Abstract
Objective
This study aimed to improve the accuracy of CT for detection of COVID-19-associated pneumonia and to identify patient subgroups who might benefit most from CT imaging.
Methods
A total of 269 patients who underwent CT for suspected COVID-19 were included in this retrospective analysis. COVID-19 was confirmed by reverse-transcription-polymerase-chain-reaction. Basic demographics (age and sex) and initial vital parameters (O2-saturation, respiratory rate, and body temperature) were recorded. Generalized mixed models were used to calculate the accuracy of vital parameters for detection of COVID-19 and to evaluate the diagnostic accuracy of CT. A clinical score based on vital parameters, age, and sex was established to estimate the pretest probability of COVID-19 and used to define low, intermediate, and high risk groups. A p-value of <0.05 was considered statistically significant.
Results
The sole use of vital parameters for the prediction of COVID-19 was inferior to CT. After correction for confounders, such as age and sex, CT showed a sensitivity of 0.86, specificity of 0.78, and positive predictive value of 0.36. In the subgroup analysis based on pretest probability, positive predictive value and sensitivity increased to 0.53 and 0.89 in the high-risk group, while specificity was reduced to 0.68. In the low-risk group, sensitivity and positive predictive value decreased to 0.76 and 0.33 with a specificity of 0.83. The negative predictive value remained high (0.94 and 0.97) in both groups.
Conclusions
The accuracy of CT for the detection of COVID-19 might be increased by selecting patients with a high-pretest probability of COVID-19.
Abbreviations: AUC, Area Under the Curve; CI, Confidence Interval; COVID-19, Coronavirus Disease 2019; CT, Computed Tomography; IQR, Interquartile Range; NPV, Negative Predictive Value; PPV, Positive Predictive Value; ROC, Receiver Operating Characteristic; RT-PCR, Reverse Transcription-Polymerase Chain Reaction; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2
Keywords: Computed tomography, Pretest probability, Prediction of COVID-19, Vital parameters
1. Introduction
Coronavirus disease 2019 (COVID-19) has challenged healthcare systems worldwide. In regions with rapidly increasing numbers of infections, resources such as hospital beds and ventilators became scarce, and healthcare providers were faced with the prospect of having to make difficult decisions about triage and allocation [1,2]. Therefore, tools for early and precise diagnosis as well as outcome predictors for therapeutic decisions and targeted use of resources are needed.
Real-time reverse-transcription-polymerase chain reaction (RT-PCR) from deep nasal or pharyngeal throat swaps is currently considered the gold standard for the diagnosis of COVID-19. However, chest computed tomography (CT) is often used in hospitals, as it can provide immediate results and has shown high diagnostic accuracy [[3], [4], [5]]. Besides, CT imaging was reported to identify COVID-19 patients at earlier stages than RT-PCR [5]. This has been attributed to possible migration of the virus from the upper respiratory tract to the lungs or faulty execution of throat swaps.
Despite its novelty, the imaging characteristics of COVID-19 are well described, most commonly including ground glass opacities in a peripheral, bilateral dissemination pattern [6., [7], [8]]. Still, the role of CT as a screening tool remains controversial due to its limited ability to distinguish between COVID-19 and other types of viral pneumonia [9,10]. In a recent statement from the Fleischner Society, it was hence recommended that only patients with a high pretest probability of disease (moderate to severe clinical features, mild symptoms with risk factors for progression, or high pretest probability only) should undergo CT imaging [11].
Our aim was therefore to evaluate the accuracy of CT as a function of clinical symptoms and to estimate pre- and posttest probabilities of COVID-19 after CT imaging for different risk groups.
2. Material and methods
We retrospectively included 269 patients who underwent diagnostic chest CT for suspected COVID-19 between March 27th and April 27th, 2020. COVID-19 was confirmed or ruled out by RT-PCR. Repeat PCRs were performed in case of a discrepancy between CT and PCR, usually on the same day, with the new test initiated immediately after both CT and initial PCR results were available. This was true for both negative PCR and positive CT and vice versa, provided that patients had not left our institution in the meantime. Similarly, repeat RT-PCR was performed, when RT-PCR results were negative or indeterminate but suspicion of COVID-19 remained. This study was approved by the Institutional Review Board of Charité Universitätsmedizin Berlin.
2.1. CT protocol
Low-dose chest CT scans for suspected COVID-19 were acquired on two different CT scanners of our hospital: an 80-slice scanner (Aquilion Prime, Canon Medical Systems Cooperation, Otowara, Japan) and a 64-slice scanner (Lightspeed VCT, General Electric, Boston, Massachusetts, United States).
For the Canon Aquilion Prime, imaging parameters were set as followed: tube voltage 100 kV, tube current modulation between 10 and 100 mA, maximum resolution time 0.27 s, noise index 27, pitch factor 1.388 and slice thickness 0.5 mm. Parameters for the Lightspeed VCT were as follows: tube voltage 100 kV, tube current modulation between 10 and 100 mA, 0.35 s maximum resolution time, noise index 39, pitch factor 1.375, and slice thickness 0.625 mm. Iterative reconstruction was used (Canon Aquilion Prime: AIDR 3D; Lightspeed VCT: ASIR) with a lung and a soft tissue kernel (Canon Aquilion Prime: Fc01 and Fc85; Lightspeed VCT: “standard” and “lung”).
2.2. Data acquisition
All CT images were evaluated manually and data on presence/absence of COVID-19 was assessed. Vital parameters (O2 saturation, respiratory rate, and body temperature) were taken from the initial admission form or request form for chest CT. Further patient characteristics, such as sex or age, were extracted from our Radiology Information System database.
2.3. Statistical analysis
All statistical analysis was conducted using the “R” statistical programming language including the “tidyverse” and “lme4” libraries [12., 13., 14.]. Variables were expressed as means +/− standard deviation if normally distributed and as median and interquartile range (IQR) if not. Categorical variables were expressed as frequencies and percentages. Normal distribution was tested using the Shapiro-Wilk test. To compare continuous variables, the Wilcoxon rank sum test was used while Pearson's chi-square test or Fisher's exact test were used to compare categorical variables. The diagnostic value of vital parameters was evaluated using receiver operating characteristic (ROC) analysis and compared using the area under the curve (AUC). Buderer's formula was employed for post-hoc power analysis of our sample size [15].
The diagnostic accuracy of CT was calculated through a generalized mixed logistic regression model, adapted from the model proposed by Coughlin et al. [16], which was extended by a random intercept to allow adjustment for different scanner types and clinical sites (formula 1).
| (1) |
Here, Yij is defined as the binary result of chest CT in each patient. Yij = 1 is suspected COVID-19 and Yij = 0 is absence of COVID-19 in CT. X represents a matrix of covariates including the result of the RT-PCR test (Xij = 1 confirmed COVID-19, Xij = 0 absence of COVID-19 as defined by RT-PCR). β0 is the intercept, β1 represents the dichotomous result of RT-PCR, and βk corresponds to Yk for (k = 1, …, k) patient characteristics for patient i on site (or scanner) j. The random variation in the intercept is denoted by δj, while εij indicates the error. 95% confidence intervals were calculated trough parametric bootstrapping with 1000 iterations. Pre- and posttest probabilities of COVID-19 were calculated using likelihood ratios. A p-value of p < 0.05 was considered statistically significant.
3. Results
A total of 269 patients was included in this retrospective study (155 males (58%) with a median age of 71 years (IQR: 24.4)). Assessment of initial vital parameters showed a median respiratory rate of 18/min (IQR = 7/min), O2-saturation of 97% (IQR: 4%), and body temperature of 37 °C (IQR: 1.4 °C). Typical CT findings of COVID-19 were observed in 79 patients, of whom 44 were male (56%) and 35 were female (44%). In the 269 patients analyzed, COVID-19 was confirmed by RT-PCR in 34 patients (12%), with males (n = 20) being slightly more often affected than females (n = 14). However, this difference was not significant. Of the 34 RT-PCR positive patients, 28 were also CT positive.
Patient characteristics are summarized in Table 1 . It can be seen, that vital parameters different between patients without COVID-19 and patients with COVID-19, with COVID-19 patients showing higher values for CRP, higher body temperature, higher respiratory rate and lower oxygen saturation.
Table 1.
Study population and vital parameters.
| Absence of COVID-19 | Presence of COVID-19 | |
|---|---|---|
| Patients (number) | 235 | 34 |
| Women (number, %) | 100 (42.6) | 14 (41.2) |
| Age | 72, IQR: 25 | 64.79, SD: 13.47 |
| Vital parameters | ||
| Body temperature (°C) | 36.9 IQR: 1.4 | 38.01 SD: 1.12 |
| Respiratory rate (breaths/min) | 18 IQR: 5 | 23.32 SD: 6.99 |
| Oxygen saturation (%) | 98 IQR: 4 | 95 IQR: 9 |
This table gives an overview of patient characteristics of the overall study population subdivided by absence and presence of COVID-19. It becomes apparent, that the vital parameters body temperature, respiratory rate and oxygen saturation differ between the groups (absence of COVID-19 versus COVID-19), with COVID-19 patients showing higher values for CRP, higher body temperature, higher respiratory rate and lower oxygen saturation. Abbreviations: IQR: Interquartile range (in case of non-normal distribution of data); SD: standard deviation (normal distribution of data).
3.1. Diagnostic accuracy of a clinical prediction model
The diagnostic value of respiratory rate, body temperature, and blood oxygen saturation was investigated to determine whether these vital parameters alone could be used for the diagnosis of COVID-19, thus reducing the need for CT.
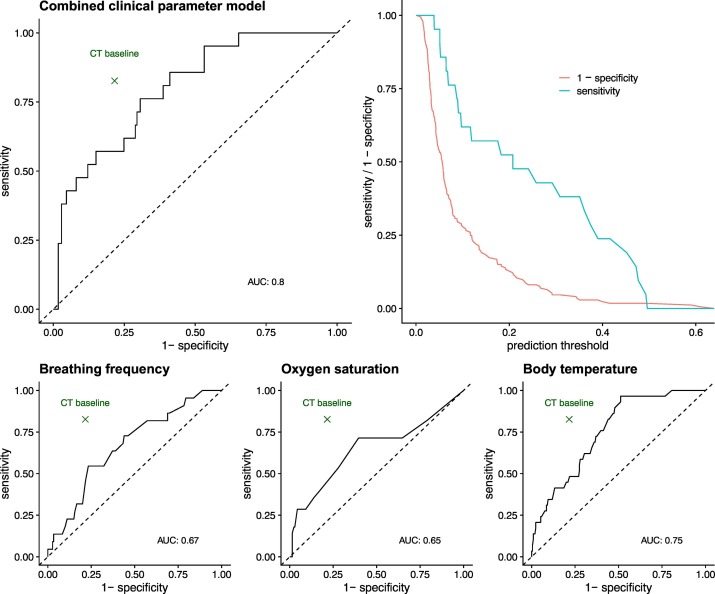
We found that the median respiratory rate and blood oxygen saturation differed significantly between patients with confirmed COVID-19 and patients with a negative RT-PCR result. The median respiratory rate of patients with confirmed COVID-19 pneumonia was 23/min (IQR: 7/min) and significantly higher than in patients without infection (18/min, IQR 5/min, Wilcoxon test (W) = 2637.5, p = 0.011). Similarly, patients with COVID-19 showed a significantly higher body temperature of 38.0 °C (IQR: 1.1 °C) and lower blood oxygen saturation of 95% (IQR 9%) compared to patients without COVID-19 (36.9 °C IQR: 1.4 °C, W = 4384, p < 0.001 and 97% IQR: 4%, W = 2029, p = 0.009). Taken together, these data indicate a potential diagnostic value of vital parameters in patients with suspected COVID-19. To compare the diagnostic accuracy of clinical parameters and CT findings, ROC curves were analyzed. The area under the curve (AUC) for respiratory rate was 0.67 (95% CI = 0.53–0.80), 0.75 (95% CI = 0.61–0.84) for body temperature, and 0.65 (95% CI = 0.60–0.75) for blood oxygen saturation. We found the largest AUC for combined clinical parameters (0.80, 95% CI = 0.68–0.90). Nevertheless, the diagnostic accuracy of the combined clinical parameters did not outperform the diagnostic accuracy of CT (see Fig. 1 ).
Fig. 1.
Predictive value of clinical parameters compared to CT.
This figure shows the diagnostic value of vital parameters alone for the diagnosis of COVID-19 and compares its accuracy with that of CT (results obtained with the baseline model). The point estimate of CT accuracy remains above the curve in all subfigures, indicating that vital parameters alone are not more accurate than a CT scan.
3.2. Diagnostic accuracy of CT
Two different models for estimating diagnostic accuracy of CT were constructed. The first, a baseline model, was only corrected for the scanner type, while the second model was also corrected for age, sex, breathing frequency, body temperature, and blood oxygen saturation. We found a sensitivity of 0.82 (95% CI 0.67–0.98) and specificity of 0.78 (95% CI 0.67–0.90) with the baseline model and a sensitivity of 0.86 (95% CI 0.71–1.00) and specificity of 0.78 (95% CI 0.57–0.98) with the models corrected for patient characteristics and initial vital parameters.
3.3. Accuracy of CT as a function of COVID-19 pretest probability
Using patient age, sex, and initial vital parameters, we propose a clinical risk score for COVID-19, which can be calculated as follows: One point is given each for a respiratory rate of ≥20/min, a body temperature ≥ 38 °C, a blood oxygen saturation ≤ 95%, patient age ≥ 65 years, and male sex. Based on this risk score, we defined three risk groups for the presence of COVID-19: low risk (0–1 points), intermediate risk (2–3 points), and high risk (4–5 points).
The pretest probability of COVID-19 was <10% in patients in the low-risk group, 10–25% in patients in the intermediate-risk group, and >25% in the high-risk group.
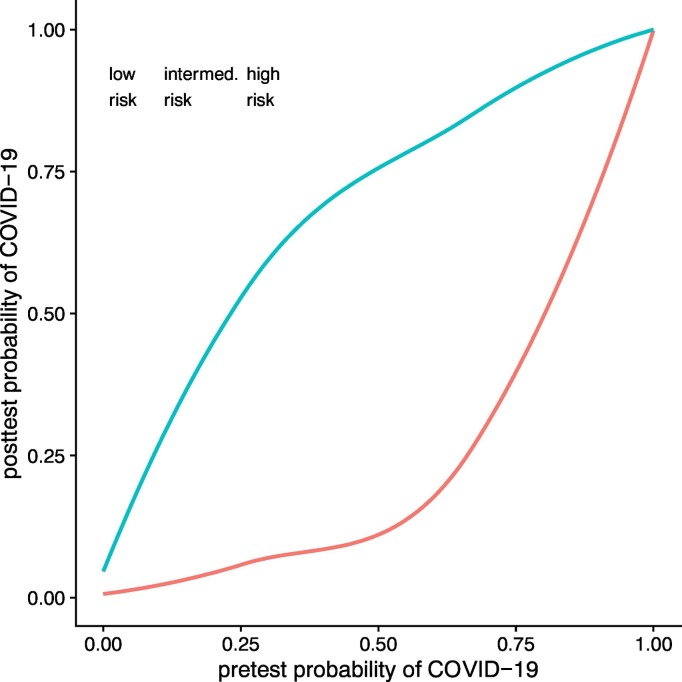
Based on the CT model, corrected for age, sex and vital parameters, for patients in the low-risk group, the positive predictive value (PPV) and negative predictive value (NPV) were 0.33 (95% CI 0.21–0.46) and 0.97 (0.95–1). In the intermediate-risk group, the PPV decreased to 0.29 (95% CI 0.15–0.44) and the NPV increased by 0.01 to 0.98 (95% CI 0.94–1). In the high-risk group, the PPV increased to 0.54 (95% CI 0.40–0.69), with the NPV only decreasing marginally to 0.94 (95% CI 0.88–1). Table 2 provides an overview of CT accuracy as a function of COVID-19 pretest probability, and Fig. 2 illustrates posttest probabilities for COVID-19 before and after a negative/positive CT result.
Table 2.
Diagnostic accuracy models of CT and CT combined with vital parameters.
| Model | Sensitivity |
Specificity |
PPV |
NPV |
|---|---|---|---|---|
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | |
| CT (baseline) | 0.82 (0.67–0.98) | 0.78 (0.67–0.90) | 0.35 (0.15–0.53) | 0.97 (0.96–1.00) |
| CT (adjusted) | 0.86 (0.71–1.00) | 0.78 (0.57–0.98) | 0.36 (0.16–0.55) | 0.97 (0.95–1.00) |
| Low-risk | 0.76 (0.61–0.91) | 0.83 (0.77–0.90) | 0.33 (0.21–0.46) | 0.97 (0.95–0.99) |
| Intermediate-risk | 0.84 (0.63–1.00) | 0.76 (0.65–0.87) | 0.29 (0.15–0.44) | 0.98 (0.94–1.01) |
| High-risk | 0.89 (0.76–1.00) | 0.68 (0.54–0.83) | 0.54 (0.40–0.69) | 0.94 (0.88–1.00) |
This tables shows the results for different models of diagnostic accuracy (baseline CT and CT adjusted for age, sex, and vital parameters) for all patients regardless of risk group, as well as the accuracy when the adjusted CT model was stratified by the pre-test probability of COVID-19.
Fig. 2.
Probability of COVID-19 after CT.
This figure shows the probability of COVID-19 before CT on the x-axis and the calculated and extrapolated posttest probabilities after a positive (blue line) or negative (red line) CT scan. It becomes apparent that for patients in the low-risk group, even after a positive scan, the probability of COVID-19 is only 50%, casting doubt on the appropriateness of using CT in this patient subset. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
To date, RT-PCR remains the reference standard for the diagnosis of COVID-19, while CT has the advantage of detecting patients with lung infiltrates fast and early. Although the role of chest CT as a screening tool for COVID-19 or other respiratory tract infections is being avidly discussed, it is currently not recommended by the Fleischner Society and the American College of Radiology [12,20].
We found that CT accuracy is influenced by the prevalence/pretest probability of COVID-19 and was highest in patients with a high pretest probability/prevalence estimate of COVID-19. Based on our results, we therefore believe that CT may have added value in the management of COVID-19, not as a screening tool, but for triaging patients in the emergency department to the best treatment path depending on the severity of the disease. In patients presenting with fever and shortness of breath, a positive CT scan might shorten the time to diagnosis and facilitate fast hospitalization and focused treatment. On the other hand, a negative CT result does not completely rule out COVID-19 and especially symptomatic patients should therefore initially continue to be isolated, with further testing to identify possible other causes that explain their symptoms, such as abdominal infections, malignancies or cardiovascular diseases.
Nevertheless, not all patients with some form of respiratory symptoms should undergo CT imaging. Especially in patients with few symptoms (e.g. young patients with normal body temperature), CT examinations are most likely not effective with a very low positive predictive value. As positive CT results may only have added value if the positive predictive value is high [11,16], we advocate our approach of calculating a fast-and-easy to use score to estimate the pre-test probability of COVID-19. Another important aspect is, that maintaining hygiene can be difficult when a larger number of patients need to have a CT scan in a short time, and CT might thus become a hub for new infections [17]. Finally, the results of the present study might be helpful to institutions without rapid access to Covid-19 tests, especially when a low radiation dose protocol for minimization of radiation burden is used.
Luo et al. previously investigated the benefit of a scoring system based on the most common imaging findings to improve CT accuracy [5,18]. They reported score-dependent specificities ranging from 0.23 to 0.95. However, the disadvantage of their approach is that their scoring system requires the patient to undergo a CT first. In our approach, we chose variables that are among the first parameters to be assessed when the patient arrives in the emergency department/hospital. This allows immediate calculation of the pretest probability of COVID-19 and subsequent further triage of patients depending on their pretest probability. As a limitation, we have not included other patient information such as medical history, other diseases (especially lung diseases and obesity), laboratory data and medication, as they were not reliably recorded on initial assessment of patients [[18], 19., [20]]. However, the inclusion of some of these information might have been beneficial, as it could also have allowed for an assessment of the potential severity of the disease course. Leung et al. have shown that chronic obstructive lung disease (COPD) is a risk factor for COVID-19, as the patients affected by both had a more severe course of the disease [19,20]. Furthermore, several laboratory examinations have were demonstrated to be associated with more severe (e.g. elevated procalcitonin) or even less severe (elevated white blood cell count) disease (21).
Some studies, which advocated CT as a screening tool, reported surprisingly high sensitivities [5,18]. Ai et al. found a very high sensitivity of 0.97 for the detection of COVID-19 (higher than our reported values), but a relatively low specificity of 0.25 [5]. In their meta-analysis Xu et al. reported similar values for sensitivity (0.92) and specificity (0.25 and 0.33) [18]. While our sensitivity was lower, our specificity was substantially higher. A possible explanation for this may be the low prevalence of confirmed COVID-19 cases in our patient population (12% versus 59% in Ai et al.) and therefore a high proportion of true negative results [5]. However, it has to be noted that the above-quoted estimates of diagnostic accuracy should be interpreted with caution, as they were obtained in patients with already suspected COVID-19 in a hospital setting and are therefore prone to substantial selection bias [11,19]. The accuracy, especially the negative predictive value of RT-PCR has been criticized in recent studies. Kucirka et al. conducted a meta-analysis and estimated the false negative rate to be 38% by the day of onset of symptoms, which decreased to 20% the third day after onset of symptoms, but then slowly started to increase again [20]. False negative results could therefore also have affected the accuracy of CT in our study, resulting in a high estimate of sensitivity and a low estimate of specificity.
Our study has several more limitations. Similar to other retrospective studies with COVID-19 data, it was performed in a hospital setting, which probably led to some selection bias. Patients presenting in the emergency department have a higher a priori probability of disease, so the accuracy of the CT is not transferable to an outpatient setting. Selection bias could be further enhanced by the fact that the parameters used for our risk score might also have been used by the physician before referring a patient for CT. The lack of patient information regarding their medical history, laboratory data and ongoing limitation has to be acknowledged as another important limitation of this study In addition, the absolute number of COVID-19 cases confirmed by RT-PCR was relatively low. This small sample size may have affected the pre-test probabilities calculated using our risk score, and estimates of diagnostic accuracy of CT might differ to some degree if repeated in larger cohorts. However, we tried to at least partially correct for this uncertainty by calculating 95% confidence intervals for all estimates of diagnostic accuracy. Furthermore, we observed an overlap for clinical data between the infected and non-infected groups, as shown by the IQR, which likely has an impact on the diagnostic performance of the model. Also, the inclusion of the pulse rate might have had a positive effect on our calculation of the pre-test probability. However, since it was not reliably documented during initial patient assessment, it could not be included in our analysis.
5. Conclusions
A simple score to estimate the pre-test probability of COVID-19 can be calculated using age, sex, O2 saturation, respiratory rate, and body temperature. Provided that only patients with a high pre-test probability undergo the examination, the accuracy of CT for the detection of COVID-19 might be increased.
Declaration of competing interest
There are no competing interests to declare.
References
- 1.Emanuel E.J., Persad G., Upshur R., Thome B., Parker M., Glickman A., et al. Fair allocation of scarce medical resources in the time of Covid-19. N Engl J Med. 2020;382:2049–2055. doi: 10.1056/NEJMsb2005114. [DOI] [PubMed] [Google Scholar]
- 2.Joebges S., Biller-Andorno N. Ethics guidelines on COVID-19 triage-an emerging international consensus. Crit Care. 2020;24(1):201. doi: 10.1186/s13054-020-02927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. 200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Tang Y., Mo Y., Li S., Lin D., Yang Z., et al. A diagnostic model for coronavirus disease 2019 (COVID-19) based on radiological semantic and clinical features: a multi-center study. Eur Radiol. 2020;30(9):4893–4902. doi: 10.1007/s00330-020-06829-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 Feb 26 doi: 10.1148/radiol.2020200642. Published online 2020 Feb 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson S, Kay FU, Abbara S, Bhalla S, Chung JH, Chung M, et al. Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. J Thorac Imaging 2020. [DOI] [PMC free article] [PubMed]
- 7.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 8.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3):200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology 2020:201343. [DOI] [PMC free article] [PubMed]
- 10.Adams H.J.A., Kwee T.C., Kwee R.M. COVID-19 and chest CT: do not put the sensitivity value in the isolation room and look beyond the numbers. Radiology. 2020 Apr 27 doi: 10.1148/radiol.2020201709. Published online 2020 Apr 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology. 2020;296:172–180. doi: 10.1148/radiol.2020201365. 201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Team RC . 2013. R: a language and environment for statistical computing. [Google Scholar]
- 13.Wickham H. 1(1) 2017. Tidyverse: easily install and load the ‘tidyverse’. R package version 2017. [Google Scholar]
- 14.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:14065823 2014.
- 15.Buderer N.M.F. Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad Emerg Med. 1996;3(9):895–900. doi: 10.1111/j.1553-2712.1996.tb03538.x. [DOI] [PubMed] [Google Scholar]
- 16.Coughlin S.S., Trock B., Criqui M.H., Pickle L.W., Browner D., Tefft M.C. The logistic modeling of sensitivity, specificity, and predictive value of a diagnostic test. J Clin Epidemiol. 1992;45(1):1–7. doi: 10.1016/0895-4356(92)90180-u. [DOI] [PubMed] [Google Scholar]
- 17.Hope M.D., Raptis C.A., Shah A., Hammer M.M., Henry T.S. A role for CT in COVID-19? What data really tell us so far. The Lancet. 2020;395(10231):1189–1190. doi: 10.1016/S0140-6736(20)30728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu B., Xing Y., Peng J., Zheng Z., Tang W., Sun Y., et al. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 2020;30(10):5720–5727. doi: 10.1007/s00330-020-06934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radiology ACo. ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. ACR website Advocacy-and Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CTfor-Suspected-COVID19-Infection Updated March 2020;22.
- 20.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173(4):262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]