Abstract
Traditional production of industrial and therapeutic proteins by eukaryotic cells typically requires large-scale fermentation capacity. As a result, these systems are not easily portable or reusable for on-demand protein production applications. In this study, we employ Bioproduced Proteins On Demand (Bio-POD), a F127-bisurethane methacrylate hydrogel-based technique that immobilizes engineered Pichia pastoris for preservable, on-demand production and secretion of medium- and high-molecular weight proteins (in this case, SEAP, α-amylase, and anti-HER2). The gel samples containing encapsulated-yeast demonstrated sustained protein production and exhibited productivity immediately after lyophilization and rehydration. The hydrogel platform described here is the first hydrogel immobilization using a P. pastoris system to produce recombinant proteins of this breadth. These results highlight the potential of this formulation to establish a cost-effective bioprocessing strategy for on-demand protein production.
Keywords: Protein production, Hydrogel, Pichia pastoris, Immobilization, Lyophilization
Graphical abstract
Highlights
-
•
Hydrogel immobilization used for first time for Pichia pastoris protein production.
-
•
Diffusion of large protein from hydrogels is not a hindrance to protein production.
-
•
Yeast-laden hydrogel samples lost minimal productivity following lyophilization.
-
•
Encapsulation-based production outperformed suspension cultures.
-
•
Bio-POD enabled portable, reusable, preservable, on-demand production of proteins.
1. Introduction
Mainstream enzyme applications ranging from laundry detergents and food processing to precision-medicine enabling antibody production (both native and heterologous) are a foundational technology of biotech and biopharma industries [[1], [2], [3], [4]]. Not surprisingly, the global markets for non-therapeutic enzymes and protein drugs are projected to reach $7.0 billion by 2023 [5] and $228.4 billion by 2021 [6], respectively. Much of this expansion has been due to recent progress in genetic tool development to manipulate the microorganisms, filamentous fungi, and mammalian cells utilized in biopharmaceutical and industrial enzyme production [[7], [8], [9]]. However, much of this industry is still encumbered with large fermentation tanks, submerged fermentation, and even roller-bottle based processes [4,[10], [11], [12]]. In the end, these processes lack much of the flexibility and portability necessary for on-demand production to meet oscillating demands for varied products as well as for small-scale products requiring on-site production due to cold-chain requirements.
Among the potential hosts for protein production, the unicellular yeast Pichia pastoris (which has recently been divided into the three species Komagataella phaffii, Komagataella pastoris and Komagataella pseudopastoris [13], with K. phaffii being utilized exclusively in this study) has unique advantages over other platform hosts (such as Escherichia coli and mammalian cells). These advantages include: (i) powerful secretory ability [13], (ii) improved protein folding and post-translational modification over prokaryotic systems [9], (iii) ease of genetic manipulation [7,14], and (iv) high-cell density cultivation with associated high expression levels and low processing costs as compared with mammalian cell lines [15,16]. In addition, the strong and tightly regulated alcohol oxidase I (AOX1) promoter derived from methylotrophic P. pastoris and its unique Crabtree-negative phenotype has enabled high cell density and high protein yield production during aerobic fermentation [7,9]. Moreover, P. pastoris is capable of protein post-translational modifications including N- as well as O-linked glycosylation without hyper-glycosylation, thereby reducing hyper-antigenicity of recombinant proteins for therapeutic use [9].
However, even with P. pastoris as a host, the use of suspension culture still predominates the field. The immobilization of microorganisms on/within various polymeric matrices offers several potential benefits over suspension fermentations, including ease of product separation in cases where there is no cell leakage from the scaffold, high cell loading capacity, reduced microbial contamination, and improved biocatalytic reusability [17]. Moreover, the proper immobilization technique could enable flexible scaffolds for small-scale, on-demand production. While immobilization techniques can improve secreted product production in S. cerevisiae when compared to traditional suspension fermentation [18], there is scant literature implementing live cell immobilization approaches for protein production, and especially utilizing P. pastoris. Furthermore, the properties of previously reported polymeric matrices are incompatible with protein production and large-scale fermentation schemes. For example, invertase production from P. pastoris PpBfrA in a calcium alginate matrix was compromised due to heat-inactivation of cells and the reversible ionic crosslinking of the matrix by charge-bearing metabolites and pH of the medium [19,20]. The use of agar beads with embedded L-alanyl-l-glutamine-producing P. pastoris GPAp was shown to be sensitive to the high immobilization temperature of 50 °C [21]. Finally, the use of magnetic nanoparticles limited recombinant protein production due to the nanoparticle compromising the cytoplasmic membrane integrity of immobilized cells, leading to undesirable effects including cell leakage and abnormal cell function [22], phenomena that were not completely investigated in a study utilizing immobilized P. pastoris GS115 Albumin for production of human serum albumin [23]. As a result, there is often a mismatch between the immobilization material and process conditions for protein production.
In recent work, we employed a temperature-responsive, shear-thinning triblock copolymer F127-bisurethane methacrylate (F127-BUM) hydrogel for on-demand biomolecule production of mostly small molecules using either mono- or co-cultures [24,25]. Moreover, we demonstrated that this unique hydrogel formulation provides preservation capacity (i.e. metabolic activity of lyophilized consortia hydrogels retained 100% efficiency after long-term storage at room temperature for 3 months) and reusability (i.e. yeast-laden hydrogels enabled ethanol production for over 1 year of repeated use) [24]. This approach was found to be suitable for production of small molecules and very small polypeptides (as with the case for colicin V production). Furthermore, the lyophilization process exhibited minimal impact on the mechanical integrity of the cell-laden hydrogel samples (i.e. the Young's moduli were identical and little change in microstructure was observed in SEM images between the pre- and post-lyophilized materials). Importantly, this hydrogel system was compatible with yeast hosts.
In this work, we demonstrate that engineered P. pastoris can be embedded within a F127-BUM hydrogel ink to fabricate extruded hydrogel filaments suitable for cell preservation and continuous production of secreted recombinant proteins of increasing size (Fig. 1). Through this approach, we enable a scheme for Bioproduced Proteins On Demand (Bio-POD). Specifically, we show that secreted proteins are able to effectively diffuse from within the printed yeast gel to the surrounding culture media which allows the product to be easily harvested and for the yeast-embedded hydrogels to be re-used. Additionally, it is possible to simply preserve these hydrogel inks through lyophilization and retain productivity that is on par with that of pre-lyophilized samples in a manner that outcompetes liquid culture with respect to consistency over multiple rounds of sample reuse. To our knowledge, this is the first study highlighting the use of a hydrogel system employing P. pastoris for robust and re-useable on-demand recombinant protein production.
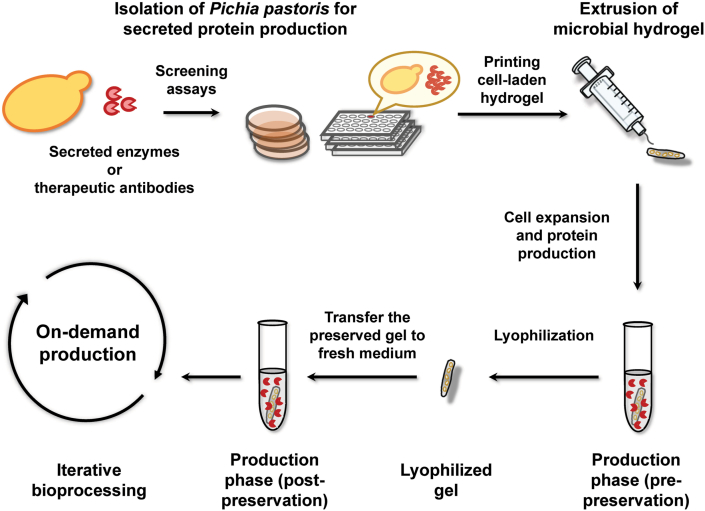
Fig. 1.
Overview of Bio-POD bioprocessing for on-demand protein production. Strain development of secreted proteins in engineered P. pastoris is achieved through auxotrophic or antibiotic selection coupled with appropriate biocatalytic screening assays. The engineered yeast cells are then encapsulated within the hydrogel and extrusion printed using syringes. The printed and UV-cured yeast-laden hydrogels are subsequently transferred to culture medium for cell expansion/enzyme production. The hydrogels can optionally proceed to lyophilization for storage after repetitive uses of the living microbial materials. Next, the preserved gels can be rehydrated in fresh medium for on-demand protein production, with iterative re-uses depending on user needs.
2. Materials & methods
2.1. Strains, media and plasmid construction
All strains, plasmids, primers and a gBlock gene fragment used in this study are listed in Supplementary Table 1, Supplementary Table 2, and Supplementary Table 3. Oligonucleotide primers used for PCR amplification were purchased from Integrated DNA Technologies (Coralville, IA). All Gibson-assembled DNA [26] were electroporated (2 mm Electroporation Cuvettes, Bioexpress) into E. coli competent cells with a BioRad Genepulser Xcell at 2.5 kV. E. coli NEB10β was used for gene cloning or propagation of all expression vectors. For propagation of pPIC9 series, NEB10β was cultivated in LB medium (1% tryptone, 0.5% yeast Extract and 1% NaCl) supplemented with 100 μg/mL ampicillin (Sigma). For propagation of pPICZα series, NEB10β was grown in low salt LB medium (1% tryptone, 0.5% yeast Extract and 0.5% NaCl, pH7.5) supplemented with 25 μg/mL zeocin (Sigma)) with 225 rpm orbital shaking at 37 °C. Starter cultures of yeast strains were routinely grown in YPD (1% yeast extract, 2% peptone and 2% glucose) medium at 30 °C. Electroporation of Pichia with an integrative expression cassette was performed according to Pichia Expression Kit's (Invitrogen) instructions. Minimal dextrose (MD) medium (1.34% yeast nitrogen base (YNB), 4 × 10−5% biotin and 2% glucose) was used in auxotrophic selection study. Buffered glycerol complex medium (BMGY) and buffered methanol complex medium (BMMY) were prepared according to the manual of EasySelect™ Pichia Expression Kit (Invitrogen) and used in recombinant protein production.
For SEAP production, an amplicon containing a human SEAP gene amplified from the plasmid TB-SIZ-SV40pA [27] using primers P1 as well as P2 was generated to replace the N-terminal human secretion signal peptide with S. cerevisiae α-factor mating signal sequence, and remove hydrophobic C-terminal signal peptide for PI-G anchor attachment [28]. The amplicon was then Gibson assembled with the PCR product amplified from pPIC9 empty vector (Invitrogen) with primers P3 and P4 to construct pPIC9-SEAP. To construct pPICZα empty vector, the gene fragment amplified from the plasmid pPICZalphaB-SapL3 (Addgene) with primer P5 and P6 was Gibson assembled with primer P7. To construct pPICZα-AmyL, the amplicon without SapL3 gene was amplified from pPICZalphaB-SapL3 using primers P5 as well as P6, and Gibson assembled with the PCR product amplified from the amylase gBlock codon-optimized for P. pastoris (Supplementary Table 3) with primer P8 and P9. To construct pPICZ-HIS4, the amplicon amplified from pPIC9 empty vector using primers P10 as well as P11 was Gibson assembled with the PCR product amplified from pPICZα using primers P12 as well as P13. For construction of pPICZ-LC, the light chain gene of anti-HER2 amplified from the plasmid AbVec-hIgK-hu4D5 [29] using primers P14 and P15 was Gibson assembled with the PCR product amplified from pPICZ-HIS4 using primers P16 and P17. To construct pPICZ-HC, the heavy chain gene of anti-HER2 amplified from the plasmid AbVec-IgG1-hu4D5 using primers P18 as well as P19 was Gibson assembled with the gene fragment amplified from pPICZα using primers P16 and P17. For anti-HER2 production, the amplicon amplified from pPICZ-HC using primers P20 as well as P22, and the PCR product amplified from pPICZ-LC using primers P21 as well as P23, were separately digested with EcoRI and BamHI restriction enzymes, and then digested products were ligated to construct pPICZ-aHER2, which contains full-length of anti-HER2.
2.2. F127-BUM hydrogel preparation
F127 is a commercially available triblock copolymer of poly (ethylene oxide)-b-poly (propylene oxide)-b-poly (ethylene oxide) that can be transformed into F127-BUM as previously described [30]. The extrusion of yeast-laden hydrogels and photocuring procedures were performed by following our previously reported hydrogel work [24]. In short, 30 wt% F127-BUM polymer solution was mixed with photo-radical initiator 2-hydroxy-2-methylpropiophenone (Sigma-Aldrich; 2.5 μL for every 1 g of hydrogel solution) to facilitate polymerization of the methacrylate functional groups upon UV exposure. Photo-initiated polymer solution was mixed with 4.5 × 107 cells before extrusion to yield robust, viable microbe-laden gels upon brief photocuring (with an exposure to 365 nm light at 0.55 mW cm−2 for 5 min using Spectroline® XX-15NF model).
2.3. SEAP secretion and production
The plasmids pPIC9 and pPIC9-SEAP harboring S. cerevisiae α-factor mating signal sequence were first digested by restriction enzyme SalI to promote insertion in the his4 locus. Then the linearized integration cassettes were separately transformed into P. pastoris GS115 by electroporation. MD agar plates and broth media were used for selection of His+ transformants. Five colonies from MD plates were picked and examined for production levels. Seeding cultures were grown in BMGY medium. Protein induction was performed using 15 mL of BMMY medium (0.5% methanol) with the initial OD600 of 0.5 in a 125 mL flask. Cultures were incubated at 30 °C with an orbital speed 225 rpm. A final concentration of 0.5% methanol was aseptically added to the flask every 24 h to maintain protein induction. To measure SEAP production, the supernatants obtained from centrifugation to pellet yeast cells were taken for analysis using the NovaBright™ Secreted Placental Alkaline Phosphatase (SEAP) Enzyme Reporter Gene Chemiluminescent Detection Kit 2.0 (Invitrogen). Finally, the highest SEAP producer was selected and designated as Pp02.
For SEAP production in hydrogels, starter cultures of Pp01 control strain and Pp02 SEAP-producing strain were first grown in YPD at 30 °C. Then 4.5 × 107 overnight cells were embedded in 0.3 g of polymer. The printed and cured cell-laden gels were subsequently incubated in 3 mL of BMGY at 30 °C for cell expansion for 24 h (we define this cell outgrowth stage as round 0, the transfer to and subsequent induction in BMMY for a first production stage as round 1 of reuse, the next transfer to new BMMY as round 2 and so on). It should be noted that the SEAP induction was not initiated at round 0. After 24 h incubation, each gel sample was transferred to 3 mL of BMMY (in the presence of the inducer 0.5% methanol) for induction of secreted SEAP expression (round 1). For the liquid culture system, 4.5 × 106 seeding yeast cells for each strain were transferred to 3 mL of BMGY media and incubated at 30 °C for 24 h (round 0). Then 4.5 × 106 cells for each strain were transferred to 3 mL of BMMY medium for protein induction (round 1). Two consecutive reuses after round 1 were performed and samples were processed via lyophilization treatment as described in the previous study [24]. Once ready for testing, the preserved samples were transferred directly into 3 mL of BMMY media for two additional repetitive uses. All reuse batches were carried out with 30 °C incubation for 48 h and 0.5% methanol was aseptically added to the culture every 24 h to maintain protein induction. Each gel sample was washed twice with 800 μL of BMMY media to ensure carry-over of secreted SEAP enzyme and metabolites from the previous batch were not being introduced to the next batch of reuse. For liquid culture system, repetitive uses were performed through sub-culturing 50 μL of yeast from previous batch to the next batch. Supernatants at 48-h timepoint were taken from each round of reuse and analyzed by luminescence measurement.
Luminescence was measured with a BioTek Citation 3 at emission detection range from 500 nm to 600 nm resulting in maximal emission at of 540 nm. SEAP standards were prepared by adding the purified SEAP enzyme (Invitrogen) to the Pp01 control strain spent medium.
2.4. α-Amylase secretion and production
The plasmids pPICZα and pPICZα-AmyL containing S. cerevisiae α-factor secretion signal sequence were first digested by restriction enzyme Pme I to promote integration into the AOX1 locus. Then the linearized integration cassettes were separately transformed into P. pastoris GS115 by electroporation. YPD agar plates supplemented with 100 μg/mL zeocin were used for selection of zeocin resistant transformants. Zeocin resistance protein binds zeocin antibiotic in a stoichiometric manner. Therefore, selection with a higher concentration of zeocin makes it easy to screen for high-copy number transformed variants. 48 colonies were picked from the agar plate and transferred to a 96-deep-well microplate containing 500 μL of YPD+100 μg/mL zeocin broth medium for each well. Through a series of selections under gradually increased selection pressure (from 100, 500–1000 μg/mL zeocin) at 30 °C, 10 clones with high growth rate were selected for testing α-amylase production. 500 μL of BMGY and BMMY (with 0.5% methanol) media were used for cell outgrowth and α-amylase expression. After 48 h fermentation, supernatants containing secreted α-amylase from the pelleted yeast cells were collected and the enzyme activities were analyzed by measuring the size of the halos forming on starch agar plates (3% agar and 5% (w/v) soluble starch (Alfa Aesar; product number A11961)) and plate-based starch-iodine assay. Finally, the highest α-amylase producer was selected and designated as Pp04.
For evaluation of α-amylase production in hydrogels, starter cultures of Pp03 control strain and Pp04 α-amylase-producing strain were first grown in YPD at 30 °C. The procedure of preparation of yeast-laden hydrogels and liquid cultures is the same as describe in the above SEAP production section, except that each round of reuse took 24 h. Liquid cultures were seeded with an initial inoculum of 4.5 × 106 or 4.5 × 107 cells for comparison of amylase production. Cell counts for the 50 μL transfers in the liquid culture system for each round was calculated by converting OD600 value to cell numbers (OD600 of 1 for P. pastoris is 5 × 107 cells/mL). To evaluate the growth between Pp01 (HIS+; complementation of GS115 the wild‐type HIS4 gene) and wild-type GS115 (HIS−) pre-lyophilization, cultures were performed using 3 mL of BMGY medium with the initial OD600 of 0.05 in a test tube, and 13-h as well as 23-h timepoint samples were taken for cell growth measurement. Subsequently, 3 × 107 cells from the 23-h pre-lyophilized samples were transferred to sterile culture tubes and processed via lyophilization treatment. The lyophilized Pp01 (HIS+) and wild-type GS115 (HIS−) were separately grown in 3 mL of BMMY methanol medium and 15-h as well as 24-h timepoint samples were taken for growth measurement. The cell growth was measured by Ultrospec 2100 Pro UV/Visible Spectrophotometer observing optical density at 600 nm.
For plate-based starch-iodine assay, assay reactions were initiated by mixing 40 μL of 5% soluble starch solution and 40 μL of B. licheniformis α-amylase standard (Sigma; product number A4551) solution prepared in Pp03 control strain spent media or 40 μL of supernatants from the pelleted yeast samples. After incubation at 37 °C for 5 min, 20 μL of HCl was added to the mixture to stop the enzymatic reaction, followed by the addition of 100 μL of iodine-KI reagent (5 mM I2 and 5 mM KI in DI water). Then 100 μL of the iodine-treated samples were transferred to a transparent flat-bottomed 96-well microplate and the absorbances at 570 nm were recorded using Tecan Infinite M Plex.
2.5. Anti-HER2 secretion and production
The pPICZ-aHER2 harboring two S. cerevisiae α-factor mating signal sequences and full-length of anti-HER2 genes was first digested by restriction enzyme XbaI to promote insertion in the his4 locus. Then the linearized integration cassette was transformed into P. pastoris GS115 by electroporation. YPD agar plate supplemented with 100 μg/mL zeocin was used for selection of zeocin resistant transformants. The zeocin selection procedure was described in section 2.4. Finally, two clones with high growth rate were selected for testing anti-HER2 production in flasks. Seeding cultures were grown in BMGY medium. Protein induction was performed using 50 mL of BMMY medium (0.5% methanol) with the initial OD600 of 0.3 in a 125 mL flask. Cultures were incubated at 30 °C with an orbital speed of 225 rpm. A final concentration of 0.5% methanol was aseptically added to the flask every 24 h to maintain protein induction. To measure anti-HER2 production, the supernatants obtained from pelleted yeast cells were taken for analysis using enzyme-linked immunosorbent assay (ELISA) described below. Supernatant from the 72-hr timepoint sample was taken for Western blotting analysis. Finally, the highest anti-HER2 producer was selected and designated as Pp05.
Full-length, soluble control anti-HER2 antibody was expressed and purified using traditional CHO expression as described [29]. Briefly, the anti-HER2 variable heavy and light chain sequences (synthesized by Life Technologies with Cricetulus griseus codon optimization) were cloned into AbVec IgG1 and IgK plamids [31] via AgeI and SalI or AgeI and BsiWI restriction sites, respectively, resulting in plasmids AbVec-IgK-h4D5 and AbVec-IgG1-hu4D5. After lipofection and transient expression in CHO–K1 cells, the antibody was purified from the media by ammonium sulfate precipitation and protein A chromatography, followed by buffer exchange into phosphate buffered saline, pH 7.4. Quantification of expression of anti-HER2 from Pichia pastoris cultures was determined by ELISA alongside the purified anti-HER2 control produced in CHO cells as described [29]. Purified HER2-Fc (R&D Systems) was coated overnight at 4 °C on high binding plates at a concentration of 0.2 μg/mL. After blocking with milk, purified anti-HER2 control antibody was serially diluted in five-fold steps starting from 1 μg/mL and added to HER2 coated and uncoated wells. Undiluted and five-fold diluted Pichia pastoris media were also added to the coated wells and uncoated wells. After a 60-min incubation at room temperature, wells were washed three times and anti-human-IgK-HRP (Southern Biotech) secondary antibody was added at 1:2500 for 60 min at room temperature. After a final wash, signal was developed with tetramethylbenzadine chromogenic substrate (TMB Substrate Kit; Thermo Scientific), followed by a 1 M hydrochloric acid quench and signal recorded as the absorbance at 450 nm (Tecan Infinite M Plex). All washes were performed with PBS-0.05% Tween-20 (designated as PBS-T) while blocking and diluent media comprised this wash buffer with non-fat dry milk (5% w/v) (designated as PBS-T-milk). For evaluation of anti-HER2 production in hydrogels, starter cultures of Pp03 control strain and Pp05 anti-HER2-producing strain were first grown in YPD at 30 °C. All procedures of preparation of yeast-laden hydrogels and liquid cultures as well as lyophilization are the same as described in the above SEAP production section, except that each round of reuse took 24 h.
2.6. SDS-PAGE analysis of secreted SEAP and α-amylase
After fermentation, SEAP or α-amylase-producing yeast cells from the suspension cultures were removed by centrifuging at 4000×g for 30 min at 4 °C. The supernatants were concentrated using Amicon® Ultra-15 centrifugal filter (Millipore) with a 30 kDa molecular weight cutoff. The concentrated samples were then analyzed by running 10% SDS-PAGE gel and stained with Coomassie Blue.
2.7. Western blotting of anti-HER2 antibody
Anti-HER2-producing Pichia cells from the cultures were removed by centrifuging at 4000×g for 30 min at 4 °C. The supernatants were then concentrated using Amicon® Ultra-15 centrifugal filter (Millipore) with a 10 kDa molecular weight cutoff. The resulting concentrated Pichia media was diluted 1:30 in PBS, and 10 μl was mixed with 2 μl 6X reducing or non-reducing sample buffer (0.375 M Tris pH 6.8, 12% SDS, 60% glycerol, 0.06% bromophenol blue for both, plus 9% β-mercaptoethanol for the reducing buffer). Reduced samples were boiled for 5 min and non-reduced samples were incubated at 42 °C for 2 min before being subjected to protein electrophoresis on a 4–20% gradient SDS-PAGE gel (Bio-Rad cat # 4,561,093) alongside a prestained marker (Fisher Scientific cat # PI-26634). The resulting protein bands were transferred to a PVDF membrane (Thermo Fisher Scientific cat # IB24001) using an iBlot 2 instrument at 25 V for 6 min. The membrane was then blocked in PBS-T-milk for 30 min and stained with a mixture of goat-anti-human-kappa-HRP (Southern Biotech cat # 2060–05) and goat-anti-human-Fc-HRP (Jackson ImmunoResearch cat # 109-035-098), each diluted 1:2000 in PBS-T-milk for 1 h at room temperature. After washing four times with PBS-T, the chemiluminescent substrate (Clarity Western ECL Substrate, #1705061) was added and the membrane was imaged on ChemiDoc Imaging System (BIO-RAD) for 5 s.
3. Results
To test the Bio-POD approach, we selected three proteins to demonstrate the feasibility and relevance of our system and to test the limits of production across a range of protein sizes. In prior research, we demonstrated that E. coli-laden hydrogels were capable of secreting a small peptide antibiotic, colicin V (around 10 kDa), in a consistent manner (even after lyophilization) over four consecutive re-uses in contrast to that of suspension culture [24]. Here, we sought to evaluate the novel use of P. pastoris within this gel-based system for secreted placental alkaline phosphatase, α-amylase, and the antibody anti-human epidermal growth factor receptor 2 (also known as anti-HER2, Herceptin® or Trastuzumab) as model proteins ranging from 60 to 150 kDa. In each case, we generated a P. pastoris cell line capable of modest-level production, printed P. pastoris-laden hydrogel inks, and tested protein production pre- and post-lyophilization with repeated use for comparison to a traditional suspension culture. At the onset, it should be mentioned that the goal of this study was to evaluate the use of this hydrogel based system and compatibility with P. pastoris-enabled protein secretion, not to evaluate the absolute amount produced as we did not perform extensive cell line engineering.
3.1. On-demand production of SEAP using the Bio-POD system
To first evaluate if the F127-BUM hydrogel-based platform with a eukaryotic expression host is suitable for production of higher molecular weight proteins, we engineered P. pastoris to produce secreted placental alkaline phosphatase (SEAP; a protein around 60 kDa), a commonly used reporter enzyme in mammalian cell studies [27]. Integration of the SEAP gene expression cassette into P. pastoris followed by screening resulted in isolating strain Pp02 capable of secreting 15.1 ± 0.4 ng/mL of SEAP protein in shake flasks 48 h post-induction (Fig. 2 A and B).
Fig. 2.
Generation of SEAP-producing P. pastoris and SEAP production in hydrogel system. (A) Comparison of SEAP production between 48 h and 72 h fermentation at 30 °C. P. pastoris Pp01 and Pp02 were used as the control and SEAP-producing strain, respectively. 0.5% methanol was added every 24 h to maintain induction (n = 3 for each strain). (B) SDS-PAGE analysis of recombinant SEAP production. The protein marker was loaded into M lane of SDS-PAGE and supernatant of pelleted yeast containing SEAP was loaded into S lane. The red arrow indicates the size of SEAP (around 60 kDa) secreted to the media. (C) SEAP production assessed both pre- and post-lyophilization (data from round 2 and round 5 of reuse respectively) from hydrogels. (D) SEAP production in hydrogels (blue bars) with repeated use compared to liquid culture (orange bars) performance. All reuse batches were carried out with 30 °C incubation for 48 h. All the samples were treated with lyophilization after round 3 of reuse. Data are mean ± s.d.; n = 6 biological replicates for hydrogels n = 3 biological replicates for liquid culture. *P < .05; **P < .01; ***P < .001 via two-sample t-test (hydrogel vs corresponding liquid culture for each round of reuse).
Next, we encapsulated the resulting Pp02 strain in the hydrogel matrix and allowed cells to proliferate for 24 h prior to SEAP induction testing (i.e. a round 0 gel use outgrowth step followed by round 1 production stage in which the gels were transferred to induction media). In this experiment, we performed a total of five repetitive use assays (rounds 1–5) as well as assessed SEAP production both pre- and post-preservation by lyophilizing the gel at the midpoint of this reuse experiment (Fig. 2D). Each round of reuse involved cultivating the gels in BMMY for 48 h (round 1 acts as a methanol adaption phase where both transcriptional de-repression and induction mechanisms occur), with media supplemented with an additional 0.5% (v/v) methanol for induction at 24 h. To showcase the robustness of the gel system, the lyophilization procedure (without addition of cryoprotectants) consisting of a 10-min freezing step in liquid nitrogen prior to lyophilization was performed as described in previous work [24]. With this simplified procedure, post-lyophilized hydrogel samples (average of round 4 and round 5) retained nearly 90% of pre-lyophilization (average of rounds 1–3) SEAP secretion capacity (Fig. 2D), with the highest post-lyophilization titer (20.2 ± 1.9 ng/mL) reaching approximately 80% of the maximum pre-lyophilization titer (25.3 ± 1.9 ng/mL) (Fig. 2C). Furthermore, the average titer of round 2–5 of hydrogel reuse (18.0 ± 5.4 ng/mL) was quite similar to the round 1 hydrogel titer (17.1 ± 4.6 ng/mL), indicating that the encapsulated cells remained metabolically active despite multiple rounds of reuse and simplified preservation procedures (Fig. 2D).
Finally, we compared these SEAP production values to those of traditional suspension cultures treated in a similar manner. Specifically, the pre- and post-lyophilization average values for suspension cultures were roughly 60% and 45% of the corresponding hydrogel values, respectively (Fig. 2D). Likewise, the highest average pre- and post-lyophilization values for suspension cultures were approximately 52% and 65% of corresponding hydrogel titers, respectively (Fig. 2D). Collectively, these results demonstrate that the F127-BUM hydrogel matrix enables sustained production and secretion of SEAP by P. pastoris cells with higher titers, more consistency over rounds of reuse, and enhanced cell viability and protein production post-preservation compared to similarly treated liquid suspension cultures.
3.2. On-demand production of α-amylase using the Bio-POD system
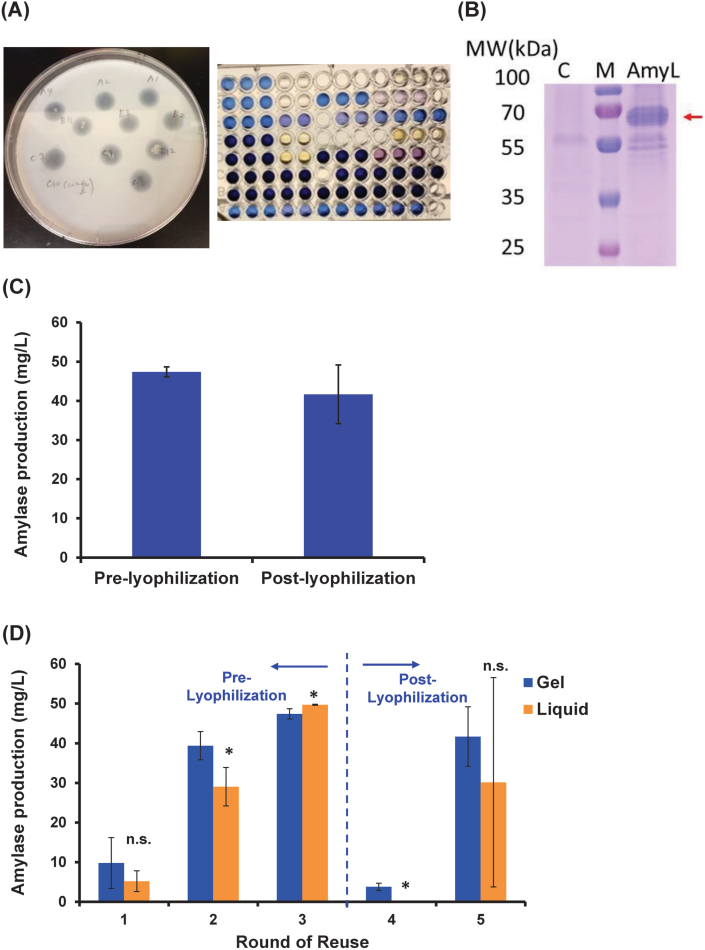
Encouraged by the results with SEAP, we next selected the production of another similarly-sized enzyme, namely α-amylase (around 60 kDa) which is a starch hydrolyzing enzyme widely employed in food, detergent and textile industries [32]. To first establish a cell line for demonstration of sustained production and secretion of α-amylase using the Bio-POD technique, we engineered P. pastoris to overproduce and secrete Bacillus licheniformis α-amylase by a similar gene integration and selection strategy [33]. The resulting strain we selected for this work, Pp04, was capable of secreting 3.2 mg/L α-amylase 24 h post-induction in a 96-deep-well microplate (Fig. 3A and B).
Fig. 3.
Generation of α-amylase-producing P. pastoris and α-amylase production in hydrogels system. (A) Zeocin-resistant transformants were cultured in a 96-deep-well microplate and selected based on the cell growth. Secreted α-amylase capacities of each isolated strains were evaluated via starch agar plate (measuring the size of the halos) and plate-based starch-iodine assay, where dark blue wells contain no amylase and lighter colored wells ranging from light blue to yellow contain increasing amounts of active amylase. (B) SDS-PAGE analysis of recombinant amylase. C lane: supernatant of pelleted Pp03 culture (control); M lane: protein marker; AmyL lane: supernatant of pelleted Pp04 culture (amylase strain). The red arrow indicates the size of amylase (around 60 kDa) secreted to the media. (C) Amylase production assessed both pre- and post-lyophilization (data from round 2 and round 5 of reuse respectively) from hydrogels. (D) Amylase production in hydrogels (blue bars) with repeated use compared to liquid culture (orange bars) performance. All reuse batches were carried out with 30 °C incubation for 24 h. All the samples were treated with lyophilization after round 2 of reuse. Data are mean ± s.d.; n = 3 biological replicates for both hydrogel and liquid culture samples. *P < .05; via two-sample t-test (hydrogel vs corresponding liquid culture for each round of reuse).
We next encapsulated Pp04 in the gel matrix, allowed cells to proliferate for 24 h prior to induction, and performed 5 repetitive uses after round 0 in a similar fashion to the SEAP production assays described in section 3.1 (Fig. 2). One deviation from the SEAP experiments was that amylase-secreting cells were grown in BMMY for only 24 h per round of reuse, as this timeframe was sufficient for amylase production to be detected via a plate-based starch-iodine assay (Fig. 3A and Supplementary Fig. 1). As with the SEAP demonstration, the P. pastoris-laden hydrogels were lyophilized mid-experiment without the addition of cryoprotectants. Post-lyophilization (average of round 4 and round 5) hydrogel samples retained around 86% of pre-lyophilization (average of rounds 1–3) amylase secretion capacity (Fig. 3D), with the highest post-lyophilization titer (41.68 ± 7.5 mg/L) approximately 88% of the maximum pre-lyophilization titer (47.40 ± 1.29 mg/L) (Fig. 3C). Compared with the SEAP demonstration, the extent of decrease in titer at round 4 (i.e. after lyophilization) was more significant likely due to the slower growth phenotype of these cells and reduced fitness of the P. pastoris GS115 HIS4 mutant strain (HIS−) [34] used in this amylase study (in contrast, complementation of HIS4 was applied for the SEAP case resulting in a HIS+ strain). Previous studies have demonstrated that a HIS− strain is more prone to osmotic stress than HIS+ strain even when culturing in a complex medium. Consistent with the previous studies, our result also showed that the HIS− strain used in this amylase study exhibited a slower growth than the HIS+ strain (Pp01) pre- and post-lyophilization (Supplementary Fig. 2). The HIS− strain displayed a slight slower growth than HIS+ strain pre-lyophilization (Supplementary Fig. 2A), while a dramatically impaired cell growth for lyophilized HIS− strain (3-fold and 11-fold lower at 15-h and 24-h timepoint, respectively) was observed when compared with that for HIS+ strain (Supplementary Fig. 2B), indicating restoring histidine prototrophy of GS115 can mitigate osmotic stress. Nevertheless, the observation of high average titer post-lyophilization confirms that the encapsulated cells remained metabolically active following multiple rounds of reuse and even in light of simplified, harsh preservation procedures.
As with SEAP, we compared amylase production values from hydrogels with those obtained from traditional suspension cultures. The pre- and post-lyophilization average value for suspension cultures were roughly 79% and 61% of the corresponding hydrogel values, respectively (Fig. 3D). Furthermore, the maximum pre- and post-lyophilization values for suspension cultures were approximately 87% and 58% of corresponding hydrogel values, respectively (Fig. 3D). The amylase production with a higher inoculum size of 4.5 × 107 cells for liquid culture system (which is the same seeding number as used in hydrogel system) was also evaluated pre- and post-lyophilization (Supplementary Fig. 3). These results showed that the liquid cultures dramatically decreased in cell viability following lyophilization (regardless of seeding level) (Supplementary Fig. 3A) and thus resulted in highly variable production levels (Supplementary Fig. 3B). As further evidence, it should be noted that the liquid culture system produced undetectable levels of protein in round 4 (i.e. after lyophilization) (Supplementary Fig. 3B) unlike the hydrogel system in this round (Fig. 3D), thus suggesting that the hydrogel indeed provides protection to the cells from preservation techniques. Taken together, these results demonstrate that the amylase-secreting P. pastoris-laden F127-BUM gels were metabolically active and were able to both produce and secrete high amounts of enzyme in a manner that outperformed liquid cultures both in terms of absolute titer and consistency during subsequent rounds of repetitive use, especially following lyophilization and subsequent rehydration.
3.3. On-demand production of an antibody using the Bio-POD system
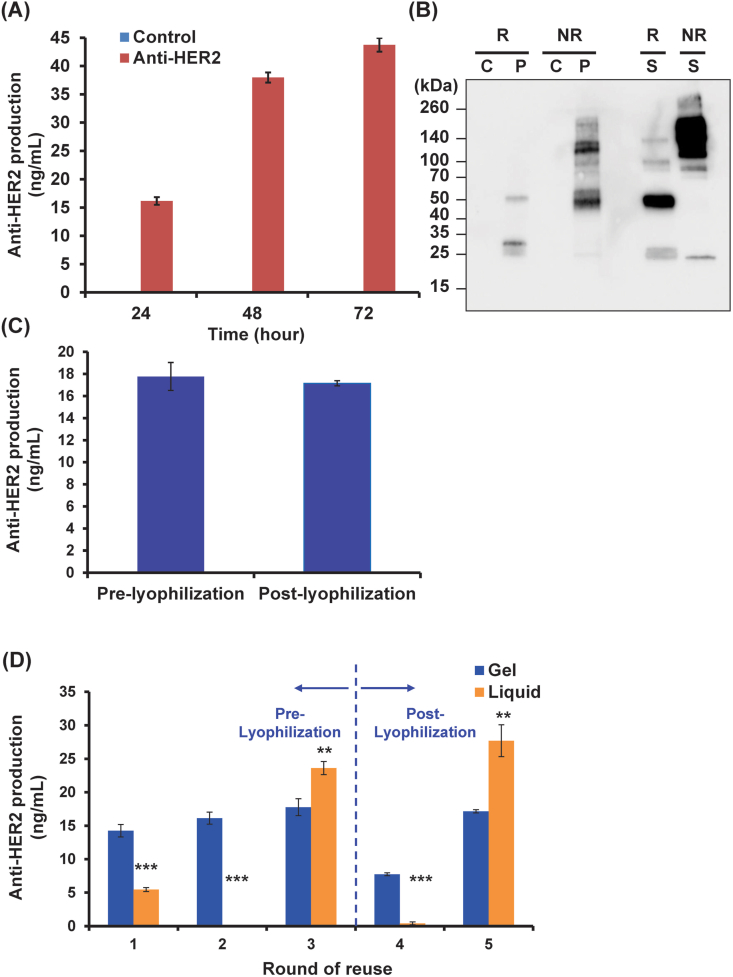
Profound successes in monoclonal antibody production technologies have been achieved across a different expression systems including bacteria, insects, plants, yeasts, and mammalian cell lines [35]. To showcase that diffusion of large products through the hydrogel matrix is possible and that our Bio-POD system may be amenable to production of complex proteins, we selected the therapeutic antibody Herceptin® (anti-HER2) as a case study. Herceptin was approved by the US Food and Drug Administration for the treatment of breast cancer in 1998 [29,36], and is a 150 kDa hetero-dimer comprised of two heavy and two light chains with 16 inter- and intra-chain di-sulfide bonds [37] and an essential glycosylation appended to residue N297 on the heavy chain. Integration of the full-length anti-HER2 expression cassette into P. pastoris followed by screening resulted in isolating a strain, Pp05, capable of secreting 37 ng/mL of anti-HER2 antibody in shake flasks at 72 h post-induction (Fig. 4A and B).
Fig. 4.
Generation of anti-HER2-producing P. pastoris and anti-HER2 production in hydrogels system. (A) Time-course profile of anti-HER2 production at 30 °C. P. pastoris Pp03 and Pp05 were used as the control and SEAP-producing strain, respectively. 0.5% methanol was added every 24 h to maintain induction (n = 3 for each strain). (B) Western blotting analysis of anti-HER2 production. R: reducing condition; NR: non-reducing condition; C: supernatant from pelleted negative control Pp03 strain; P: supernatant from pelleted anti-HER2-producing strain Pp05; S: anti-HER2 control purified from CHO cells. (C) Anti-HER2 production assessed both pre- and post-lyophilization (data from round 3 and round 5 of reuse respectively) from hydrogels. (D) Anti-HER2 production in hydrogels (blue bars) with repeated use compared to liquid culture (orange bars) performance. All reuse batches were carried out with 30 °C incubation for 24 h. All the samples were treated with lyophilization after round 3 of reuse. Data are mean ± s.d.; n = 3 biological replicates for hydrogels n = 3 biological replicates for liquid culture. *P < .05; **P < .01; ***P < .001 via two-sample t-test (hydrogel vs corresponding liquid culture for each round of reuse).
We next encapsulated the resulting Pp05 in the F127-BUM matrix, performed three continuous re-use cycles followed by a lyophilization process and additional two subsequent re-uses (a similar scheme as described in section 3.1) to evaluate the reusability and preservation capacity for on-demand production of anti-HER2. Since a 24 h induction time was sufficient to detect anti-HER2 production by a plate-based ELISA assay (Fig. 4A), we utilized this timepoint for growing the anti-HER2-secreting cells in BMMY for each round of reuse. At the onset, these results were able to demonstrate relatively sustained production of anti-HER2 from the embedded cells, as well as secretion out of the hydrogel matrix and into the supernatant. Anti-HER2 production was also observed post-lyophilization and re-use. While a mild decline in average titer was observed for post-lyophilized hydrogel samples (12.5 ± 0.0 ng/mL for round 4–5 samples) compared to pre-lyophilized samples (16.1 ± 0.8 ng/mL for rounds 1–3 gels) (Fig. 4C), the highest post-lyophilization titer (17.2 ± 0.2 ng/mL) was quite similar to the maximum pre-lyophilization titer (17.8 ± 1.3 ng/mL) (Fig. 4D). Furthermore, the average titer of round 2–5 hydrogel reuse (14.7 ± 0.5 ng/mL) was nearly on par with the round 1 hydrogel titer (14.3 ± 0.9 ng/mL). These results demonstrate that the yeast-laden gels remained metabolically active and that immobilization of cells in the F127-BUM polymeric matrix resulted in minimal loss of protein production capacity during iterative re-uses and after lyophilization.
As with SEAP and amylase productions described in section 3.1, 3.2, we finally compared the anti-HER2 production between the embedded yeast cells using the Bio-POD platform and suspension yeast cultures. In this comparison, we found an approximately 1.3-fold improvement in anti-HER2 production (average of round 1–5 was 14.6 ± 0.5 ng/mL) for yeast-laden F127-BUM-based hydrogels when compared to liquid cultures (average of round 1–5 was 11.4 ± 1.0 ng/mL) (Fig. 4D). While a mild decrease in production level for post-lyophilized gel samples was observed (average of round 4–5 was 12.5 ± 0.0 ng/mL) compared to the corresponding suspension culture titers (14.1 ± 1.5 ng/mL), the pre-lyophilized cell-laden gels displayed a nearly 1.7-fold higher production (average of round 1–3 was 16.1 ± 0.8 ng/mL) than that of corresponding liquid culture titer (9.7 ± 0.5 ng/mL). Moreover, the anti-HER2 production levels by the suspension system was variable across repeated subculture cycles, a trend not observed in the hydrogel-based immobilized cell scheme (Fig. 4D). Specifically, no anti-HER2 production was observed for the suspension system during the second round of re-use, and the antibody activity was dramatically reduced after lyophilization (fourth round of reuse). It is interesting to note that our prior results with production of colicin V also presented with inconsistent production in liquid cultures and these problems were resolved using the gel-based system [24], however the detailed mechanism of this phenomenon is still unclear. In sum, the Bio-POD hydrogel system presented here demonstrated the capacity for robust reusability and strong preservation capacity of embedded P. pastoris secreting anti-HER2 as compared to suspension cultures, thus underscoring the ability of using this immobilization technique to effectively produce and diffuse the large therapeutic proteins.
4. Discussion
On-demand protein production enables small batches of biologic products to be produced for precision medicine treatments [38] as well as in off-the-grid scenarios such as active military missions and in developing nations where existing large-scale manufacturing infrastructure is scarce and storage occurs at variable climates without clear access to cold-chain conditions. While cell immobilization and preservation techniques provide a viable path toward this vision, efforts prior to this report have been limited for P. pastoris. One major hurdle has been the incompatibility of encapsulation materials with the bioprocessing goals of viable cells with high protein titers. In this regard, ionically-crosslinked gels (such as calcium alginate gels), while commonly utilized in many biomedical applications, can hinder diffusion and protein quality/export can vary with protein isoelectric point [39]. In other cases, the high extrusion temperatures of polymers can damage cells [40], which limits processing options for material-cell mixtures. In contrast to these approaches, the hydrogel approach presented here allows for both ease of material processing and protein production.
The work presented here demonstrates that encapsulation of P. pastoris cells within an F127-BUM hydrogel matrix establishes a platform for on-demand, sustained production of high molecular weight recombinant proteins (demonstrated up to 150 kDa). This system also bypasses the challenges associated with other materials, as the aqueous F127-BUM polymer mixture is a shear-thinning hydrogel at ambient temperature that undergoes a reversible gel-to-sol transition at ~17 °C, thus facilitating the processing to obtain homogenous viable cell distributions [24]. This study focuses on investigating the reusability and preservability of the gel system for protein on-demand production. We conducted a side-by-side comparison with liquid cultures in a test tube scale to demonstrate that similar titers could be achieved by using the same strain in gel and liquid culture systems. For all three protein products tested, the hydrogel platform displayed a higher absolute average titer, a better consistency in round-to-round re-use, and better preservation traits compared to liquid suspension cultures.
For complex therapeutic proteins such as Herceptin, the therapeutic efficacy largely depends on the folding and glycosylation patterns [41]. While these factors were not the main focus in this work, they could likewise be incorporated with further strain engineering. For example, P. pastoris protein glycoengineering that has been employed to improve the conformational stability of protein pharmaceuticals [42,43] could be applied to this production strain in the future. In this work, the Bio-POD platform yielded significantly more functional protein post-lyophilization (round 4) for all three protein cases compared with liquid culture system. This difference can be attributed to the robustness of the F127-BUM polymer in providing protection from preservation techniques (i.e. both macro- and micro-structure of hydrogel have been previously demonstrated to remain nearly unchanged after lyophilization, and confocal microscopy images indicated that the yeast cell distribution were not significantly affected by preservation [24]). The ability to preserve protein-secreting cells within a lyophilized matrix that enables rehydration for rapid production demonstrates the portability and robustness of our platforms (especially compared to simple glycerol stocks of strains) to enable on-demand protein production in point-of-care settings without need for cold-chain requirements. The sustained production of these distinct protein products from the gels at titers on par with traditional culturing processes demonstrates that diffusion of large products through the gel matrix was not a substantial impediment in this platform as we were able to detect protein in the supernatant at levels near/above a liquid culture. It should be noted that we did not utilize the codon-optimized version of genes for Pichia for some human protein cases in the current study, and thus the production level was expected at ng/mL or mg/L. To this end, this work demonstrated that the F127-BUM hydrogel material can serve to preserve protein production function under variable conditions.
Quantification of cell number within these hydrogels remains analytically unreliable using this particular hydrogel formulation despite trying mechanical disruption (slicing and lyophilization) of the cell-laden gels and RNA extraction for qPCR experiment. Specifically, the results were unreliable (triplicate with a high variance) due to the technical difficulty of completely breaking down the entire gel. Likewise, the use of viability dyes suffered from challenges in z-stack measurements for microscopy. These challenges unfortunately impede the accuracy of cell number and viability quantification in these particular hydrogels. Despite the current formulation of F127-BUM hydrogel requiring a short exposure to long-wave ultraviolet light (365 nm) for photocuring, it is possible that mutagenesis of embedded cells could occur, but these effects have not been apparent from production titers acquired in this work. Nevertheless, the Bio-POD approach presented in this work provides a major feature for use in point-of-care settings that is not feasible with traditional liquid cultures. Specifically, the lyophilized gels without addition of cryoprotectants could be stably stored at room temperature without the need of −80 °C freezer [24], thus enabling a field-deployable platform.
5. Conclusion
Taken together, these results demonstrate that the Bio-POD technique enables repeated use and preservation for the on-demand production of recombinant proteins using an engineered P. pastoris platform. Based on the reusability and preservation capacity exhibited here, we believe that this hydrogel platform can provide for the portable, reusable, and stable production of proteins for on-demand applications without a need for cold-chain requirements. Looking forward, this platform could be coupled with simple, miniaturized downstream protein purification modules [[44], [45], [46], [47]] to yield a field-deployable platform for on-demand production of protein products.
CRediT authorship contribution statement
Shuo-Fu Yuan: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Sierra M. Brooks: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Annalee W. Nguyen: Investigation, Resources, Writing - review & editing. Wen-Ling Lin: Investigation, Resources. Trevor G. Johnston: Resources. Jennifer A. Maynard: Writing - review & editing, Supervision. Alshakim Nelson: Writing - review & editing, Supervision, Funding acquisition. Hal S. Alper: Conceptualization, Methodology, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
None of the authors have any conflicts of interest with the work presented.
Acknowledgements
This work was supported by the Camille and Henry Dreyfus Foundation (H.A.). A.N. acknowledges both UW CoMotion and Royalty Research Fund for supporting this work. We thank Dr. Hung-Wen (Ben) Liu and Dr. Nathaniel A. Lynd for access to lyophilizers.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.01.019.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Singh R., Kumar M., Mittal A., Mehta P.K. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 2016;6:174. doi: 10.1007/s13205-016-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 3.Rieder L., Teuschler N., Ebner K., Glieder A. Wiley Online Library; 2019. Chapter 1.3 - Eukaryotic Expression Systems for Industrial Enzymes, Industrial Enzyme Applications; pp. 47–69. [Google Scholar]
- 4.Singhania R.R., Patel A.K., Thomas L., Goswami M., Giri B.S., Pandey A. Chapter 13 - industrial enzymes. In: Pandey A., Höfer R., Taherzadeh M., Nampoothiri K.M., Larroche C., editors. Industrial Biorefineries & White Biotechnology. Elsevier; Amsterdam: 2015. pp. 473–497. [Google Scholar]
- 5.Dewan S.S. BCC Research; 2018. Global Markets for Enzymes in Industrial Applications.https://www.bccresearch.com/market-research/biotechnology/global-markets-for-enzymes-in-industrial-applications.html [Google Scholar]
- 6.Dewan S.S. BCC Research; 2017. Global Markets for Bioengineered Protein Drugs.https://www.bccresearch.com/market-research/biotechnology/bioengineered-protein-drugs-report.html [Google Scholar]
- 7.Vieira Gomes A.M., Souza Carmo T., Silva Carvalho L., Mendonça Bahia F., Parachin N.S. Comparison of yeasts as hosts for recombinant protein production. Microorganisms. 2018;6:38. doi: 10.3390/microorganisms6020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baghban R., Farajnia S., Rajabibazl M., Ghasemi Y., Mafi A., Hoseinpoor R., Rahbarnia L., Aria M. Yeast expression systems: overview and recent advances. Mol. Biotechnol. 2019;61:365–384. doi: 10.1007/s12033-019-00164-8. [DOI] [PubMed] [Google Scholar]
- 9.Juturu V., Wu J.C. Heterologous protein expression in Pichia pastoris: latest research progress and applications. Chembiochem. 2018;19:7–21. doi: 10.1002/cbic.201700460. [DOI] [PubMed] [Google Scholar]
- 10.Clapp K.P., Castan A., Lindskog E.K. Chapter 24 - upstream processing equipment. In: Jagschies G., Lindskog E., Łącki K., Galliher P., editors. Biopharmaceutical Processing. Elsevier; 2018. pp. 457–476. [Google Scholar]
- 11.Srivastava N., Srivastava M., Mishra P.K., Gupta V.K., Molina G., Rodriguez-Couto S., Manikanta A., Ramteke P.W. Applications of fungal cellulases in biofuel production: advances and limitations. Renew. Sustain. Energy Rev. 2018;82:2379–2386. doi: 10.1016/j.rser.2017.08.074. [DOI] [Google Scholar]
- 12.Buyel J.F., Twyman R.M., Fischer R. Very-large-scale production of antibodies in plants: the biologization of manufacturing. Biotechnol. Adv. 2017;35:458–465. doi: 10.1016/j.biotechadv.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Fischer J.E., Glieder A. Current advances in engineering tools for Pichia pastoris. Curr. Opin. Biotechnol. 2019;59:175–181. doi: 10.1016/j.copbio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Kang Z., Huang H., Zhang Y., Du G., Chen J. Recent advances of molecular toolbox construction expand Pichia pastoris in synthetic biology applications. World J. Microbiol. Biotechnol. 2016;33:19. doi: 10.1007/s11274-016-2185-2. [DOI] [PubMed] [Google Scholar]
- 15.Katla S., Karmakar B., Tadi S.R.R., Mohan N., Anand B., Pal U., Sivaprakasam S. High level extracellular production of recombinant human interferon alpha 2b in glycoengineered Pichia pastoris: culture medium optimization, high cell density cultivation and biological characterization. J. Appl. Microbiol. 2019;126:1438–1453. doi: 10.1111/jam.14227. [DOI] [PubMed] [Google Scholar]
- 16.Kim H., Yoo S.J., Kang H.A. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 2015;15:1–16. doi: 10.1111/1567-1364.12195. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-García J., García-Martínez T., Mauricio J.C., Moreno J. Yeast immobilization systems for alcoholic wine fermentations: actual trends and future perspectives. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najafpour G., Younesi H., Syahidah Ku Ismail K. Ethanol fermentation in an immobilized cell reactor using Saccharomyces cerevisiae. Bioresour. Technol. 2004;92:251–260. doi: 10.1016/j.biortech.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Martínez D., Menéndez C., Echemendia F.M., Pérez E.R., Trujillo L.E., Sobrino A., Ramírez R., Quintero Y., Hernández L. Complete sucrose hydrolysis by heat-killed recombinant Pichia pastoris cells entrapped in calcium alginate. Microb. Cell Factories. 2014;13:87. doi: 10.1186/1475-2859-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheetham P.S.J., Blunt K.W., Bocke C. Physical studies on cell immobilization using calcium alginate gels. Biotechnol. Bioeng. 1979;21:2155–2168. doi: 10.1002/bit.260211202. [DOI] [Google Scholar]
- 21.Li Y.-M., Gao J.-Q., Pei X.-Z., Du C., Fan C., Yuan W.-J., Bai F.-W. Production of l-alanyl-l-glutamine by immobilized Pichia pastoris GS115 expressing α-amino acid ester acyltransferase. Microb. Cell Factories. 2019;18:27. doi: 10.1186/s12934-019-1077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansari F., Grigoriev P., Libor S., Tothill I.E., Ramsden J.J. DBT degradation enhancement by decorating Rhodococcus erythropolis IGST8 with magnetic Fe3O4 nanoparticles. Biotechnol. Bioeng. 2009;102:1505–1512. doi: 10.1002/bit.22161. [DOI] [PubMed] [Google Scholar]
- 23.Taghizadeh S.-M., Ebrahiminezhad A., Ghoshoon M.B., Dehshahri A., Berenjian A., Ghasemi Y. Magnetic immobilization of Pichia pastoris cells for the production of recombinant human serum albumin. Nanomaterials. 2020;10:111. doi: 10.3390/nano10010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston T.G., Yuan S.-F., Wagner J.M., Yi X., Saha A., Smith P., Nelson A., Alper H.S. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation. Nat. Commun. 2020;11:563. doi: 10.1038/s41467-020-14371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha A., Johnston T.G., Shafranek R.T., Goodman C.J., Zalatan J.G., Storti D.W., Ganter M.A., Nelson A. Additive manufacturing of catalytically active living materials. ACS Appl. Mater. Interfaces. 2018;10:13373–13380. doi: 10.1021/acsami.8b02719. [DOI] [PubMed] [Google Scholar]
- 26.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Iii, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343. doi: 10.1038/nmeth.1318. https://www.nature.com/articles/nmeth.1318#supplementary-information [DOI] [PubMed] [Google Scholar]
- 27.Cheng J.K., Alper H.S. Transcriptomics-guided design of synthetic promoters for a mammalian system. ACS Synth. Biol. 2016;5:1455–1465. doi: 10.1021/acssynbio.6b00075. [DOI] [PubMed] [Google Scholar]
- 28.Heimo H., Palmu K., Suominen I. Human placental alkaline phosphatase: expression in Pichia pastoris, purification and characterization of the enzyme. Protein Expr. Purif. 1998;12:85–92. doi: 10.1006/prep.1997.0808. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen A.W., Le K.C., Maynard J.A. Identification of high affinity HER2 binding antibodies using CHO Fab surface display, Protein Engineering. Design and Selection. 2018;31:91–101. doi: 10.1093/protein/gzy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millik S.C., Dostie A.M., Karis D.G., Smith P.T., McKenna M., Chan N., Curtis C.D., Nance E., Theberge A.B., Nelson A. 3D printed coaxial nozzles for the extrusion of hydrogel tubes toward modeling vascular endothelium. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab2b4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith K., Garman L., Wrammert J., Zheng N.-Y., Capra J.D., Ahmed R., Wilson P.C. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta D., Satyanarayana T. Bacterial and archaeal α-amylases: diversity and amelioration of the desirable characteristics for industrial applications. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J.-R., Li Y.-Y., Liu D.-N., Liu J.-S., Li P., Chen L.-Z., Xu S.-D. Codon optimization significantly improves the expression level of α -amylase gene from Bacillus licheniformis in Pichia pastoris. BioMed Res. Int. 2015 doi: 10.1155/2015/248680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brady J.R., Whittaker C.A., Tan M.C., Kristensen D.L., II, Ma D., Dalvie N.C., Love K.R., Love J.C. Comparative genome-scale analysis of Pichia pastoris variants informs selection of an optimal base strain. Biotechnol. Bioeng. 2020;117:543–555. doi: 10.1002/bit.27209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ministro J., Manuel A.M., Goncalves J. Therapeutic antibody engineering and selection strategies. In: Silva A.C., Moreira J.N., Lobo J.M.S., Almeida H., editors. Current Applications of Pharmaceutical Biotechnology. Springer International Publishing; Cham: 2020. pp. 55–86. [Google Scholar]
- 36.Ecker D.M., Jones S.D., Levine H.L. The therapeutic monoclonal antibody market. mAbs. 2015;7:9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H., May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolsten M., Søgaard M. Precision medicine: an approach to R&D for delivering superior medicines to patients. Clin. Transl. Med. 2012;1:7. doi: 10.1186/2001-1326-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wawrzyńska E., Kubies D. Alginate matrices for protein delivery - a short review. Physiol. Res. 2018;67:S319–S334. doi: 10.33549/physiolres.933980. [DOI] [PubMed] [Google Scholar]
- 40.Ngo T.D., Kashani A., Imbalzano G., Nguyen K.T.Q., Hui D. Additive manufacturing (3D printing): a review of materials, methods, applications and challenges. Compos. B Eng. 2018;143:172–196. doi: 10.1016/j.compositesb.2018.02.012. [DOI] [Google Scholar]
- 41.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 42.Solá R.J., Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharmacol. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C.-P., Tsai T.-I., Cheng T., Shivatare V.S., Wu C.-Y., Wu C.-Y., Wong C.-H. Glycoengineering of antibody (Herceptin) through yeast expression and in vitro enzymatic glycosylation. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:720. doi: 10.1073/pnas.1718172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bareither R., Pollard D. A review of advanced small-scale parallel bioreactor technology for accelerated process development: current state and future need. Biotechnol. Prog. 2011;27:2–14. doi: 10.1002/btpr.522. [DOI] [PubMed] [Google Scholar]
- 45.Baumann P., Hubbuch J. Downstream process development strategies for effective bioprocesses: trends, progress, and combinatorial approaches. Eng. Life Sci. 2017;17:1142–1158. doi: 10.1002/elsc.201600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millet L.J., Lucheon J.D., Standaert R.F., Retterer S.T., Doktycz M.J. Modular microfluidics for point-of-care protein purifications. Lab Chip. 2015;15:1799–1811. doi: 10.1039/C5LC00094G. [DOI] [PubMed] [Google Scholar]
- 47.Crowell L.E., Lu A.E., Love K.R., Stockdale A., Timmick S.M., Wu D., Wang Y., Doherty W., Bonnyman A., Vecchiarello N., Goodwine C., Bradbury L., Brady J.R., Clark J.J., Colant N.A., Cvetkovic A., Dalvie N.C., Liu D., Liu Y., Mascarenhas C.A., Matthews C.B., Mozdzierz N.J., Shah K.A., Wu S.-L., Hancock W.S., Braatz R.D., Cramer S.M., Love J.C. On-demand manufacturing of clinical-quality biopharmaceuticals. Nat. Biotechnol. 2018;36:988–995. doi: 10.1038/nbt.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.