Abstract
Coxiella burnetii is an obligate intracellular zoonotic bacterium that causes Q fever. Ruminants, including cattle, are broadly known to be reservoirs for this bacterium. Since 2006, many research groups have evaluated the herd-level prevalence of C. burnetii in cattle by molecular techniques on composite milk samples. This study explored the global C. burnetii herd-level prevalence from studies done on bovine bulk-tank milk (BTM) samples using PCR-based analysis. Also, moderators were investigated to identify sources of heterogeneity. Databases (CAB Abstracts, Medline via Ovid, PubMed, Web of Science and Google Scholar) were searched for index articles on C. burnetii prevalence in BTM samples by PCR published between January-1973 and November-2018. Numerous studies (1054) were initially identified, from which seventeen original publications were included in the meta-analysis based on the pre-defined selection criteria. These studies comprised 4031 BTM samples from twelve countries. A random-effects model was used because of considerable heterogeneity (I2 = 98%) to estimate the herd-level prevalence of C. burnetii as 37.0%(CI95%25.2–49.5%). The average herd size appeared to account for a high level of the heterogeneity. No other moderators (geographic location, gross national income or notification criteria for Q fever) seemed to be determinant. This systematic evaluation demonstrated a high molecular prevalence of C. burnetii in BTM samples both in European and non-European countries, evidencing a widespread herd-level circulation of this agent in bovine dairy farms around the world. Meta-regression showed herd size as the most relevant moderator with the odds of a BTM sample testing positive doubling with every unit increase.
Keywords: Q fever, Coxiella burnetii, Coxiellosis, Meta-prevalence, PCR, IS1111
Graphical abstract
Highlights
-
•
First meta-analysis of the PCR-based prevalence of C. burnetii in bovine milk
-
•
Results showed a high molecular prevalence of C. burnetii in bulk-tank milk samples.
-
•
C. burnetii is widely distributed in dairy farms in Europe and the wider world.
-
•
Current results reinforce the need for further investigations on this zoonosis.
1. Introduction
Coxiella burnetii the intracellular Gram-negative bacterium responsible for the zoonotic disease Q fever [1] has many reservoirs, including ruminants, that represent the primary source of environmental contamination and of infection in people [2]. This agent causes fertility disorders and metritis in cattle and is implicated in bovine abortion [[3], [4], [5]]. It often leads to abortion in small ruminants when a pregnant dam is infected, as C. burnetii exhibits a specific tropism for the trophoblast cells in placental cotyledons [6].
Coxiella burnetii has a complex epidemiological pattern and characteristics that make its control challenging. It is widely disseminated in nature and infects a large number of species, including mammals, birds, reptiles and fish [7]. There are two maintenance cycles in nature, one involving domestic species, and another including wild animal species and their ectoparasites. Ticks may be involved in the transmission of C. burnetii between wildlife and domestic species [8]. Additionally, the agent is extremely resistant remaining viable in the environment over extended periods [8]. Coxiella burnetii can also undergo air-borne transmission by contaminated dust particles, which can be facilitated by hot and dry weather conditions [9,10].
A large human outbreak of Q fever reported in the Netherlands (2007–2010), comprising more than 4000 cases, emphasised the need for robust surveillance campaigns and highlighted its importance as a threat to public health [9,11]. Transmission to people is principally by the inhalation of aerosolised contaminated animal placenta and birth fluids during abortions or the birth of normal offspring [12]. Practices such as the assistance of calving, handling of birth products, and manure spreading may present a high risk for C. burnetii transmission to humans [[13], [14], [15]]. There is no consensus about the importance or effectiveness of the digestive route of infection by the consumption of raw milk and dairy product [6,[16], [17], [18]]. Nevertheless, respiratory exposure to aerosols produced during milking of animals should not be underestimated [19].
The level of bacterial load by the different routes differs among ruminants [6]. While parturition products are the primary source of shedding in small ruminants, milk seems to play a central role as a shedding route of C. burnetii in dairy cattle [20,21]. Even asymptomatic animals [20] or seronegative cattle [22] have been identified as C. burnetii milk shedders. Coxiella burnetii can be excreted in milk for up-to 13 months [9,23], although this may be intermittent [6]. Two patterns of shedding have been identified in dairy cows which can be persistent heavy shedders or sporadic shedders [20].
Based on these heterogeneous shedding patterns, composite samples such as bulk-tank milk (BTM) constitute useful and easily accessible specimens for large scale epidemiological investigation. A positive result provides robust evidence for the identification of infected herds. Bulk-tank milk testing is the preferred diagnostic approach for disease notification in many countries [24] and has epidemiological value for the monitoring of infection status over time in follow-up evaluations [25].
Recent large human Q fever outbreaks in the Netherlands, Spain, France and Germany have increasingly focussed attention on coxiellosis in many European countries where strategies including mandatory notification of the disease have been implemented. We systematically review studies of the herd prevalence of C. burnetii in dairy cattle using PCR on BTM samples, conduct a meta-analysis to determine the overall European and global prevalences and assess geographic region, average herd size, local legislation for coxiellosis and per capita income in each country where studies were conducted as potential moderators.
2. Material and methods
2.1. Literature search and study selection
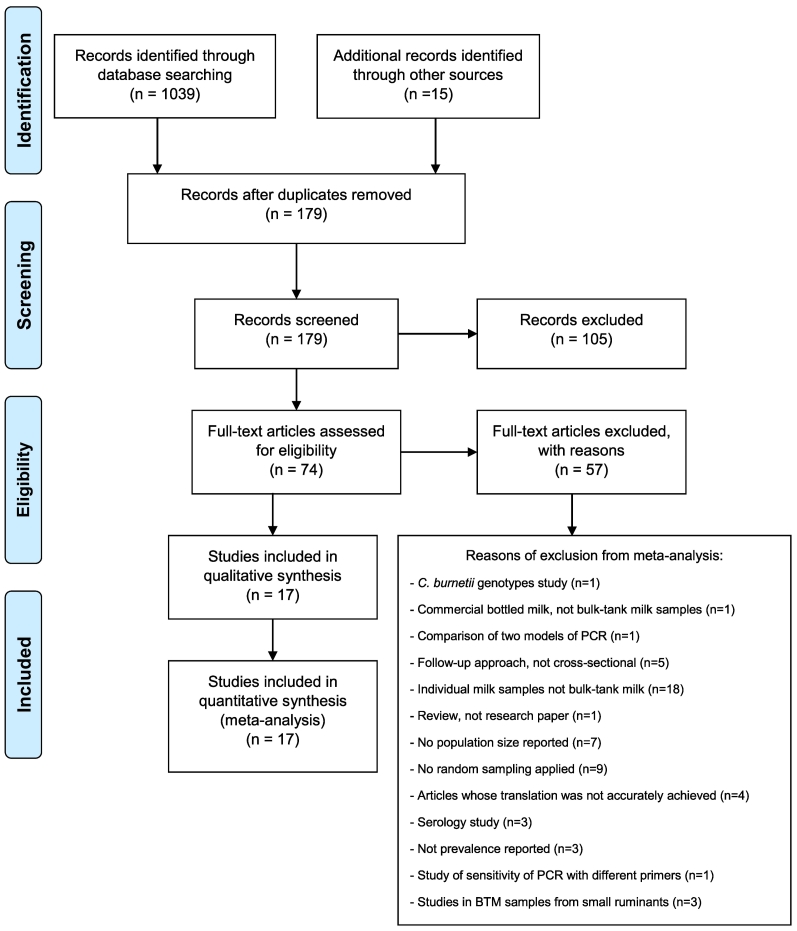
The systematic review and meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [26] (Fig. 1). The search strategy identified publications reporting the prevalence of C. burnetii on BTM samples analysed by molecular studies. The following electronic databases were used to identify studies published from January 1973 up to November 2018 (week 43 of 2018): CAB Abstracts, Medline, PubMed, Web of Science, Scopus, Science Direct and Google Scholar. The literature search comprised the terms: “Coxiella burnetii” or “Q fever” or “coxiellosis” and “PCR” or “qPCR” or “real-time PCR” or “molecular diagnosis” and “BTM” or “milk”, with no language restriction. No constraint in study designs was applied at this phase. Additional publications were identified by cross checking references included in the articles. Duplicates were identified by reference management software (Mendeley) and manually removed.
Fig. 1.
PRISMA flow diagram describing the study design process for the systematic review and meta-analysis of the molecular prevalence of Coxiella burnetii in bulk-tank milk from bovine dairy herds.
2.2. Eligibility - inclusion criteria
Publications on studies fulfilling all the following criteria were eligible for inclusion: (i) molecular investigation of C. burnetii by PCR, (ii) random sampling, (iii) composite single test-day samples obtained from the bulk storage tank located on a dairy cattle farm, (iv) primary studies, but not reviews, (v) cross-sectional studies reporting prevalence. Authors of articles not stating the total number of dairy cattle herds from which the sample was drawn were contacted to provide this missing data. Publications were examined by two independent reviewers (AR and MF) to ensure they matched the inclusion criteria. Discrepancies between the two reviewers on eligibility were discussed with the rest of authors until reaching agreement.
2.3. Data extraction and meta-analysis
Studies were screened by title, and abstract and irrelevant publications were excluded. The remaining studies were full-text checked against the inclusion criteria described above. Articles that did not fulfil all these criteria were excluded. The number of publications excluded are shown in Fig. 1. Data were systematically extracted from all the studies that satisfied the inclusion criteria, including: the first author identity, year of publication, study title, journal title, country, study methodology (duration of sampling, herd size, sample size, the number of positives herds and/or prevalence, randomisation), molecular technique and target gene used. When available, information about the factors associated with the C. burnetii infection was also reported.
The C. burnetii herd prevalence determined in BTM samples (dependant variable) was considered as the effect size for the studies included in the meta-analysis. This meta-analysis of proportions was performed as outlined by Wang [27]. The heterogeneity among studies was first investigated by Cochran's Q (X2) that tests the null hypothesis of homogeneity, and then quantified by the Higgins' I2 statistic [28]. The heterogeneity was measured to select the model for the overall weighted C. burnetii herd prevalence estimation. As the level of heterogeneity was high, a random-effects model was first used to address both within-study variance (the sampling error) and the between-studies variance (τ2). Possible sources of heterogeneity were investigated through the analysis of moderators. The evaluated moderators included: i) geographic region: Europe vs non-Europe; ii) average herd size; iii) local legislation for Q fever: mandatory notification vs non-mandatory notification [[29], [30], [31], [32], [33], [34], [35], [36], [37]], and iv) gross national income (GNI) per capita classification from the year the study was conducted, based on the Atlas method [38]. A subgroup analysis was performed for the categorical moderators. Categorical moderators were analysed using a mixed-effects model. The statistical significance of the moderators was evaluated by an omnibus test (QM) within the mixed-effects model [39]. The proportion of heterogeneity accounted for by each moderator was explored by the R2 index. Meta-regression was also utilised to explore heterogeneity among the studies. All the moderators and their interactions were entered in the initial model and non-significant terms were then dropped stepwise (from lowest R2 to highest R2) [40]. The odds ratio (OR) for loge average herd size was additionally investigated. Association among moderators was assessed by the Pearson correlation coefficient (r). Results from the meta-analysis with the corresponding 95% confidence intervals were summarized using forest plots. Egger's test was used to test for the possibility of a publication bias for studies with low or high effect sizes [41]. All the assessments were conducted using open RStudio software (Boston, MA) with metafor package, mvmeta package and metaprop commands [39,42].
3. Results
3.1. Description of the studies
After removal of duplicates, a total of 179 studies were identified initially (Fig. 1). Seventeen studies from twelve different countries (Belgium, Colombia, Hungary, Iran [2 studies], Italy [3 studies], Latvia, Netherlands [2 studies], Portugal, Spain, South Korea, UK and USA [2 studies]) were eligible for the meta-analysis based on the inclusion criteria. Six of those studies were conducted in non-European countries and 11 in European countries; 10 were conducted in countries where Q fever is a notifiable disease, while 7 were from countries where it is not. The study conducted in the Basque Country was included in the subgroup with mandatory notification, although this is the only Spanish province where the notification for Q fever is compulsory. Finally, 3 studies were conducted in upper-middle-income countries and 14 studies were in high income countries. The seventeen selected articles are summarized in Table 1 and included test results for a total of 4031 BTM samples collected over 9 years (2006 to 2015). Studies employed either conventional PCR (n = 5), quantitative PCR (n = 9) or nested PCR (n = 3). The transposon-like repetitive region of the bacterial genome (IS1111) was the gene most frequently used as the target in these PCRs (n = 14), followed by com1 (n = 2), icd (n = 1) and 16S rRNA genes (n = 1) (Table 1).
Table 1.
Characteristics and main results of the eligible studies ordered by molecular prevalence of Coxiella burnetii in bulk-tank milk samples.
| Author | Year | Country | Study area | Average herd size | Period of study | Risk factor analysis | Gross national income per capita [38] | Is Q fever a mandatory notifiable disease? | Molecular approach | Target gene | N herds in study area | Percentage of herds sampled | BTM(i) samples tested | Positive BTM samples | Prevalence | 95% CI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Boroduske et al. [43] | 2017 | Latvia | Nationwide | 8.6 | 2015 | Yes | High-income | Yes | qPCR | IS1111 | 5040 | 5 | 252 | 27 | 10.7 | 7.2 | 14.9 |
| Kargar et al. [23] | 2013 | Iran | Johrom | 3.7 | – | Yes | Upper-middle-income | Yes | nPCR | com1 | 3000 | 3.3 | 100 | 11 | 11 | 5.5 | 18.0 |
| Seo et al. [44] | 2018 | South Korea | Gyeongsang | 74 | 2015 | No | High-income | Yes | nPCR | 16S rRNA | 869 | 69.9 | 607 | 108 | 17.8 | 14.8 | 20.9 |
| Rahimi et al. [45] | 2010 | Iran | Chaharmahal and Bakhtiari | 48 | 2008 | No | Upper-middle-income | Yes | nPCR | com1 | 95 | 29.5 | 28 | 5 | 17.9 | 5.5 | 34.5 |
| van Engelen et al. [46] | 2014 | Netherlands | Nationwide | 71.7 | 2009–2011 | Yes | High-income | Yes | qPCR | IS1111 | 20,746 | 1.5 | 309 | 58 | 18.8 | 14.6 | 23.3 |
| Anastácio et al. [47] | 2016 | Portugal | Nationwide | 21.7 | 2009–2013 | Yes | High-income | No | PCR | IS1111 | 1712 | 2.6 | 45 | 9 | 20 | 10.9 | 33.8 |
| Velasova et al. [48] | 2017 | UK | Nationwide | 133 | 2014–2015 | No | High-income | No | qPCR | icd/IS1111 | 10,491 | 2.1 | 220 | 57 | 25.9 | 20.3 | 31.9 |
| Czaplicki et al. [49] | 2012 | Belgium | Wallonia | 28.5 | 2006 | Yes | High-income | No | qPCR | IS1111 | 5086 | 1 | 50 | 15 | 30 | 8.7 | 51.3 |
| Magnino et al. [50] | 2009 | Italy | Cremona, Montova and Pavia | 180 | 2007–2008 | No | High-income | No | PCR | IS1111 | 3550 | 11.2 | 400 | 161 | 40.2 | 35.5 | 45.1 |
| Valla et al. [51] | 2014 | Italy | Nationwide | 42.5 | 2011–2013 | No | High-income | No | PCR | IS1111 | 30,000 | 1.1 | 344 | 138 | 40.1 | 35.0 | 45.4 |
| Contreras et al. [37] | 2015 | Colombia | Monteria | 150–600 | 2012 | No | Upper-middle-income | No | PCR | IS1111 | 3341 | 0.3 | 11 | 5 | 45.5 | 16.7 | 75.8 |
| Astobiza et al. [52] | 2012 | Spain | Bizkaia | 46.1 | 2009–2010 | No | High-income | No / Yes(ii) | qPCR | IS1111 | 178 | 100 | 178 | 92 | 51.7 | 44.4 | 59 |
| Muskens et al. [25] | 2011 | Netherlands | Nationwide | 65.7 | 2007 | No | High-income | Yes | qPCR | IS1111 | 21,313 | 1.6 | 341 | 193 | 56.6 | 50.7 | 61.9 |
| Vicari et al. [34] | 2013 | Italy | Lombardy | 182 | 2011 | No | High-income | No | PCR | IS1111 | 5750 | 5 | 287 | 173 | 60.3 | 54.5 | 65.9 |
| Bauer et al. [53] | 2015 | USA | Indiana | 145.3 | 2011 | No | High-income | Yes | qPCR | IS1111 | 1225 | 25.8 | 316 | 193 | 61.1 | 55.6 | 66.4 |
| Gyuranecz et al. [54] | 2012 | Hungary | Nationwide | 14.5 | 2010–2011 | No | High-income | Yes | qPCR | IS1111 | 17,172 | 0.1 | 15 | 10 | 66.7 | 40.5 | 88.7 |
| APHIS [55] | 2007 | USA | 18 states(iii) | 162.6 | 2007 | No | High-income | Yes | qPCR | IS1111 | 54,100 | 1 | 528 | 406 | 76.9 | 73.2 | 80.4 |
(i): BTM: bulk-tank milk samples, one per herd; PCR: conventional PCR; qPCR: real-time PCR; nPCR: nested PCR. (ii) mandatory notification in Basque Country. (iii) California, Idaho, Indiana, Iowa, Kentucky, Michigan, Minnesota, Missouri, New Mexico, New York, Ohio, Pennsylvania, Texas, Vermont, Virginia, Washington, Wisconsin.
3.2. The estimated overall meta-prevalence of Coxiella burnetii in BTM samples
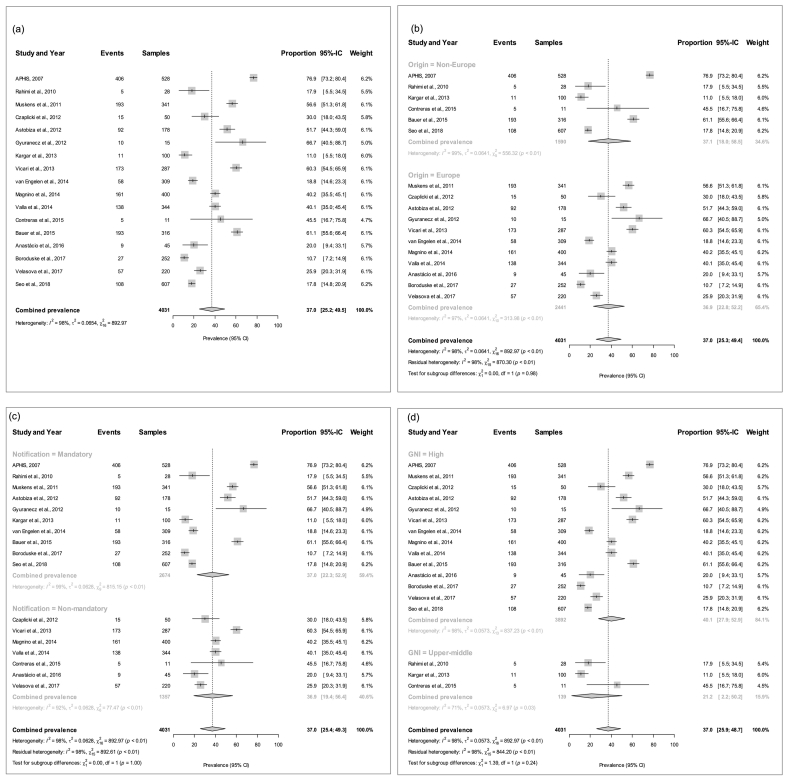
The median size of the eligible studies was 252 BTM samples. Of the total 4031 BTM samples, 1661 were diagnosed positive by molecular techniques. The percentages of positive BTM samples among the studies ranged from 10.7 to 76.9%. The overall weighted prevalence of C. burnetii in the random-effects meta-analysis was estimated at 37.0% (CI95%25.2–49.5%). The I2 value of 98.0% (CI95%95.9–99.0) suggested high heterogeneity, with a τ2 of 0.0654 (CI95%0.3296–1.4997), and an X2 statistic of 892.97 (P < 0.0001). The overall meta-analysis is shown in a forest plot (Fig. 2a). No obvious evidence of publication bias was detected in the meta-analysis on the basis of Egger's test (P = 0.599).
Fig. 2.
Forest plot for the meta-analysis of herd-level Coxiella burnetii prevalence based on bulk-tank milk samples from the seventeen studies that matched the inclusion criteria in the systematic review. (a) All studies. (b) European and non-European country subgroups. (c) Grouped by mandatory and non-mandatory notification. (d) Grouped by the per capita Gross National Income (GNI) level.
3.3. The meta-prevalence of Coxiella burnetii and moderator analyses
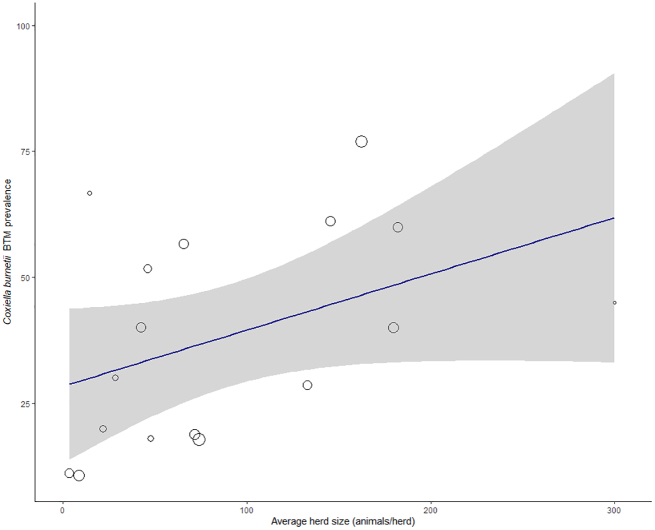
The weighted average prevalence was similar within each of the two geographic subgroups (36.9% in European countries and 37.1% in non-European countries; (I2 = 98%; X2 = 870.29, P < 0.01; QM (df = 1) = 0.002, P = 0.98), albeit with differing 95% confidence intervals of 22.8%– 52.2% in the former and 18.0%–58.5% in the latter group of countries (Fig. 2b). Similarly, countries with mandatory and non-mandatory notification of Q fever had a prevalence around 37.0% (CI95%22.3–52.9% and CI95%19.4–56.4%, respectively; (I2 = 98%; X2 = 892.61, P < 0.01; QM (df = 1) = 0.010, P = 1.00) (Fig. 2c). In the subgroup analysis based on the GNI per capita (Fig. 2d), the prevalence was 40.1% (CI95%27.9–52.9%) in high-income countries and 21.2% (CI95%2.2–50.2%) in upper-middle-income countries (I2 = 98%; R2 = 3.10%; X2 = 844.20, P < 0.01; QM (df = 1) = 1.39, P = 0.24). None of the three factors above appeared to contribute meaningfully to the observed level of heterogeneity based on the subgroup analysis. The meta-regression revealed that average herd size accounted for a significant proportion of the heterogeneity (I2 = 97%; R2 = 33.01%; X2 = 552.23, P < 0.01; QM = 4.55, P = 0.03). As a significant moderator, high-size herds presented a higher herd-level C. burnetii BTM prevalence (Fig. 3). The odds ratio for the loge of herd size was 2.00 (CI95%1.24–3.52; P = 0.02). A strong positive correlation was found between countries being located in Europe and high GNI per capita income (r = 0.633, P < 0.05), but between location in Europe and compulsory disease notification (r = −0.239, P = 0.24), and between high GNI per capita and notification (r = −0.076, P = 0.82) correlations were weak and negative. Herd size was not meaningfully correlated with the origin of the studies (r = −0.468, P = 0.12), notification (r = −0.428, P = 0.16), or with GNI per capita (r = −0.444, P = 0.14).
Fig. 3.
Bubble plot for meta-regression of herd-level Coxiella burnetii prevalence based on bulk-tank milk with average herd size as continuous covariate. Points represent the seventeen studies that matched the inclusion criteria in the systematic review. Bubble size is in relation to the weight of each primary study.
4. Discussion
Global serological or molecular prevalences from pathogens as diverse as Toxoplasma gondii and Helicobacter pylori have been estimated by meta-analyses following a systematic review of the published body of studies [56,57]. We conducted a comprehensive keyword-based systematic review of the literature on the global molecular prevalence of C. burnetii in bovine BTM samples and data from those studies matching the inclusion criteria was extracted and included in a meta-analysis. For the purpose of this review, only adequately randomised studies with a cross-sectional design were included.
Heterogeneity among studies was first investigated by Higgins' I2 statistic which indicates the proportion of heterogeneity not due to chance. A high level of heterogeneity (≥75%) indicates another source of variability besides the random error. The high I2 value (98%) led to the choice of a random-effects model for estimating the overall weighted C. burnetii herd-level prevalence among eligible articles, which makes no assumption that the prevalence is constant across the studies. The meta-analysis shows that C. burnetii is widely distributed in dairy farms around twelve countries from 3 continents (America, Europe, and Asia). The best estimate of global C. burnetii herd-level prevalence, based on the studies matching the current inclusion criteria, was 37.0%. While there was no obvious evidence of publication bias based on Egger's test, this test has limited power and the possibility of bias cannot be altogether excluded [58].
Bulk tank milk samples are a widely used approach for studying infectious diseases of dairy livestock at the population level, despite that dry cows and unhealthy animals are not included and hence BTM only provides a partial representation of the herd sanitary status. The analysis of BTM samples represents a suitable and convenient approach for the investigation of C. burnetii, not only for initial farm-level screening in situations where their disease status is unknown, but also for repeated analyses during monitoring programmes or after sanitary interventions such as antibiotic administration [59] or vaccination [60,61]. A positive BTM result confirms herd exposure to C. burnetii.
The molecular diagnostic methods of studies included in this meta-analysis targeted different regions of the bacterial C. burnetii genome. The repetitive element IS1111 was selected in most of the published studies as this multiple copy gene is presumed to increase the sensitivity of the test [62]. Other studies used PCRs targeting com1, icd and 16S rRNA genes. The com1 element is frequently used for accurate quantification, as this is a single-copy gene [63]. Additionally, the analysis of 16S RNAs may reveal the prevalence of Coxiella as a genus, by the identification of both C. burnetii and Coxiella-like organisms [44].
The overall weighted C. burnetii prevalence found in bovine dairy herds was higher than the 5.1% to 22.1% range reported for BTM samples from sheep dairy flocks [47,64,65]. This difference could be explained by the primary route of bacterial transmission in each species. A higher C. burnetii prevalence might be expected in bovine milk, which is the predominant route of shedding for cows (and with a longer duration), whereas milk is less important for transmission from goats and sheep [9,23].
Two nationwide studies in Dutch dairy herds revealed markedly different prevalence levels in 2011 (56.6%) and 2014 (18.8%) [25,46], when using the same molecular approach in a similar number of herds. The lower prevalence in 2014 might be related to compulsory control measures applied in dairy goat farms after the large human Q fever outbreak in 2007–2010 [11,66]. There is some albeit limited evidence that the same outbreak strain may affect both cattle and goats in the Netherlands [67], and measures applied to goat farms might have indirectly helped to reduce prevalence in bovine herds. Similarly, three studies conducted in Italian herds in 2013 and 2014 also reported differences in C. burnetii prevalence. Valla et al. (2014) [51] revealed a nationwide prevalence of 40.0%, while Vicari et al. (2013) [34] found a higher prevalence of 60.0% in the northwest region of Lombardy, where almost half of Italian cows' milk is produced [68]. The molecular prevalence of C. burnetii found in Lombardy represented a marked increase compared to a previous two-year study (2007–2008) conducted in the same region (40.0%) [50].
Differences in the bacterial shedding patterns among ruminants and uncertainty about the importance of milk-borne infection may result in emphasis on different control measures depending on the species. In small ruminants, the identification of high-risk dams before parturition is important in avoiding zoonotic risk [69]. In cattle where milk is the primary shedding route, pre-partum monitoring may not be as appropriate [69]. Identification of chronic C. burnetii milk shedding cattle may be more effective in preventing environmental contamination, decreasing the risks of transmission among animals and preventing the spread of the bacterium.
Only five of the seventeen selected articles included analysis of factors associated with C. burnetii infection. Herd size, cattle density and purchasing replacement animals from external sources were all linked with C. burnetii infection [43,46]. Additionally, the presence of ticks on cattle was associated with BTM PCR positivity [46].
For both cattle and small ruminants, a positive correlation between herd size and herd prevalence of C. burnetii has been reported [70,71]. The association between herd size, density of animals and an enhanced risk of C. burnetii infection has been well demonstrated [10,72]. Close contact between cows is an intrinsic characteristic of dairy herd management systems, and larger herds offer even greater chances for contact and transmission. Densely populated farms are prone to a higher risk of transmission of the pathogen within the herd after C. burnetii is introduced into the farm. Additionally, high animal density leads to greater bacterial load and thus higher environmental contamination [73], which may represent an increased risk of transmission to either cattle or people. This meta-analysis showed that elevated prevalence of C. burnetii is associated with large-sized herds, where the odds of a BTM sample testing positive double with every unit increase in loge herd size (odds ratio CI95%1.24–3.52). Accordingly, of the moderators analysed, average herd size had the largest effect, accounting 33.0% of the observed level of heterogeneity among studies.
While Q fever has been studied in both European and non-European countries, these two contexts have not previously been contrasted. The overall prevalence of C. burnetii infection was remarkably similar in European and non-European studies (both 37%). The greater variability among non-European studies (CI95% 18.0%–58.5%) than among European studies (CI95% 22.8%–52.2%) could be accounted for by the differences in the numbers of studies and herds investigated.
The mandatory notification of a disease should be helpful not only for early identification of outbreaks but also to enable evaluation of the effectiveness of control strategies. For instance, legislation implemented by the Dutch government in the face of the largest Q fever outbreak ever recorded included compulsory notification of coxiellosis [66]. In the current meta-analysis, a remarkable similarity was noted between overall weighted prevalence of C. burnetii in BTM samples from countries with mandatory (37.0%, CI95%22.3–52.9%) and non-mandatory (36.9%, CI95%19.4–56.4%) notification legislation.
In our meta-analysis, the GNI per capita seems to have a minor effect as a moderator of the prevalence of C. burnetii in BTM samples. When the studies were stratified according to this indicator of economic development, high-income countries had twice the overall weighted prevalence of upper-middle income countries, albeit that this difference was not statistically significant (P = 0.24). All publications matching the inclusion criteria were conducted in high and upper-middle income countries. None of the studies conducted in low-middle and low-income countries that were identified in the initial search fulfilled the inclusion criteria and were rejected from the meta-analysis. For instance, an ineligible study carried out in Egypt reported a 22% molecular prevalence of C. burnetii in individual milk samples [74] and one carried out in Bangladesh reported 15.6% seroprevalence in herd milk specimens [75]. These findings suggest that further field studies could prove rewarding. The overall prevalence in low-middle and low-income countries remains unknown. There is evidence of extensive ruminant infection with C. burnetii throughout African countries where the threat of human exposure and significant economic impact are possibly underestimated [76].
Some heterogeneity might have resulted from methodological variation among nine of the 17 studies that used qPCR to detect the IS1111 target. Four of these [25,46,49,52] used the TaqVet Coxiella burnetii LSI kit and followed the same manufacturer's instructions for the amplification reaction and for the interpretation of the results. These four studies considered samples as positive with a cycle threshold (Ct) < 40. Two further studies used threshold Ct values of 36.5 [53] and 36.95 [54], while the remaining three studies using qPCR to detect IS1111 did not report threshold Ct values.
Moreover, whereas the IS1111 transposon-like element is a multi-copy gene [77], the 16S rRNA target used in South Korean study [44] and the com1 target used in the two studies in Iran [23,45] are both single copy genes. The assays used in these studies might have had lower sensitivity and indeed, the studies using the single copy assays had three of the four lowest prevalence values. All three of these studies were in non-European countries where the disease is notifiable, and the two Iranian studies were in an upper-middle income country, which may have introduced a degree of bias in the analysis.
Although the moderator analysis identified average herd size as one source, most of the heterogeneity remained unexplained (residual heterogeneity I2 = 97.0%; P < 0.01). It is quite possible that other factors, not currently addressed, influence the C. burnetii herd-level prevalence. Unsurprisingly, two of the moderators were highly correlated; studies in European and in high-income countries showed a significant and positive correlation (r = 0.627, P < 0.01). Awareness of the relationships between moderators that may potentially induce bias in the analysis should be considered when drawing conclusions [78].
5. Conclusion
This meta-analysis reports a high overall global prevalence of C. burnetii in BTM samples of 37.0% (CI95%25.2–49.5%), showing widespread herd-level circulation of this agent in bovine dairy farms. These results should be of interest not only for European countries where C. burnetii is a well-known health threat, but also in countries where epidemiological investigations have been limited, its importance as a zoonosis may be underestimated and prevention strategies may need to be implemented. Information on local biosecurity practices and environmental conditions would be valuable for a full understanding of C. burnetii prevalence globally, but such descriptions were lacking in most of the publications considered in the meta-analysis. While this study has shown the global herd prevalence of C. burnetii in dairy cattle to be high, in many countries, including high-income countries such as Belgium, Italy, Portugal and UK, the disease is not currently notifiable, and control is not mandatory. To make it so might represent an additional burden on dairy farmers and would require justification on economic or public health grounds for which further study might be required. The high herd-level circulation of C. burnetii in bovine dairy farms in several countries showed by this study reinforces the need for further investigations on this globally important zoonosis.
Author contributions
AR and MCE conceptualised the study. AR and MF performed the systematic review, including data collection and screening the retrieved records. AR, MCE and LGC conducted the data-analysis. All authors (AR, MF, LGC, KT, FRC and MCE) made contributions to the interpretation of results. All authors participated in the manuscript drafting and reviewing.
Funding
The first author is supported by a PhD scholarship (POS_EXT_2015_1_123804) from Agencia Nacional de Innovación e Investigación (ANII), Uruguay.
Declaration of Competing Interest
None declared.
Acknowledgements
We thank Emma Place from the School of Veterinary Sciences (University of Bristol) for assistance with literature review strategies. The first author is supported by a PhD scholarship (POS_EXT_2015_1_123804) from Agencia Nacional de Innovación e Investigación (ANII), Uruguay. The graphical abstract was created at Biorender.com.
References
- 1.Porter S.R., Czaplicki G., Mainil J., Guattéo R., Saegerman C. Q fever: current state of knowledge and perspectives of research of a neglected zoonosis. Int. J. Micro. 2011;13:248418. doi: 10.1155/2011/248418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woldehiwet Z. Q fever (coxiellosis): epidemiology and pathogenesis. Res. Vet. Sci. 2004;77(2):93–100. doi: 10.1016/j.rvsc.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Lang G.H. Coxiellosis (Q fever) in animals. Q Fever. 1990;1:23–48. [Google Scholar]
- 4.To H, Htwe K.K., Kako N., Kim H.J., Yamaguchi T., Fukushi H., Hirai K. Prevalence of Coxiella burnetii infection in dairy cattle with reproductive disorders. J. Vet. Med. Sci. 1998;60(7):859–861. doi: 10.1292/jvms.60.859. [DOI] [PubMed] [Google Scholar]
- 5.Bildfell R.J., Thomson G.W., Haines D.M., McEwen B.J., Smart N. Coxiella burnetii infection is associated with placentitis in cases of bovine abortion. J. Vet. Diagn. Investig. 2000;12(5):419–425. doi: 10.1177/104063870001200505. [DOI] [PubMed] [Google Scholar]
- 6.Rodolakis A., Berri M., Hechard C., Caudron C., Souriau A., Bodier C.C., Blanchard B., Camuset P., Devillechaise P., Natorp J.C., Vadet J.P., Arricau-Bouver N. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J. Dairy Sci. 2007;90(12):5352–5360. doi: 10.3168/jds.2006-815. [DOI] [PubMed] [Google Scholar]
- 7.Sawyer L.A., Fishbein D.B., McDade J.E. Q fever: current concepts. Rev. Infect. Dis. 1987;9:935–946. doi: 10.1093/clinids/9.5.935. [DOI] [PubMed] [Google Scholar]
- 8.Aitken I.D., Bogel K., Cracea E., Edlinger E., Houwers D., Krauss H., Rady M., Rehacek J., Schiefer H.G., Schmeer N. Q fever in Europe: current aspects in aetiology, epidemiology, human infection, diagnosis and therapy. Infection. 1987;15:323–327. doi: 10.1007/BF01647731. [DOI] [PubMed] [Google Scholar]
- 9.Roest H.I., Tilburg J.J., Van der Hoek W., Vellema P., Van Zijderveld F.G., Klaassen C.H., Raoult D. The Q fever epidemic in the Netherlands: history, onset, response and reflection. Epidemiol. Infect. 2011;139(1):1–2. doi: 10.1017/S0950268810002268. [DOI] [PubMed] [Google Scholar]
- 10.Nusinovici S., Frössling J., Widgren S., Beaudeau F., Lindberg A. Q fever infection in dairy cattle herds: increased risk with high wind speed and low precipitation. Epidemiol. Infect. 2015;143(15):3316–3326. doi: 10.1017/S0950268814003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneeberger P.M., Wintenberger C., Van der Hoek W., Stahl J.P. Q fever in the Netherlands–2007–2010: what we learned from the largest outbreak ever. Med. Mal. Infect. 2014;44(8):339–353. doi: 10.1016/j.medmal.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Roest H.J., van Gelderen B., Dinkla A., Frangoulidis D., van Zijderveld F., Rebel J., van Keulen L. Q fever in pregnant goats: pathogenesis and excretion of Coxiella burnetii. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berri M., Rousset E., Champion J.L., Arricau-Bouvery N., Russo P., Pepin M., Rodolakis A. Ovine manure used as garden fertiliser as a suspected source of human Q fever. Vet. Rec. 2003;153:269–270. doi: 10.1136/vr.153.9.269. [DOI] [PubMed] [Google Scholar]
- 14.Sun W.W., Cong W., Li M.H., Wang C.F., Shan X.F., Qian A.D. Coxiella burnetii seroprevalence and risk factors in cattle farmers and farm residents in three North-eastern provinces and inner Mongolia autonomous region, China. Biomed. Res. Int. 2016;2016 doi: 10.1155/2016/7059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard H., Brockmann S.O., Kleinkauf N., Klinc C., Wagner-Wiening C., Stark K., Jansen A. High seroprevalence of Coxiella burnetii antibodies in veterinarians associated with cattle obstetrics, Bavaria, 2009. Vector Borne Zoonot. Dis. 2012;12(7):552–557. doi: 10.1089/vbz.2011.0879. [DOI] [PubMed] [Google Scholar]
- 16.Fishbein D.B., Raoult D. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am. J. Trop. Med. Hyg. 1992;47(1):35–40. doi: 10.4269/ajtmh.1992.47.35. [DOI] [PubMed] [Google Scholar]
- 17.Rodolakis A. Q fever in dairy animals. Ann. N. Y. Acad. Sci. 2009;1166:90–93. doi: 10.1111/j.1749-6632.2009.04511.x. [DOI] [PubMed] [Google Scholar]
- 18.Eldin C., Mélenotte C., Mediannikov O., Ghigo E., Million M., Edouard S., Mege J.L., Maurin M., Raoult D. From Q fever to Coxiella burnetii infection: a paradigm change. Clin. Microbiol. Rev. 2017;30:115–190. doi: 10.1128/CMR.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loftis A.D., Priestley R.A., Massung R.F. Detection of Coxiella burnetii in commercially available raw milk from the United States. Foodborne Pathog. Dis. 2010;7(12):1453–1456. doi: 10.1089/fpd.2010.0579. [DOI] [PubMed] [Google Scholar]
- 20.Guatteo R., Beaudeau F., Joly A., Seegers H. Coxiella burnetii shedding by dairy cows. Vet. Res. 2007;38(6):849–860. doi: 10.1051/vetres:2007038. [DOI] [PubMed] [Google Scholar]
- 21.Keshavamurthy R., Singh B.B., Kalambhe D.G., Aulakh R.S., Dhand N.K. Prevalence of Coxiella burnetii in cattle and buffalo populations in Punjab, India. Prev. Vet. Med. 2019;166:16–20. doi: 10.1016/j.prevetmed.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Barberio A., Badan M., Busa A., Ceglie L., Capello K., Comin A., Zuliani F., Gioia G., Natale A. Association between serological response and shedding of Coxiella burnetii in milk in dairy cattle. Large Anim. Rev. 2014;20(1):3–8. [Google Scholar]
- 23.Kargar M., Rashidi A., Doosti A., Ghorbani-Dalini S., Najafi A. Prevalence of Coxiella burnetii in bovine bulk milk samples in southern Iran. Comp. Clin. Pathol. 2013;22(3):331–334. [Google Scholar]
- 24.Van der Hoek W., Dijkstra F., Schimmer B., Schneeberger P.M., Vellema P., Wijkmans C., ter Schegget R., Hackert V., van Duynhoven Y. Q fever in the Netherlands: an update on the epidemiology and control measures. Eurosurveillance. 2010;15(12):19520. [PubMed] [Google Scholar]
- 25.Muskens J., Van Engelen E., Van Maanen C., Bartels C., Lam T.J. Prevalence of Coxiella burnetii infection in Dutch dairy herds based on testing bulk tank milk and individual samples by PCR and ELISA. Vet. Rec. 2011;168(3):79. doi: 10.1136/vr.c6106. [DOI] [PubMed] [Google Scholar]
- 26.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 27.Wang N. How to Conduct a Meta-analysis of Proportions in R: A Comprehensive Tutorial Conducting Meta-Analyses of Proportions in R. John Jay Coll Crim Justice, pp. 0–62.
- 28.Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. Wiley-Blackwell; Chichester: 2009. Introduction to Meta-Analysis. [Google Scholar]
- 29.Sidi-Boumedine K., Rousset E., Henning K., Ziller M., Niemczuck K., Roest H.I.J., Thiery E. Development of harmonised schemes for the monitoring and reporting of Q-fever in animals in the European Union. EFSA Support. Public. 2010;7(5):48E. [Google Scholar]
- 30.Dorsett-Martin W.A. Considering Q fever when working with laboratory sheep. Lab. Anim. 2010;39(3):86. doi: 10.1038/laban0310-86. [DOI] [PubMed] [Google Scholar]
- 31.Kwak W., Chu H., Hwang S., Park J.H., Hwang K.J., Gwack J., Choi Y.S., Youn S.K., Park M.Y. Epidemiological characteristics of serologically confirmed q fever cases in South Korea, 2006–2011. OPHRP. 2013;4(1):34–38. doi: 10.1016/j.phrp.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulyok K.M., Kreizinger Z., Hornstra H.M., Pearson T., Szigeti A., Dán Á., Balla E., Keim P.S., Gyuranecz M. Genotyping of Coxiella burnetii from domestic ruminants and human in Hungary: indication of various genotypes. BMC Vet. Res. 2014;10(1):107. doi: 10.1186/1746-6148-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanzo J.M., Garcia-Calabuig M.A., Audicana A., Dehesa V. Q fever: prevalence of antibodies to Coxiella burnetii in the Basque Country. Int. J. Epidemiol. 1993;22(6):1183–1188. doi: 10.1093/ije/22.6.1183. [DOI] [PubMed] [Google Scholar]
- 34.Vicari N., Faccini S., Ricchi M., Garbarino C., Decastelli L., Boldini M., Rosignoli C., Dalmasso A., Bronzo V., Fabbi M. Occurrence of Coxiella burnetii in bulk tank milk from northwestern Italy. Vet. Rec. 2013;172(26):687. doi: 10.1136/vr.101423. [DOI] [PubMed] [Google Scholar]
- 35.Anon . Q Fever Annual Epidemiological Report for 2015. In. Stockholm. 2017. European centre for disease prevention and control. [Google Scholar]
- 36.Esmaeili S., Mobarez A.M., Khalili M., Mostafavi E. High prevalence and risk factors of Coxiella burnetii in milk of dairy animals with a history of abortion in Iran. Comp. Immunol. Microbiol. Infect. Dis. 2019;63:127–130. doi: 10.1016/j.cimid.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Contreras V., Máttar S., González M., Álvarez J., Oteo J.A. Coxiella burnetii in bulk tank milk and antibodies in farm workers at Montería, Colombia. Rev. Colomb. Cienc. Pec. 2015;28(2):181–187. [Google Scholar]
- 38.The World Bank Data and Statistics World Bank Country and Lending Groups. 2019. https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bankcountry-and-lending-groups Available from:
- 39.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36(3):1–48. [Google Scholar]
- 40.Li X., Dusseldorp E., Su X., Meulman J.J. Multiple moderator meta-analysis using the R-package Meta-CART. Behav. Res. Methods. 2020:1–17. doi: 10.3758/s13428-020-01360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta- analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.RStudio Team . RStudio, PBC; Boston, MA: 2020. RStudio: Integrated Development for R.http://www.rstudio.com/ URL. Available online. (accessed on 26 June 2020) [Google Scholar]
- 43.Boroduske A., Trofimova J., Kibilds J., Papule U., Sergejeva M., Rodze I., Grantina-Ievina L. Coxiella burnetii (Q fever) infection in dairy cattle and associated risk factors in Latvia. Epidemiol. Infect. 2017;145(10):2011–2019. doi: 10.1017/S0950268817000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo M.G., Ouh I.O., Kwak D. Herd prevalence and genotypes of Coxiella burnetii in dairy cattle bulk tank milk in Gyeongsang provinces of South Korea. Trop. Anim. Health Prod. 2018;50(6):1399–1404. doi: 10.1007/s11250-018-1564-0. [DOI] [PubMed] [Google Scholar]
- 45.Rahimi E., Doosti A., Ameri M., Kabiri E., Sharifian B. Detection of Coxiella burnetii by nested PCR in bulk milk samples from dairy bovine, ovine, and caprine herds in Iran. Zoonoses Publ. Hlth. 2010;57(7–8):e38–e41. doi: 10.1111/j.1863-2378.2009.01289.x. [DOI] [PubMed] [Google Scholar]
- 46.Van Engelen E., Schotten N., Schimmer B., Hautvast J.L., Van Schaik G., Van Duijnhoven Y.T. Prevalence and risk factors for Coxiella burnetii (Q fever) in Dutch dairy cattle herds based on bulk tank milk testing. Prev. Vet. Med. 2014;117(1):103–109. doi: 10.1016/j.prevetmed.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Anastácio S., Carolino N., Sidi-Boumedine K., Da Silva G.J. Q fever dairy herd status determination based on serological and molecular analysis of bulk tank milk. Transbound. Emerg. Dis. 2016;63(2):e293–e300. doi: 10.1111/tbed.12275. [DOI] [PubMed] [Google Scholar]
- 48.Velasova M., Damaso A., Prakashbabu B.C., Gibbons J., Wheelhouse N., Longbottom D., van Winden S., Green M., Guitian J. Herd-level prevalence of selected endemic infectious diseases of dairy cows in Great Britain. J. Dairy Sci. 2017;100(11):9215–9233. doi: 10.3168/jds.2016-11863. [DOI] [PubMed] [Google Scholar]
- 49.Czaplicki G., Houtain J.Y., Mullender C., Porter S.R., Humblet M.F., Manteca C., Saegerman C. Apparent prevalence of antibodies to Coxiella burnetii (Q fever) in bulk tank milk from dairy herds in southern Belgium. Vet. J. 2012;192(3):529–531. doi: 10.1016/j.tvjl.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 50.Magnino S., Vicari N., Boldini M., Rosignoli C., Nigrelli A., Andreoli G., Pajoro M., Fabbi M. Rilevamento di Coxiella burnetii nel latte di massa di alcune aziende bovine lombarde. Large Anim. Rev. 2009;15(1):3–6. [Google Scholar]
- 51.Valla G., Bizzarri D., Ferrari G., Bussacchini M. Prevalenza di Coxiella burnetii nel latte di massa in allevamenti di bovine da latte italiani e possibile correlazione con problemi riproduttivi. Large Anim. Rev. 2014;20:51–56. [Google Scholar]
- 52.Astobiza I., Ruiz-Fons F., Pinero A., Barandika J.F., Hurtado A., Garcia-Perez A.L. Estimation of Coxiella burnetii prevalence in dairy cattle in intensive systems by serological and molecular analyses of bulk-tank milk samples. J. Dairy Sci. 2012;95(4):1632–1638. doi: 10.3168/jds.2011-4721. [DOI] [PubMed] [Google Scholar]
- 53.Bauer A.E., Olivas S., Cooper M., Hornstra H., Keim P., Pearson T., Johnson A.J. Estimated herd prevalence and sequence types of Coxiella burnetii in bulk tank milk samples from commercial dairies in Indiana. BMC Vet. Res. 2015;11(1):186. doi: 10.1186/s12917-015-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gyuranecz M., Dénes B., Hornok S., Kovács P., Horváth G., Jurkovich V., Horváth G., Jurkovich V., Varga T., Hajtós I., Szabó R., Magyar T., Vass N., Hofmann-Lehmann R., Erdélyi K., Bhide M., Dán A. Prevalence of Coxiella burnetii in Hungary: screening of dairy cows, sheep, commercial milk samples, and ticks. Vector Borne Zoonot. 2012;12(8):650–653. doi: 10.1089/vbz.2011.0953. [DOI] [PubMed] [Google Scholar]
- 55.APHIS Veterinary Service Centers for Epidemiology and Animal Health. Prevalence of Coxiella burnetii in Bulk-tank Milk on U.S. Dairy Operations, 2007. 2011. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/monitoring-and-surveillance/nahms Available from:
- 56.Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., Chan F.K.L., Sung J.J.Y., Kaplan G.G., Ng S.C. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 57.Montazeri M., Mikaeili Galeh T., Moosazadeh M., Sarvi S., Dodangeh S., Javidnia J., Sharif M., Daryani A. The global serological prevalence of Toxoplasma gondii in felids during the last five decades (1967–2017): a systematic review and meta-analysis. Parasit. Vectors. 2020:1–10. doi: 10.1186/s13071-020-3954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sterne J.A.C., Sutton A.J., Ioannidis J.P.A., Terrin N., Jones D.R., Lau J., Carpenter J., Rücker G., Harbord R.M., Schmid C.H., Tetzlaff J., Deeks J.J., Peters J., Macaskill P., Schwarzer G., Duval S., Altman D.G., Moher D., Higgins J.P.T. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 59.Taurel A.F., Guatteo R., Lehebel A., Joly A., Beaudeau F. Vaccination using phase I vaccine is effective to control Coxiella burnetii shedding in infected dairy cattle herds. Comp. Immunol. Microb. 2014;37(1):1–9. doi: 10.1016/j.cimid.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Astobiza I., Barandika J.F., Juste R.A., Hurtado A., García-Pérez A.L. Evaluation of the efficacy of oxytetracycline treatment followed by vaccination against Q fever in a highly infected sheep flock. Vet. J. 2013;196(1):81–85. doi: 10.1016/j.tvjl.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 61.Boarbi S., Mori M., Rousset E., Sidi-Boumedine K., Van Esbroeck M., Fretin D. Prevalence and molecular typing of Coxiella burnetii in bulk tank milk in Belgian dairy goats, 2009–2013. Vet. Microbial. 2014;170:117–124. doi: 10.1016/j.vetmic.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 62.Kargar M., Rashidi A., Doosti A., Najafi A., Ghorbani-Dalini S. The sensitivity of the PCR method for detection of Coxiella burnetii in the milk samples. Zahedan J. Res. Med. Sci. 2015;17(6):29–32. [Google Scholar]
- 63.Kersh G.J., Wolfe T.M., Fitzpatrick K.A., Candee A.J., Oliver L.D., Patterson N.E., Self J.S., Priestley R.A., Loftis A.D., Massung R.F. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl. Environ. Microbiol. 2010;76(13):4469–4475. doi: 10.1128/AEM.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.García-Pérez A.L., Astobiza I., Barandika J.F., Atxaerandio R., Hurtado A., Juste R.A. Investigation of Coxiella burnetii occurrence in dairy sheep flocks by bulk-tank milk analysis and antibody level determination. J. Dairy Sci. 2009;92(4):1581–1584. doi: 10.3168/jds.2008-1672. [DOI] [PubMed] [Google Scholar]
- 65.Marenzoni M.L., Luca S.D., Stefanetti V., Castelli L., Bietta A., Coletti M., Passamonti F., Ranucci D., Proietti P.C. Investigation of Coxiella burnetii occurrence in dairy sheep flocks by molecular analysis in Umbrian area: a pilot study. Large Anim. Rev. 2013;19(5):225–229. [Google Scholar]
- 66.Schimmer B., Morroy G., Dijkstra F., Schneeberger P.M., Weers-Pothoff G., Timen A., Wijkmans C., van der Hoek W. Large ongoing Q fever outbreak in the south of the Netherlands, 2008. Eurosurveillance. 2008;13(31):18939. [PubMed] [Google Scholar]
- 67.Roest H.I., van Solt C.B., Tilburg J.J., Klaassen C.H., Hovius E.K., Roest F.T., Vellema P., van den Brom R., van Zijderveld F.G. Search for possible additional reservoirs for human Q fever, The Netherlands. Emerg. Infect. Dis. 2013;19(5):834. doi: 10.3201/eid1905.121489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zucali M., Tamburini A., Sandrucci A., Bava L. Global warming and mitigation potential of milk and meat production in Lombardy (Italy) J. Clean. Prod. 2017;153:474–482. [Google Scholar]
- 69.Lucchese L., Capello K., Barberio A., Zuliani F., Stegeman A., Ceglie L., Guerrini E., Marangon S., Natale A. IFAT and ELISA phase I/phase II as tools for the identification of Q fever chronic milk shedders in cattle. Vet. Microbiol. 2015;179(1–2):102–108. doi: 10.1016/j.vetmic.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 70.Schimmer B., Lenferink A., Schneeberger P., Aangenend H., Vellema P., Hautvast J., van Duynhoven Y. Seroprevalence and risk factors for Coxiella burnetii (Q fever) seropositivity in dairy goat farmers’ households in the Netherlands, 2009–2010. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0042364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCaughey C., Murray L.J., McKenna J.P., Menzies F.D., McCullough S.J., O’Neill H.J., Wyatt D.E., Cardwell C.R., Coyle P.V. Coxiella burnetii (Q fever) seroprevalence in cattle. Epidemiol. Infect. 2010;138(1):21–27. doi: 10.1017/S0950268809002854. [DOI] [PubMed] [Google Scholar]
- 72.Agger J.F., Paul S. Increasing prevalence of Coxiella burnetii seropositive Danish dairy cattle herds. Acta Vet. Scand. 2014;56(1):46. doi: 10.1186/s13028-014-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suman P., Uddin A.S.S., Md-Tariqul I., Nayan B., Jens A. Coxiella burnetii infection (Q fever) in humans and animals: a general overview. Ann. Vet. Anim. Sci. 2016;3(1):1–19. [Google Scholar]
- 74.Amin W.F., Ahmed S.O. Detection of Coxiella burnetii in bovine milk samples using polymerase chain reaction. Assiut. Vet. Med. J. 2009;55(123):23–31. [Google Scholar]
- 75.Rahman M., Alam M., Islam M., Bhuiyan A.K., Rahman A.K.M. Serological and molecular evidence of Q fever in domestic ruminants in Bangladesh. Vet. Med. Int. 2016:1–8. doi: 10.1155/2016/9098416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanderburg S., Rubach M.P., Halliday J.E., Cleaveland S., Reddy E.A., Crump J.A. Epidemiology of Coxiella burnetii infection in Africa: a One Health systematic review. PLOS NTDs. 2014;8(4):e2787. doi: 10.1371/journal.pntd.0002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoover T.A., Vodkin M.H., Williams J.C. A Coxiella burnetii repeated DNA element resembling a bacterial insertion sequence. J. Bacteriol. 1992;174(17):5540–5548. doi: 10.1128/jb.174.17.5540-5548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lipsey M.W. Those confounded moderators in meta-analysis: good, bad, and ugly. Ann. Am. Acad. Pol. Soc. Sci. 2003;587(1):69–81. [Google Scholar]