Abstract
Background
Exposure to viral or bacterial pathogens increases the number of neutrophils with a relative decrease in lymphocytes, leading to elevated neutrophil to lymphocyte ratio (NLR). This study aimed to investigate whether differences in NLR among real-time polymerase chain reaction (PCR)-positive and -negative patients presenting with a prediagnosis of COVID-19 pneumonia could be useful in the differential diagnosis.
Methods
The study included 174 patients admitted because of suspected COVID-19 infection between March and April 2020. Patients were divided into two groups: PCR-negative and PCR-positive. Hemogram, NLR, urea, creatinine, aspartate aminotransferase, alanine aminotransferase, bilirubin, ferritin, D-dimer, C-reactive protein, procalcitonin, troponin, and coagulation parameters were analyzed.
Results
On comparison of laboratory parameters between both groups at presentation, PCR-positive patients were significantly more likely to have leukopenia (p < 0.001), thrombocytopenia (p = 0.006), neutropenia (p < 0.001), lymphopenia (p = 0.001), and increased NLR (p = 0.003). Furthermore, PCR-positive patients showed significant elevations of ferritin (p = 0.012) and procalcitonin (p = 0.038) and significant lower potassium levels (p = 0.05).
Conclusion
COVID-19 pneumonia has become a major global health problem. Early diagnosis and treatment of these patients are crucial, as COVID-19 pneumonia shows a rapid progression in most cases. Thus, leukopenia, thrombocytopenia, elevated NLR, and elevated ferritin may be useful as supplementary diagnostic tests in these patients, which may allow early initiation of treatment and may contribute to preventing progression in patients with abnormal results.
Keywords: COVID-19, NLR, Lymphopenia, PCR
1. Introduction
Coronavirus disease 2019 (COVID-19) is a respiratory infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel emergent virus that was first recognized in Wuhan, China, and since then has rapidly spread worldwide [1]. Although most patients infected with COVID-19 are either asymptomatic or only have a mild disease, approximately one-third of patients have more severe disease caused by an acute respiratory distress syndrome, which may lead to the need for mechanical ventilation (MV) and admission to an intensive care unit or even death [2]. According to the current data of the World Health Organization, the number of infected people and deaths has increased to 63,921,921 and 1,480,622, respectively (December 2019-November 2020) [3].
The nucleic acid real-time polymerase chain reaction (PCR) test has become the standard method for the diagnosis of SARS-CoV-2 infection [3]. This process can take up to 2–3 h. The reported diagnostic accuracy of real-time PCR is between 60% and 80% [4]. These real-time PCR test kits have several limitations. Despite the increasing demand for PCR testing, diagnostic delays remain a challenging problem in patients with COVID-19 pneumonia because of increasing numbers of patients, prolonged testing process, high cost, need for certified laboratories, and the need for experienced personnel. In addition, the reported false negativity rates may be as high as 20% [4]. Therefore, considering the current lack of a rapid and reliable testing method, particularly in countries where test availability is low, supportive laboratory tests should be used to establish a diagnosis if possible. The rapid diagnosis of COVID-19 infection using simple laboratory tests will initiate treatment and break the transmission chain [4], [5]. Several supportive laboratory tests may help in the diagnosis of COVID-19 infection, including leukopenia, lymphopenia, neutrophil to lymphocyte ratio (NLR) elevation, D-dimer elevation, and ferritin elevation [6], [7], [8]. A study has reported that neutralizing antibodies play a crucial role in identifying COVID-19 infection [9]. Another study claimed that serum S100B protein could be a crucial marker in the diagnosis of severe COVID-19 [10]. However, Zeng et al. reported that serum interleukin (IL)-2 receptor, IL-6, IL-8, IL-10, tumor necrosis factor-alpha (TNF-α), ferritin, procalcitonin lactate dehydrogenase, and high-sensitive C-reactive protein (hsCRP) were higher in critically ill patients than moderate and severe patients [11].
Leukocytes play a fundamental role in defense of the immune system during bacterial or viral infection. When the body encounters a viral or bacterial infection, leukocytes provide an immune response. It causes the release of free oxygen radicals that destroy the cell wall of the virus and disrupt its structure by producing a high number of neutrophils. Whereas lymphocytes, the cornerstone of immunity to viral infections, increase the release of CD8 + T lymphocytes and reduce the release of CD4 + T lymphocytes. Thus, it suppresses cellular immunity and lymphopenia, thus increasing NLR [8], [12]. In severe patients, proinflammatory cytokine stimulates Th1, leading to an excessive immune response and uncontrolled release of cytokines and chemokines such as IL-6 [13].
In this study, we aimed to compare inflammatory parameters such as hemogram, CRP, procalcitonin, and ferritin between real-time PCR-positive and -negative patients admitted to our hospital.
2. Materials and method
2.1. Patients
This retrospective single-center study included 174 patients admitted because of suspected COVID-19 infection between March and April 2020. Permissions required for the scientific research project endorsed by the ministry were obtained from the Ministry of Health. Patients were assigned into two groups: PCR positive and PCR negative with typical tomography findings. Demographic characteristics, symptoms, radiological findings, current medications, comorbid conditions, and duration of hospital stay were recorded. Moreover, complete blood count, NLR, urea, creatinine, aspartate aminotransferase, alanine aminotransferase, bilirubin, ferritin, D-dimer, CRP, procalcitonin, troponin, and prothrombin time, and active partial thromboplastin time were analyzed. NLR was defined as the ratio between the absolute neutrophil count and lymphocyte count in the blood. After admission, nasopharyngeal samples for PCR analysis were obtained by a physician. The amplification and analysis of the DNA and RNA were performed using a CFX96 real-time PCR detection system (CFX96; Bio-Rad, USA).
Inclusion criteria: Patients above the age of 18 years, patients who were PCR positive and negative, and having typical tomography findings. Exclusion criteria: patients below the age of 18 years and patients without typical symptoms or absence of tomographic findings compatible with pneumonia.
Patients with an oxygen saturation of <88%, respiratory rate of >30 per minute, pulse rate of >110 beats per minute, blood pressure of <90/60 mmHg, and bilateral pneumonia were admitted to the intensive care unit, whereas other patients were treated in the normal ward. Patients were discharged in case of clinical improvement, regression of radiological findings, and a negative control PCR test after the fifth day following admission.
2.2. Treatment
Treatment with hydroxychloroquine was initiated after admission. Favipiravir was added to this regimen in patients with radiological progression, persistent fever, and the need for intensive care admission occurring within the first 72 h. If the need for oxygen persisted despite hydroxychloroquine and favipiravir or if no clinical improvement occurred, tocilizumab was added. Both groups received similar treatments.
2.3. Radiologic evaluation
The radiological assessment and classification of the patients were based on the criteria proposed by the Radiological Society of North America Expert Consensus [14]. Thus, bilateral ground-glass and reverse halo appearance were considered typical radiological findings; unilateral ground-glass or consolidation or nonperipheral perihilar ground-glass appearance were considered possible radiological findings; isolated consolidation, cavitary lung lesions, or pleural effusions were considered atypical radiological findings; and absence of any pathological findings suggestive of pneumonia was considered a negative radiological finding. According to the Radiological Society of North America Expert Consensus, thoracic tomography was not typical, and PCR-negative patients were excluded.
2.4. Statistical analysis
The study data were analyzed using the Statistical Package for Social Sciences for Windows, 22.0 software pack. Continuous variables were expressed as appropriate means and standard deviations or medians and interquartile ranges. Categorical variables were summarized as counts and percentages for each category. The normal distribution of the parametric data was evaluated using the Shapiro-Wilk test. Categorical variables between PCR-positive and -negative groups were compared using the chi-square test. Nonparametric data were compared between the two groups using the Mann-Whitney U test. The optimal cutoff values of the continuous NLR, leukocyte, neutrophil, and lymphocyte counts were calculated using the receiver operating curve (ROC) analysis. For statistical results, a p-value of less than 0.05 was considered significant, with corresponding 95% confidence intervals.
3. Results
3.1. Demographic features
A total of 174 patients were included in the study. Patients were assigned into two groups: PCR-positive (n = 109; 64 males [58.7%]) and PCR-negative patients (n = 65; 38 males [58.5%]) with typical tomography findings. The gender distribution was similar between the groups (p = 0.974) (Table 1 ). When the two groups were examined for age, the mean age of the PCR-positive group was 44.85 ± 18.41 years, and that of the PCR-negative group was 42.29 ± 18.06 years (p = 0.373; Table 1). In the PCR-positive group, the number of patients with hypertension (HT), diabetes mellitus (DM), asthma, chronic renal failure (CRF), chronic obstructive pulmonary disease (COPD), cerebrovascular disease (CVD), and chronic heart failure (CHF) was 14 (12.8%), 11 (10%), 5 (4.5%), 5 (4.5%), 3 (2.7%), 3 (2.7%), and 1 (0.9%), respectively. In the PCR-negative group, the number of patients with HT, DM, asthma, COPD, CHF, and CVD was 4 (6.1%), 3 (4.6%), 2 (3%), 2 (3%), 1 (1.5%), and 1 (1.5%), respectively. There was a statistically significant difference between the two groups in comorbidity (p = 0.049; Table 1).
Table 1.
Demographic characteristic of patients.
| PCR |
p-value | |||
|---|---|---|---|---|
| Positive (n = 109) |
Negative (n = 65) |
|||
| Age (year) (Mean ± SD) | 44.85 ± 18.41 | 42.29 ± 18.06 | 0.373 | |
| Gender | Male | 64 (58.7%) | 38 (58.5%) | 0.974 |
| Female | 45 (41.3%) | 27 (41.5%) | ||
| Oxygen status | Without oxygen | 97 (89%) | 61 (93.8%) | 0.276 |
| With oxygen | 8 (7.3%) | 4 (6.2%) | ||
| Mechanical ventilation | 4 (3.7%) | 0 (0%) | ||
| Hospitalization time (Day) (Mean ± SD) | 9.77 ± 4.98 | 6.49 ± 2.76 | <0.001* | |
| Unit | Service | 96 (88.1%) | 61 (93.8%) | 0.215 |
| Intensive care | 13 (11.9%) | 4 (6.2%) | ||
| Discharge status | Discharge | 104 (95.4%) | 63 (96.9%) | 0.624 |
| Death | 5 (4.6%) | 2 (3.1%) | ||
| Additional disease | Present | 37 (33.9%) | 13 (20%) | 0.049* |
| Absent | 72 (66.1%) | 52 (80%) | ||
p < 0.05: is statistically significant
p < 0.001
3.2. Evaluation of symptoms
The most common symptoms in both groups were cough, fever, and shortness of breath. Other common symptoms included sore throat, sputum, and loss of taste and odor. In addition, weakness, headache, nausea, and vomiting were observed rarely. No significant difference was observed between the two groups for symptoms (Table 2 ).
Table 2.
The difference in symptoms between groups.
| Symptoms | PCR |
p-value | |
|---|---|---|---|
| Positive (N = 109) |
Negative (N = 65) |
||
| Cough | 77 (70.6%) | 54 (83.1%) | 0.06 |
| Fewer | 49 (45%) | 24 (36.9%) | 0.29 |
| Dyspnea | 42 (38.5%) | 21 (32.3%) | 0.40 |
| Sputum | 2 (1.8%) | 1 (1.7%) | 0.88 |
| Throat Ache | 13 (11.9%) | 7 (10.8%) | 0.81 |
| Loss Of Taste And Smell | 8 (7.3%) | 5 (7.7%) | 0.93 |
| Other Symptoms* | 32 (29.4%) | 13 (20%) | 0.88 |
p < 0.05 : is statistically significant
: weakness, malaise, headache, nausea, vomiting
3.3. Evaluation of treatments
In wards where PCR-positive and PCR-negative patients were hospitalized, 96 (88.1%) PCR-positive patients were admitted to the service, and 13 (11.9%) patients were admitted to the intensive care unit. In the other group, 61 (93.8%) patients received service, and 4 (6.2%) patients were followed up in the intensive care unit (Table 1). Moreover, 97 (89%) patients in the PCR-positive group did not require oxygen therapy, whereas 8 (7.3%) required oxygen therapy, and 4 (3.7%) required MV. In the PCR-negative group, 61 (93.8%) patients did not require oxygen therapy, whereas 4 (6.2%) required oxygen therapy. None of the patients in this group required MV (p = 0.276; Table 1).
3.4. Evaluation of mortality
A total of 104 (95.4%) patients were discharged, and 5 (4.6%) died in the PCR-positive group, whereas 63 (96.9%) were discharged and 2 (3.1%) died in the other group, with no significant difference between the two groups (p = 0.624; Table 1). The mean duration of hospitalization was 9.77 ± 4.98 days and 6.49 ± 2.76 days in the PCR-positive and -negative patients, respectively, with a statistically significant difference between the two groups (p < 0.001; Table 1).
3.5. Evaluation of radiological findings
Radiological images of the patients in the PCR-positive group were classified according to the Radiological Society of North America Expert Consensus. The PCR test was negative in 30 (27.5%) patients, and the radiological appearance was typical in 64 (58.7%) patients, probable in 11 (10.1%), and atypical in four (3.7%).
3.6. Evaluation of laboratory parameters
The two groups were transmitting in the laboratory at the time of admission to the hospital. A significant increase was observed in leukopenia (p < 0.001), thrombocytopenia (p = 0.006), neutropenia (p < 0.001), lymphopenia (p = 0.001), and NLR (p = 0.003) in the PCR-positive group. However, a significant decrease was observed in ferritin (p = 0.012), procalcitonin (p = 0.038), and potassium (p = 0.05) levels in the PCR-positive group (Table 4). No significant differences were observed in lactate dehydrogenase (p = 0.182), albumin (p = 0.094), D-dimer (p = 0.949), troponin (p = 0.394), and CRP (p = 0.322) levels (Table 3 ).
Table 4.
The effects of laboratory parameters on PCR positivity.
| White blood cell (WBC) | Platelet | Lymphocyte | Neutrophil | |
|---|---|---|---|---|
| Cut-off | 6.67 | 208 | 1.51 | 4.425 |
| Sensitivity | 68% | 55% | 61% | 72% |
| Specificity | 71% | 72% | 65% | 67% |
| Positive predictive value | 79% | 77% | 74% | 79% |
| Negative predictive value | 56% | 49% | 49% | 59% |
Table 3.
Comparison of laboratory parameters with PCR positivity.
| PCR |
p-value | ||
|---|---|---|---|
| Positive (n = 109) Median (interquartile range) |
Negative (n = 65) Median (interquartile range) |
||
| WBC (White Blood Cells) | 5.70 (3.15 to15.25) | 8.13 (3.07–17.07) | <0.001* |
| Platelets | 204 (72–732) | 239 (78–462) | 0.006* |
| Neutrophils | 3.61 (0.98–12.97) | 5.24 (1.20–14.26) | <0.001* |
| Lymphocytes | 1.40 (0.39–4.57) | 1.86 (0.58–5.15) | 0.001* |
| PDW (Platelet Distribution Width) | 11.80 (8.20–19.80) | 12.00 (9.40–21.40 | 0.366 |
| RDW (Red Cell Distribution Width) | 12.60 (10.90–20.00) | 12.60 (11.40–12.80) | 0.717 |
| MPV (Mean Platelet Volume) | 10.50 (8.80–13.50) | 10.60 (9.10–12.80) | 0.821 |
| NLR (Neutrophils/Lymphocytes) | 3.73 (1.16–16.01) | 3.17 (0.75–6.25) | 0.003* |
| CRP (C-reactive protein) | 8.00 (0.04–266.60) | 9.90 (0.10–81.80) | 0.322 |
| Procalcitonin | 0.02 (0.01–32.60) | 0.01 (0.00–4.24) | 0.038* |
| D-dimer | 409.00 (96.00–3750,000) | 386.00 (123.00–13,400) | 0.949 |
| Troponin | 2.50 (2.00–8989.15) | 2.50 (2.10–64.57) | 0.394 |
| Albumin | 4.30 (2.80–5.10) | 4.40 (3.00–5.00) | 0.94 |
| K+ (potassium) | 3.90 (3.10–5.40) | 4.10 (2.80–5.60) | 0.05* |
| Ferritin | 180.00 (23.20–3142.50) | 134.90 (5.50–600.00) | 0.012* |
| LDH (Lactate Dehydrogenase) | 239.00 (156.00–645.00) | 258.00 (98.00–640.00) | 0.182 |
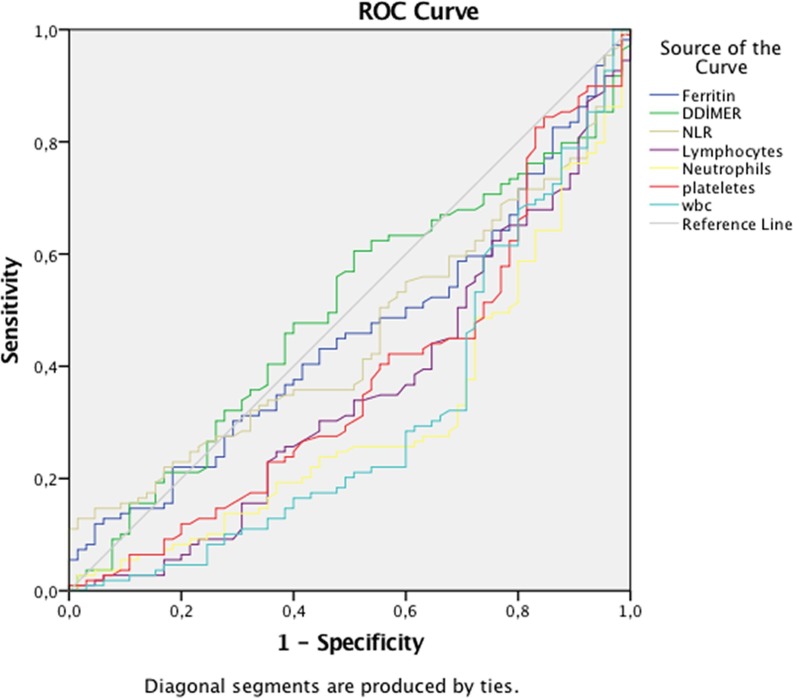
In addition, a ROC curve analysis was performed on the PCR-positive group. The PCR test was significantly positive for leukopenia, lymphopenia, thrombocytopenia, and neutropenia (Fig. 1 ). The cutoff value, sensitivity, and specificity for leukopenia were 667 thousand, 68%, and 71%, respectively. The cutoff value, sensitivity, and specificity for lymphopenia were 151 thousand, 61%, and 65%, respectively. The cutoff value, sensitivity, and specificity for neutropenia were 4.425, 72%, and 67%, respectively. The cutoff value, sensitivity, and specificity for thrombocytopenia were 208 thousand, 55%, and 72%, respectively (Table 4).
Fig. 1.
ROC curve analysis of NLR, D-dimer, platelets, ferritin, lymphocyte, neutrophil, and leukocyte count. Area under the curve of lymphocyte: 356; p < 0.001, area under the curve of neutrophils: 307; p < 0.001, area under the curve of platelets: 375; p = 0.006, area under the curve of white blood count: 310; p < 0.001.
4. Discussion
COVID-19 infection has become a crucial problem all over the world. In this study, we studied parameters and searched for findings that may help in early diagnosis and treatment. PCR positivity was found to be higher in patients with leukopenia, lymphopenia, thrombocytopenia, increased NLR, and increased ferritin.
COVID-19 is a disease that affects several systems in the body, including the lungs. It can be transmitted very quickly from person to person or through droplets and can spread rapidly in this manner. It is more common in men. Although all age groups can contract COVID-19, 15–49 years of age range is less likely to be affected. Cough and fever are among the common symptoms [15]. Li et al. reported that COVID-19 infection is more common in men. Moreover, the most frequent presentation was a cough, and it is more common in the same market between the ages of 15 and 49 years. In addition, fever was seen in 98% of the cases, and the mortality rate was higher in intensive care patients.[1], [6]. It occurred more often in men than in women. Moreover, fever and cough were common symptoms, with a mortality rate reported as 11% [16]. In this study, although the disease was more prevalent among males, it did not affect the young population. In addition, the most common symptoms were cough and fever, with a mortality rate of 4.6%.
A leukocyte count of less than 4000 µL is termed as leukopenia. Leukocytes provide an immune response when the body encounters a viral or bacterial infection. After coming in contact with the virus, it affects all cells and releases a high number of cytokines, as previously seen in SARS-COV and MERS-COV infection [17]. Thus, it can directly kill the virus or trigger humoral immunity and cause a cytokine storm by releasing IL1, IL6, and TNF-α. On the other hand, lymphocytes, the cornerstone of the immunity of viral infections, increase the release of CD8 + T lymphocytes, and decrease the release of CD4 + T lymphocytes. Thus, it suppresses cellular immunity, and lymphopenia occurs. Coronavirus has been shown to affect the bone marrow and the release of leukocytes. Zeng et al. reported that IL-2, IL-6, IL-8, IL-10, and TNF-α secretion increased in critical patients with COVID-19 [11]. In one study, it was reported that IL-6 release increased after the imbalanced response of the immune system in critically ill patients with COVID-19 [13]. A study reported that leukopenia was detected in patients with COVID-19 and these patients were more likely to die [17]. In another study, >80% of the patients had leukopenia [18]. Similarly, other studies have shown that leukopenia was more common among patients with COVID-19 [6], [16]. In this study, the number of leukocytes significantly decreased in the PCR-positive group. Therefore, we believed that the presence of leukopenia is a supportive laboratory parameter for diagnosis in patients with COVID-19.
In PCR-positive patients, the virus could have caused lymphopenia by affecting lymphocytes. Liu et al. suggested that the coronavirus may affect lymphocytes, leading to a decrease in the absolute lymphocyte count. Therefore, it has been reported that the virus may consume defense cells and cause the disease to progress more severely [12]. In another study, Henry et al. suggested that lymphopenia was observed in critically ill patients and in cases with high mortality, and this may lead to a decreased immune response [17]. Moreover, lymphopenia can be observed in severe patients with MERS-COV infection [19]. Another study showed that lymphopenia may help in identifying disease severity in patients with COVID-19 [7]. A study conducted in Wuhan has shown that the observation of lymphopenia may help in the early diagnosis of COVID-19 [20]. In a study conducted by Zeng et al., the presence of high hsCRP and lymphopenia was emphasized in critical patients with COVID-19 [11]. In this study, lymphopenia was significantly observed in patients with PCR positivity. Therefore, lymphopenia can be used as an auxiliary laboratory parameter in the diagnosis of PCR-positive patients.
When the body encounters a viral infection, it stimulates antigen-presenting CD8 + T lymphocytes and causes a cytokine storm by releasing a high number of neutrophils. This increase NLR by causing a decrease in the absolute lymphocyte count in viral infections [21], [22]. In a study of 237 patients in China, it was reported that NLR is an independent risk factor affecting mortality in influenza infections such as H7N9. In another study, the mortality rate of patients with an NLR > 19.94 was found to be higher [23]. A study by Yang et al. showed that the increase in NLR can be used as an independent prognostic biomarker in patients with COVID-19 [8]. In a study conducted in China with 116 COVID-19 pneumonia patients, it was shown that the increase in NLR may be associated with the severity of COVID-19 disease [7]. Another study showed that the increase in NLR in patients with COVID-19 may help in early diagnosis [20]. In this study, an increase in NLR at the time of diagnosis was more pronounced in PCR-positive patients. We believed that increase in NLR in patients with COVID-19 should be recognized earlier and treated.
Platelet is one of the crucial factors in coagulation and an essential part of systemic inflammation. In the case of active inflammation, it is triggered by the immune system and can lead to thrombosis. In addition, as chronic inflammation continues, it interacts with white blood cells and triggers cytokine release. Platelet is the origin of several immune mediators and inflammatory structures. In a viral infection, platelets stimulate the immune system, increasing the release of IL1, IL6, and IL1b and creating an immune response. COVID-19 infection affects the bone marrow and causes thrombocytopenia by inhibiting the synthesis of hematopoietic cells after entering the cells [24]. Studies have shown that the rate of thrombocytopenia in COVID-19 ranges from 40% to 60%. A study reported that the mortality rate of patients with thrombocytopenia was high in patients with COVID-19 [25]. Another study showed that low platelet levels were observed in patients diagnosed with COVID-19, which affected mortality [26]. Similarly, other studies have shown that thrombocytopenia supports the diagnosis and affects mortality in COVID-19 patients [6], [27]. In this study, we found that the number of platelets significantly decreased in patients with PCR positivity. Thus, thrombocytopenia may be a parameter that can help in early diagnosis, apart from the radiological appearance of the patients.
Ferritin is an intracellular iron storage protein and a crucial part of natural immunity, which activates macrophages. Ferritin levels increase in viral or bacterial infections. Moreover, studies have reported high ferritin levels in COVID-19 pneumonia. Ferritin is believed to trigger the macrophage activation syndrome at extremely high levels because of excessive macrophage release. This may be the cause of the cytokine storm that occurs in COVID-19 infection. In a study, patients with COVID-19 had an elevated ferritin level with a significant effect on mortality [27]. Another study found that 63% of patients had high ferritin levels [12]. Another study reported that the S100B protein could be used as a biomarker in severe COVID-19 patients and correlated with ferritin [10]. In this study, a significant increase was observed in ferritin levels in PCR-positive patients. As ferritin levels increase in COVID-19 infection, we believed that it may be a crucial laboratory marker for the diagnosis of the infection.
Several notable limitations were observed in this study. First, data were obtained from a single clinical research center. Second, the experimental data were limited. Furthermore, the conclusion of this study may differ from that of other studies at home and abroad and must be further improved in clinical cases. Third, in PCR-negative patients, samples were taken only from the nasopharynx and oropharynx. We were unable to examine PCR in bronchoalveolar lavage or deep tracheal aspirate in these patients because of the heavy patient load. Finally, a small number of patients were included in the study because of time constraints.
In conclusion, COVID-19 pneumonia has become a major disease worldwide. As COVID-19 pneumonia progresses rapidly, early diagnosis and treatment are crucial. Radiological imaging and PCR are a crucial part of the diagnosis. Because of the higher cost, prolonged test results, unavailability of PCR laboratories in each site, shortage of microbiologists experienced in PCR, and false negativity of the PCR test, there is an unmet need for rapid laboratory tests. It is crucial to diagnose and start treatment early with clinical, radiology, and rapid hemogram testing. We believed that leukopenia, lymphopenia, thrombocytopenia, NLR increase, and ferritin increase are crucial parameters in PCR positivity.
Ethics committee approval for the study was obtained from the University of Harran Ethics Committee with number HRU/20.15.20.
CRediT authorship contribution statement
M. Kabak: Conceptualization, Methodology, Software, Data curation, Writing - original draft, Validation, Writing - review & editing. B. Çil: Visualization, Investigation. I. Hocanlı: Supervision.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk Factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan. China, JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.who.int/dg/speeches/detail/who-director-general-s-opening (accessed 2 January 2021).
- 4.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in WuhanChina. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S., Cai X., Wang H., He G., Lin Y., Lu B., Chen C., Pan Y., Hu X. Abnormalities of peripheral blood system in patients with COVID-19 in WenzhouChina. Clin. Chim Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang A.P., Liu J.P., Tao W.Q., Li H.M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore J.P., Klasse P.J. COVID-19 vaccines: “warp speed” needs mind melds, not warped minds. J. Virol. 2020;94(17) doi: 10.1128/JVI.01083-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aceti A., Margarucci L.M., Scaramucci E., Orsini M., Salerno G., Di Sante G., Gianfranceschi G., Di Liddo R., Valeriani F., Ria F., Simmaco M., Parnigotto P.P., Vitali M., Romano Spica V., Michetti F. Serum S100B protein as a marker of severity in Covid-19 patients. Sci. Rep. 2020;10(1):18665. doi: 10.1038/s41598-020-75618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Z., Yu H., Chen H., Qi W., Chen L., Chen G., Yan W., Chen T., Ning Q., Han M., Wu D. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Crit. Care. 2020;24(1):525. doi: 10.1186/s13054-020-03255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., Gao G.F. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gozalbo-Rovira R., Gimenez E., Latorre V., Frances-Gomez C., Albert E., Buesa J., Marina A., Blasco M.L., Signes-Costa J., Rodriguez-Diaz J., Geller R., Navarro D. SARS-CoV-2 antibodies, serum inflammatory biomarkers and clinical severity of hospitalized COVID-19 patients. J. Clin. Virol. 2020;131:104611. doi: 10.1016/j.jcv.2020.104611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne D., O'Neill S.B., Muller N.L., Silva Muller C.I., Walsh J.P., Jalal S., Parker W., Bilawich A.M., Nicolaou S. RSNA expert consensus statement on reporting chest CT findings related to COVID-19: interobserver agreement between chest radiologists. Can. Assoc. Radiol. J. 2020 doi: 10.1177/0846537120938328. 846537120938328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. I. China novel coronavirus, T. research, a novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 18.Chen L., Liu H.G., Liu W., Liu J., Liu K., Shang J., Deng Y., Wei S. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E005. doi: 10.3760/cma.j.issn.1001-0939.2020.0005. [DOI] [PubMed] [Google Scholar]
- 19.Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., AlHajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan China. Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hufford M.M., Kim T.S., Sun J., Braciale T.J. The effector T cell response to influenza infection. Curr. Top Microbiol. Immunol. 2015;386:423–455. doi: 10.1007/82_2014_397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Z., Chen Z., Li X., Xu L., Guan W., Cao Y., Hu Y., Zhang J. Host immunological response and factors associated with clinical outcome in patients with the novel influenza A H7N9 infection. Clin. Microbiol. Infect. 2014;20(8):O493–O500. doi: 10.1111/1469-0691.12505. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Zou P., Gao H., Yang M., Yi P., Gan J., Shen Y., Wang W., Zhang W., Li J., Liu P., Li L. Neutrophil-lymphocyte ratio as an early new marker in AIV-H7N9-infected patients: a retrospective study. Ther. Clin. Risk Manag. 2019;15:911–919. doi: 10.2147/TCRM.S206930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X., Yang Q., Wang Y., Wu Y., Xu J., Yu Y., Shang Y. Thrombocytopenia and its association with mortality in patients with COVID-19. J. Thromb. Haemost. 2020;18(6):1469–1472. doi: 10.1111/jth.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. U.K. Hlh across speciality collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]