentration of 50 μM or GA4+7 (Gibberellin 4+7, Duchefa) at a final concentration of 100 μM for 24 and 48 hours depending on the experiment (see legend).
GUS histochemical assay
β-Glucuronidase activity of transgenic lines carrying the GUS enzyme was assayed essentially as described in Moubayidin et al.12 using the β-glucoronidase substrate X-GlcA, (5-Bromo-4-chloro-3-indolyl-β-D-glucuronic acid, Duchefa) dissolved in DMSO. X-GlcA solution: 100 mM Na2HPO4, 100 mM NaH2PO4, 0.5 mM K3 K3 Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100 and 1 mg/ml X-GlcA. Seedlings were incubated at 37°C in the dark for an appropriate time allowing tissue staining depending on the GUS line assayed. Imaging was done using the Axio Imager A2 (Zeiss) microscopy. For each line and time-point, at least 20 roots were analyzed and the percentages of phenotypes were evaluated.
RNA isolation, reverse-transcription and qRT-PCR
Total RNA was extracted from 5 dpg or 8 dpg old roots using the NucleoSpin RNA Plus (Macherey-Nagel). The cDNA was retro-transcribed using the SuperScript III First-Strand VILO cDNA Synthesis Kit (ThermoFisher Scientific). Quantitative RT-PCR (qRT-PCR) analysis were performed using the gene-specific primers listed in Table S2. All primers are given in the 5′-to-3′ direction. All the primers were tested for their qPCR efficiency of 2-fold amplifications per cycle by qRT-PCR with the Standard curve method. PCR amplifications were carried out using the SensiFast SYBR Lo-Rox (Bioline) mix. Amplification was monitored in real time with a 7500 Real Time PCR System (Applied Biosystems). Amplification of ORNITHINE TRANSCARBAMYLASE (OTC) and GLYCERALDEIDE-PHOSPHATE-DEHIDROGENASE (GAPDH) served as housekeeper controls. Data are expressed in 2-ΔΔct value. Three technical replicates of qRT-PCR were performed on two independent RNA batches. Results were comparable in all the experiments and with both housekeepers. Student’s t test was performed to assess the significance of the differences between each sample and the control sample. In figures are reported data normalized to OTC. In Figures 1P and 4A the normalization base is 5 dpg.
MIR165a/166a Fluorescence Quantification
The fluorescence value of MIR165A::GFP and MIR166A::GFP (Figure S1) was obtained as reported in Di Mambro and Sabatini.48 The plugin MeasureRGB of the software ImageJ (https://imagej.nih.gov/ij/) quantify the Σ of pixels of the channel (raw intensity density—RawIntDen). RawIntDen values of GFP channel of confocal microscope images were obtained taking into consideration the same area for 5 and 8 dpg of MIR165A::GFP and MIR166A::GFP, respectively, starting from the QC, keeping the same acquisition setting for both 5 and 8 dpg. Student’s t test was used to determine the statistical significance (https://graphpad.com:443/quickcalcs/ttest2.cfm) as reported in the relative figure legend.
ChIP-qPCR analysis
ChIP was conducted following modified protocols from Lawrence et al.50 and Kaufmann et al.51 on 2-3 biological replicates of PHB::GFP (Col-0), as control, and PHB-GFP (Col-0) roots at 8 dpg.
0.8-1.5g of roots were harvested in 50ml collection tubes and cooled on ice. Tubes were covered with Miracloth (Merk Millipore) and tissues were rinsed twice with 40ml of ddH2O. Plant material was fixated with 37ml of ddH2O and 1ml of 37% (w/v) formaldehyde on ice. Then, vacuum was applied for ten minutes. Vacuum was slowly released and material was mixed inverting the tubes gently. After five minutes vacuum was re-applied for another ten minutes. This step was repeat three times. To quench crosslinking, 2.5ml (1.25M stock) of glycine (Biorad) was added and vacuum was applied for five minutes. The vacuum was released slowly and plant material was rinsed twice with ddH2O. The plant material was dried between two tissue layers and quick-frozen in liquid nitrogen. Then plant material was ground to a fine powder and placed into a pre-cooled 50ml tube.
30 mL of ice-cold Extraction Buffer 1 (0.4M sucrose, 10 mM TRIS-HCl pH 8.0, 10 Mm MgCl2, 5 mM DDT, protease inhibitor cocktail) was added to the material and immediately vortexed until a homogeneous mixture was obtained. Tubes were kept on ice on 30 minutes. The solution was filtrated twice through Miracloth (Merk Millipore) and centrifuged for 15 minutes (4000 rpm, 4°C). The supernatant was gently discarded and the pellet was re-suspended in 1ml of Extraction Buffer 2 (0.25 sucrose, 10 mM TRIS-HCl pH 8.0, 10 Mm MgCl2, 0.15% Triton X-100, 5 mM DTT, protease inhibitor cocktail). Samples were centrifuged for 12 minutes (10000 rpm, 4°C). The pellet was re-suspended in 300 μl of Extraction buffer 2. 300 μl of Extraction Buffer 3 (1.7 M sucrose, 10 mM TRIS-HCl pH 8.0, 2 mM MgCl2, 0.15% Triton X-100, 5 mM DTT, protease inhibitor cocktail) was added and pellet was carefully layered upon it. Tubes were centrifuged for one hour (13000 rpm, 4°C) and the supernatant was carefully removed. This step permits the separation of two phases were nuclei are suspended in the pellet. The pellet was re-suspended in 500 μl of Nuclei Lysis Buffer (50 mM TRIS-HCl pH 8.0, 10 mM EDTA, 1% (v/v) SDS, protease inhibitor cocktail). Chromatin was sonicated with a probe sonicator (Brenson) for 3 cycles of 5 s of a 25% of power and 5 s of cooling between the pulses. Samples were cooled for 4 minutes and re-sonicated for 6 cycles for 5 s of 28% of power and 5 s of cooling between the pulses. Tubes were placed on ice the whole time. The sonication allows to obtain fragments of approximately 600 bp. The tubes were centrifuged for ten minutes (13000 rpm, 4°C) and the supernatant was transferred into new 2 mL safe lock tubes. This step was repeated and the supernatant (0.4 ml) was transferred to 15 mL falcon tubes containing 3.6 mL of ChIP Dilution Buffer (1% (v/v) Triton X-100, 1.2 mM EDTA, 16.7 mM TRIS-HCl (pH 8.0), 167 mM NaCl) (1:10 dilution). 120 μl of sample was set aside as input DNA control.
To preclear chromatin, first of all, 30 μl of blocked agarose beads (Chromoteck, Planegg-Martinsried, Germany) were suspended in 10 mL of ChIP Dilution Buffer ad mixed. The tubes were centrifuged for two minutes (2500 rpm, 4°C) and the supernatant was discarded. This wash step was repeated twice. At this point, chromatin was mixed with the blocked agarose beads and incubated at 4°C for one hour. In the meantime, 40 μl of GFP-trap agarose beads and 40 μl of blocked agarose beads (Chromotek, Planegg- Martinsried, Germany) were suspended, separately, in 1 mL of ChIP Dilution Buffer into new 2 mL safe lock tubes. The beads were centrifuged for three minutes (2500 rpm, 4°C) and supernatant was carefully discarded. This wash step was repeated twice. Afterward, chromatin was centrifuged for three minutes (2500 rpm, 4°C) and the supernatant was carefully transferred to a new pre-cooled 15 mL falcon tube, taking care to transfer any beads. This step was repeated and the supernatant was transferred to a new pre-cooled 15 mL tubes. At this point the resulting 4 mL of samples were divided into 2 mL aliquots: one aliquot was added to the GFP-trap agarose beads and the other to the blocked agarose beads, as a negative control. The samples were incubated for the IP at 4°C overnight on a rotating wheel.
Samples were centrifuged for three minutes (2500 rpm, 4°C) and the supernatant was removed. Then, 1 mL of Low salt Buffer (150 Mm NaCl, 0.1% SDS, 1% (v/v) Triton X-100, 2 mM EDTA, 20 mM TRIS-HCl pH 8.0) was added in the tubes and beads were incubated at 4°C on a rotating wheel for seven minutes and, then, centrifuged for three minutes at 2500 g at 4°C. This wash step was repeated utilizing, in order, the following buffer: High Salt Buffer (500 mM NaCl, 0.1% SDS, 1% (v/v) Triton X-100, 2 mM EDTA, 20 mM TRIS-HCl pH 8.0) and TE Buffer (10 mM TRIS-HCl pH 8, 1 mM EDTA). The TE Buffer wash was performed three times.
To elute the protein-DNA complex from the beads 100 μl of cold Elution Buffer (0.1 M glycine, 0.5 M NaCl, 0.05% Tween-20, pH was adjusted to 2.8) was added to the samples, which were immediately vortexed and incubated for one minutes at 37°C while shaking vigorously. Tubes were centrifuged for one minutes (13000 rpm, room temperature) and the supernatant (eluate) was transferred to a new safe lock 2 mL tubes, where 50 μl of TRIS-HCL (1 M stock, pH 9.0) was added to neutralize it. The elution step was repeated incubating tubes, with remaining protein-DNA complex, for two minutes at 37°C while shaking vigorously. Tubes were centrifuged for one minute (13000 rpm, room temperature) and supernatant were transferred to the eluate of the first elution. 50 μl of TRIS-HCl (1 M stock, pH 9.0) was added to neutralize. The elution step was repeated incubating tubes at 37°C while shaking vigorously and spinning for one minutes (13000 rpm, room temperature). The supernatant was combined with previously eluates. 50 μl of TRIS-HCl (1 M stock, pH 9.0) was added to neutralize, obtaining a final volume of 450 μl. The samples were centrifuged for two minutes (13000 rpm, room temperature) and the supernatant was transferred to new 2 mL safe lock tubes, taking care to disintegrate any pellet that may have been formed.
12.5 μl of proteinase K (18 mg/ml stock; final concentration should be 0.5 mg/ml) was added to the eluates, which were incubated at 37°C overnight to reverse crosslinking. A second aliquot of proteinase K (same amount) was added to the samples and the tubes were incubated at 65°C for six hours.
DNA was purified with the MinElute PCR purification kit (Quiagen, Venlo, NL). The total volume (472.5 μl) of the eluted DNA was split in two aliquots and each of them was mixed with 1181,25 μl of Binding Buffer PB, provided by the kit, and 30 μl of sodium acetate (3 M stock, pH 5.0). At this point, kit instructions were followed. The elution step was performed incubating for five minutes DNA with 25 μl of ddH2O. Then, DNA was centrifuged for 30 s (13000 rpm, room temperature). This step was repeated. The total volume (50 μl) was diluted with 50 μl of ddH2O to perform qRT-PCR analysis.
qRT-PCR was performed using the 7500 Real Time PCR System (Applied Biosystems). Primers (Table S3), spannig three region of GA2ox2 promoter, were tested for their qPCR efficiency of 2-fold amplifications per cycle by Standard curve method. PCR amplifications were carried out using the SensiFast SYBR Lo-Rox (Bioline) mix. Analysis was performed in triplicates from 2-3 independent chromatin immunoprecipitations. The fold enrichment of fragments was obtained as ratio of anti-GFP IP to control beads IP of each independent replicate. UBQ10 (UBQ10-F 5′-GGCCTTGTATAATCCCTGATGAATAAG-3′, UBQ10-R 5′- AAAGAGATAACAGGAACGGAAACATAGT −3′) was used for normalization. Primers are given in the 5′-to-3′ direction.
Seeds sterilization protocol for ChIP
In 50 mL falcon tubes seeds were mixed for five minutes with a solution composed by bleach 50% and Tween 10%. Seeds were centrifuged for one minute (410 g, room temperature) and supernatant was discarded. H2O was added to seeds. They were mixed for five minutes and centrifuged for one minute (410 g, room temperature). This wash step was repeated for other four times (five total washes).48 Seeds were dried under sterile flux and they were stratified with agarose 0.1% in darkness at 4°C for three days. Then, they were grown on solid 1/2 MS medium at 22°C in long-day conditions (16-h-light/8-h-dark cycle).
Quantification and Statistical Analysis
Statistical analysis was performed using GraphPad (https://www.graphpad.com/scientific-software/prism/). In all plots, error bars represent standard deviations (SD). The significance of the data was evaluated using the Student’s t test (∗p < 0,05, p ∗∗ < 0,01, p∗∗∗ < 0,005, NS Not Significant). For the statistical analysis of the MC frequency percentage was performed a one-way ANOVA analysis with post hoc Dunnet testing. Significantly different groups of samples are indicated using lower case letters.
GFP fluorescence signal intensity was measured and quantified with the ImageJ (https://imagej.nih.gov/ij/) software.
All experiments have been performed in at least three replications, using enough number of samples to ensure statistical significance.
Acknowledgments
We are grateful to M. Tsiantis, R. Heidstra, R. Sozzani, K. Nakajima, and N. Harbered for providing material. We thank M. Del Bianco for useful comments and I. Sbrocca, C. Mittelberger, M.M. Moretti, and E. Rattalino for technical help. This work was supported by a FIRB (Futuro in Ricerca 2013) project grant from the Ministero dell’Istruzione, dell’Università e della Ricerca (FIRB2013-RBFR13DCDS to R.D.I.), by a mobility grant from the Autonomous Province of Bozen/Bolzano (SENSE2GROW to S.J.U.), and a European Research Council grant (260368 to S.S.). We thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs.
Author Contributions
Conceptualization, R.D.I.; Methodology, G.B., S.J.U., V.R., D.S., E.S., N.S., F.L.S., R.D.M., and R.D.I.; Formal Analysis, G.B., S.J.U., V.R., D.S., E.S., N.S., R.D.M., P.V., and R.D.I.; Investigation, G.B., S.J.U., V.R., D.S., E.S., N.S., R.D.M., P.V., S.S., and R.D.I.; Data Curation, G.B., S.J.U., V.R., D.S., E.S., N.S., R.D.M., P.V., and R.D.I.; Writing – Original Draft Preparation, R.D.I.; Writing – Review & Editing, S.S., P.V., P.C., and R.D.I.; Supervision, R.D.I.; Project Administration, R.D.I.; Funding Acquisition, S.J.U., S.S., and R.D.I.
Declaration of Interests
The authors declare no competing interests.
Published: November 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.10.038.
Supplemental Information
References
- 1.Choi J.W., Lim J. Control of asymmetric cell divisions during root ground tissue maturation. Mol. Cells. 2016;39:524–529. doi: 10.14348/molcells.2016.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Ruocco G., Di Mambro R., Dello Ioio R. Building the differences: a case for the ground tissue patterning in plants. Proc. Biol. Sci. 2018;285:20181746. doi: 10.1098/rspb.2018.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paquette A.J., Benfey P.N. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum S.F., Dubrovsky J.G., Rost T.L. Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am. J. Bot. 2002;89:908–920. doi: 10.3732/ajb.89.6.908. [DOI] [PubMed] [Google Scholar]
- 5.Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A., Benfey P.N. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S.A., Jang S., Yoon E.K., Heo J.O., Chang K.S., Choi J.W., Dhar S., Kim G., Choe J.E., Heo J.B. Interplay between ABA and GA modulates the timing of asymmetric cell divisions in the Arabidopsis root ground tissue. Mol. Plant. 2016;9:870–884. doi: 10.1016/j.molp.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Cui H. Middle cortex formation in the root: an emerging picture of integrated regulatory mechanisms. Mol. Plant. 2016;9:771–773. doi: 10.1016/j.molp.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Carlsbecker A., Lee J.Y., Roberts C.J., Dettmer J., Lehesranta S., Zhou J., Lindgren O., Moreno-Risueno M.A., Vatén A., Thitamadee S. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyashima S., Koi S., Hashimoto T., Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 10.Di Mambro R., Sabatini S., Dello Ioio R. Patterning the axes: a lesson from the root. Plants. 2018;8:8. doi: 10.3390/plants8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Ruocco G., Bertolotti G., Pacifici E., Polverari L., Tsiantis M., Sabatini S., Costantino P., Dello Ioio R. Differential spatial distribution of miR165/6 determines variability in plant root anatomy. Development. 2018;145:dev153858. doi: 10.1242/dev.153858. [DOI] [PubMed] [Google Scholar]
- 12.Dello Ioio R., Galinha C., Fletcher A.G., Grigg S.P., Molnar A., Willemsen V., Scheres B., Sabatini S., Baulcombe D., Maini P.K., Tsiantis M. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr. Biol. 2012;22:1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Heidstra R., Welch D., Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., Chen X., Tang G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heo J.O., Chang K.S., Kim I.A., Lee M.H., Lee S.A., Song S.K., Lee M.M., Lim J. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA. 2011;108:2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacifici E., Polverari L., Sabatini S. Plant hormone cross-talk: the pivot of root growth. J. Exp. Bot. 2015;66:1113–1121. doi: 10.1093/jxb/eru534. [DOI] [PubMed] [Google Scholar]
- 18.Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverstone A.L., Ciampaglio C.N., Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koizumi K., Hayashi T., Wu S., Gallagher K.L. The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl. Acad. Sci. USA. 2012;109:13010–13015. doi: 10.1073/pnas.1205579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book. 2008;6:e0103. doi: 10.1199/tab.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plackett A.R., Powers S.J., Fernandez-Garcia N., Urbanova T., Takebayashi Y., Seo M., Jikumaru Y., Benlloch R., Nilsson O., Ruiz-Rivero O. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 26.Thomas S.G., Phillips A.L., Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 28.Li C., Zheng L., Wang X., Hu Z., Zheng Y., Chen Q., Hao X., Xiao X., Wang X., Wang G., Zhang Y. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 2019;285:1–13. doi: 10.1016/j.plantsci.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Torres-Garcia J., Latrasse D., Benhamed M., Schilderink S., Zhou W., Kulikova O., Hirt H., Bisseling T. Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number. Plant Cell. 2017;29:2183–2196. doi: 10.1105/tpc.17.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow C.N., Zheng H.Q., Wu N.Y., Chien C.H., Huang H.D., Lee T.Y., Chiang-Hsieh Y.F., Hou P.F., Yang T.Y., Chang W.C. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016;44(D1):D1154–D1160. doi: 10.1093/nar/gkv1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima K., Sena G., Nawy T., Benfey P.N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 32.Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 2010;20:1138–1143. doi: 10.1016/j.cub.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 33.Singh A., Roy S., Singh S., Das S.S., Gautam V., Yadav S., Kumar A., Singh A., Samantha S., Sarkar A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017;7:3408. doi: 10.1038/s41598-017-03632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong X., Flores-Vergara M.A., Hong J.H., Chu H., Lim J., Franks R.G., Liu Z., Xu J. SEUSS integrates gibberellin signaling with transcriptional inputs from the SHR-SCR-SCL3 module to regulate middle cortex formation in the Arabidopsis root. Plant Physiol. 2016;170:1675–1683. doi: 10.1104/pp.15.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark N.M., Fisher A.P., Berckmans B., Van den Broeck L., Nelson E.C., Nguyen T.T. Protein complex stoichiometry and expression dynamics of transcription factors modulate stem cell division. Proc. Natl. Acad. Sci. USA. 2020;117:15332–15342. doi: 10.1073/pnas.2002166117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koizumi K., Hayashi T., Gallagher K.L. SCARECROW reinforces SHORT-ROOT signaling and inhibits periclinal cell divisions in the ground tissue by maintaining SHR at high levels in the endodermis. Plant Signal. Behav. 2012;7:1573–1577. doi: 10.4161/psb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koizumi K., Gallagher K.L. Identification of SHRUBBY, a SHORT-ROOT and SCARECROW interacting protein that controls root growth and radial patterning. Development. 2013;140:1292–1300. doi: 10.1242/dev.090761. [DOI] [PubMed] [Google Scholar]
- 38.Hay A.S., Pieper B., Cooke E., Mandáková T., Cartolano M., Tattersall A.D., Ioio R.D., McGowan S.J., Barkoulas M., Galinha C. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J. 2014;78:1–15. doi: 10.1111/tpj.12447. [DOI] [PubMed] [Google Scholar]
- 39.Boccaccini A., Santopolo S., Capauto D., Lorrai R., Minutello E., Belcram K., Palauqui J.C., Costantino P., Vittorioso P. Independent and interactive effects of DOF affecting germination 1 (DAG1) and the Della proteins GA insensitive (GAI) and Repressor of ga1-3 (RGA) in embryo development and seed germination. BMC Plant Biol. 2014;14:200. doi: 10.1186/s12870-014-0200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleck B., Harberd N.P. Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J. 2002;32:935–947. doi: 10.1046/j.1365-313x.2002.01478.x. [DOI] [PubMed] [Google Scholar]
- 41.Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005;15:1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 42.Levesque M.P., Vernoux T., Busch W., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Lohmann J.U. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Mambro R., Svolacchia N., Dello Ioio R., Pierdonati E., Salvi E., Pedrazzini E., Vitale A., Perilli S., Sozzani R., Benfey P.N. The lateral root cap acts as an auxin sink that controls meristem size. Curr. Biol. 2019;29:1199–1205.e4. doi: 10.1016/j.cub.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 44.Pierdonati E., Unterholzner S.J., Salvi E., Svolacchia N., Bertolotti G., Dello Ioio R., Sabatini S., Di Mambro R. Cytokinin-dependent control of GH3 group II family genes in the Arabidopsis root. Plants. 2019;8:94. doi: 10.3390/plants8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dill A., Sun T. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 48.Di Mambro R., Sabatini S. Developmental analysis of Arabidopsis root meristem. Methods Mol. Biol. 2018;1761:33–45. doi: 10.1007/978-1-4939-7747-5_3. [DOI] [PubMed] [Google Scholar]
- 49.Ursache R., Andersen T.G., Marhavý P., Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 51.Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-seq) or hybridization to whole genome arrays (ChIP-CHIP) Nat. Protoc. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.