Abstract
The actin fold is found in cytoskeletal polymers, chaperones, and various metabolic enzymes. Many actin-fold proteins, like the carbohydrate kinases, do not polymerize. We found that Glk1, a Saccharomyces cerevisiae glucokinase, forms two-stranded filaments with unique ultrastructure, distinct from other cytoskeletal polymers. In cells, Glk1 polymerized upon sugar addition and depolymerized upon sugar withdrawal. Polymerization inhibits enzymatic activity; the Glk1 monomer-polymer equilibrium sets a maximum rate of glucose phosphorylation regardless of Glk1 concentration. Mutation eliminating Glk1 polymerization alleviated concentration-dependent enzyme inhibition. Yeast containing non-polymerizing Glk1 were less fit when growing on sugars and more likely to die when refed glucose. Glk1 polymerization arose independently from other actin-related filaments and may allow yeast to rapidly modulate glucokinase activity as nutrient availability changes.
One-sentence summary
Yeast glucokinase activity is limited by its polymerization, which is critical for cell viability during glucose refeeding.
The Actin ATPase clan (1) is a diverse group of structurally similar protein families found in all domains of life (2). Several of the Actin ATPase families form polymers, but the metabolic enzymes, such as hexose kinases, do not (3). It is unclear if polymerization evolved several times within this clan or if these polymerizing families descend from a single, ancient, polymerizing ancestor, a hypothesis suggested by phylogenetic (4) and structural studies (5).
Cells use several mechanisms to change enzyme activity in response to environmental changes, including allosteric and post-translational regulation. Enzymes can also change their physical state, assembling into filaments or gels, which serves as a sensitive, tunable way to control activity. Enzyme polymerization can regulate flux through pathways (6), store enzymes during starvation (7), and measure and signal cellular states (8).
Hexokinases and glucokinases of fungi are from a single family (the hexokinase family) within the Actin ATPase clan. Fungal glucokinases phosphorylate glucose, mannose, and glucosamine, while the fungal hexokinases also phosphorylate fructose. The glucokinases have a higher substrate affinity and lower Vmax than the hexokinases (9). S. cerevisiae has three hexokinase family proteins: a glucokinase (Glk1) and two hexokinases (Hxk1 and Hxk2). Hxk2 is expressed in glucose-rich environments and regulates the expression of the other two enzymes; Hxk1 and Glk1 repression is alleviated without glucose (10) (Fig. S1).
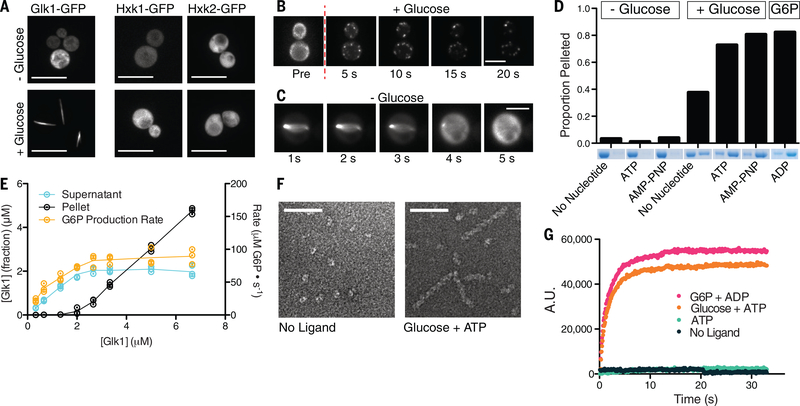
To probe the cell biology of these isozymes we made monomeric-superfolder-GFP (msfGFP) fusions to each at their native loci and examined their behavior in cells. Without glucose, Glk1-msfGFP was diffuse. When glucose-starved cells were refed glucose, Glk1-msfGFP formed filamentous structures, which rapidly disassembled upon glucose removal (Fig. 1A–C; Movies S1–2). Glk1-msfGFP also polymerized upon refeeding other Glk1 substrates: mannose or glucosamine (Fig. S2; S3). Hxk1-msfGFP and Hxk2-msfGFP did not oligomerize (Fig. 1A).
Fig 1. Glk1 forms filaments in response to its substrates at high enzyme concentration.
A) Fluorescence images of stationary phase Glk1-msfGFP (left), Hxk1-msfGFP (middle), and Hxk2-msfGFP (right) cells before (top) or after (bottom) glucose addition. Scale bars: 10 μm. B, C) Fluorescence images Glk1-msfGFP cells from a time-lapse video as glucose is added (top) or removed (bottom). Puncta in B eventually coalesce into a single, filamentous structure as in A and C. Scale bars: 5 μm. D) Purified Glk1 was ultracentrifuged at 436k x g for 30 min with different ligand combinations. The supernatant (left) and pellet (right) of each condition were subjected to SDS-PAGE. E) Purified Glk1 concentration was varied in saturating glucose and ATP and assayed for enzyme activity (glucose-6-phosphate production rate) or ultracentrifuged. The concentration of Glk1 in the supernatant and pellet was measured. N = 3. Means are connected by lines. F) Electron micrographs of negative-stained samples: 7.5 μM Glk1 in the absence of ligands (left) or in saturating glucose and ATP (right). Scale bar: 50 nm. G) 90-degree light scattering of 7.5 μM Glk1 mixed with different ligand combinations.
To understand what regulates Glk1 polymerization, we studied it in vitro. Other enzymes form condensates in low pH (7), however Glk1 did not (Fig. S4). Rather, Glk1 polymerized in the presence of its substrates (ATP and glucose, mannose, or glucosamine) or its products (ADP and sugar-6-phosphate). Modest polymerization also occurred with Glk1 inhibitors (Fig. 1D; S5; S6) (9). Although fructose and galactose induced Glk1 polymerization in vivo, they did not in vitro, suggesting in vivo polymers assemble when cells convert these sugars into glucose-6-phosphate (G6P) (Fig. S2; S3; S7) (11, 12).
Like actin, Glk1 exhibits a critical concentration (CC) for polymerization. Beneath 2 μM Glk1 there was no polymerization. Above 2 μM the concentration of Glk1 polymer increased, while unpolymerized Glk1 remained constant (Fig. 1E). This is consistent with the lack of polymers in fermenting cells where Glk1 expression is suppressed by glucose. Indeed, Glk1-msfGFP polymers were observed in glucose when Glk1-msfGFP was expressed from a strong promoter (Fig. S9). Consistent with the rapid polymerization observed in vivo, in vitro Glk1 polymerization reached steady state in a matter of seconds (Fig. 1G).
Polymerization can either activate or inhibit enzyme activity (6, 13), so we measured Glk1 activity as we varied Glk1’s concentration. Beneath Glk1’s CC, G6P production rate increased with Glk1. Above the CC the rate of product formation was constant (Fig. 1E). Thus, polymerization inhibits Glk1 activity, and the monomer-polymer equilibrium caps monomer concentration, thereby keeping net enzymatic activity constant.
We used electron microscopy of negative stained samples to examine Glk1 oligomers. Glk1 formed helical filaments in the presence of substrates (Fig. 1F). The micron-scale structures seen in vivo are likely driven by crowding (14), filament binding proteins (15, 16), or dimerization of the fluorescent tag (17). Similar polymers were observed when Glk1 was fused to other “monomeric” fluorescent proteins (Fig. S8).
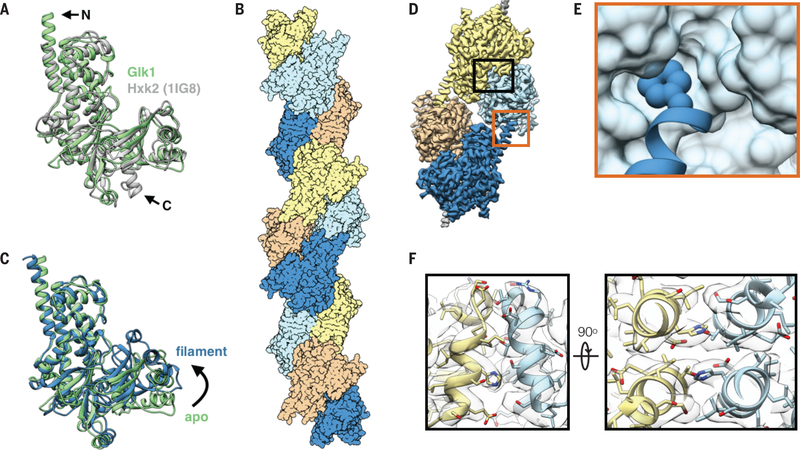
To investigate why Glk1 polymerizes and Hxk1 and Hxk2 do not, we solved the crystal structure of Glk1 (Table S1) (18). Comparing this to the S. cerevisiae Hxk2 structure (19) revealed differences in key regions: the N- and C-termini and in two loops (Fig. 2A; S10; S11). We then used cryo-electron microscopy to determine the structure of Glk1 filaments (Table S2) (20). This 3.8 Å resolution structure (Fig. 2B) revealed that Glk1 formed two-stranded, antiparallel filaments. Similar to other actin-like filaments (4), subunits were in the closed state and ligand bound (Fig. 2C; S13) (21), but unlike actin, did not flatten. Glk1 homologs alternate between open and closed states during their catalytic cycle (21), thus Glk1 inhibition likely arises from their inability to transition between states.
Fig 2. Glk1 forms anti-parallel, two-stranded filaments in its closed state.
A: Superimposition of Glk1 crystal structure (green) (residues 1–500) with Hxk2 (PDB ID: 1IG8) (19) (white) (residues 18–486). The N-terminal helix of Glk1 extends further than that of Hxk2 (arrow, N), while the C-terminal helix of Hxk2 extends further (arrow, C). B: Surface representation of a model of Glk1 filaments reconstructed from cryo-EM (3.8 Å resolution). Glk1 filaments are two-stranded, anti-parallel helices. Subunits along each strand are either orange/yellow or blue/cyan. C: Superimposition of the Glk1 crystal structure (green) with the Glk1 filament conformation from the cryo-EM reconstruction (blue). The crystal structure is not ligand bound and is in the open state while the filament form is ligand bound and in the closed state. D: Cryo-EM map of four subunits in the Glk1 filament colored by subunit. Longitudinal contact is boxed in orange, and a lateral contact is boxed in black. E: Closeup of longitudinal contact with Phe3 represented as van der Waals spheres and the next subunit represented as a surface model. Phe3 of one subunit inserts into the hydrophobic pocket near the next subunits C-terminus. F: Two orthogonal close-ups of lateral filament contact. The helix-loop-helix from residue 371–393 of one subunit (yellow) binds antiparallel to the same region of the adjacent subunit (blue). Cryo-EM map is transparent grey.
The interactions between Glk1 subunits along a strand differ from the conserved interactions in other Actin ATPase clan polymers (5) (Fig. S14). Along the strand, the N-terminal, solvent-exposed Phe3 of one subunit inserts into the hydrophobic pocket at the C-terminus of the next (Fig. 2D–E; S15). Lateral associations between strands are mediated by the helix-loop-helix between residues 371–393, which binds antiparallel to the same region on the adjacent subunit (Fig. 2F).
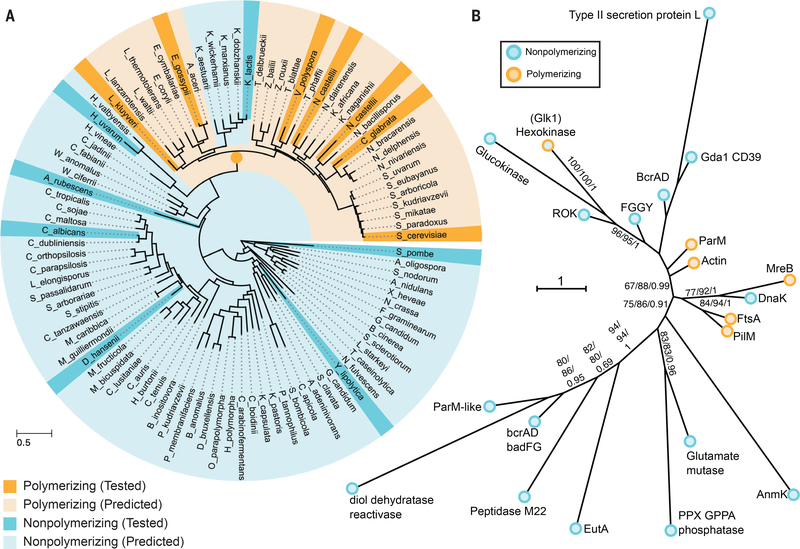
Phylogenetically, the fungal glucokinases and hexokinases form separate clades. The group of yeast containing S. cerevisiae (the Saccharomyceteceae) arose ~200 million years ago. The Glk1 homologs in most Saccharomyceteceae contain four conserved motifs missing in both the other Glk1 homologs and all Hxk1/2 homologs. These motifs are at or near filament contacts: the N- and C-termini, loop 230–243, and loop 438–444. To test if these motifs predict polymerization, we purified different Glk1and Hxk1/2 homologs and tested their ability to polymerize. Only Glk1 homologs that contained all four motifs polymerized (Fig. S10; S16). Phylogenetic logistic regression showed these motifs correlate significantly with polymerization (P=0.018). These results suggest that Glk1 polymerization arose ~200 million years ago. One ascomycete lineage, the Kluveromycetes, lost this ability (Fig. 3A) (22, 23).
Fig 3. Glucokinase polymerization evolved independently of other actin-related polymers.
A) Tree of ascomycetes as calculated by (23) indicating which species Glk1 homologs do (dark orange) and do not (dark cyan) polymerize. Species whose Glk1 homologs are predicted to polymerize based on conserved motifs (pale orange) and those predicted to not polymerize (pale cyan) are also indicated. Dark orange node marks the likely origin of Glk1 polymerization. B) Phylogeny of Actin ATPase families, summarizing phylogenetic analysis of 802 sequences from Actin ATPase protein families. A maximum likelihood tree was inferred under the LG+C20 substitution model in IQ-Tree (28). This displays the backbone structure of that ML tree with each family collapsed. Support values indicated are ultrafast bootstrap / SH-LRT / aBayes. Much of the backbone is uncertain; bootstrap supports shown when SH-LRT (middle value) > 70. This tree suggests the hexokinase family, which contains Glk1, forms a clade with ROK kinases and glucokinases, and is only distantly related to other polymer forming actin families. Families that do not polymerize are cyan while families that do polymerize are orange. The full phylogeny is available online.
The hexokinase family, which contains Glk1, segregates from the known polymer forming Actin ATPase families (Fig. 3B). This pattern is consistent with pairwise similarity between Hidden Markov Models of each family (Fig. S17; Table S3), and several lines of evidence suggest that Glk1 polymerization evolved independently of other actin-fold polymers: A) polymerizing Glk1 sequences form a subclade within the hexokinase family (Fig. 3A); B) that broader hexokinase family is monophyletic in the global actin-fold tree ( Fig. 3B, 100% bootstrap, 100% SH-aLRT, 1 aBayes); C) hexokinases robustly group with the non-polymerizing glucokinases and ROK kinases (96%, 95%, 1), and D) the monophyly of Glk1s with other polymerizing actins was rejected by an AU-test (P = 0.00106).
Next we examined how disrupting Glk1’s polymerization affected enzymatic activity and cell physiology. To create non-polymerizing Glk1 (NonPol-Glk1), we mutated the N-terminal phenylalanine involved in inter-subunit contacts to serine (Glk1-F3S). This mutation eliminated polymerization both in vitro, and in vivo (Fig. S18A–B). NonPol-Glk1 was enzymatically active but lacked the concentration-dependent inhibition of wild-type Glk1 (Fig. S18C).
To distinguish between the cellular effects of a lack of inhibition and the absence of polymers, we mutated the catalytic lysine (K182A) (Fig. S13B) (24) to create catalytically dead Glk1 (CatDead-Glk1). We combined these mutations (F3S/K182A) to create non-polymerizing, catalytically dead Glk1 (NonPolCatDead-Glk1). CatDead-Glk1 formed polymers in vivo and in vitro but did not generate G6P. NonPolCatDead-Glk1 neither polymerized nor produced G6P (Fig. S18A–C).
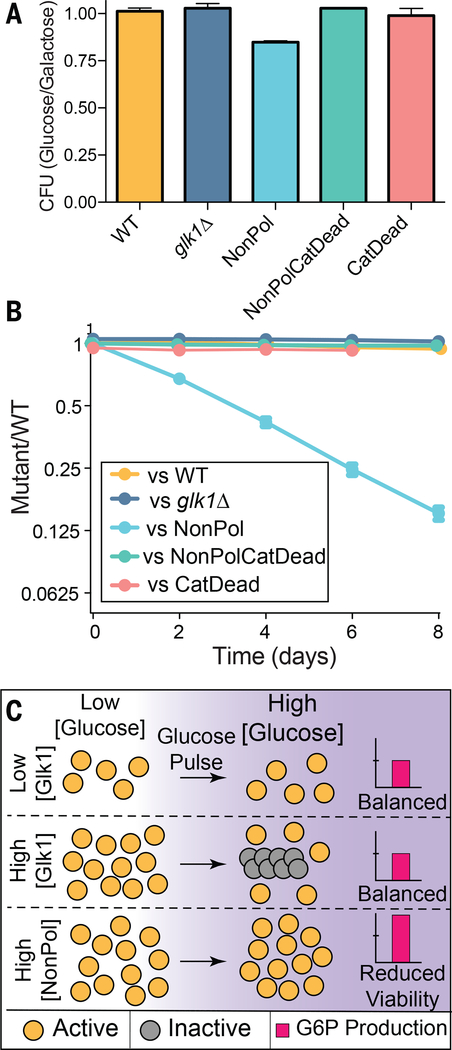
When starved yeast are refed glucose, excess sugar kinase activity is toxic due to an imbalance between the early steps of glycolysis, which consume ATP, and the late steps, which generate ATP (25). Hxk1 and Hxk2 activity is inhibited by trehalose-6-phosphate, a transiently-accumulating metabolite (26). Because Glk1 is not inhibited by trehalose-6-phosphate, we hypothesized that Glk1 polymerization limits activity when starved cells are refed glucose. Consistent with this model, when cells grown in galactose were refed glucose, 15% of NonPol-Glk1 cells die (Fig. 4A). This death was caused by unregulated Glk1 activity and not lack of Glk1 polymers: glk1Δ, CatDead-Glk1, or NonPolCatDead-Glk1 behaved indistinguishably from wild-type cells. Thus, Glk1 polymerization limits the rate of glucose phosphorylation during glucose refeeding.
Fig 4. Elimination of Glk1 polymerization reduces fitness.
A) Cells were pre-conditioned in citrate buffered synthetic (CBS) medium with galactose and refed either glucose or galactose. The ratio of the resulting colonies are reported here. Mean +/− SD (N = 4). B) Wild-type cells expressing mCherry were competed against cells expressing GFP with different Glk1 genotypes through growth and dilution cycles in synthetic medium with glucose. The proportion of strains was measured after dilution by flow cytometry. Mean +/− SD (N = 5). C) Schematic of how Glk1 polymerization affects glucokinase activity. When Glk1 concentration is high and glucose increases, Glk1 polymerizes until the monomer concentration equals the CC. Glk1 polymers lack enzyme activity: regardless of Glk1 concentration, the concentration of active enzyme is the same after glucose addition. When Glk1’s ability to polymerize is disrupted, it’s glucokinase activity is unconstrained, leading to fitness and viability defects.
Unregulated Glk1 activity is detrimental to fitness over the entire growth cycle. We used differential fluorescent labeling to compare the fitness of wild-type cells against each mutant. We grew mixed cultures to saturation, diluted them into fresh medium every 48 hours, and measured the proportion of strains by flow cytometry. NonPol-Glk1 cells had a substantial fitness defect in these conditions, averaging to a fitness cost of 6% (Fig. 4B) (27). In contrast, no growth defect was observed when mixed cultures were maintained in a glucose-rich environment (Fig. S19), suggesting that environmental changes are required to expose the growth defects of NonPol-Glk1 cells. Similar effects were observed competing these strains on other sugars (Fig. S20). Cells lacking Glk1 activity (glk1Δ, CatDead-Glk1, NonPolCatDead-Glk1) showed minor growth defects in acetate. Thus, Glk1 activity is important for growth on non-sugar sources and Glk1 polymerization prevents toxic overactivity during refeeding.
Glk1 polymerization governs its bulk rate of catalysis, with the Glk1 CC setting the upper limit of flux through the entire Glk1 pool (Fig. 4C). This mode of self-regulation is robust to growth state and cell-to-cell variation in protein level and it allows rapid adaptation to transient perturbations. In this sense, Glk1 polymerization behaves as a molecular surge protector, protecting the cell against nutrient spikes.
Supplementary Material
Acknowledgments:
Special thanks to Quincey Justman for her gift of strains and extended discussions of this work. We thank Rachelle Gaudet for sharing beamtime and resources and for crystallographic advice, along with Christina Zimanyi, Lukas Bane, and Elizabeth May. We thank Jim Wilhelm for helpful discussions. We thank the Arnold and Mabel Beckman Cryo-EM Center at the University of Washington for access to electron microscopes. This work used NE-CAT beamlines (GM124165) at the APS (DE-AC02-06CH11357).
Funding: This work was supported by NIH grants DP2AI117923-01 to ECG, R01GM043987 to AWM, R01GM118396 to JMK, R01GM123089 to FD, F31GM116441 to PRS, and the NSF-Simons Center for Mathematical & Statistical Analysis of Biology at Harvard (#1764269) and the Harvard Quantitative Biology Initiative. ECG and PRS were also supported by Wellcome grant 203276/Z/16/Z, and support from the Volkswagen Foundation. TAW is supported by a Royal Society University Research Fellowship.
Footnotes
Competing Interests
The authors have no competing interests.
Data and Materials Availability
Atomic coordinate files of the Glk1 crystal structure (PDB ID 6P4X) and the cryoEM filament structure (PBD ID 6P4X) are hosted online at the RCSB PDB (www.rcsb.org). The EM map of the Glk1 cryoEM filament structure is hosted online at the EM Data Resource (https://www.emdataresource.org/) under the accession number EMD-20309. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.El-Gebali S et al. , The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickstead B, Gull K, The evolution of the cytoskeleton. J. Cell Biol 194, 513–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bork P, Sander C, Valencia A, An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A 89, 7290–7294 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoddard PR, Williams TA, Garner E, Baum B, Evolution of polymer formation within the actin superfamily. Mol. Biol. Cell 28, 2461–2469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J, Bacterial actin MreB forms antiparallel double filaments. Elife. 3, e02634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry RM et al. , Large-scale filament formation inhibits the activity of CTP synthetase. Elife. 3, e03638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrovska I et al. , Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife. 3, 6063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzmann TM et al. , Phase separation of a yeast prion protein promotes cellular fitness. Science. 359, eaao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Maitra PK, A glucokinase from Saccharomyces cerevisiae. J. Biol. Chem 245, 2423–2431 (1970). [PubMed] [Google Scholar]
- 10.Rodríguez A, De La Cera T, Herrero P, Moreno F, The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J 355, 625–631 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey PA, The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10, 461–470 (1996). [PubMed] [Google Scholar]
- 12.Green JB, Wright AP, Cheung WY, Lancashire WE, Hartley BS, The structure and regulation of phosphoglucose isomerase in Saccharomyces cerevisiae. Mol. Gen. Genet 215, 100–106 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Lynch EM et al. , Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol 24, 507–514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilitski F et al. , Measuring cohesion between macromolecular filaments one pair at a time: depletion-induced microtubule bundling. Phys. Rev. Lett 114, 138102 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Griffith LM, Pollard TD, Cross-linking of actin filament networks by self-association and actin-binding macromolecules. J. Biol. Chem 257, 9135–9142 (1982). [PubMed] [Google Scholar]
- 16.Takatsuki H, Bengtsson E, Månsson A, Persistence length of fascin-cross-linked actin filament bundles in solution and the in vitro motility assay. Biochim. Biophys. Acta 1840, 1933–1942 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J, Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9, 480–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoddard PR, Garner EC, Murray AW, PDBID: 6P4X, Crystal Structure of the S. cereviseae glucokinase, Glk1. [Google Scholar]
- 19.Kuser PR, Krauchenco S, Antunes OA, Polikarpov I, The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem 275, 20814–20821 (2000). [DOI] [PubMed] [Google Scholar]
- 20.PDBID: 6PDT.
- 21.Kuettner EB et al. , Crystal structure of hexokinase KlHxk1 of Kluyveromyces lactis: a molecular basis for understanding the control of yeast hexokinase functions via covalent modification and oligomerization. J. Biol. Chem 285, 41019–41033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagman A, Säll T, Piskur J, Analysis of the yeast short-term Crabtree effect and its origin. FEBS J. 281, 4805–4814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X-X et al. , Reconstructing the Backbone of the Saccharomycotina Yeast Phylogeny Using Genome-Scale Data. G3 (Bethesda) 6, 3927–3939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J et al. , Lys169 of human glucokinase is a determinant for glucose phosphorylation: implication for the atomic mechanism of glucokinase catalysis. PLoS ONE. 4, e6304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Heerden JH et al. , Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science. 343, 1245114–1245114 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Blázquez MA, Lagunas R, Gancedo C, Gancedo JM, Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 329, 51–54 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Duveau F et al. , Fitness effects of altering gene expression noise in Saccharomyces cerevisiae. Elife. 7, 173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ, IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchuk NA et al. , Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32, e19–19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiestl RH, Gietz RD, High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet 16, 339–346 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Malakhov MP et al. , SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5, 75–86 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Gibson DG, Enzymatic assembly of overlapping DNA fragments. Meth. Enzymol 498, 349–361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ et al. , Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K, Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SHW, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Song Y et al. , High-resolution comparative modeling with RosettaCM. Structure. 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frenz B, Walls AC, Egelman EH, Veesler D, DiMaio F, RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods 14, 797–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RY-R et al. , Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife. 5, 352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D, Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boratyn GM et al. , BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chojnacki S, Cowley A, Lee J, Foix A, Lopez R, Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddy SR, Accelerated Profile HMM Searches. PLoS Comput. Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinegger M et al. , HH-suite3 for fast remote homology detection and deep protein annotation. bioRxiv, 560029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, Godzik A, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T, trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS, UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu A, Romero R, Petty HR, An enzymatic colorimetric assay for glucose-6-phosphate. Anal. Biochem 419, 266–270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnston M, Flick JS, Pexton T, Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol 14, 3834–3841 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bharat TAM, Murshudov GN, Sachse C, Löwe J, Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 523, 106–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Behrmann E et al. , Structure of the rigor actin-tropomyosin-myosin complex. Cell. 150, 327–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Löwe J, He S, Scheres SHW, Savva CG, X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization. Proc. Natl. Acad. Sci. U.S.A 113, 13396–13401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinate files of the Glk1 crystal structure (PDB ID 6P4X) and the cryoEM filament structure (PBD ID 6P4X) are hosted online at the RCSB PDB (www.rcsb.org). The EM map of the Glk1 cryoEM filament structure is hosted online at the EM Data Resource (https://www.emdataresource.org/) under the accession number EMD-20309. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.