Abstract
Objectives
To examine severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant replacement in association with containment capacity and changes in case fatality at country level.
Methods
Altogether, 69 571 full SARS-CoV-2 genomes collected globally within the first 6 months of the pandemic were examined. The correlation between variant replacement and containment capacity was examined by logistic regression models using the WHO International Health Regulation (IHR) score, the Oxford COVID-19 Government Response Tracker (OxCGRT) and the vulnerability index INFORM as proxies, while correlation with changes in monthly crude case fatality ratios was examined by a mixed effect model.
Results
At the global level, variant lineage G∗, characterized by the S-D614G mutation, replaced the older lineages L and S in March 2020. European countries—including Finland, France and Italy—were the first to reach a 50% increment of G∗, whereas only Singapore and South Korea had non-G∗ persisting throughout the first 6 months. Countries with higher IHR scores (β-coefficient –0.001, 95%CI –0.016, –0.001; p 0.034) and higher stringency indexes (OxCGRT) (β-coefficient –0.011, 95%CI –0.020, –0.001; p 0.035) were associated with lower levels of G∗ replacement, whereas higher vulnerability indexes (INFORM) (β-coefficient 0.049, 95%CI 0.001, 0.097; p 0.044) were associated with higher replacement levels. Crude case fatality ratio showed a positive correlation with G∗ replacement (β-coefficient: 0.034, 95%CI 0.011, 0.058; p 0.004), even after adjusting for testing capacity and other country-specific characteristics.
Conclusions
SARS-CoV-2 variant lineage G∗ (S-D614G) replaced older lineages more efficiently in countries with lower containment capacity, and its possible association with increased disease severity deserves further investigation.
Keywords: Containment, Fatality, Lineage, Replacement, SARS-CoV-2, Variant
Introduction
An outbreak of unexplained pneumonia was recognized in December 2019 in Wuhan, China [1]. On 30th January 2020 the World Health Organization (WHO) declared the outbreak a Public Health Emergency of International Concern [2]. European countries then became another epicentre, and the WHO declared the novel coronavirus disease 2019 (COVID-19) a pandemic on 11th March 2020 [3].
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is expected to mutate and evolve during the course of adaptation in humans [4]. The Global Initiative on Sharing All Influenza Data (GISAID) classified variants into six high-level phylogenetic groups from the early split of S and L, to the further evolution of L into V and G, and later of G into GH and GR [5]. A recent report suggested that a mutation of the spike protein, S-D614G, could be associated with increased infectivity [6]. To explore the implications of SARS-CoV-2 evolution, we analysed the replacement of SARS-CoV-2 variants during the first 6 months of the pandemic in association with containment capacity and case fatality from a global perspective.
Methods
Variant identification and replacement pattern
Complete SARS-CoV-2 genome sequences collected within the first 6 months of the pandemic (on or before 30th June 2020) were downloaded from GISAID [5]. All sequences were called for open reading frames (ORFs) using blastn v2.10.1 [7] and bedtools v2.29.2 [8], and concatenated based on aligned ORFs using mafft v7.471 [9]. Sequences of poor quality—including those missing more than 5000 bp or having more than ten ambiguous variations—were filtered. A tree inferred from all sequences was prepared using FastTree v2 [10] to select representative genomes from main topology branches for calling single nucleotide polymorphisms (SNPs) and for constructing a maximum likelihood phylogenetic tree using RAxML v8.2.12 [11]. With reference to GISAID, variants were assigned to four lineages: L (reference sequence, nucleotide T28144, ORF8 amino acid L84), S (nt T28144C, ORF8 aa L84S), V (nt G26144T, ORF3a aa G251V) and G∗ (nt A23403G, S gene aa D614G). The lineage G∗ was further divided into three clades, including G, GH (G25563T, ORF3a aa Q57H) and GR (G28883C, N gene aa G204R).
The geographic source and date of collection were retrieved from GISAID to construct the dispersion and replacement of SARS-CoV-2 variants at global, continent and country levels.
Containment capacity and implementation of public health measures
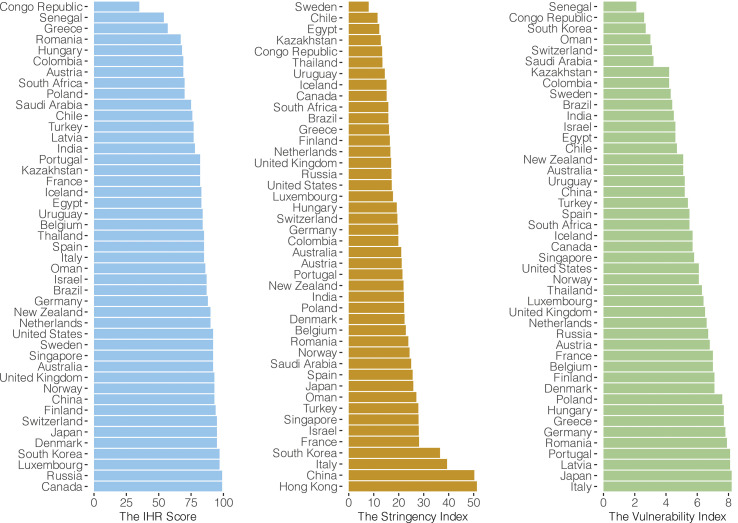
We hypothesized that the degree of spread of, and hence the replacement with, newly emerged variants could be related to the containment capacity. Three internationally recognized indexes—namely the WHO International Health Regulation (IHR) score, the stringency index, and the vulnerability index—were used as proxies of containment capacity and implementation.
The IHR score
Members of the IHR report to the WHO annually on the implementation of capacity required by the regulations to sustain public health response and surveillance. These regulations are legal instruments designed to develop the capacity of all members for preventing, detecting, assessing, notifying, and responding to internationally concerned public health events. The IHR score includes 13 IHR capacity items assessed by 24 indicators [12].
The stringency index
We use the Oxford COVID-19 Government Response Tracker (OxCGRT) developed by the University of Oxford to reflect the stringency of government responses to the pandemic across regions over time [13]. The constructs of OxCGRT consist of workplace closure, school closure, public transport closure, public event cancellation, staying-at-home requirements, restrictions on gathering size, restrictions on internal movement, restrictions on international travel, and public information campaigns. This index has previously been shown to be a good predictor for COVID-19 pandemic control [14].
The vulnerability index
The composite index INFORM for risk management was developed by the European Commission to indicate the vulnerability of regions at risk of crisis that could overwhelm its response capacity [15]. It ranks regions according to their needs for global assistance, and provides a risk profile for each region. This index has been validated by a panel of experts [16].
We performed three separate multivariable linear regression models to examine the association between variant replacement and containment capacity. At the global level, we observed that the new variant lineage G∗ started to replace the older variants in March 2020. Therefore, we took March as a critical switching point, where the proportion of G∗ lineage among new sequences detected in March was treated as a continuous outcome variable, and the most updated IHR score (2019), stringency index (the average of all OxCGRT scores on or before 31st March 2020) and vulnerability index (INFORM scores at 2018) as the variables to test for associations. All the three models were controlled for Gross Domestic Product (GDP) per capita from the World Bank [17], Human Development Index (HDI) from the United Nations [18], and the population density of each country from the World Population Review 2020 [19].
Case fatality
We obtained the numbers of cumulative incidences of and deaths from COVID-19 from the WHO dashboard [20], and determined the crude case fatality ratios (CFRs) by dividing the cumulative number of deaths by the cumulative number of reported cases for each country. A scatterplot was generated to explore the association between CFRs and the proportion of variants belonged to the G∗ lineage from January to June 2020. To account for between-country variability, we employed linear mixed effect models to examine the association between CFR and proportion of variants belonging to the G∗ lineage in each month. As the testing capacity is highly correlated with the number of cases detected, the country-specific numbers of tests per 1000 population by months and other country-specific characteristics (i.e. proportion of population aged ≥65 years, gross domestic product per capita, population density, and number of hospital beds per 1000 population) were adjusted in the model [21]. A random intercept term was used to adjust for between-country variations from repeated measurements. Suppose y ij is the monthly CFR on month j in country i, the full model form is:
where β 0 is the grand intercept, x ij is the monthly proportion of variants belonged to lineage G∗ with regression coefficient β 1, w ij is the monthly testing capacity with regression coefficient β 2 for country i on month j, and z i (p) is the p-th country-specific characteristic variable with regression coefficient β p. The country-specific random effect is modelled as α i which followed a normal distribution with mean 0 and variance σ α 2 on top of the random error (ε ij) within country i over time.
To examine whether the observation was affected by the extreme CFRs from the countries whose healthcare facilities were overwhelmed, a separate analysis excluding countries outside the regression prediction interval was conducted. A subgroup analysis for low and high testing capacity using a median cut-off was also conducted.
Results
Variant replacement—global level
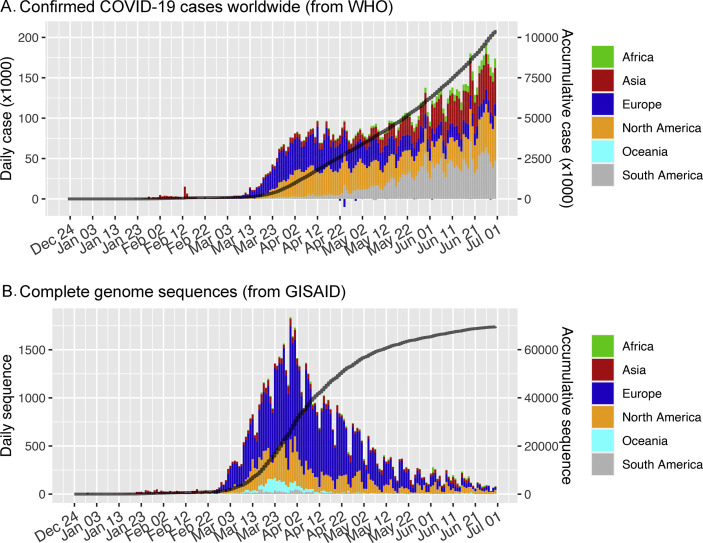
By 30th June 2020, at least 10 450 456 confirmed COVID-19 cases had been reported to the WHO [22] (Fig 1 A), with 69 571 high-quality complete genome sequences deposited in GISAID fulfilling the criteria to be included in this study (Fig. 1B). These sequences were from six continents (62.9% Europe, 23.0% North America, 7.5% Asia, 3.7% Oceania, 1.6% South America, 1.4% Africa) (Supplementary Material Table S1) and 100 countries/cities.
Fig. 1.
Number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections and available full genome sequences in the first 6 months of the pandemic. (A) Daily coronavirus disease 2019 (COVID-19) infections reported to the World Health Organization (WHO). (B) Complete genome sequences available in GISAID (Global Initiative on Sharing All Influenza Data) according to the date of collection.
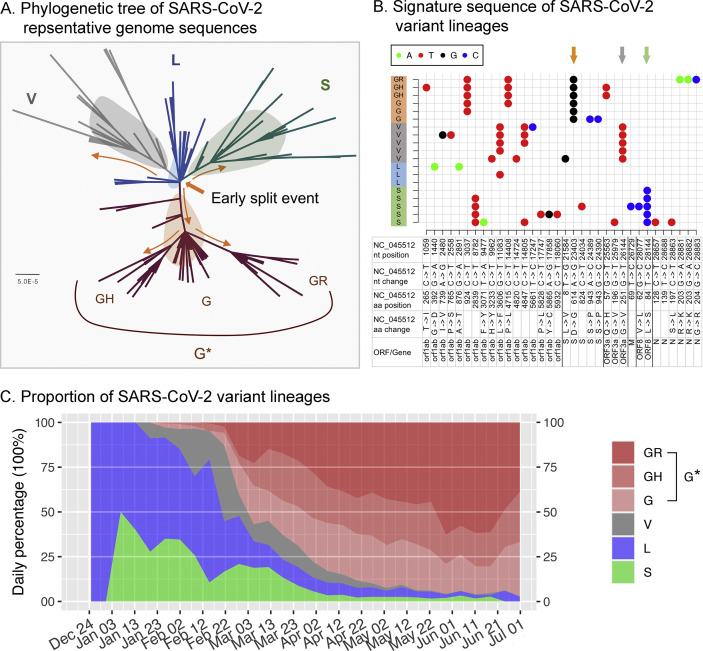
The phylogenetic tree topology revealed four major lineages (L, S, V and G∗); lineage G∗ is further divided into three clades (G, GH and GR) (Fig. 2 A). The sequence signature patterns of each lineage are shown in Fig. 2B, with S-D614G consistently detected from clades G, GR and GH of lineage G∗.
Fig. 2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant lineages. (A) A maximum likelihood tree based on representative SARS-CoV-2 complete genome sequences. Yellow arrows indicate estimated evolutionary events of variant split. The lineage G∗ comprises clades G, GH and GR. (B) Signature sequence patterns of SARS-CoV-2 variant lineages. Arrows highlight lineage-specific sequence variations. (C) The change in proportion of SARS-CoV-2 variants in the first 6 months of the pandemic. The x-axis indicates the collection date of the isolates.
When the outbreak was first recognized in December 2019, all sequenced isolates (before 1st January 2020) were from China and belonged to lineage L, whereas lineage S was detected from early January in China (Fig. 2C). Then, from January to mid-February, both lineages L and S were detected and contributed to the majority of sequenced isolates.
Isolates belonging to lineage V were first reported from China on 21st January, and were soon followed by another newly detected lineage, G∗, on 24th January, also from China. Subsequently, two clades (GH and GR) of lineage G∗ were first reported on 27th January and 16th February, respectively; both were from the United Kingdom. From the global perspective, lineage G∗ appeared from late January, and quickly replaced L and S starting from early March. Lineage V remained as a minor fraction until early May, and was then rarely detected (Fig. 2C).
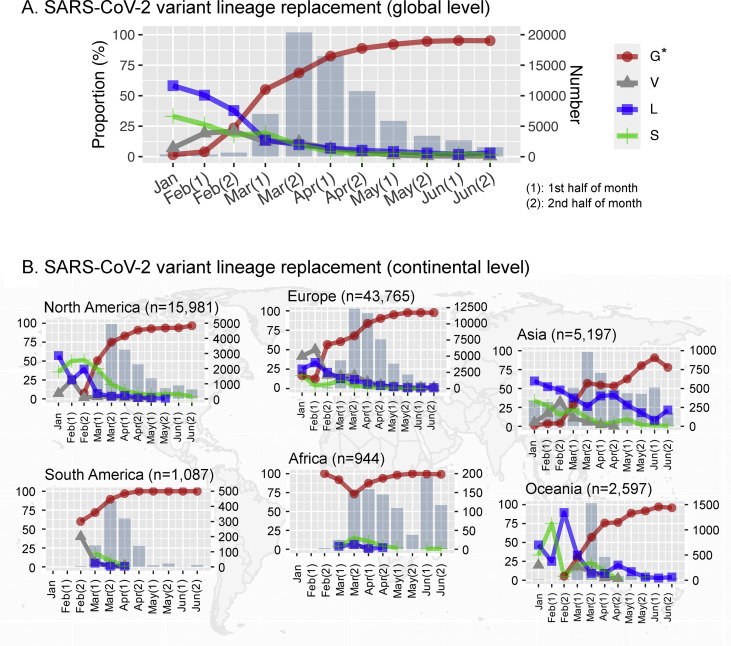
Replacement of lineage G∗ as observed at the global level (Fig. 3 A) was reproduced in most continents, including Europe, North America, Oceania and Asia (Fig. 3B). While lineage G∗ also predominated in South America and Africa, early sequences from these two continents were not available to make a clear distinction between replacement by or persistent of lineage G∗ right from the beginning.
Fig. 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant lineage replacement over the first 6 months of the pandemic. The proportion of variant lineages identified over half-month periods from the earliest available sequence to June 2020 is shown at (A) global and (B) continental levels. Jan, the complete genomes isolated on January 2020 and before. ‘1’ and ‘2’ represent the first and second half of the month, respectively.
Variant replacement—country level
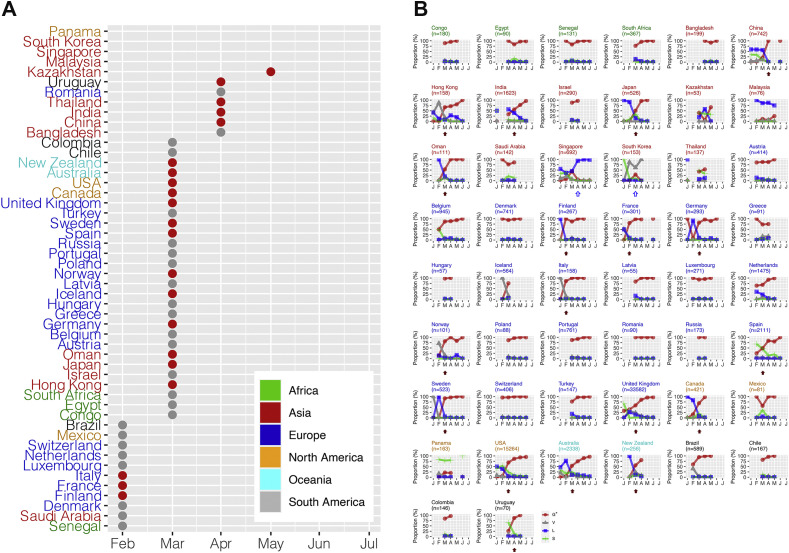
Globally, replacement by lineage G∗ occurred in March 2020, but with variations in the timing of replacement at country level; some countries did not exhibit substantial replacement throughout the first 6 months of the pandemic. We included countries with more than 50 high-quality full-genome sequences collected within the first 6 months of the pandemic for country-level analysis. Hong Kong, a city of China, also provided more than 50 high-quality full genomes, and was included in the analysis. Fig. 4 A shows the time to reach 50% increase, i.e., 50% of the newly identified isolates belonged to lineage G∗. Finland, France and Italy were the first countries to reach a 50% increase in February, whereas others—e.g. Panama, Malaysia, Singapore and South Korea—did not reach 50% increase during our study period (until the end of June). In some countries—such as Senegal and Saudi Arabia, Denmark, Luxembourg, Netherlands, Switzerland Mexico and Brazil—lineage G∗ was predominant right from the beginning.
Fig. 4.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant lineage replacement over the first 6 months of the pandemic at country level. Countries with >50 complete genome sequences available before 30th June 2020 were included. Hong Kong, a city of China, with >50 complete genome sequences available, was also included. (A) Red/grey dots indicate the time when lineage G∗ accounted for 50% or more of the new isolates. Red dots indicate countries with non-G∗ lineages predominating in the early phase, then replaced by lineage G∗. Grey dots indicate countries in which lineage G∗ predominated right from the beginning. (B) Solid arrows indicate countries showing a clear pattern of variant replacement by lineage G∗. Empty arrows indicate countries showing persistence of a non-G∗ lineage.
Fig. 4B shows the change in proportions of the four lineages at country level. First, a clear pattern of variant displacement by lineage G∗ was observed in China, Hong Kong, India, Japan, Oman, Finland, France, Germany, Italy, Norway, Spain, Sweden, the United Kingdom, Canada, the USA, Australia, New Zealand and Uruguay. In these countries, non-G∗ lineage(s) predominated in the earlier phase, and were then replaced by lineage G∗. Second, there was a clear pattern of persistence with a non-G∗ lineage; this pattern was exhibited by only two countries, Singapore and South Korea. The third pattern represents countries in which lineage G∗ was predominant at a later stage, but there weren't enough earlier sequences to confirm replacement.
Variant replacement and containment capacity
The association between variant replacement and containment capacity was examined at country level. We first included 45 countries with at least one of the three proxy parameters to indicate containment capacity (Supplementary Material Fig. S1). Multivariate regression analyses showed that countries with higher IHR scores (β-coefficient: –0.001, 95%CI –0.016, –0.001; p 0.034) and higher stringency indexes (β-coefficient: –0.011, 95%CI –0.020, –0.001; p 0.035) were associated with lower levels of lineage G∗ replacement, whereas higher vulnerability indexes (β-coefficient: 0.049, 95%CI 0.001, 0.097; p 0.044) were associated with higher levels of variant replacement.
Variant replacement and disease severity
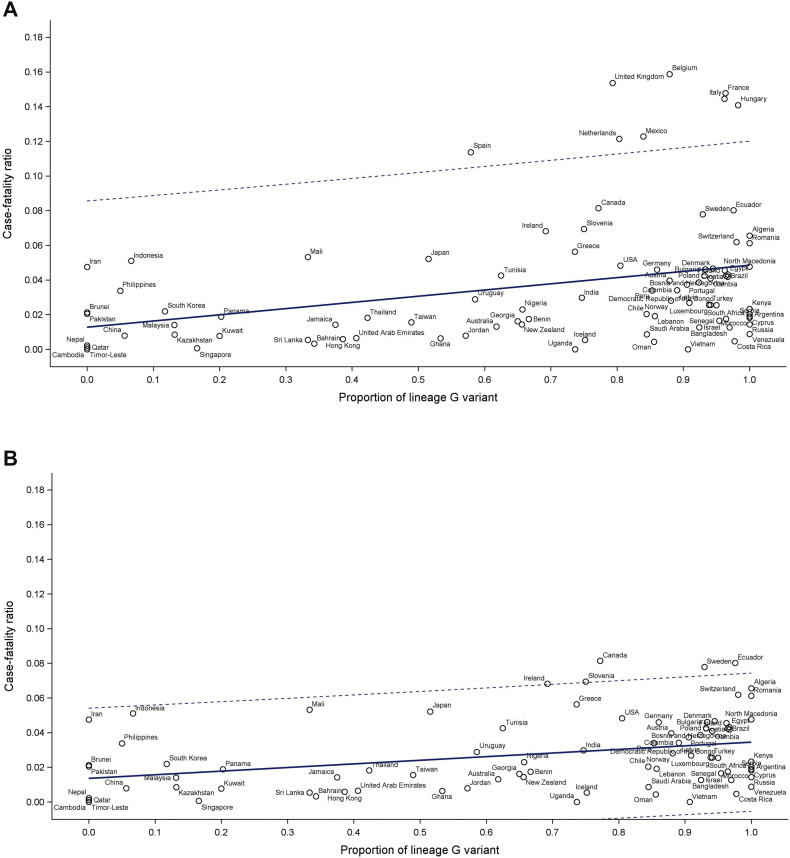
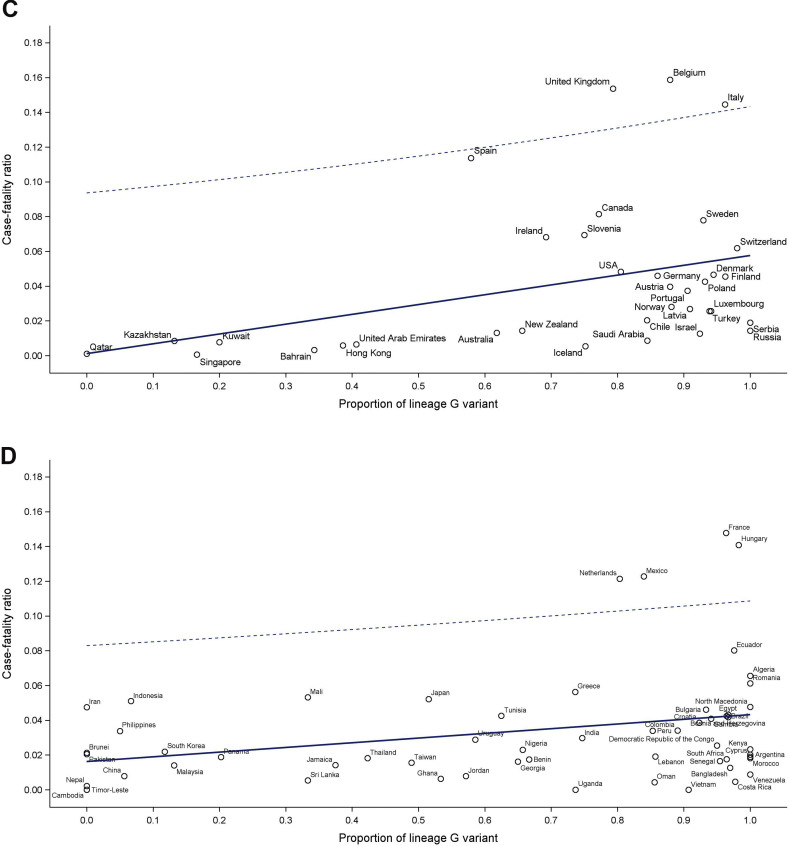
Fig. 5A shows the association between CFRs and proportion of variant lineage G∗ for each country over the first 6 months of the pandemic. The mixed effect model analysis accounting for monthly measurements, testing capacity, and country-specific characteristics revealed a significant association between CFR and variant replacement (β-coefficient: 0.034, 95%CI 0.011, 0.058; p 0.004). While some countries experienced an overwhelming healthcare demand, and thus their CFRs were inflated, the association between CFR and proportion of variant lineage G∗ was still statistically significant (β-coefficient: 0.024, 95%CI 0.006, 0.041; p 0.008) when countries with high CFRs (including Spain, Hungary, The Netherlands, Italy, France, the United Kingdom, Belgium, and Mexico) were excluded (Fig. 5B). The association was also robust among countries with higher (>40) or lower (≤40) cumulated numbers of SARS-CoV-2 tests performed per 1000 population (Figs. 5C,D).
Fig. 5.
Relationship between crude case fatality ratios and proportions of lineage G variant. (A) All countries. (B) Countries with case fatality outside the predicted level excluded. (C) Countries having >40 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) tests performed per 1000 population. (D) Countries having ≤40 SARS-CoV-2 tests performed per 1000 population.
Discussion
The S-D614G mutation is a signature of variants of lineage G∗, including clades G, GH and GR, which could induce conformational modification that facilitates exposure of the cleavage domain to proteases [23]. While a higher infectivity of this variant has been suggested, its implication for disease severity and the fate of the pandemic is unknown [24].
The phenomenon of global replacement by lineage G∗ (S-D614G variants) as documented in this study could be due to its survival benefits or simply a founder effect. Since we did not examine replication or transmission efficiency directly, we can only hypothesize based on the changes in proportion of different lineages over time. For a majority of counties, lineage(s) L/S was/were present in the early period and then replaced by lineage G∗, suggesting lineage G∗ has survival benefits [6]. In a few countries, including Korea and Singapore, lineage G∗ did not outgrow the others. Thus, survival benefits were not exhibited in these populations. For a group of countries, notably South America and Africa, in which lineage G∗ predominated right from the beginning, it is consistent with a founder effect. Taken together, it is possible that both survival benefits and founder effect have accounted for the global predominance of lineage G∗ (S-D614G variants).
We observed that S-D614G replacement started in late February 2020, and was followed by the exponential upsurge in reported cases 2 weeks later in mid-March 2020. We thus use March 2020 as the ‘critical variant replacement period’ to examine its association with containment capacity and response. The results showed that countries with higher containment and public health response capacity had delayed lineage replacement, probably due to their success in suppressing importation and/or delaying local spread of this newly emerging variant. Our findings support using the IHR score, OxCGRT and INFORM to reflect the stringency of government responses and the vulnerability of a country in the context of pandemic. Of note, two countries in Asia, Singapore and South Korea, exhibited a clear persistence of the older lineages, L and V respectively, suggesting that their later waves were due mainly to continuous circulation and upsurge of local infections, rather than to importation of new variants. Strategies to suppress importation in these two countries could be of learning value to others.
We examined the changes in population-level case fatality ratio in association with the changes in proportion of lineage G∗ in an attempt to understand its effect on disease severity. It is anticipated that comparing crude fatality ratios at country level is subject to biases and confounders. We tried a few alternative analyses by excluding countries with extremely high case fatality, and by stratifying countries according to their testing capacity. We also adjusted for country-specific demographic features. While a significant association between lineage G∗ replacement and increased disease severity was observed from our mixed effect model, one should interpret this cautiously as it may not represent a causative association. For instance, information on the infected population in each country, such as age distribution and comorbidity status, was not available for a more robust analysis.
Our study has limitations. First, the availability of genome sequences is subject to biases such as sequencing capacity, sampling location and timing. Second, deaths and infections due to SARS-CoV-2 are bound to be underreported and linked to complex confounders. Nevertheless, the variant replacement revealed in this study is no doubt a genuine observation, and its possible association with increased disease severity should be further verified using appropriate patient cohorts and biological models.
Author contributions
PKSC conceived and supervised the study. ZC, KCC, MCSW and JH collected and analysed data. SSB, MHW, RWYN, CKCL interpreted data and prepared the manuscript.
Transparency declaration
Dr Maggie H. Wang is one of the shareholders in Beth Bioinformatics Co. Ltd. All other authors declare no conflicts of interest. This study was supported by the Health and Medical Research Fund Commissioned Research on the Novel Coronavirus Disease (COVID-19) (reference no. COVID190103) from the Food and Health Bureau, Hong Kong SAR Government; and the Project Impact Enhancement Fund (Project number PIEF/Ph2/COVID/11) from the Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China.
Acknowledgements
We gratefully acknowledge the authors and the originating and submitting laboratories of the sequences from GISAID's EpiFlu™ Database (https://www.epicov.org/) on which this research is based.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.01.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization 2019-nCoV outbreak is an emergency of international concern 2020. http://www.euro.who.int/en/health-topics/health-emergencies/international-health-regulations/news/news/2020/2/2019-ncov-outbreak-is-an-emergency-of-international-concern Available from:
- 3.World Health Organization WHO Director-General’s opening remarks at the media briefing on COVID-19—11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Available from:
- 4.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Nat Acad Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Global initiative on sharing all influenza data. https://www.gisaid.org/ Available from: [DOI] [PMC free article] [PubMed]
- 6.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh K., Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization E-SPAR State Party Annual Report. https://extranet.who.int/e-spar Available from:
- 13.The University of Oxford Variation in Government responses to COVID-19. https://www.bsg.ox.ac.uk/research/publications/variation-government-responses-covid-19 Available from:
- 14.Wong M.C., Huang J., Teoh J., Wong S.H. Evaluation on different non-pharmaceutical interventions during COVID-19 pandemic: An analysis of 139 countries. J Infect. 2020;81:e70–e71. doi: 10.1016/j.jinf.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marin-Ferrer M., Vernaccini L., Poljansek K. EUR; 2017. Index for Risk Management INFORM Concept and Methodology Report—Version 2017. [Google Scholar]
- 16.Wong M.C., Teoh J.Y., Huang J., Wong S.H. The potential impact of vulnerability and coping capacity on the pandemic control of COVID-19. J Infect. 2020 Nov;81:816–846. doi: 10.1016/j.jinf.2020.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.United Nations Development Programme, Human Development Reports 2019 Human Development Index Ranking. http://hdr.undp.org/en/content/2019-human-development-index-ranking Available from:
- 18.The Economist Intelligence Unit World Bank and Central Intelligence Agency World Factbook. https://www.cia.gov/library/publications/the-world-factbook/geos/we.html Available from:
- 19.Countries by density by population 2020 World Population Review. https://worldpopulationreview.com/countries/countries-by-density/ Available from:
- 20.World Health Organization Coronavirus disease (COVID-19) outbreak situation. Dashboard. 2020 https://covid19.who.int/ Available from: [Google Scholar]
- 21.Our World in Data, Statistics and Research Coronavirus (COVID-19) Testing 2020. https://ourworldindata.org/coronavirus-testing Available from:
- 22.Johns Hopkins University COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) https://coronavirus.jhu.edu/map.html Available from:
- 23.Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.