Abstract
We are still facing a Covid-19 pandemic these days and after the aggressively infection control measures taken by the governments in the whole world, there is a need of a rapid pharmaceutical solution in order to control this crisis. The computer aided chemistry and molecular docking is a rapid tool for drug screening and investigation. Moreover, more metal-based drugs are tested daily by research institutes for their antiviral activity. Here, we make use of theoretical studies on previously published biological active complex molecules of vanadium as an example of evaluating possible drug candidates before entering the laboratory. We used DFT calculation studies for structural elucidation and optimization of the molecules and molecular docking studies on several Covid-19 related proteins. Our findings suggest that drug discovery should always be computer -aided. Additionally, it is found that Vtocdea and VXn molecules are seem to be good candidates for further studies as antiviral agents.
Keywords: Docking, Vanadium, Covid-19, Drug, Complexes, Computational
1. Introduction
Nowadays the scientific community faces a unique situation due to the Covid–19 disease. The World Health Organization (WHO), declared the coronavirus disease a global pandemic in March 2020 [1]. Generally speaking, Covid–19 is a member of Beta coronoviruses, the former human coronaviruses SARS and MERS [2]. SARS-CoV–2 belongs to the + RNA virus family that utilize single-stranded positive-sense RNA molecules as genomes. SARS-CoV–2, a positive strand RNA virus encodes four structural proteins the matrix (M), small envelope (E), spike (S) and nucleocapsid phosphoprotein (N). Moreover, in a research study, sixteen non-structural proteins (nsp1–16), Covid-19 related, indicated the link between nucleocapsid phosphoprotein N and the viral genome to the viral membrane with its N-terminal RNA binding domain (N-NTD) that captures the RNA genome [3].
The effective options of drug therapy and vaccination are currently under evaluation and our societies are trying to suppress the disease with aggressive infection control measures [4]. Having in mind these facts we are underlying the need of a rapid evaluation of possible drug candidates. This will save money and time to the drug industry in order to shift the research in a synthetic path. As far as now, several studies, discussed bioinformatics [5], [30], [31] and computer aided techniques as a more efficient way to fight the Covid–19 pandemic disease. In addition, Shi et al. [6] developed a molecular docking web-based server to discover new drug targets, or active compounds. Yan et al. [7] conclude that a structure-based rational design of binders with enhanced affinities the S protein of the coronaviruses may facilitate development of decoy ligands or neutralizing antibodies for suppression of viral infection, and Lung et al. [8] suggested a potential SARS‐CoV‐2 RdRp inhibitor for further study using molecular docking studies. Several other research groups proposed the combination of computational and experimental approaches for investigation on pharmaceutical molecules [34], [35], [36]. Molecular docking has several strengths, among which the method’s ability to screen large compound databases at low cost compared to experimental techniques [9], [10] and Open source software development provides many advantages to users of modelling applications, not the least of which is that the software is free and completely extendable [11], [12].
On the other hand, there is an increase interest in the development of metal-based drugs [13], [14] and some of them exhibited interest activities against viruses [15], [16]. Vanadium complexes are in use because of the low toxicity of vanadium metal in comparison with other transition metals. Pharmaceuticals based on vanadium complexes have attracted the interest of scientists due to the biological activity of vanadium molecules, antidiabetic properties, anticancer activity and their low toxicity [17].
Herein, we describe the use of five vanadium (III/IV/V) complexes as potential candidates in drug screening with computer aided chemistry for Covid–19. In particular, we used the example of β-tocDEA (V5+) and β-tocDPA (V4+) as reported in [17], the binuclear V5 + bdeah [18], and binuclear V3 + bicah complex molecule [19]. The amphiphilic vanadium complexes exhibit enhanced hydrolytic stability. All compounds found to be cytotoxic for cancer cells exhibiting activity similar or higher to cis-platin. Finally, based on the theoretical studies we designed for the first time the V5 + Xn complex molecule, a vanadium complex with xanthohumol which is a prenylated chalcone. We performed molecular dynamics studies, DFT studies and molecular docking studies against BSA (transport protein), 6M03 (Covid–19 main protease), 6M71 (SARS-COV–2 RNA-dependent polymerase) and 6YI3 the N-terminal RNA-binding domain of the SARS-CoV–2 nucleocapsid phosphoprotein.
2. Materials and methods
2.1. Materials
To create 2D and 3D structures of the ligand molecules, Chem3d Pro 12.0 and AVOGADRO 1.90.0 software programmes were used. Ligand optimisations were done by ORCA 4.1.1 software while ligands and receptor preparation studies were done by Chimera 1.14. Molecular docking studies were carried out by Autodock 4.2 and docking results were analysed by Chimera 1.14 program. Additionally, calculations on docking studies performed using iGEMDOCK 2.1 software. BSA, 6M03, 6M71 and 6YI3 coded crystal structures were selected from Protein Data Bank (www.rcsb.org). Ligand molecules were collected by literature [17], [18], [19].
2.2. Methods
2.2.1. Computational chemistry
Geometric optimization calculations were performed in accordance with DFT method [20]. The quantum mechanical wavefunction contains, in principle, all the information about a given system. Optimization and vibration frequency calculations were made at B3LYP level with ORCA version 4.0.1 program. B3LYP is a hybrid density functional theory method. ORCA input files were created by AVOGADRO version 1.2.0 software. Frequency calculations were performed to obtain thermodynamic properties and to verify that each optimization achieved an energy minimum. The quantum chemical descriptors extracted directly from the ORCA output file were Huckel charge, electronic density, dipole moment, the energy of the highest occupied molecular orbital (HOMO), and the energy of the lowest unoccupied molecular orbital (LUMO) [21], [22].
2.2.2. Receptor preparation
All water molecules were removed, hydrogen atoms were added, the end residues were repaired, and energy minimization was performed on the protein. The protein structure was then prepared by assigning the hydrogen atoms, charges and energy minimization using CHIMERA software. Docking calculations were performed through Autodock generation using the ligand, the protein was fixed while the ligand was flexible, and the default parameters were used as described [23].
2.2.3. Docking studies
Docking studies were performed with a number of 150 individuals in population, maximum energy evaluations of 2,500,000, and maximum generation of 54,000 to result in 100 docking poses [24]. Using iGEMDOCK 2.1 software we doubled checked our docking scores. The coded crystal structures were selected from the Protein Data Bank (www.rcsb.org). The energy minimization was performed using 500 steepest descent steps with 0.02 Å step size and an update interval of 10. Before performing the molecular docking of ligand and receptor, the ligands were optimized by addition of hydrogens using CHIMERA software and the optimized structures were obtained by DFT studies using ORCA.
2.2.4. Method validation
Two targets in relation to Plasmodium falciparum glutamate dehydrogenase (PDBID:2BMA) and HCV H77 NS5B polymerase (PDBID: 2XI3) were used as control targets, and docking showed that interaction varies with GScore ranging from −3 to −8.3. Among candidates, the highest interaction was shown by Remdesivir showed GScore of −8.35 with HCV H77 NS5B polymerase. In the present study, to compare the selected antiviral drugs with already reported anti-SARS-CoV-2 drugs, lopinavir [32] and remdesivir triphosphate [33] were selected as positive inhibitors of SARS-CoV-2 for in silico analysis in the present study.
3. Results and discussion
3.1. Theoretical chemistry
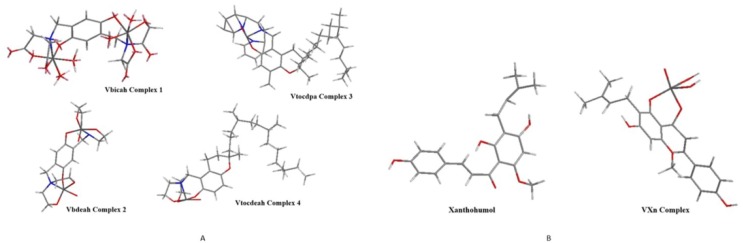
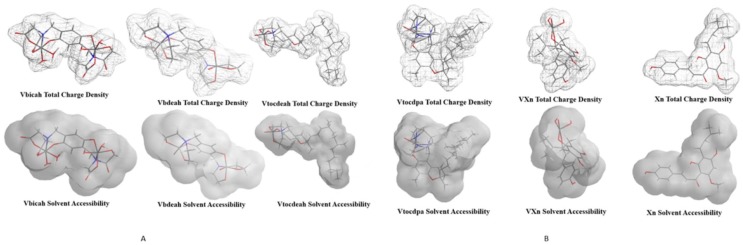
The optimized structures of the studied complex molecules are depicted in Fig. 1 a. The DFT results are within the same range as MP2. For Vbicah and Vbdea we can say that the theoretical studies agreed with the experimentally x-ray structures from the literature. The structures of xanthohumol molecule and complexed with vanadium structure of the molecule can be found in Fig. 1b. The Huckel charges and the calculated molecular geometry parameters can be found in Tables A1, A2 and B respectively in the Supplementary material. Calculated geometry parameters for Vbicah and Vbdeah are in conformity with experimental ones [18], [19]. It is widely known that DFT calculation results strongly depend on used functionals and to some extend on used solvents in model [25], [26], [27]. The molecular electrostatic potential and the solvent accessibility of all the molecules studied in this paper are shown in Fig. 2 a and b. These were in good correlation with Huckel charges values.
Fig. 1.
a. Optimized calculated structures of the Vanadium complexed molecules. b. Optimized calculated structures of xanthohumol and its design vanadium complexed structure.
Fig. 2.
a. Total charge densities and solvent accessibilities for Vbicah, Vbdeah and Vtocdea complex molecules. b. Total charge densities and solvent accessibilities for Vtocdpa, VXn complex molecules and Xanthohumol molecule.
As can be seen from Table 1 . The calculated total energies of Vtocdea, Vtocdpa and VXn are in order from lowest to highest Vtocdea.
Table 1.
Calculated minimized energies for complex molecules of Vanadium with natural phenolic ligands.
| Calculated Values | Vtocdea | Vtocdpa | VXn |
|---|---|---|---|
| Stretch | 32.54 | 81.37 | 45.28 |
| Bond | 84.91 | 215.51 | 182.21 |
| Stretch-Bond | 1.27 | −14.20 | 0.26 |
| Torsion | 18.15 | 22.38 | −1.47 |
| Non-1.4 VDW | −19.89 | −23.76 | −0.87 |
| 1.4 VDW | 36.08 | 37.86 | 20.33 |
| Dipole-Dipole | 2.15 | ------- | 0.65 |
| Total Energy | 156.2 Kcal/mol | 357.79 Kcal/mol | 246.43 Kcal/mol |
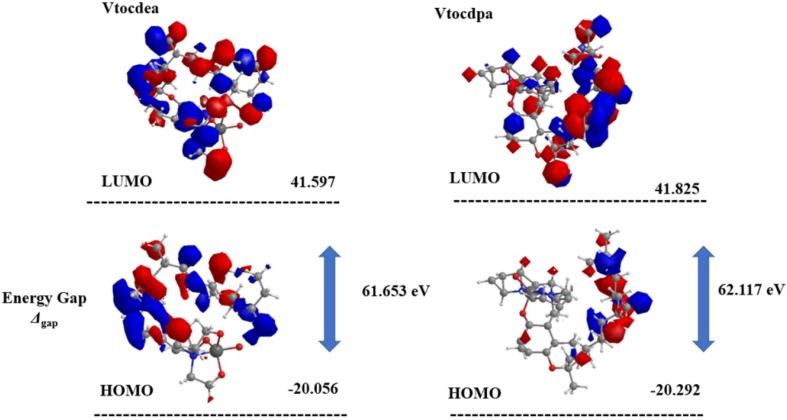
The value of the energy difference between HOMO and LUMO as well as the highest occupied molecular orbital (EHOMO) and lowest unoccupied molecular orbital (ELUMO) energies plays a very important role in stability and reactivity of molecules. The EHOMO energies of molecules show the molecule’s ability to give electrons. On the other hand, ELUMO characterizes the ability of the compound to accept electrons. The energy gap of Vtocdea and Vtocdpa is similar, a fact that it is corelated with the experimental values [17], showing that the vanadium ion in Vtocdea is in oxidation state + 5 while in Vtocdpa the vanadium ion is in oxidation state + 4 (Fig. 3 ). The electronic correlation effects play an essential role in the stability of these type of systems, and therefore, it will affect the equilibrium distances and the interaction energies.
Fig. 3.
HOMO and LUMO molecular orbital of Vtocdea and Vtocdpa.
Electronegativity (χ) is a measure of the power of an atom to attract a bonding pair of electrons.
According to this theorem is expressed as follows. Based on Eq. (1) larger Δgap always indicates lower chemical reactivity and higher kinetic stability of the investigated species. The chemical reactivity of molecules is caused by the simultaneous effect of different parameters.
| (1) |
The distribution and energy of HOMO is an important parameter to explain the antioxidant potential of phenolic antioxidants. The electron-donating capacity of the molecule can be predicted by looking at the energy values of HOMO. The value of the energy difference between HOMO and LUMO as well as the highest occupied molecular orbital (EHOMO) and lowest unoccupied molecular orbital (ELUMO) energies plays a very important role in stability and reactivity [24].
As we can see in Table 2 . Vtocdea and Vtocdpa have similar reactivities and kinetic stability. The same is expected for VXn which is a complex molecule designed based on these two. Based on these results we have used the geometry of the model Tocopherol to design VXanthohumol (both are vanadium complexes with natural derived ligands) and the results confirmed that.
Table 2.
Quantum chemical parameters for studied molecules.
| Complex Molecule | Metal Oxidation State | HOMO | LUMO | Δgap |
|---|---|---|---|---|
| Vbicah | V3+ | −13.043 | 0.526 | 13.569 |
| Vbicah | V3+ | −15.036 | 36.811 | 51.847 |
| Vbdea | V5+ | −14.557 | 32.555 | 47.112 |
| Vbdea | V5+ | −15.298 | 36.063 | 51.361 |
| Vtocdpa | V5+ | −20.056 | 41.397 | 61.453 |
| Vtocdpa | V4+ | −20.292 | 41.825 | 62.117 |
| VXn | V4+ | −18.801 | 43.473 | 62.274 |
3.2. Molecular docking
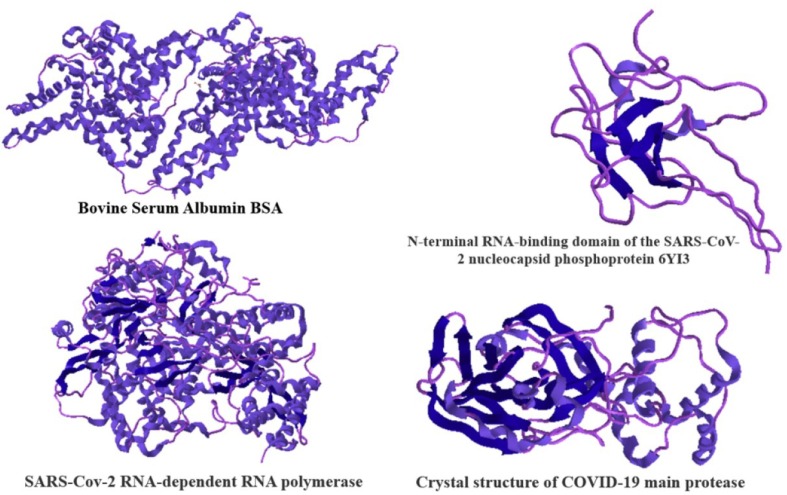
Molecular docking method is one of the frequently referred structure-based design methods [28], [29]. We decided that because the ligand molecules would be screened as a drug candidate for Covid-19, to perform studies with several related proteins to Covid-19 as we discussed in introduction, and at least with one plasma transport protein. In Fig. 4 we can see the crystal structures that we used for molecular docking studies.
Fig. 4.
The crystal structures of the proteins that studied theoretically.
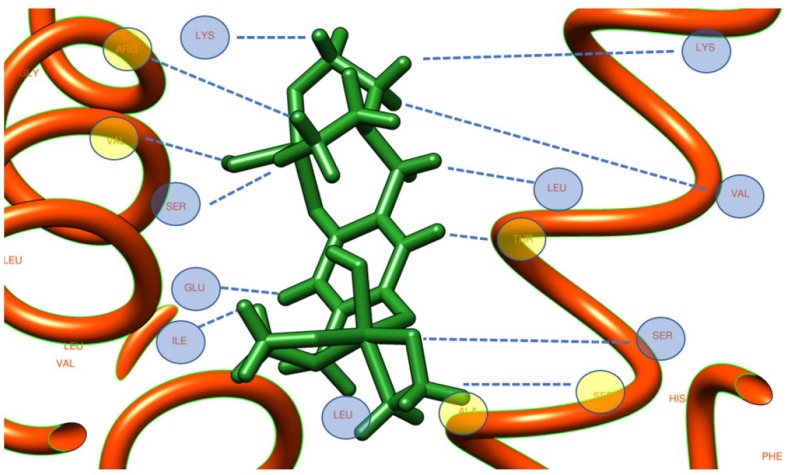
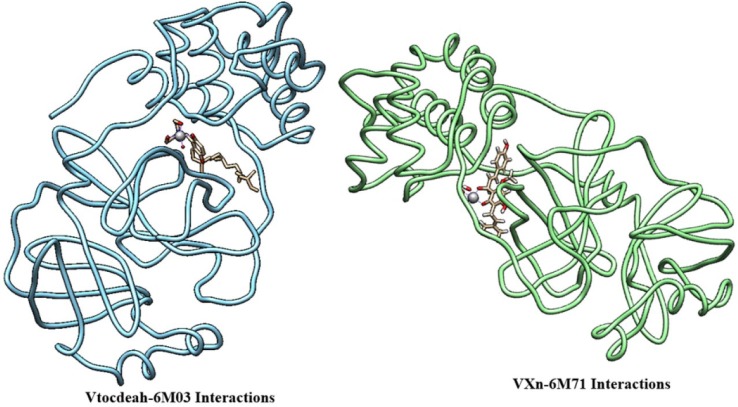
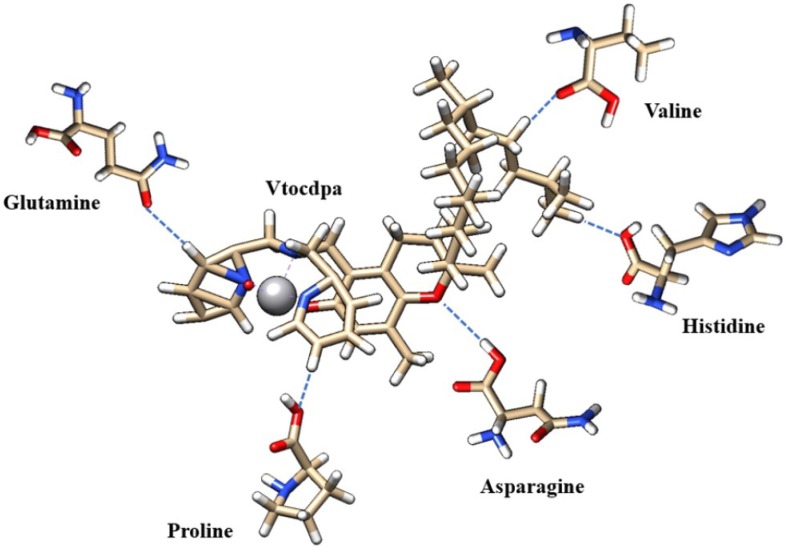
The Vtocdea molecule interaction with 6YI3 is depicted in Fig. 5 while, in Fig. 6 we can see the relatively position of Vtocdea and VXn molecules on 6M03 and 6M71 respectively. Moreover, the hydrogen bonding interactions of amino acid residue of structure 6M71 with Vtocdpa molecule, are shown in Fig. 7 .
Fig. 5.
The amino acid binding residue of Vtocdea molecule with 6YI3.
Fig. 6.
Binding structures of Vtocdeah-6M03 and VXn-6M71 interactions.
Fig. 7.
The interactions of Vtocdpa with 6M71 amino acid residues.
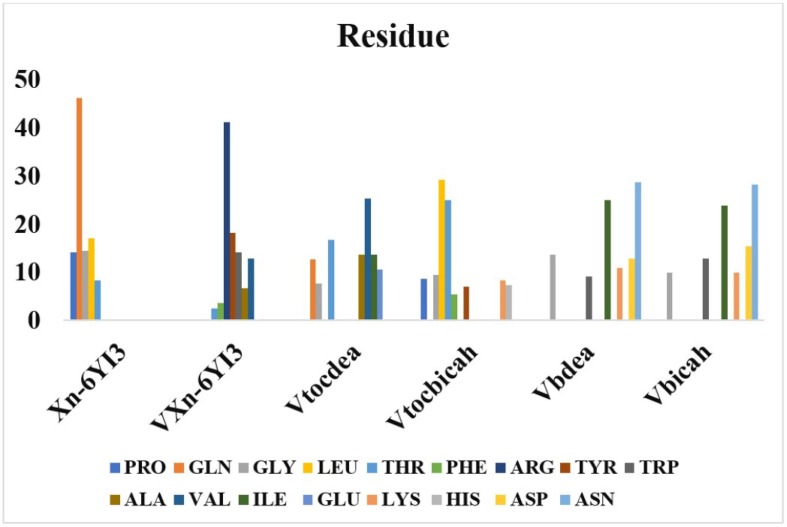
Docking analyses showed that VXn coded ligand has the best binding score among all ligands with BSA protein, followed by Xn with a binding free energy of −10.872 kcal/mol. It makes hydrogen bonds with ASP108, PRO146, TYR147 and ILE455 (four H bonds), and also interacts with BSA with Van der Waals forces with a binding free energy of −9.323 kcal/mol. Regarding the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein (6YI3) the molecule Vbicah has a binding free energy of −10.888 kcal/mol, followed by VXn with −10.523 kcal/mol. In Fig. 8 we are seeing the percentage interaction energies of the amino acid’s residue of 6YI3 on the studied molecules. In particular, the highest energy interaction of Xn molecule with 6YI3 is related with the amino acid glutamine, while for VXn the highest energy related interaction with the same protein comes from the amino acid arginine. Interestingly both Vbdeah and Vbicah interact more with the amino acid asparagine of the 6YI3.
Fig. 8.
The percentage interaction energies of the responsible amino acids of 6YI3 on the different moleculesstudied in this work.
Continuing with SARS-COV-2 RNA-dependent polymerase (6M71) starting with the ligand molecule with the highest affinity to the protein, Vbicah is the first with −10.266 kcal/mol followed by Vtocdea −9.891 Kcal/mol, Vtocdpa −9.706 Kcal/mol, Xn −9.603 Kcal/mol, VXn −9.364 Kcal/mol and finally Vbdea with −8.647 Kcal/mol. Finally, for Covid-19 main protease (6M03), Vtocdea and VXn are amongst the highest molecules in terms of binding affinity with the specific protein. Vtocdea interacts through hydrogen bonds with SER255 and PRO322 (2H bonds) whereas, VXn interacts through hydrogen bonds with the amino acids GLN110 and THR111. All the detailed information about the energy amounts of the ligand molecules through the different proteins and the hydrophobic interactions of the amino acids with these molecules can be found in Table 3 and Table 4 .
Table 3.
ΔGbind energies and energy bond distribution of the molecule’s interaction with BSA, 6YI3, 6M71 and 6M03.
| Complex (Protein-Ligand) | ΔG (Kcal/mol) | Van der Waals contribution (Kcal/mol) | Hydrogen Bond contribution (Kcal/mol) |
|---|---|---|---|
| Remdesivir-6 M03 Control 1 | −7.800 | ------- | ------- |
| Lopinavir-6 M03 Control 2 [37] | −7.300 | ------- | ------- |
| Vbicah-BSA | −8.581 | −7.151 | −1.430 |
| Vbicah-6YI3 | −10.888 | −7.963 | −2.925 |
| Vbicah-6 M71 | −10.266 | −6.550 | −3.716 |
| Vbicah-6 M03 | −10.662 | −4.726 | −5.937 |
| Vbdea-BSA | −8.566 | −5.744 | −2.822 |
| Vbdea-6YI3 | −8.678 | −6.663 | −2.015 |
| Vbdea-6 M71 | −8.647 | −6.528 | −2.119 |
| Vbdea-6 M03 | −8.735 | −7.116 | −1.618 |
| Vtocdea-BSA | −8.552 | −8.160 | −0.350 |
| Vtocdea-6YI3 | −7.858 | −7.858 | 0 |
| Vtocdea-6 M71 | −9.891 | −9.341 | −0.555 |
| Vtocdea-6 M03 | −8.601 | −7.632 | −0.998 |
| Vtocdpa-BSA | −9.914 | −9.443 | −0.633 |
| Vtocdpa-6YI3 | −9.679 | −9.329 | −0.350 |
| Vtocdpa-6 M71 | −9.706 | −8.912 | −0.500 |
| Vtocdpa-6 M03 | −8.724 | −8.432 | −0.274 |
| VXn-BSA | −10.872 | −9.323 | −1.550 |
| VXn-6YI3 | −10.523 | −7.436 | −3.087 |
| VXn-6 M71 | −9.364 | −8.06 | −1.303 |
| VXn-6 M03 | −9.183 | −7.941 | −1.242 |
| Xn-BSA | −10.332 | −9.404 | −0.928 |
| Xn-6YI3 | −8.740 | −7.041 | −0.170 |
| Xn-6 M71 | −9.603 | −7.454 | −2.149 |
| Xn-6 M03 | −8.046 | −6.277 | −0.177 |
Table 4.
Amino acid residue of each protein that interacted with the studied molecules.
| Complex (Protein-Ligand) | Hydrogen Bonded Amino Acid Residue | Hydrophobic Interacted Amino Acid Residue |
|---|---|---|
| Vbicah-BSA | LEU304, HIS304 | LEU301, PRO302, PRO303, LEU304, TYR333, ARG336, HIS 337, PHE373 |
| Vbicah-6YI3 | ASP23, LYS25, ASN86, GLY89, TRP92 | ASP88, ILE90, TRP92 |
| Vbicah-6 M71 | TYR129, HIS133, SER709, THR710, ASP711, LYS714, ASN781 | TYR32, LYS780 |
| Vbicah-6 M03 | HIS41, SER46, LEU141, ASN142, GLY143, SER144, CYS145, HIS163, GLU166 | HIS41, MET49, ASN142, HIS163, GLU166 |
| Vbdea-BSA | HIS145, SER192, SER428, ARG458 | LEU189, SER192, ARG196 |
| Vbdea-6YI3 | ASP23, LYS25, GLY89, ILE90 | ASN86(M), ASP88, ILE91, TRP92 |
| Vbdea-6 M71 | LYS47, ASN138, ASP140, THR141, LEU42 | TYR32, LYS47, ALA130, ASP140, THR141 |
| Vbdea-6 M03 | GLY143, SER144, CYS145, HIS163 | HIS41, SER46, MET49, LEU141, ASN142, MET165, GLN189 |
| Vtocdea-BSA | ASN161 | ASP129, GLU130, LYS131, TRP 134, ASN158, ASN161 |
| Vtocdea-6YI3 | ------- | THR14, ALA116, ILE117, VAL118, GLN120, GLU134, GLY135 |
| Vtocdea-6 M71 | SER255, PRO322 | TYR265, LYS267, TRP268 |
| Vtocdea-6 M03 | GLN110, ASN203 | GLY109, GLN110, VAL202, ASN203, GLU240, PRO241, HIS246, ILE249 |
| Vtocdpa-BSA | ARG144 | ASP111, LEU112, PRO113, LYS114, LEU115, ARG144, ARG185, GLU424, ILE522 |
| Vtocdpa-6YI3 | LEU149 | HIS19, LEU121, THR126, LEU127, PRO128, LYS129, GLY130, PHE131, TYR132 |
| Vtocdpa-6 M71 | ASP761 | HIS439, SER549, ARG555, TRP617, GLU811, CYS813, SER814, ARG836 |
| Vtocdpa-6 M03 | HI246 | GLN107, PRO108, GLY109, VAL202, GLU240, HIS246 |
| VXn-BSA | ASP108, PRO146, TYR147, ILE455 | HIS145, LEU189, SER192, ALA193, ARG196, ARG458 |
| VXn-6YI3 | PHE26, ARG28, TYR83, TRP92, THR95 | ARG28, TYR83, TRP92, VAL93 |
| VXn-6 M71 | ARG29, THR252 | THR246, LEU247, ARG249, THR319, LEU460, PRO461 |
| VXn-6 M03 | GLN110, THR111 | GLY109, GLN110, THR292, PHE294 |
| Xn-BSA | ARG458 | ASP108, HIS145, LEU189, SER192, ALA193, ARG196, ARG458 |
| Xn-6YI3 | PRO122, GLN123, GLY124 | GLN120, LEU121, PRO122, GLN123, GLY124, THR126 |
| Xn-6 M71 | LYS621, ARG624 | ARG553, ASP618, TYR619, PRO620, LYS621, ASP623, ARG624, ASP760 |
| Xn-6 M03 | ASP33, ASP34, SER81, LYS88 | ASP33, VAL35, LYS88, LEU89, LYS90, TYR101 |
The presence of the electrostatic forces on those interactions was limited. Electrostatic interactions can only be seen on the interaction between Vtocdea with BSA (−0.042 Kcal/mol), Vtocdea with 6M03 (−0.029 Kcal/mol), Vtocdpa with BSA (−0.162 Kcal/mol), Vtocdpa with 6M71 (−0.294 Kcal/mol) and Vtocdpa with 6M03 (−0.018 Kcal/mol). The electrostatic interactions of Vtocdea and Vtocdpa are in correlation with literature [17], indicating that these molecules are having anticancer activity through the disruptions of electron transfer cycle in mitochondria. Having in mind the theoretical studies we suggest that molecules such as Vtocdea and VXn are good candidates to tested in vitro and in vivo, against the Covid-19 disease.
4. Conclusions
In this study we performed several theoretical studies on vanadium complexes with biological activity from the literature. Based on that, we designed a new one the VXn molecule that showed similar activity with Vtocdea. We used computational chemistry methods to take valuable information of the molecules and performed molecular docking studies on Covid-19 related structures. Knowing from literature that Vtocdea exhibits the highest biological activity having at the same time the least toxicity in healthy cells, we conclude that these techniques should be used by the scientific community to save time and money on the fight against Covid-19 and take this valuable information in order to discover novel drug therapies with high effectiveness and the least toxicity. Additionally, it is found that Vtocdea and VXn molecules are seem to be good candidates for further studies as antiviral agents.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comtox.2021.100157.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 Coronavirus (SARS-CoV-2) - an update on the status. Infect. Genet. Evol. 2020;83(April) doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80-.) 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Zhang B., Jin Z., Yang H., Rao Z. The crystal structure of COVID-19 main protease in complex with an inhibitor N3. PDB Release. 2020;119(February):17–20. doi: 10.2210/PDB6LU7/PDB. [DOI] [Google Scholar]
- 6.Shi Y., Zhang X., Mu K., Peng C., Zhu Z., Wang X., Yang Y., Xu Z., Zhu W. D3Targets-2019-NCoV: a webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm. Sin. B. 2020;xxx doi: 10.1016/j.apsb.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinesh D.C., Chalupska D., Silhan J., Veverka V., Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. bioRxiv. 2020;2020(04) doi: 10.1101/2020.04.02.022194. pp. 02.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Liu H.Te, Wu C.Y. The potential chemical structure of Anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira L., dos Santos R., Oliva G., Andricopulo A. Molecular docking and structure-based drug design. Strategies. 2015;20 doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luque F. Frontiers in computational chemistry for drug discovery. Molecules. 2018;23(11):2872. doi: 10.3390/molecules23112872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirhadi S., Sunseri J., Koes D.R. Open source molecular modeling. J. Mol. Graph. Model. 2016;69:127–143. doi: 10.1016/j.jmgm.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M.A. Miteva, S. Violas, M. Montes, D. Gomez, P. Tuffery, B.O. Villoutreix. FAF-Drugs: Free ADME/Tox Filtering of Compound Collections. Nucleic Acids Res., 2006, 34 (WEB. SERV. ISS.), 738–744. DOI:10.1093/nar/gkl065. [DOI] [PMC free article] [PubMed]
- 13.De Clercq E. Antiviral metal complexes. Met. Based. Drugs. 1997;4(3):173–192. doi: 10.1155/MBD.1997.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biju S., Parac-Vogt T.N. Recent advances in lanthanide based nano-architectures as probes for ultra high-field magnetic resonance imaging. Curr. Med. Chem. 2018;27(3):352–361. doi: 10.2174/0929867325666180201110244. [DOI] [PubMed] [Google Scholar]
- 15.Ghaffari Hadi, Tavakoli Ahmad, Moradi Abdolvahab, Tabarraei Alijan, Bokharaei-Salim Farah, Zahmatkeshan Masoumeh, Farahmand Mohammad, Javanmard Davod, Kiani Seyed Jalal, Esghaei Maryam, Pirhajati-Mahabadi Vahid, Monavari Seyed Hamidreza, Ataei-Pirkooh Angila. Inhibition of H1N1 influenza virus infection by zinc oxide nanoparticles: another emerging application of nanomedicine. J. Biomed. Sci. 2019;26(1) doi: 10.1186/s12929-019-0563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langland J., Jacobs B., Wagner C.E., Ruiz G., Cahill T.M. Antiviral activity of metal chelates of caffeic acid and similar compounds towards herpes simplex, VSV-Ebola Pseudotyped and Vaccinia Viruses. Antiviral Res. 2018;160:143–150. doi: 10.1016/j.antiviral.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Hadjiadamou Ioanna, Vlasiou Manolis, Spanou Smaragda, Simos Yannis, Papanastasiou George, Kontargiris Evangelos, Dhima Irida, Ragos Vasilios, Karkabounas Spyridon, Drouza Chryssoula, Keramidas Anastasios D. Synthesis of Vitamin E and aliphatic lipid Vanadium(IV) and (V) complexes, and their cytotoxic properties. J. Inorg. Biochem. 2020;208:111074. doi: 10.1016/j.jinorgbio.2020.111074. [DOI] [PubMed] [Google Scholar]
- 18.Drouza C., Vlasiou M., Keramidas A.D. Vanadium(IV/V)-p-dioxolene temperature induced electron transfer associated with ligation/deligation of solvent molecules. Dalton Trans. 2013;42(33):11831–11840. doi: 10.1039/c3dt50619c. [DOI] [PubMed] [Google Scholar]
- 19.Drouza C., Vlasiou M., Keramidas A.D. Synthesis, characterization of Dinuclear Vanadium(III) hydroquinonate- iminodiacetate complexes. Inorg. Chim. Acta. 2014;420:103–111. doi: 10.1016/j.ica.2013.12.033. [DOI] [Google Scholar]
- 20.Wakui N., Yoshino R., Yasuo N., Ohue M., Sekijima M. Exploring the selectivity of inhibitor complexes with Bcl-2 and Bcl-XL: a molecular dynamics simulation approach. J. Mol. Graph. Model. 2018;79:166–174. doi: 10.1016/j.jmgm.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 21.J. A. Rocha, N.C.S. Rego, B.T. Carvalho, F.I. Silva, J.A. Sousa, R.M. Ramos, I.N.G. Passos, J. De Moraes, J.R.S.A. Leite, F.C.A. Lima. Computational Quantum Chemistry, Molecular Docking, and ADMET Predictions of Imidazole Alkaloids of Pilocarpus Microphyllus with Schistosomicidal Properties. PLoS One, 2018, 13(6), 1–23. DOI:10.1371/journal.pone.0198476. [DOI] [PMC free article] [PubMed]
- 22.Hayakawa Daichi, Sawada Nahoko, Watanabe Yurie, Gouda Hiroaki. A molecular interaction field describing nonconventional intermolecular interactions and its application to protein-ligand interaction prediction. J. Mol. Graph. Model. 2020;96:107515. doi: 10.1016/j.jmgm.2019.107515. [DOI] [PubMed] [Google Scholar]
- 23.Y. Yin, Y. Sun, L. Zhao, J. Pan, Y. Feng. Medicinal Chemistry Based Amides as Potent S6K1 Inhibitors †. 2020. DOI:10.1039/c9md00537d. [DOI] [PMC free article] [PubMed]
- 24.Ercan S., Şenses Y. Design and molecular docking studies of new inhibitor candidates for EBNA1 DNA binding site: a computational study. Mol. Simul. 2020;46(4):332–339. doi: 10.1080/08927022.2019.1709638. [DOI] [Google Scholar]
- 25.Çakmak E., Özbakır Işın D. A theoretical evaluation on free radical scavenging activity of 3- styrylchromone derivatives: the DFT study. J. Mol. Model. 2020;26(5) doi: 10.1007/s00894-020-04368-7. [DOI] [PubMed] [Google Scholar]
- 26.Tian Y., Chen W., Zhao Z., Xu L., Tong B. Interaction and selectivity of 14-Crown-4 Derivatives with Li+, Na+, and Mg2+ metal ions. J. Mol. Model. 2020;26(4) doi: 10.1007/s00894-020-4325-8. [DOI] [PubMed] [Google Scholar]
- 27.Mendizabal Fernando, Miranda-Rojas Sebastián, Castro-Latorre Pablo. Quantum chemistry simulation of the electronic properties in [Au(NH3)2]NO3 and [Au(NCH)2][AuCl4] extended unsupported complexes. Mol. Simul. 2020:521–529. doi: 10.1080/08927022.2020.1735634. [DOI] [Google Scholar]
- 28.Praski A., Jaworska M., Lodowski P. Structure and electronic spectra of neutral and protonated forms of anticonvulsant drug lamotrigine. J. Mol. Model. 2020;26(3):019-4266-2. doi: 10.1007/s00894-. [DOI] [PubMed] [Google Scholar]
- 29.Mohd Amin S.N., Md Idris M.H., Selvaraj M., Mohd Amin S.N., Jamari H., Kek T.L., Salleh M.Z. Virtual screening, ADME study, and molecular dynamic simulation of chalcone and flavone derivatives as 5-lipoxygenase (5-LO) inhibitor. Mol. Simul. 2020;46(6):487–496. doi: 10.1080/08927022.2020.1732961. [DOI] [Google Scholar]
- 30.Marek Prachar, Sune Justesen, Daniel Bisgaard Steen-Jensen, Stephan Thorgrimsen, Erik Jurgons, Ole Winther, and Frederik Otzen Bagger, COVID-19 Vaccine Candidates: Prediction and Validation of 174 SARS-CoV-2 Epitopes, DOI:10.1101/2020.03.20.000794. [DOI] [PMC free article] [PubMed]
- 31.Carly G. K. Ziegler et al., SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues, Cell Press, DOI:10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed]
- 32.Hall Donald C., Ji Hai-Feng. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect. Dis. 2020;35:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon Calvin J., Egor X., Tchesnokov P., Emma Woolner X., Jason X., Perry K., Feng Joy Y., Porter Danielle P., Götte Matthias. Remdesivir is a direct-acting antiviral that inhibits RNA dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna Murthy P., Sheena Mary Y., Shyma Mary Y., Yohannan Panicker C., Suneetha V., Armakovi S., Armakovi S.J., Van Alsenoy C., Suchetan P.A. Synthesis, crystal structure analysis, spectral investigations, DFT computations and molecular dynamics and docking study of 4-benzyl-5-oxomorpholine-3-carbamide, a potential bioactive agent. J. Mol. Struct. 2017:25e39. [Google Scholar]
- 35.El-Azab Adel S., Shyma Mary Y., Sheena Mary Y., Yohannan Panicker C., Abdel-Aziz Alaa A.-M., Mohamed Menshawy A., Armakovi Stevan, Armakovi Sanja J., Van Alsenoy Christian. Spectroscopic and reactive properties of a newly synthesized quinazoline derivative: combined experimental, DFT, molecular dynamics and docking study. J. Mol. Struct. 2017:863e881. [Google Scholar]
- 36.Y. Sheena Mary, Pankaj B. Miniyar, Y. Shyma Mary, K.S. Resmi, C. Yohannan Panicker, Stevan Armakovi, Sanja J. Armakovi, Renjith Thomas, B. Sureshkumar, Synthesis and spectroscopic study of three new oxadiazole derivatives with detailed computational evaluation of their reactivity and pharmaceutical potential, J. Mol. Struct. 1173 (2018) 469e480.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.