Abstract
Prompt diagnosis, patient isolation, and contact tracing are key measures to contain the coronavirus disease 2019 (COVID-19). Molecular tests are the current gold standard for COVID-19 detection, but are carried out at central laboratories, delaying treatment and control decisions. Here we describe a portable assay system for rapid, onsite COVID-19 diagnosis. Termed CODA (CRISPR Optical Detection of Anisotropy), the method combined isothermal nucleic acid amplification, activation of CRISPR/Cas12a, and signal generation in a single assay, eliminating extra manual steps. Importantly, signal detection was based on the ratiometric measurement of fluorescent anisotropy, which allowed CODA to achieve a high signal-to-noise ratio. For point-of-care operation, we built a compact, standalone CODA device integrating optoelectronics, an embedded heater, and a microcontroller for data processing. The developed system completed SARS-CoV-2 RNA detection within 20 min of sample loading; the limit of detection reached 3 copy/μL. When applied to clinical samples (10 confirmed COVID-19 patients; 10 controls), the rapid CODA test accurately classified COVID-19 status, in concordance with gold-standard clinical diagnostics.
Keywords: Biosensor, COVID-19, CRISPR/Cas, Isothermal amplification, Fluorescence polarization, Point-of-care
1. Introduction
The 2019 coronavirus (COVID-19) pandemic underscores the need for rapid, point-of-care (POC) diagnostic tests (Kilic et al., 2020; Weissleder et al., 2020). At about one year post-initial outbreak, the United States alone has had over 24 million COVID-19 cases with more than 400,000 deaths; other countries, who have managed to prevent the initial spread, now experience a “second wave” of COVID-19 outbreaks (Wang et al., 2020; Zhu et al., 2020). While COVID-19 vaccines are slowly deployed over the world, the primary response to COVID-19 is still containment, i.e., widespread implementation of diagnostic testing, isolation and contact tracing (Kilic et al., 2020; Shim et al., 2020; Weissleder et al., 2020). Among many different assays reported for COVID-19 detection, nucleic acid amplification tests (NAATs), in particular reverse transcription polymerase chain reaction (RT-PCR), are the de facto diagnostic standard. They enable highly accurate identification of SARS-CoV-2, the causative pathogen (Jiang et al., 2020; Tahamtan and Ardebili, 2020; Weissleder et al., 2020). Most PCR tests, however, are carried out in centralized laboratories, limited by lengthy assay time (1–2 h) and requiring bulky instrumentation (Petralia and Conoci, 2017). This in turn causes logistic overheads (e.g., sample transfer, protection from degradation) and long turnaround times to obtain results (2–3 days).
Isothermal nucleic acid (NA) amplification is a promising technology for onsite NAATs. Using specialized DNA polymerases with the capacity of strand displacement, these methods amplify NA at a fixed temperature, thereby simplifying device design (Zhang et al., 2012; Zhao et al., 2015; Bi et al., 2017). Combining isothermal amplifications with clustered regularly interspaced short palindromic repeats (CRISPR) has been shown to further improve assay sensitivity and specificity (Dronina et al., 2021). Upon recognition of its target strand, a CRISPR-associated (Cas) protein and guide RNA complex can indiscriminately cleave single-stranded NAs (Chen et al., 2018). This property has been exploited to amplify analytical signal i) through the collateral cleavage of non-target NA probes that have a fluorescent dye and quencher pair (Gootenberg et al., 2017; Kellner et al., 2019; Aman et al., 2020) or ii) through the release of nanoprobes that are initially anchored by non-target NA probes (Zeng et al., 2021). CRISPR systems have demonstrated a promising potential for COVID-19 diagnostics (Broughton et al., 2020; Huang et al., 2020). Technical challenges, however, limit the practical point-of-care (POC) use of these assays: i) most CRISPR tests proceed with sequential NA amplification and detection, requiring separate preparation and introduction of CRISPR reagents into samples; and ii) signal readout, typically performed with a dipstick-type lateral flow device, also incurs extra manual steps and produces subjective, qualitative results.
Here, we report on a rapid, quantitative, and streamlined COVID-19 test in a compact system. Termed CODA (CRISPR Optical Detection of Anisotropy), the system seamlessly combined a one-pot CRISPR test with a robust detection modality, fluorescence anisotropy (FA). Specifically, we adopted a one-pot assay scheme wherein isothermal NA-target amplification and CRISPR-based target recognition take place simultaneously. The activated CRISPR/Cas then cleaves fluorescent DNA reporters, changing the FA readout. The CODA approach offered practical advantages: i) the entire reaction, including reverse transcription, NA amplification, and signal detection, were conveniently carried out in a single tube and at constant temperature (42 °C); ii) the FA measurement, unlike conventional fluorescent intensity detection, was ratiometric and robust against common noises (e.g., intensity fluctuations), which led to reliable analytical signal even at low NA concentrations (LiCata and Wowor, 2008); and iii) FA reporters, which required only a fluorescent dye conjugation, were simpler and cheaper than common CRISPR/Cas probes. These features enabled us to advance a portable CODA system integrating low-noise optical detection, precise temperature control, and on-board data processing and display. With the integrated CODA system, we detected SARS-CoV-2 RNA within 20 min in a ‘sample-to-result’ manner and achieved a sensitivity down to 3 copies/μL. We further applied the system in a pilot clinical test, assessing 20 clinical samples; the results showed an excellent accordance with those by a clinical laboratory.
2. Materials and methods
2.1. Materials
All oligonucleotides used in this study were synthesized by Bioneer® (Daejeon, Korea). The sequences of the oligonucleotides are listed in Table S1. Plasmid controls of SARS-CoV-2, human RPP30, MERS-CoV, and SARS-CoV and gBlocks® gene fragments of HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 were purchased from Integrated DNA Technologies, Inc. (Coralvile, IA, USA). TwistAmp® Basic kit and RevertAid reverse transcriptase (RTase) were purchased from TwistDx™ and ThermoFisher Scientific (Waltham, MA, USA), respectively. EnGen® Lba Cas12a (Cas12a), RNase inhibitor (Murine) (RI), DNase I, HiScribe™ T7 High Yield T7 RNA synthesis kit, and Monarch® RNA cleanup kit were purchased from New England BioLabs (Beverly, MA, USA). KAPA HiFi HotStart ReadyMix PCR kit and TOPreal™ One-step RT qPCR kit were purchased from Roche (Basel, Switzerland) and Enzynomics (Daejeon, Korea), respectively. DEPC-DW, purchased from Bioneer®, was used in all experiments. All other chemicals were of analytical grade and used without further purification.
2.2. Device construction
The CODA device was modeled in computer-aided design software (Solidworks, 2019) and fabricated via computer numerical control machining (6061 aluminum) and 3-dimensional photopolymer resin printing (Formlabs). The main body housed a light source subassembly and two identical FA detectors. i) The light source consisted of an LED (Thorlabs M470D2), a linear polarizer (Thorlabs LPVISE100-A), and a convex focusing lens (Thorlabs LB1092-A). Components were concentrically aligned inside a standard tube mount (Thorlabs SM1A6, CP4S, SM1L03). The LED was attached to a metal heatsink through a thermal compound (Arctic Silver 5) and driven by a constant current amplifier (Meanwell LDD-1000L). ii) Each FA detector subassembly consisted of a convex lens (Thorlabs LB1092-A), a linear polarizer (Thorlabs LPVISE050-A), a band-pass filter (Thorlabs FLH05532), and a photodiode (Hammamatsu S1223). Optical components were assembled in slotted lens tubes (Thorlabs SM05L20C); the two detectors were oriented to have orthogonal polarizations. iii) FA signals were amplified using a configuration of precision amplifiers (LF356, AD54 CE9JHZ) and a lock-in amplifier (AD63 CE0JNZ). A 1-kHz carrier wave was generated by the integrated digital-analog converter (DAC) of an ARM Cortex M4 microcontroller (PJRC Teensy 3.2). Iv) For sample heating, a metal holder was machined in aluminum. An integrated 10 W ceramic heater was driven by a power transistor (TIP120) attached to an 18 V supply. Temperature was monitored by a 10 kΩ thermistor (Thorlabs HT10KR) and controlled to within 0.2 °C by a proportional-integral-derivative (PID) feedback loop running on the microcontroller unit (MCU). V) For the graphical user interface (GUI), a real-time dashboard was programmed using the Qt GUI framework (Fig. S1), which communicated with the MCU over the USB serial port. The MCU firmware was written in C++ (Arduino).
2.3. CODA signal processing
The oscillating fluorescence signals were captured by a photodiode; amplified 10-fold by an analog negative-feedback amplifier locked-in to the 1 kHz source signal; low-pass-filtered and amplified again by 30-fold; and converted to a digital signal by a pair of 12-bit analog-to-digital converters inside the MCU. The integrated real-time monitoring GUI displayed raw fluorescence readings from both parallel (I parallel) and perpendicular (I perpendicular) channels in real-time (every 0.1 s). The anisotropy (r) was computed according to the equation r = F·(I parallel – I perpendicular)/(I parallel + 2·I perpendicular), where F was a scaling factor (F = 4259) to match CODA values with those measured by a plate reader (Sapphire 2, TECAN). Noise fluctuations arising from both raw and computed data streams were precisely measured as the rolling standard deviation of the most recent 40 samples, to confirm that the system's measurement was reliable (see Fig. S2 for the flow of signal processing).
2.4. CODA assay
The CODA master mix was prepared by combining 240 nM gene-specific RPA primers, 100 nM reporter probe, 160 nM gene-specific Cas12a gRNAs, 2 U/μL RevertAid RTase, 0.8 U/μL RI, 640 nM Cas12a, 0.2 × NEBuffer 2.1 (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 100 μg/mL BSA, pH 7.9 at 1 × concentration), and reconstituted RPA mix. Upon addition of viral RNA and 14 mM MgOAc, the CODA mix was incubated at 42 °C for 20 min. FA was measured during the reaction. In the specificity test, plasmid controls and gene fragments, which are double-stranded DNAs, were used as a target and RTase was excluded.
2.5. In-vitro RNA preparation
PCR mix for in-vitro transcription (IVT) (20 μL) was prepared to contain 500 nM IVT primers, 1 × KAPA HiFi HotStart ReadyMix, and 104 copy/μL SARS-CoV-2 plasmid control. Thermocycling was performed with following steps: 95 °C, 2min; 95 °C, 20 s, 60 °C, 39 s, 35 cycles; 60 °C, 5 min. After agarose gel extraction, 1 μg of IVT PCR product was applied to HiScribe™ T7 High Yield T7 RNA synthesis mix with recommended composition of T7 RNA polymerase mix, rNTPs, and reaction buffer. IVT was then carried out at 37 °C for 3 h, followed by DNase I treatment for 30 min to prevent DNA contamination. Finally, the IVT RNA product was purified with Monarch® RNA cleanup kit per manufacturer's protocol and its concentration and purity were determined using NanoDrop™ 2000c (ThermoFisher Scientific, Waltham, MA, USA).
2.6. SYBR green-based qRT-PCR and melting curve analysis
For comparative analyses, qRT-PCR was performed. Viral RNA of SARS-CoV-2 was added to qRT-PCR mix composed of 250 nM primers and 1 × TOPreal™ One-step RT-qPCR reaction mix. qRT-PCR was then conducted on the ViiA 7 Real-Time PCR system (Life Technologies) with the following steps: 50 °C, 30 min for RT; 95 °C, 10 min; 95 °C, 5 s, 60 °C, 30 s, fluorescence measurement, 45 cycles. After denaturing and ramping down the PCR product to 40 °C, a melting curve analysis was carried out to validate the qRT-PCR result. As the temperature is elevated (0.05 °C/s), fluorescence is accordingly measured. Threshold cycle (C t) and melting temperature were automatically determined by the system software (Fig. S3).
2.7. Clinical sample collection and analyses
The study was approved by the Institutional Review Board of Chonnam National University Hospital. Clinical samples of nasopharyngeal and oropharyngeal swabs and sputum were collected to the universal transport medium (UTM) (Asan Pharmaceutical, Seoul, Korea) and transported to Chonnam National University Hospital Laboratory which Korea Centers for Disease Control and Prevention (KCDC) approved as biosafety level-2 (BL-2) facility in accordance with institutional biosafety requirements. Total RNA was extracted from each sample, using AdvanSure™ E3 System (LG chem, Seoul, Korea) per manufacturer's protocol. For the clinical diagnoses, extracted RNA was amplified (40 cycles) using a commercial qRT-PCR kit (PowerChek™ 2019-nCoV Real-time PCR Kit; KogeneBiotech, Seoul, Korea) and a detection system (CFX96™ Real-time PCR detection system; Bio-Rad, Hercules, CA, USA). A positive qRT-PCR result was defined as a C t ≤ 35.
3. Results and discussion
3.1. CODA assay
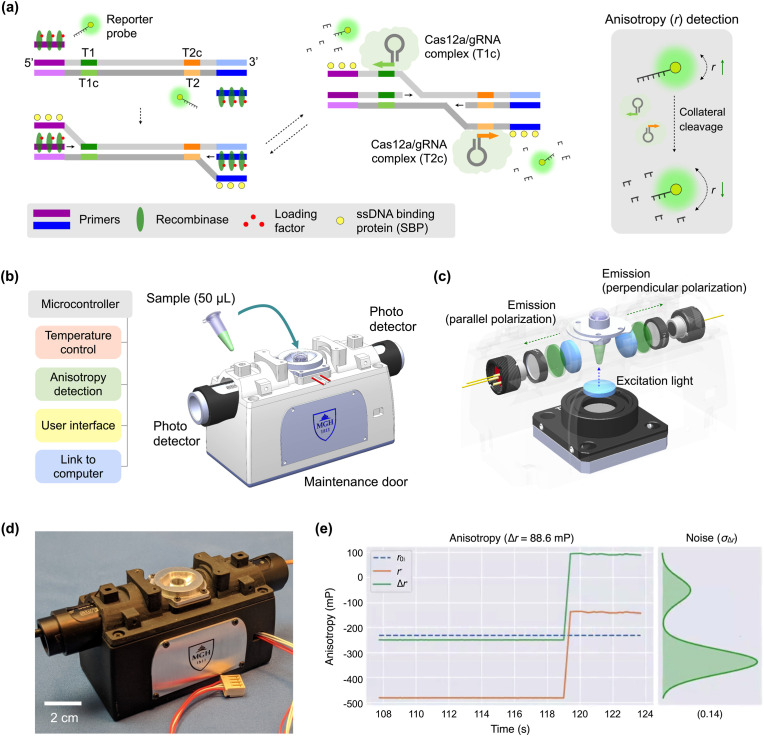
The CODA assay (Fig. 1 a) combines isothermal NA amplification and CRISPR/Cas12a detection. In the presence of the target NA sequence, reverse transcription and recombinase polymerase amplification (RT-RPA) takes place with gene-specific RPA primers. During this process, single strand DNAs downstream to the primer binding sites (purple and blue) are exposed by the strand displacement activity of DNA polymerase. When gRNAs recognize the specific sites (green and orange) at both ends of displaced single strands, Cas12a/gRNA complexes get activated to cleave DNA (Chen et al., 2018). Note that most cleavage actions would be limited to fluorescent reporter DNA (trans-cleavage). The protospacer adjacent motif (PAM) sequence is absent downstream to the recognition sites, which makes it unlikely that activated Cas12a invades and cuts target DNA (Jeon et al., 2018). Furthermore, RPA synthesizes complementary sequences on displaced single strands, effectively keeping Cas12a (RuvC nuclease domain) from cleaving single strands in target DNA.
Fig. 1.
CODA system for COVID-19 diagnosis. (a) Assay schematic. When target viral RNA is present, RT-RPA and CRISPR/Cas detection take place together. DNA polymerase recognizes target sequences and displaces double-stranded DNA. Cas12a/gRNA complexes then bind to specific sites (green and orange) in the exposed single strand, get activated, and start to cleave nearby reporter probes. This cleaving process is amplified, as RT-RPA reaction proceeds. As a result, the fluorescence anisotropy (r) of the sample decreases (right). (b) CODA device configuration. A compact device integrates rapid sample heating, precision signal processing, and real-time polarization anisotropy detection. A sample tube (50 μL) is inserted into a heated metal block, whereby two photodetectors detect fluorescence light. (c) CODA optics. A linearly polarized light illuminates a sample tube from its bottom side. Two photodetectors measure orthogonal polarization of fluorescence light emitted by the sample. (d) Photograph of the portable CODA device for onsite application. The enclosure was made of a lightweight photopolymer. Optical mounts and the sample holder were machined in aluminum. (e) A partial screenshot of an extended user interface. The CODA device communicates with a computer to present real-time data. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As exponential NA amplification proceeds, the trans-cleaving activity is reinforced, degrading a large number of fluorescent reporters. Accordingly, the overall molecular weight of fluorescent reporters decreases, which yields a lower FA value (r) due to increased Brownian motion (Fig. 1a, right). The isothermal, dual amplification of target NA and FA signal allows for rapid, one-step, and one-pot assay (20 min). Minimal intervention as well as a ratiometric FA readout make CODA robust to external interferences.
To enable onsite CODA assay, we have constructed a portable device (Fig. 1b) for CODA reaction and FA detection. Specifically, the device maintained a constant reaction temperature (42 °C) through a feedback control, measured FA values using compact optics (Fig. 1c), and processed raw data to obtain r values. The prototype device (Fig. 1d) had a form factor of 13 × 4.5 × 5 cm3. A sample container (a PCR tube with diameter 6.2 mm) was to be inserted into a form-fitting metal sleeve for heating. The device also communicated with a computer running real-time analysis software (Fig. 1e, Fig. S1) that displayed and stored data.
3.2. CODA device
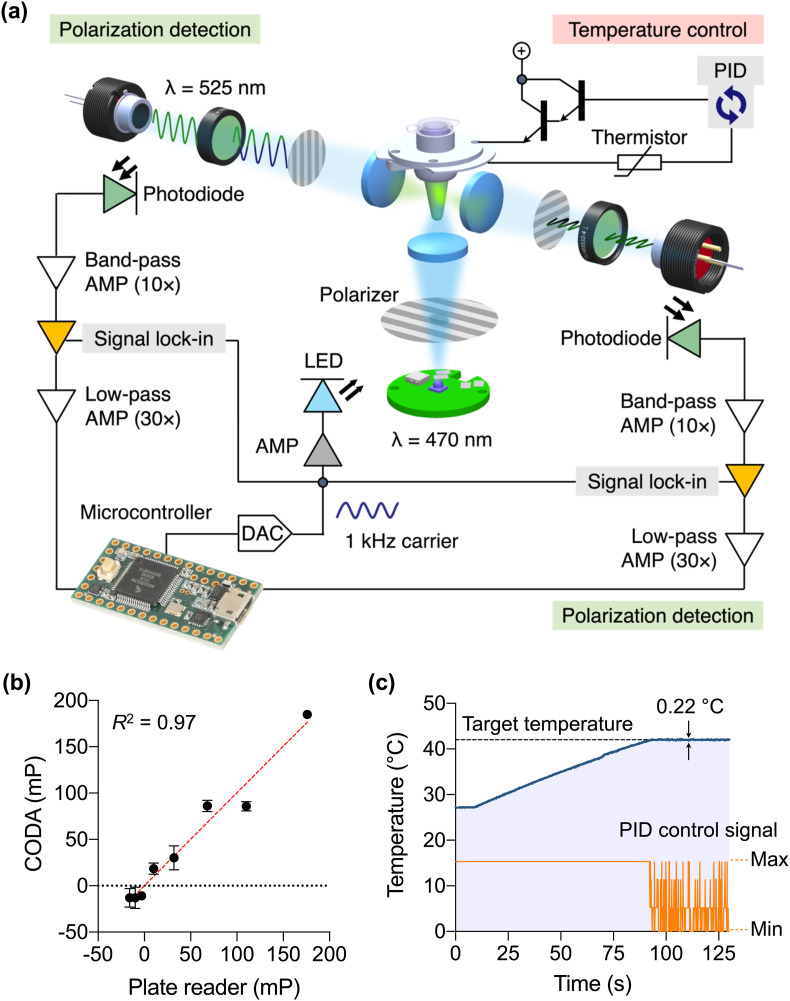
Fig. 2a shows the schematic of optoelectronics inside the CODA device. We adopted an optical lock-in method for low-noise measurements. An LED generated modulated blue light at varying intensity (ƒ = 1 kHz) which passed through a linear polarizer onto the sample. The sample emitted green light (525 nm) oscillating in intensity at the same modulation frequency (1 kHz) with polarization components that depend on the molecular weight of FA probes. Two detectors captured both parallel (I parallel) and perpendicular (I perpendicular) polarization components. Each detector consisted of a photodiode and a convex lens. An additional polarizer and a bandpass filter were used to reject the source (exciter signal) and unwanted polarization angles. Once the signal had been captured, optical lock-in amplifiers were utilized to reject common noise that may arise due to i) ambient light leaking into the device's optical path, ii) external electrical noise due to currents flowing through ground impedances, iii) electromagnetic interference from nearby electronic devices, and iv) other internal electrical noise.
Fig. 2.
Optical design and validation of CODA device. (a) Optical and electrical schematic. A light emitting diode (LED) illuminates a sample with linearly polarized light oscillating at 1 kHz. Fluorescence is captured by two photodetectors, each consisting of a photodiode, a 525-nm bandpass filter and a linear polarizer. The signal is processed by a sequence of integrated filtering/amplification steps: 10 × bandpass, lock-in, and 30 × lowpass. The cleaned signals are finally captured by a microcontroller. Temperature is controlled through a feedback control. AMP, amplifier; DAC, digital-to-analog converter; PID, proportional–integral–derivative. (b) The CODA system was benchmarked against a conventional plate reader. Samples were prepared in triplicate through the serial dilution of glycerol in an aqueous buffer, varying the viscosity. All samples contained the same amount of fluorescein (240 nM). An excellent correlation was observed between these two systems. (c) Sample heating curve. The system reached the target temperature (42 °C) within 90 s. This temperature was maintained within ±0.2 °C variations.
We compared CODA's optical performance against that of a benchtop plate reader (Sapphire 2, TECAN). We prepared standard samples of varying fluorescent anisotropy; different amounts of glycerol were added into a fluorescein solution, which changed the solution's viscosity. For CODA, parallel (I parallel) and perpendicular (I perpendicular) polarization intensities were measured every 0.1 s, and 40 readings from each channel were averaged to reduce sampling noise. The fluorescence anisotropy (r) was then computed by the onboard MCU. The measured r values showed a good match (R 2 = 0.97) between the CODA device and the plate reader (Fig. 2b), which confirmed CODA's accuracy in optical detection. Because the computed anisotropy (r) is essentially a normalized difference between polarization channels, we indeed observed that the anisotropy measurements were far more stable than their raw intensity components. Combining noise-monitoring with tightly controlled exposure times reduced the variability between measurements and increased the overall repeatability of experiments.
We next monitored CODA's temperature stability. The goal was to keep the temperature of the metal sleeve holding a sample tube at 42 °C (T set), the optimal condition for the CODA assay. The MCU checked the temperature T actual every 0.1 s by measuring the resistance across an embedded thermistor and then used the error term (T set – T actual) for the feedback control. The system rapidly reached the target temperature, within 1.4 min of heater activation, and maintained it with variations ≤0.2 °C (Fig. 2c).
3.3. CODA assay optimization
We optimized the CODA assay condition for COVID-19 detection. We designed detection probes to target two regions in nucleocapsid gene (N1 and N2 genes) of SARS-CoV-2 RNA (Table S1). A third probe targeted human ribonuclease P subunit 30 (RPP30) gene as a sample quality check. For each target gene, Cas12a gRNAs were constructed to recognize sequences near the RPA primer binding sites. This would enable Cas12a gRNAs to bind to exposed single strand DNAs upon strand displacement, activating Cas12a′s cleavage activity.
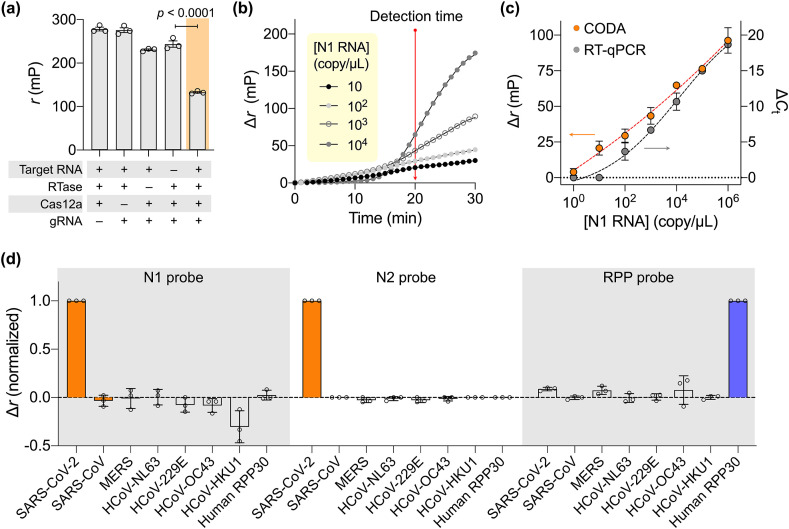
Using SARS-CoV-2 N1 RNA as a model target, we first determined the optimal FA reporter condition (Fig. S4). Probe lengths and concentrations were varied and the FA signal after Cas12a cleavage was measured. The maximal FA signal was observed with a nine-base reporter at 0.1 μM. We next validated the assay feasibility by monitoring the FA signal under various reaction conditions (Fig. 3 a). The r values were high (i.e., high anisotropy) without effective cleavage of fluorescent reporters. The highest r was observed when RT-RPA took place but not Cas12a activation. This result was presumable due to the increase of viscosity from the RPA reaction (Rust et al., 2013, 2017). With all CODA components present, however, FA decreased significantly. We defined the analytical metric as Δr = r 0 – r, where r 0 (control) was measured from the CODA assay in the absence of the target gene.
Fig. 3.
CODA assay characterization. (a) Assay mechanism validation by varying the reaction condition. FA remained high when any single assay component was missing. The value significantly decreased when all key components were present. The target N1 RNA concentration was 106 copy/μL. RTase, reverse transcriptase. (b) Temporal evolution of FA signal among serially diluted N1 RNA samples. A sample without target RNA was used as a control. The difference Δr (= r0 – r) was defined, where r0 was FA from the control sample. The net signal (Δr) increased and became commensurate with RNA concentration in 20 min. (c) Serially diluted N1 RNA samples were analyzed by CODA (20 min) and conventional RT-qPCR (80 min). CODA achieved wider dynamic range and higher sensitivity than RT-qPCR. The limit of detection was 3 copy/μL for CODA. ΔCt = Ct,0– Ct, where Ct,0 and Ct were threshold cycle values for control and RNA samples, respectively. (d) Specificity assessment. SARS-CoV-2 N1, N2, and human RPP30 probes were tested for cross reactivity with other compounding genes. Each probe generated hight analytical signal only when its corresponding target was present. Gene concentration was set to 103 copy/μL in all samples. Data are displayed as means ± s. e.m. From triplicate measurements.
We assessed the CODA assay kinetics. Samples with varying N1 RNA concentrations were prepared, and their FA signals were monitored as the CODA reaction proceeded (Fig. 3b). Within 5 min of the reaction start, we observed that signals had already risen above the background. We set the detection time to 20 min post-reaction; at this point, the signal level was commensurate with the RNA concentration, allowing for quantitative measurements.
3.4. Assay characterization
Applying these assay settings, we carried out a titration experiment with serially diluted N1 RNA samples (Fig. 3c). Based on 3σ/slope, where σ is the standard deviation at the lowest concentration in the linear range, the limit of detection was determined to be ~3 copy/μL. Notably, the CODA assay yielded a robust signal even at low RNA concentrations, achieving wider dynamic range than conventional RT-PCR. This merit can be attributed to a high signal-to-noise ratio inherent in the FA measurement: the fluorescence light entering photodetectors remained strong in the FA measurement, largely set by the initial reporter probe concentration. In RT-PCR, however, the fluorescence intensity at low RNA concentrations would be weak and competing with the detectors’ intrinsic noise.
We further tested the specificity of prepared probes (N1, N2, RPP30) against most common human coronaviruses (HCoV-NL63, HCoV-229E, HCoV-OC43, HCoV-HKU1) and zoonotic ones (SARS-CoV, MERS-CoV) (Fig. 3d). The N1 and N2 CODA assay was highly specific to SARS-CoV-2 with negligible cross-reactivity with SARS-CoV whose genome sequence is highly homologous (79.6%) to that of SARS-CoV-2 (Marra et al., 2003; Zhou et al., 2020).
3.5. Clinical sample analysis with CODA diagnostics
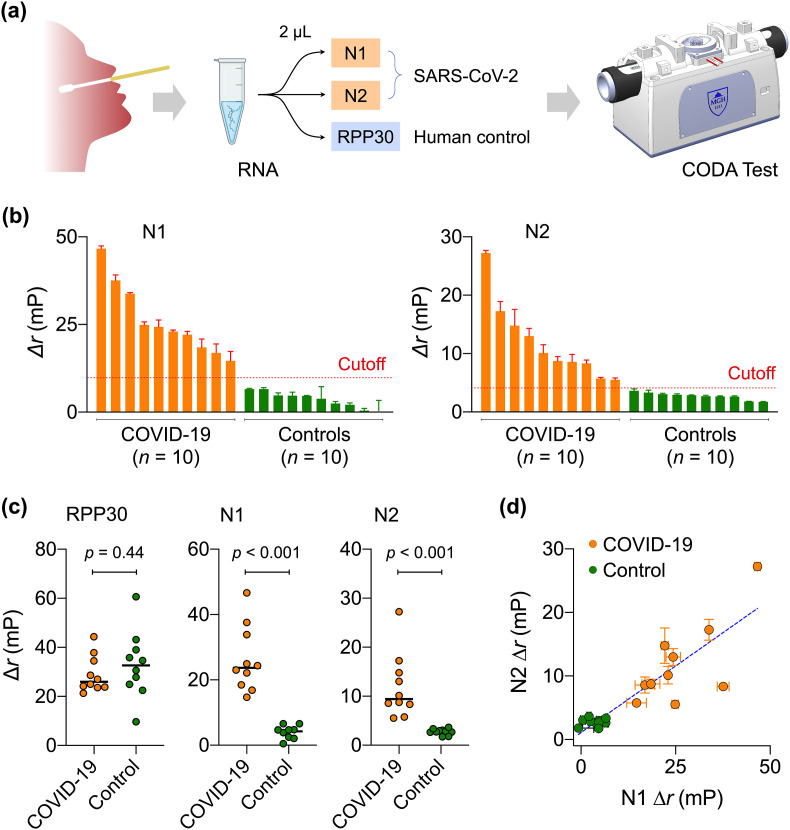
We finally applied the CODA platform to clinical COVID-19 diagnostics. Specimens (nasopharyngeal or oropharyngeal swabs, or sputum) were obtained from COVID-19 suspected individuals at Chonnam National University Hospital (CNUH, Republic of Korea). COVID-19 infection status was independently confirmed at CNUH Clinical Diagnostic Laboratory. We used ten laboratory-confirmed COVID-19 positive samples and ten confirmed negative samples (Table S2 for patient information). For each patient, we extracted total RNA from UTM. Three 2-μL aliquots were prepared from the RNA extract and analyzed for N1, N2, and RPP30 (Fig. 4 a).
Fig. 4.
COVID-19 clinical sample test with CODA platform. (a) Overall workflow of COVID-19 diagnosis with CODA. By integrating the assay and real-time signal measurement, processing, and display into the a compact device, CODA enabled onsite COVID-19 diagnosis from one-time sample loading. (b) Waterfall plot of CODA results. Clinically confirmed COVID-19 samples showed higher Δr than non-COVID-19 control samples. The cutoff values (dashed lines) were equal to 5·σT, where σT was the standard deviation of signal without target gene. (c) RPP30 levels were statistically identical between COVID-19 and control groups, passing the human sample quality check. N1 and N2 levels, however, were significantly different (two-tailed t-test, p < 0.001) between patient and control groups. (d) N1 and N2 signals of CODA, which are proportional to viral loads, were positively correlated (Pearson r = 0.76). Data are displayed as means ± s. e.m. From triplicate measurements.
Fig. 4b and Fig. S5 summarize the CODA assay results. For each gene (T = N1, N2, RPP30), we set the cutoff for positivity as Δr T = 5·σ T, where σ T is the standard deviation without the target gene. Applying these criteria, we observed that all samples passed the quality check (i.e., RPP30 positive) with no statistical difference between COVID-19 patient and control samples. In contrast, N1 and N2 were positive only in COVID-19 patient samples (Fig, 4c), matching with clinical test results (100% concordance for the current samples). Furthermore, N1 and N2 signal levels, which would be proportional to viral loads, were positively correlated (Pearson correlation coefficient, r = 0.7643; Fig. 4d).
4. Conclusions
The developed CODA platform distinguishes itself from other one-pot CRISPR/Cas assays (Ding et al., 2020). By concurrently executing isothermal amplification and CRISPR/Cas detection in a single device, CODA eliminated extra hands-on steps, completing assays within a single sample loading workflow. Measuring fluorescent anisotropy (FA) also offered practical advantages: i) the signaling probe was simpler and cheaper with CODA (i.e., fluorescent DNA) than with other methods that use DNA with a pair of fluorescence dyes and a quencher; and ii) the ratiometric nature of FA measurements made the assay robust against common noise, such as fluctuations of fluorescent intensities. The current study proved the concept by adapting CODA for POC COVID-19 diagnosis. We built a compact device integrating tight temperature control and low-noise optical measurement capability, and also established probes specific to SARS-CoV-2. Using this device, we achieved high detection sensitivity (limit of detection, 3 RNA copy/μL) and completed the entire assay within 20 min of the one-time sample loading step. Overall, CODA's assay performance was equal or superior to those of other molecular tests (see Table S3 for comparison).
Future improvements would accelerate CODA's field use. Incorporating RNA extraction into the assay system would realize a true “sample-in and answer-out” test. One promising strategy is to integrate solid-phase extraction (e.g., silica beads) (Hong et al., 2017) in a detachable cartridge. Increasing the assay throughput is also necessary, particularly for COVID-19 diagnostics, to detect all three targets (N1, N2, RPP30) at the same time. Due to its simple structure (e.g., single temperature, compact optics), the CODA device would be readily scalable for such parallelization. On the clinical side, we need to test larger patient cohorts to obtain rigorous assay statistics. Also intriguing to explore is the use of saliva as a test specimen. Not only easily collectable, saliva has also been shown to contain comparable viral loads as nasopharyngeal swabs (Wyllie et al., 2020). These efforts will help us to reach game-changing assay turnaround times, which are critical to mounting a prompt and effective response to emerging infectious diseases.
CRediT authorship contribution statement
Chang Yeol Lee: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Ismail Degani: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Jiyong Cheong: Investigation. Jae-Hyun Lee: Investigation, Writing - review & editing. Hyun-Jung Choi: Investigation, Resources, Writing - review & editing. Jinwoo Cheon: Supervision, Writing - review & editing. Hakho Lee: Conceptualization, Supervision, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the the US National Institutes of Health (R01CA229777, R21DA049577, U01CA233360, R01CA239078, R01CA237500); the US Department of Defense (W81XWH1910199, W81XWH1910194); MGH Scholar Fund; and the Korea Institute for Basic Science (IBS-R026-D1).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113049.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aman R., Mahas A., Mahfouz M. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- Bi S., Yue S., Zhang S. Chem. Soc. Rev. 2017;46:4281–4298. doi: 10.1039/c7cs00055c. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronina J., Bubniene U.S., Ramanavicius A. Biosens. Bioelectron. 2021;175:112867. doi: 10.1016/j.bios.2020.112867. [DOI] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Park K.S., Weissleder R., Castro C.M., Lee H. Chem. Commun. 2017;53:2134–2137. doi: 10.1039/c6cc09359k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Choi Y.H., Jang Y., Yu J., Goo J., Lee G., Jeong Y.K., Lee S.H., Kim I.S., Kim J.S., Jeong C., Lee S., Bae S. Nat. Commun. 2018;9:2777. doi: 10.1038/s41467-018-05245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Deng L., Zhang L., Cai Y., Cheung C.W., Xia Z. J. Gen. Intern. Med. 2020;35:1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. iScience. 2020;23:101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiCata V.J., Wowor A.J. Methods Cell Biol. 2008;84:243–262. doi: 10.1016/S0091-679X(07)84009-X. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Petralia S., Conoci S. ACS Sens. 2017;2:876–891. doi: 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- Rust P., Cereghetti D., Dual J. Lab Chip. 2013;13:4794–4799. doi: 10.1039/c3lc50857a. [DOI] [PubMed] [Google Scholar]
- Rust P., Cereghetti D., Dual J. Sens. Actuators, B. 2017;239:1118–1123. [Google Scholar]
- Shim E., Tariq A., Choi W., Lee Y., Chowell G. Int. J. Infect. Dis. 2020;93:339–344. doi: 10.1016/j.ijid.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M.J. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Wang W., Chen M., Wan Q., Wang C., Knopp D., Tang D. Nanomater. Energy. 2021;82:105711. [Google Scholar]
- Zhang B., Liu B., Tang D., Niessner R., Chen G., Knopp D. Anal. Chem. 2012;84:5392–5399. doi: 10.1021/ac3009065. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chen F., Li Q., Wang L., Fan C. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China N.C.I.A.R.T. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.