Abstract
Understanding and predicting the changes of protein structure and function upon mutation and their relationship to human health is a critical element to translate the genomic revolution into actionable interventions. Therefore, it is pertinent to explore how mutations result in structural changes leading to pathogenic proteins, but due to the protein structural knowledge gap, experimental approaches are lacking. Protein structure prediction methods, such as I-TASSER, have made it possible to predict the structure of a given amino acid sequence, thus opening a new way to explore protein structure changes upon mutations when experimental information is not available. Using known mutations from the Catalogue of Somatic Mutation in Cancer (COSMIC) and ClinVar databases, we compare predicted structure-derived properties from wild type (WT) and mutated proteins and find differences between the local and global 3D protein structures of the WT and the mutants. The studies in this relatively small sample reveal that the structural changes are quite diverse.
Keywords: Oncogenic missense variants, Protein structure prediction, Structural classification
Oncogenic Missense Variants; Protein Structure Prediction, Structural Classification
1. Introduction
Understanding and predicting the changes of protein structure and function upon mutation and their relationship to human health is a critical task to translate the genomic revolution into actionable interventions [1]. However, since the effects of mutations on structural properties of proteins are multifactorial, it is unclear which features of proteins structural changes are important in leading to pathogenicity [2]. Moreover, because there is very limited experimental structural data for proteins in general, and much less for the mutated species [3, 4], very little is known about how protein structures and their properties change upon mutation [5, 6]. It is also poorly understood if these changes can predict pathogenicity or provide ways to develop mechanistic hypotheses of pathogenicity. Using computational methods to study this problem appears to be a viable manner to explore this. This approach is also strongly supported by recent advances in computational resources and methods to predict 3D protein structures [7]. While a great deal of progress has been made in predicting variant pathogenicity [1, 8, 9, 10], the most accurate methods do not provide insights on the structural changes leading to the alteration of the protein functionality. We have demonstrated that careful comparison between wild type (WT) and mutated structures can provide insights on pathogenicity [11, 12], but general principles remain elusive. In this paper, we explore the structural changes induced by 26 known oncogenic missense mutations from the Catalogue of Somatic Mutation in Cancer (COSMIC) [13] and ClinVar [14] databases. We explored the questions proposed above by comparing the predicted 3D structures of WT and mutated proteins. We visualized the changes in the structures upon mutation, analyzing both overall and local structural changes due to the mutations. We also extracted structural features from both the 3D predicted structures and sequences for the wild types (WT) and mutants. Lastly, we analyzed them for statistical significance, and by using unsupervised machine learning, we attempted to identify common structural changes leading to oncogenesis.
2. Methods
A set of oncogenic substitution missense mutations were selected based on their classification found in COSMIC [13] and ClinVar [14] databases. While the selection of these proteins is somehow arbitrary, the selected oncogenic proteins represent a wide range of size and function while keeping its number reasonablly low to allow computational times within achievable limits. The complete description of the variants and their WTs is shown in Table 1. FASTA files for each of the WT sequences were retrieved from UniProt [15]; the mutated sequences were obtained manually by modifying the WTs according to the missense mutation annotations from the COSMIC [13] and ClinVar [14] databases. To consider ensemble effects [16], five structures corresponding to the centroids of structure clusters computed by I-TASSER [17, 18, 19] were produced per run, and two runs were performed per protein, which were retained for analysis. This approach was used because previous work [14] showed that by performing more than one I-TASSER prediction, it is possible to explore a larger conformational space, which could be more representative of the actual structures in living organisms and provide a larger data set for the clustering analysis attempted in this paper. In the interest of having structural variability in the dataset used for the clustering analysis and to avoid I-TASSER's tendency to produce consistently similar structures despite mutation, the structure predictions were performed using both default settings for one set and duplicates generated using homolog exclusion (>30% similarity). All the structure predictions were performed using the University of Utah CHPC (https://www.chpc.utah.edu/) clusters using nodes with 12 cores with 2.8 GHz Intel Xeon (Westmere X5660) processors, 24 GB memory, and Mellanox QDR Infiniband interconnect on the Ember cluster. The visual analysis and manipulation of the structures was done using 3D structures using Chimera, [20]. The features used in the classification work (see Table 2) were extracted from the predicted 3D structures using DSSP [21], I-TASSER [17, 18, 19], TM-align [22], FoldX [23], RW and RWPlus [24, 25, 26], DFIRE and dDFIRE [25], for calculating solvent accessible surface area (SASA), C scores, TM (template modeling) scores and RMSD (between reference wild types and mutant), total Gibbs free energy, and Pair-wise distance-dependent energy, side-chain orientation-dependent energy, distance-related energy, and angle-related energy, respectively. RW, RWPlus, DFIRE, and dDFIRE calculated for both wild type and mutant predicted structures were previously used in the STRUM stability change predictor by Quan et al. as structure-derived features [27]. Statistical testing (Mann-Whitney-Wilcoxon) was performed on the dataset [28], which was separated into wild type and mutant features that represented each protein structure, and p-value correction (Benjamini-Hochberg) was applied [29]. A clustering analysis (k-means clustering) was subsequently performed to evaluate the structure of the dataset using Rstudio [30].
Table 1.
Set of the 17 proteins that were investigated using 26 pathogenic substitution missense mutations selected from COSMIC [13] and ClinVar [14] databases.
| Protein | UniProt ID | PDB ID | Clinvar or COSMIC ID | AA Change | Associated Cancer |
|---|---|---|---|---|---|
| CADH1 | P12830 | 2O72 | |||
| CADH1, Mutant 1 | COSM19822 | Ala634Val | Breast Carcinoma | ||
| CADH1, Mutant 2 | NM_004360.4(CDH1):c.1008G > T (p.Glu336Asp) | Glu336Asp | Hereditary Diffuse Gastric Cancer | ||
| CBL | P22681 | 2Y1M | |||
| CBL, Mutant 1 | NM_005188.3(CBL):c.1186T > C (p.Cys396Arg) | Cys396Arg | Noonan syndrome-like disorder with juvenile myelomonocytic leukemia, Rasopathy | ||
| CBL, Mutant 2 | COSM4385831 | Gln367Pro | Genital germ cell tumor | ||
| CBL, Mutant 3 | COSM34052 | Tyr371His | Acute myeloid leukemia, juvenile myelomonocytic leukemia, chronic myelomonocytic leukemia | ||
| RET | P07949 | 4CKJ | |||
| RET, Mutant 1 | NM_020975.4(RET):c.1465G > A (p.Asp489Asn) | Asp489Asn | Multiple endocrine neoplasia, type 2, not specified, Hereditary cancer-predisposing syndrome, Hirschsprung disease | ||
| RET, Mutant 2 | COSM967 | Cys609Tyr | Pheochromocytoma | ||
| RET, Mutant 3 | COSM87267 | Cys618Ser | Carcinoma of the thyroid | ||
| RET, Mutant 4 | COSM966 | Cys634Arg | Pheochromocytoma, Carcinoma of the thyroid | ||
| RET, Mutant 5 | NM_020975.4(RET):c.1336G > C (p.Gly446Arg) | Gly446Arg | Multiple endocrine neoplasia, type 2 | ||
| RET, Mutant 6 | COSM1666596 | Gly691Ser | Acute myeloid leukemia | ||
| VHL | P40337 | 4WQO | |||
| VHL, Mutant 1 | COSM14400 | Ser65Leu | Clear cell renal cell carcinoma | ||
| VHL, Mutant 2 | NM_000551.3(VHL):c.292T > C (p.Tyr98His) | Tyr98His | Von Hippel Lindau syndrome | ||
| TP53 | P40337 | 3Q01 | |||
| TP53, Mutant 1 | COSM11066 | His193Leu | Squamous cell carcinoma of the esophagus | ||
| NF2 | P35240 | 1H4R | |||
| NF2, Mutant 1 | COSM23876 | Glu463Lys | Neurofibromatosis Type 2 | ||
| RB1 | P06400 | 4ELJ | |||
| RB1, Mutant 1 | COSM1636647 | Ser634Pro | Retinoblastoma | ||
| CALR | P27797 | 3POW | |||
| CALR, Mutant 1 | COSM1290873 | Phe46Tyr | Lymphoid Neoplasms | ||
| FANCF | Q9NPI8 | 2IQC | |||
| FANCF, Mutant 1 | COSM4521389 | Gly370Asp | Squamous cell carcinoma of head and neck | ||
| MUTYH | Q9UIF7 | 3N5N | |||
| MUTYH, Mutant 1 | COSM1645292 | Arg200Cys | Carcinoma of Colon | ||
| HRAS | P01112 | 3K8Y | |||
| HRAS, Mutant 1 | COSM490 | Gly13Asp | Chronic Myelogenous Leukemia | ||
| PPP2R1A | P30153 | 1B3U | |||
| PPP2R1A, Mutant 1 | COSM51253 | Arg183Gln | Endometrial Carcinoma | ||
| NT5C2 | P49902 | 2J2C | |||
| NT5C2, Mutant 1 | COSM4011341 | Arg413His | Gastric Adenocarcinoma | ||
| FH | P07954 | 3E04 | |||
| FH, Mutant 1 | COSM906408 | Asp179Asn | Colonic Adenocarcinoma | ||
| MAP2K1 | Q02750 | 3EQI | |||
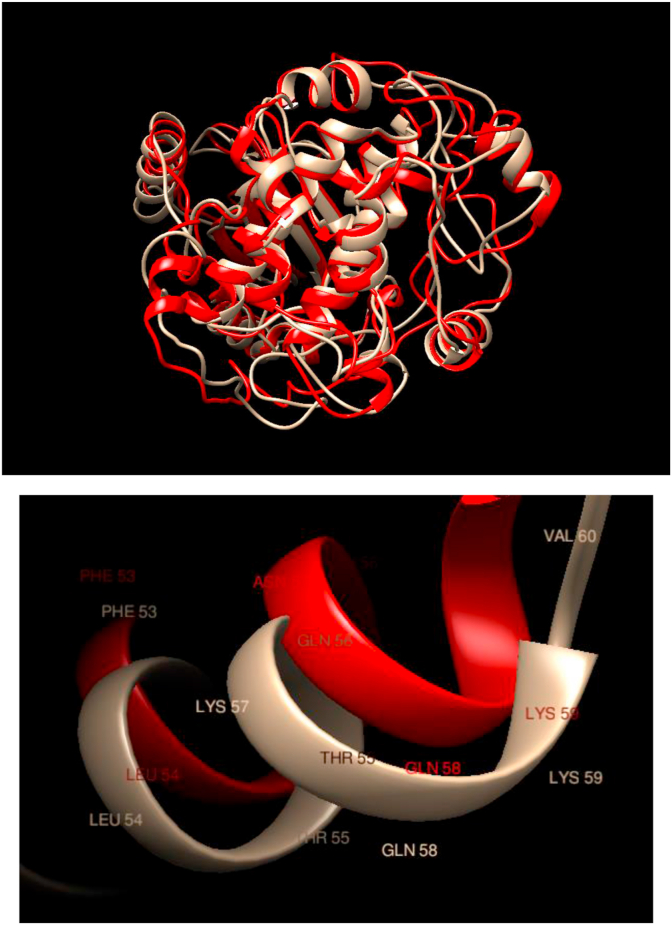
| MAP2K1, Mutant 1 | COSM1235478 | Lys57Asn | Pulmonary Adenocarcinoma | ||
| PIK3CA | P42336 | 4TV3 | |||
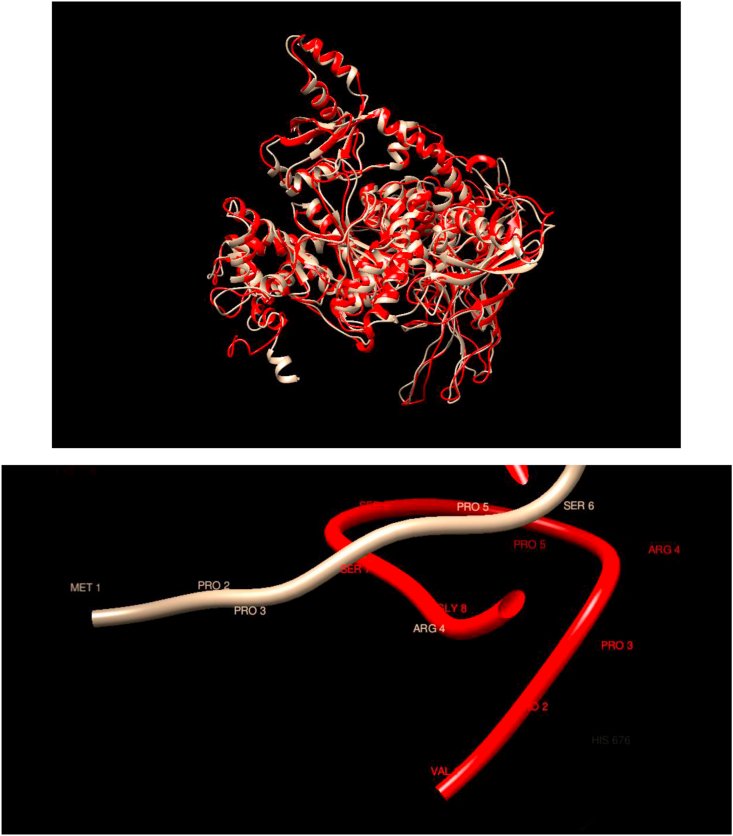
| PIK3CA, Mutant 1 | COSM1041443 | Met1Val | Glioma | ||
| MAP2K4 | P45985 | 3ALN | |||
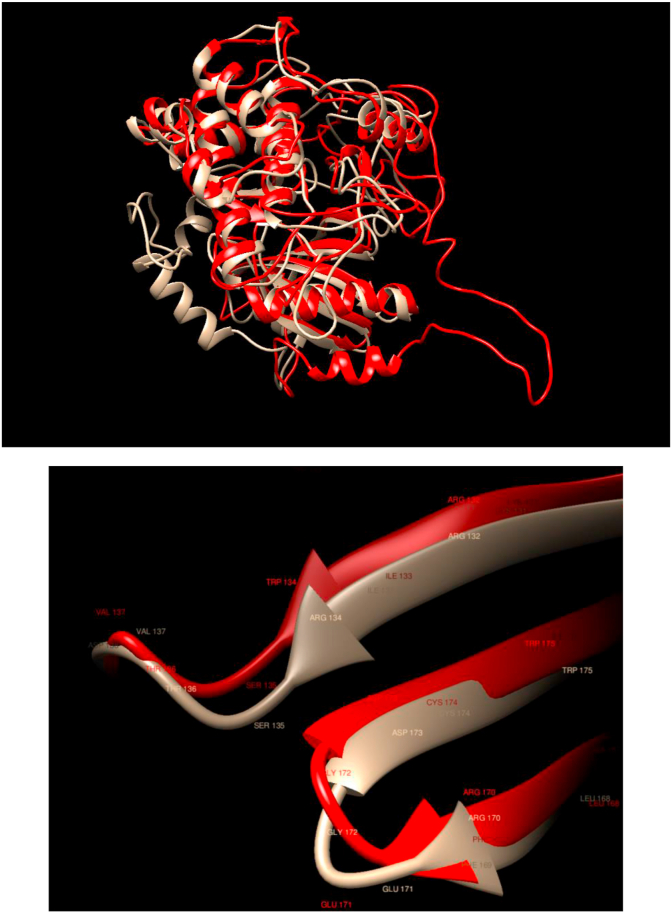
| MAP2K4, Mutant 1 | COSM137092 | Arg134Trp | Colonic Adenocarcinoma |
Table 2.
Structural features used for statistical and cluster analysis.
| Feature | Description | Software used |
|---|---|---|
| RMSD | Root mean square deviation of atomic position between reference and predicted structures for WT and mutated structures respectively. | UCSF Chimera [20] |
| Cscore | Confidence score from structure prediction | I-TASSER [17, 18, 19] |
| TMscore | Template modeling score | TMAlign [22] |
| SASA | Solvent accessible surface area of protein structure | DSSP [21] |
| Energy (kcal/mol) | Total stability energy | FoldX [23] |
| calRW (kcal/mol) | Pair-wise distance-dependent energy | calRW [26] |
| calRWPlus (kcal/mol) | Side-chain orientation-dependent energy | calRWPlus [26] |
| DFIRE2 | Distance-related energy | DFIRE2 [25] |
| dDFIRE | Angle-related energy | dDFIRE [25] |
3. Results and discussion
Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17 depict the comparison of the overall and local 3D predicted structures for a representative I-TASSER models for the WT and the different mutations from Table 1. While in some cases experimental structures are available for only short portions of the sequence, we are reasonably confident in the structure predictions and results for this study for two reasons: our structure prediction method, i.e, I-TASSER, and using 10 predicted structures per entry. Since I-TASSER uses template-based threading as part of its structure prediction, even if the proteins have not been fully experimentally resolved, we can rely on homologous structures to make reasonable predictions for the missing fragments. Furthermore, if no templates can be found, these are assembled using ab initio folding. Since we performed 10 structure predictions for each entry, we can average out biased predictions in order to make more reasonable comparisons across the features we studied and conformations we visualized. Taken together, we are reasonably confident that the structural analysis has been performed correctly.
Figure 1.
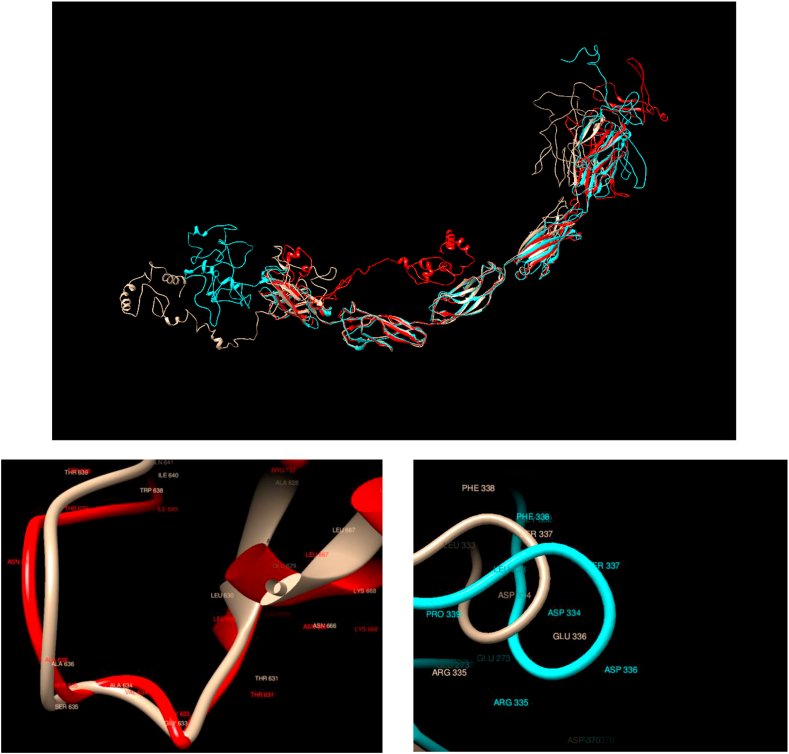
Comparison of WT and Mutated Structures for CADH1: WT: Gold; Mutant 1: Red (Ala634Val); Mutant 2: Cyan (Glu336Asp). CADH1 encodes a classical cadherin of the cadherin superfamily. Loss of function of this gene is thought to contribute to cancer progression by increasing proliferation, invasion, and/or metastasis. It is observed that both mutations lead to noticeable structural rearrangements for both flexible ends of the protein. On the other hand, the local structure in the proximity of the mutations does not change in any noticeable way.
Figure 2.
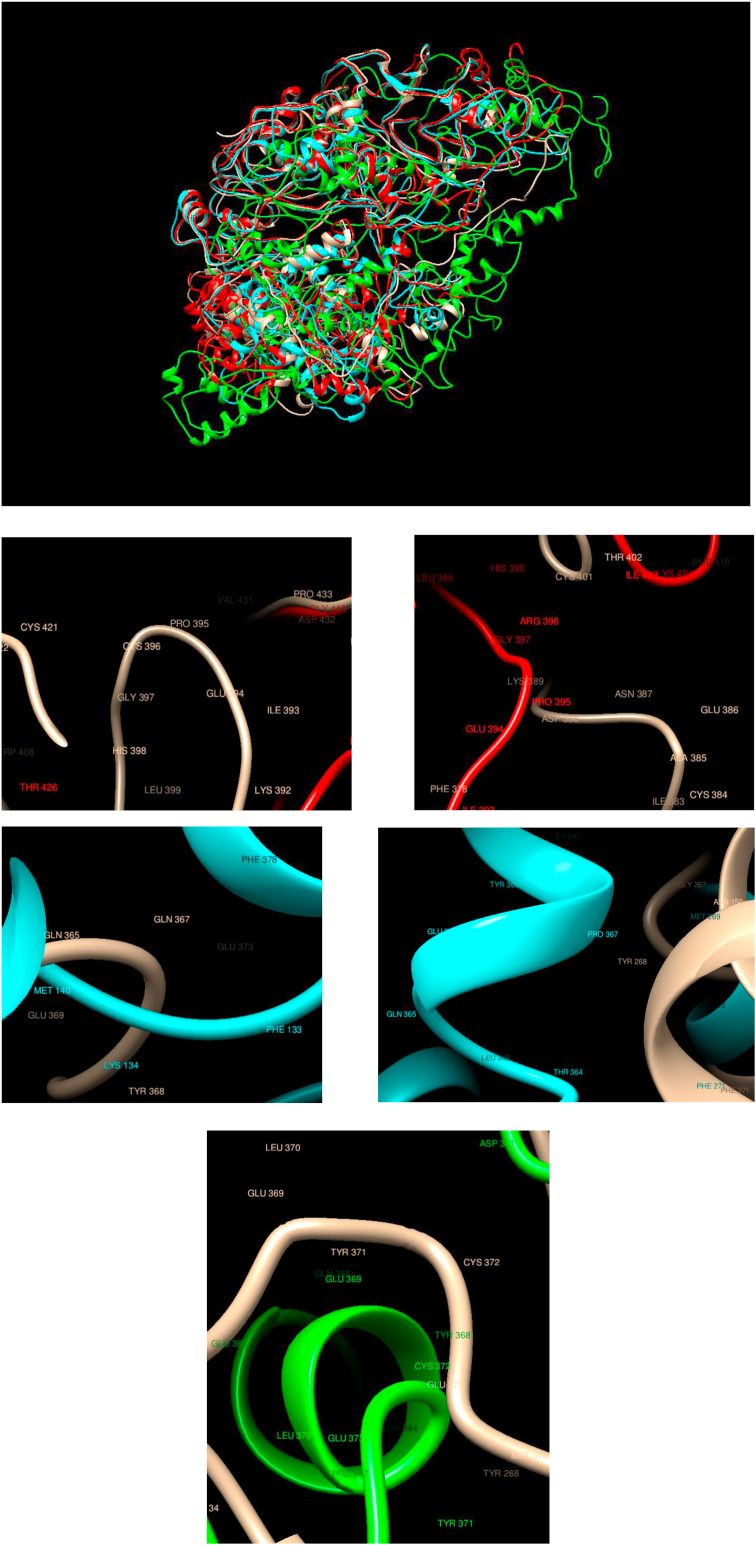
Comparison of WT and Mutated Structures for CBL: WT: Gold; Mutant 1: Red (Cys396Arg); Mutant 2: Cyan (Gln367Pro); Mutant 3: Green (Tyr371His). This gene is a proto-oncogene that encodes a RING finger E3 ubiquitin ligase. This protein mediates the transfer of ubiquitin from ubiquitin-conjugating enzymes (E2) to specific substrates. The comparison of the mutated structures shows noticeable changes, in particular for the Tyr371His variant. Also, noticeable changes can be seen for all the mutations considered here, in many cases, even showing a different secondary structure.
Figure 3.
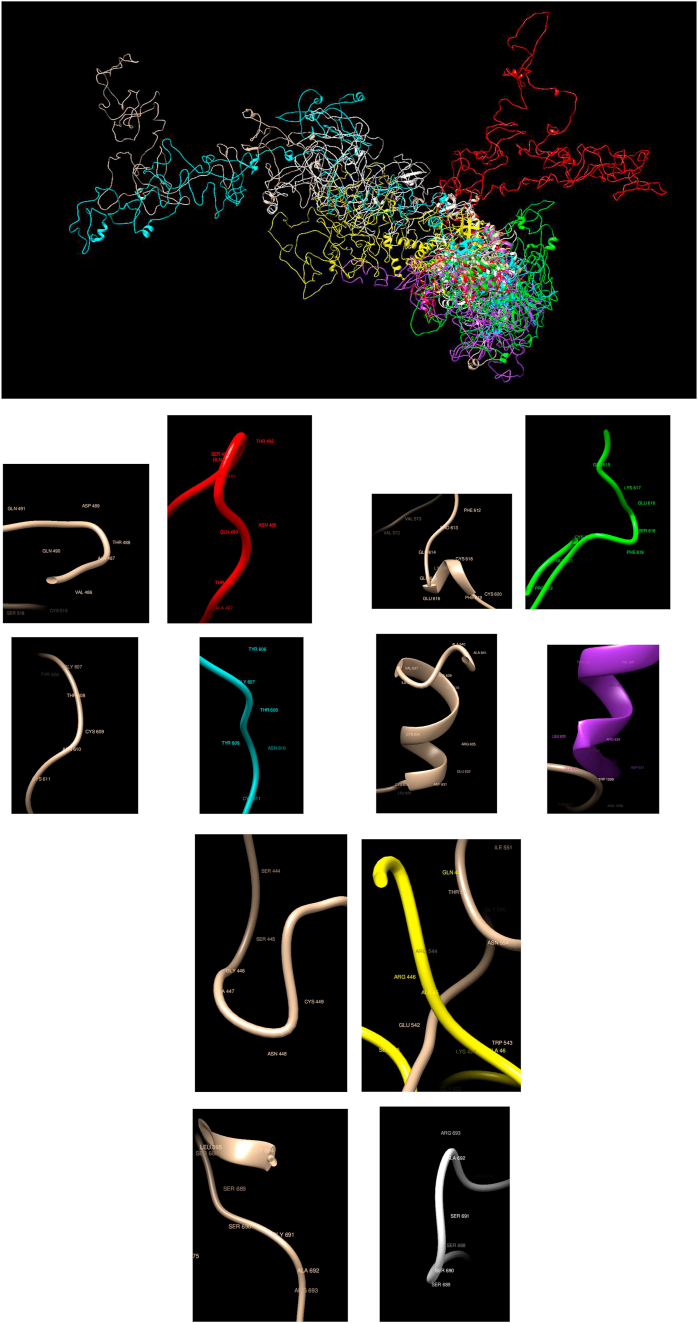
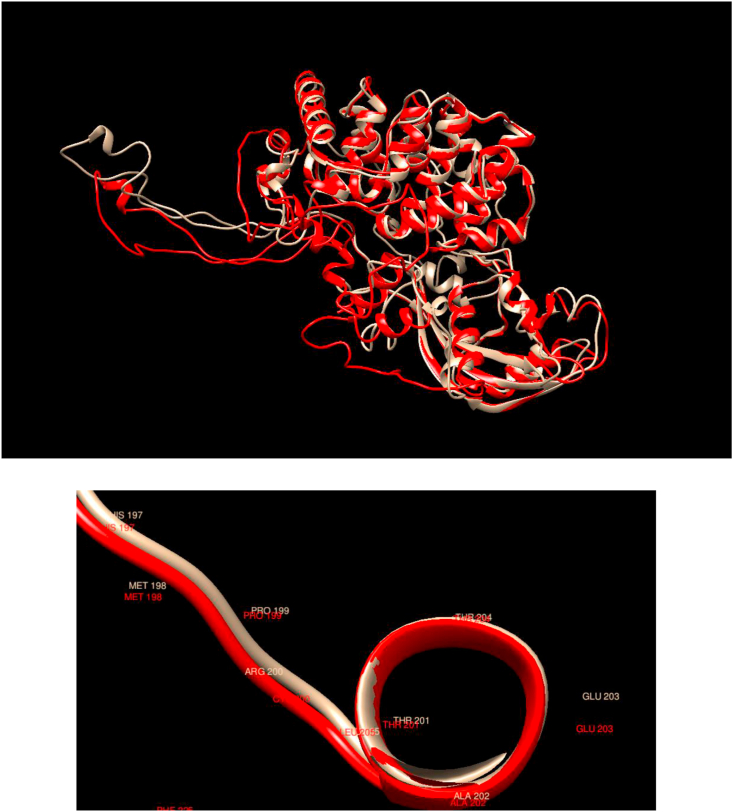
Comparison of WT and Mutated Structures for RET: WT: Gold; Mutant 1: Red (Asp489Asn); Mutant 2: Cyan (Cys609Tyr); Mutant 3: Green (Cys618Ser); Mutant 4: Purple (Cys634Arg); Mutant 5: Yellow (Gly446Arg); Mutant 6: White (Gly691Ser). This gene encodes a transmembrane receptor and member of the tyrosine-protein kinase family of proteins. Binding of ligands such as GDNF (glial cell-line derived neurotrophic factor) and other related proteins to the encoded receptor stimulates receptor dimerization and activation of downstream signaling pathways that play a role in cell differentiation, growth, migration, and survival. It is observed here that the mutated structures are quite different than the WT; in particular, the structures associated with mutations Cys618Ser and Cys634Arg are much more compact than the WT and other mutated ones. The Asp489Asn mutation appears in a different region but retains the same secondary structure, as with Cys609Tyr, Cys634Arg, Gly446Arg, and Gly691Ser, but Cys618Ser changes the local secondary structure.
Figure 4.
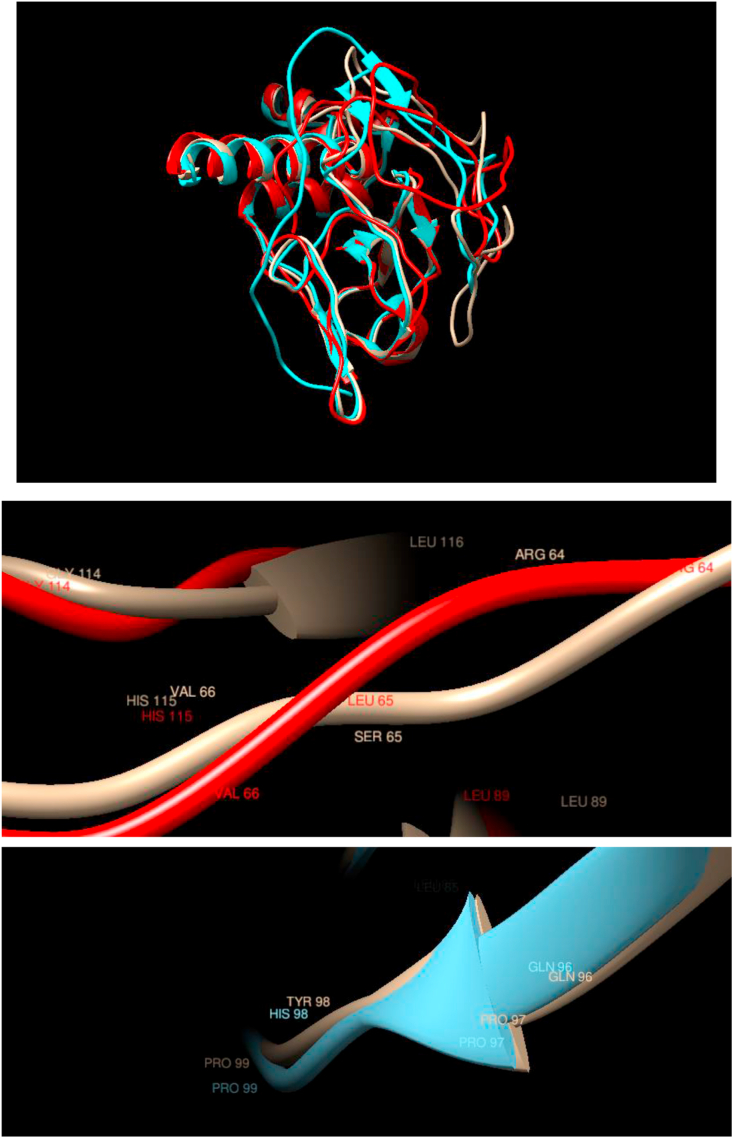
Comparison of WT and Mutated Structures for VHL: WT: Gold; Mutant 1: Red (Ser65Leu); Mutant 2: Cyan (Tyr98His). Mutations in this protein are associated with Von Hippel-Lindau syndrome (VHL), which is a dominantly inherited familial cancer syndrome predisposing to a variety of malignant and benign tumors. The mutation considered here does not change the overall structure of the protein in a noticeable way or the local structure in the neighborhood of the Ser65Leu or Tyr98His mutations, which show almost perfect overlap.
Figure 5.
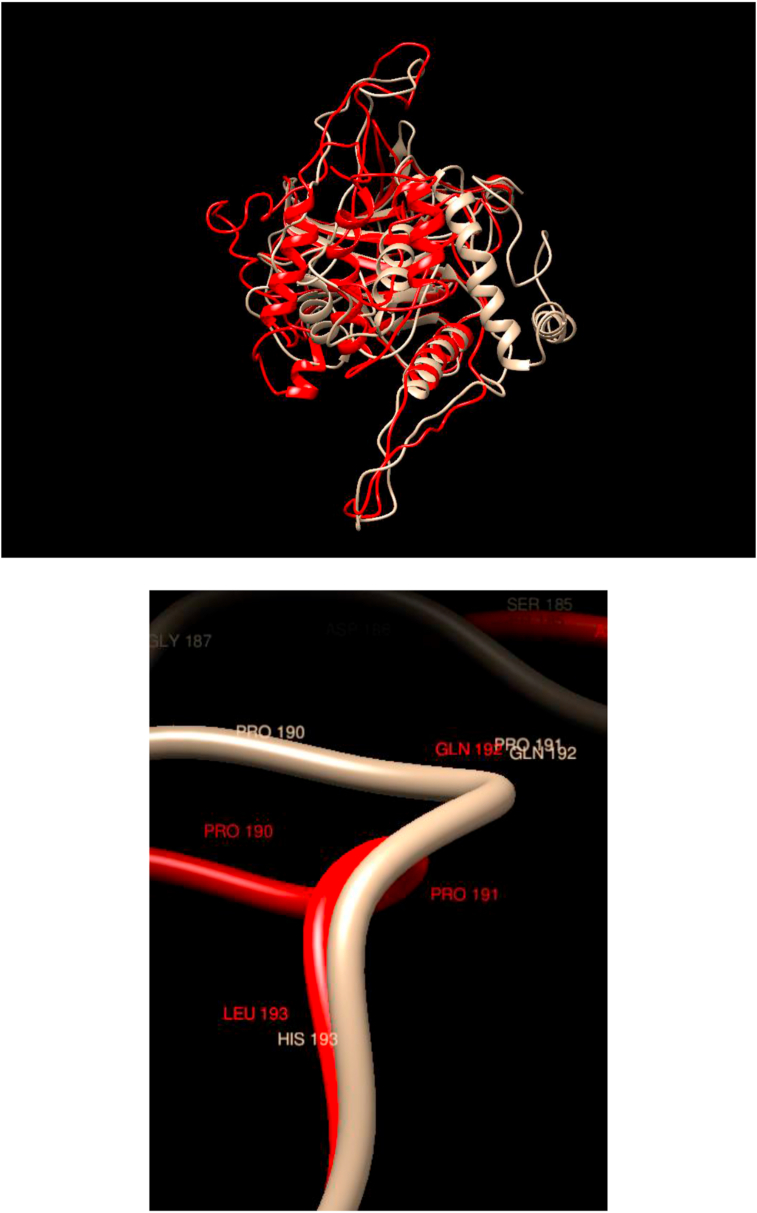
Comparison of WT and Mutated Structures for TP53: WT: Gold; Mutant 1: Red (His193Leu). This gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization. It is apparent that this mutation does not change in any noticeable way either the overall (minor the rearrangement of exposed loops) or the local structure in the neighborhood of the mutations.
Figure 6.
Comparison of WT and Mutated Structures for NF2: WT: Gold; Mutant 1: Red Glu463Lys). This gene encodes a protein that is similar to some members of the ERM (ezrin, radixin, moesin) family of proteins that are thought to link cytoskeletal components with proteins in the cell membrane. This mutation does not change in any noticeable way, either the overall or the local structure in the neighborhood of the mutations as both the overall and local structure of the WT and mutated proteins overlap almost perfectly.
Figure 7.
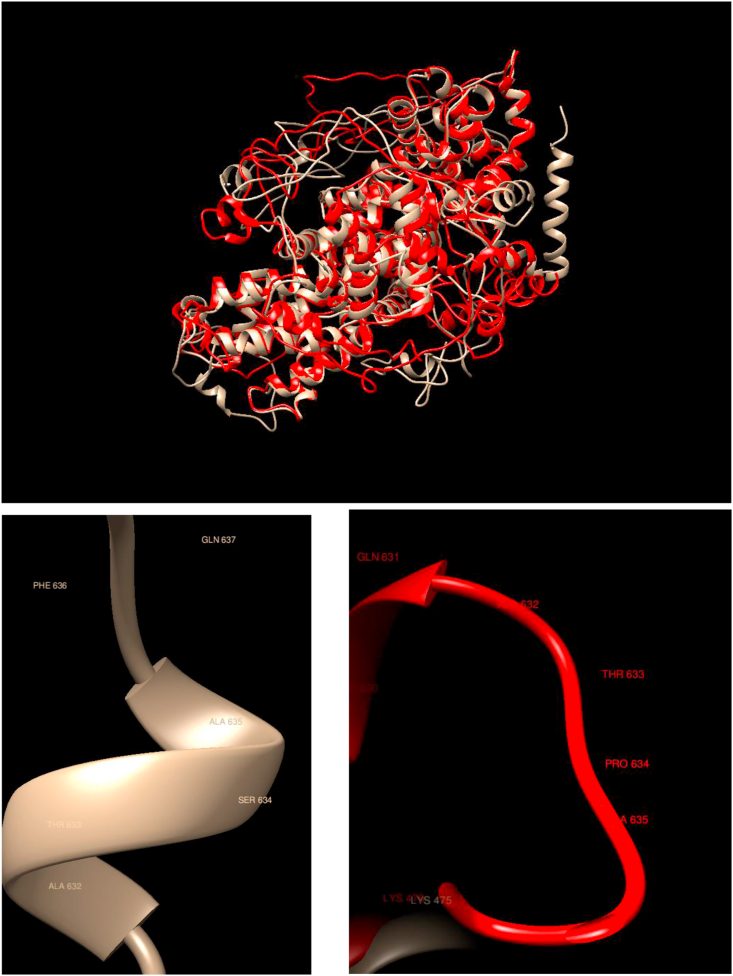
Comparison of WT and Mutated Structures for RB1: WT: Gold; Mutant 1: Red (Ser634Pro). This protein is associated with the gene of the same name that was the first tumor suppressor gene found. The encoded protein also stabilizes constitutive heterochromatin to maintain the overall chromatin structure. The changes in the overall structure upon mutation are relatively modest, but there are some considerable changes observed at the terminal helix, which is not exposed in the mutated protein. The local structure at the mutation point is substantial, as it shows a different secondary structure.
Figure 8.
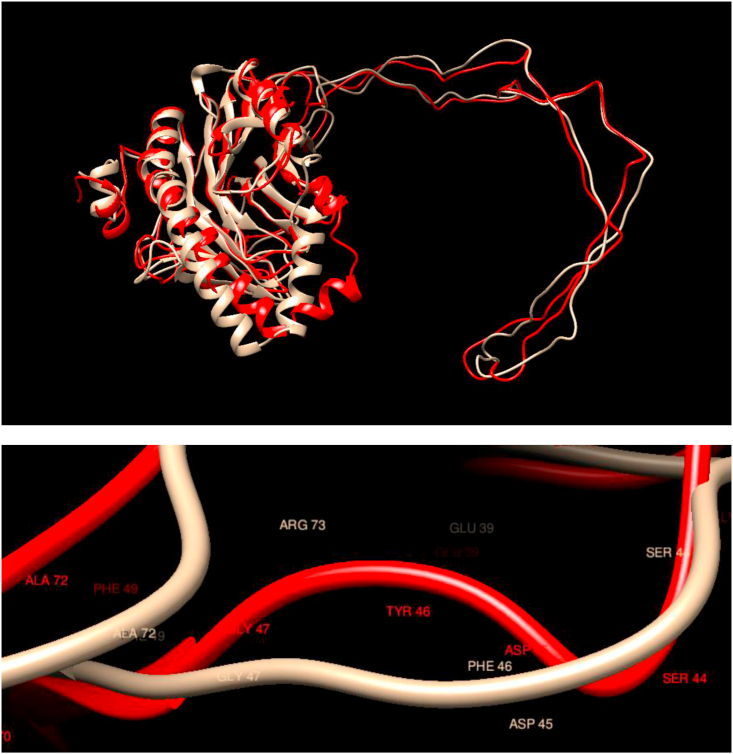
Comparison of WT and Mutated Structures for CALR: WT: Gold; Mutant 1: Red (Phe46Tyr). Calreticulin is a multifunctional protein that acts as a major Ca(2+)-binding (storage) protein in the lumen of the endoplasmic reticulum. The amino terminus of calreticulin interacts with the DNA-binding domain of the glucocorticoid receptor and prevents the receptor from binding to its specific glucocorticoid response element. From the comparison of the structures, it is apparent that neither the overall structure nor the local structure in the neighborhood of the mutations are affected in this case.
Figure 9.
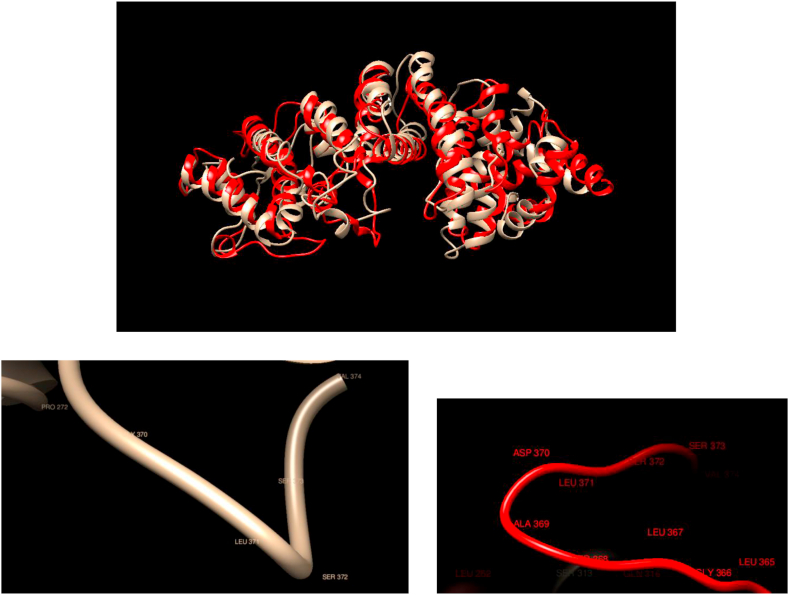
Comparison of WT and Mutated Structures for FANCF: WT: Gold; Mutant 1: Red (Gly370Asp). This protein is part of the Fanconi anemia complementation group (FANC). The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group F. The mutations do not affect the overall structure of the protein in a substantial way, except for some small changes in the exposed regions. While the structure of the mutant predicts the mutant residue in a different place, it does not appear that there is any change in secondary structure or accessibility.
Figure 10.
Comparison of WT and Mutated Structures for MUTYH: WT: Gold; Mutant 1: Red (Arg200Cys). This gene encodes a DNA glycosylase involved in oxidative DNA damage repair. It is observed a very good overlap between the WT and mutated structures, except for small changes in the exposed regions. The overlap in the neighborhood of the mutation is almost perfect, showing no local differences due to amino acid substitution.
Figure 11.
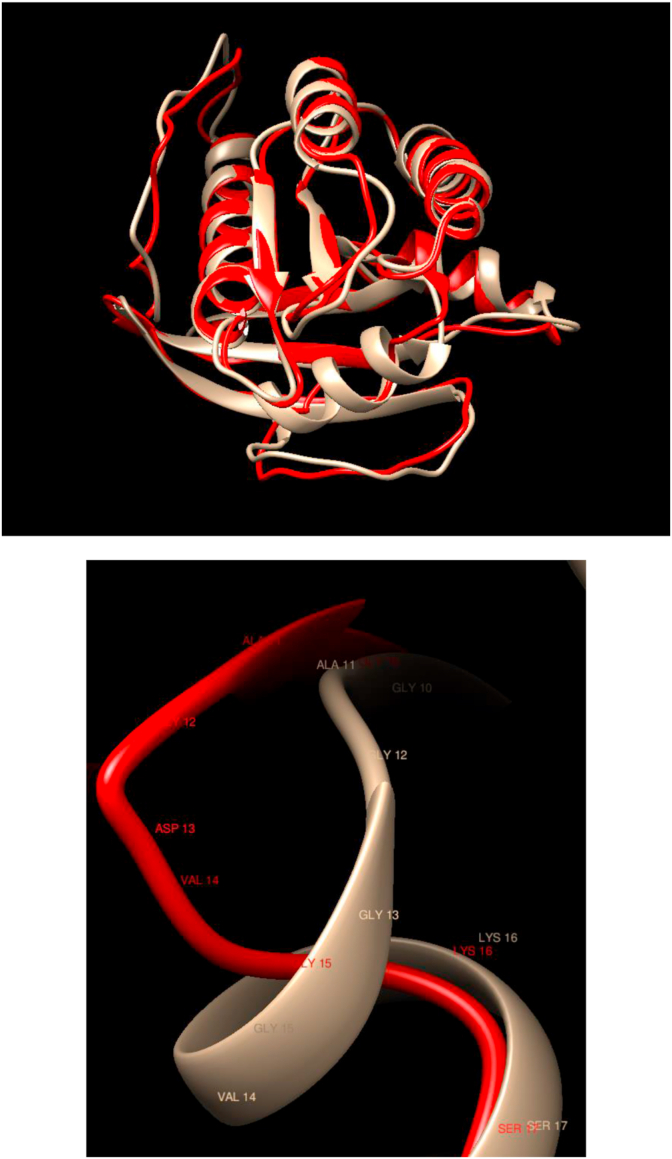
Comparison of WT and Mutated Structures for HRAS: WT: Gold; Mutant 1: Red (Gly13Asp). This gene belongs to the Ras oncogene family, whose members are related to the transforming genes of mammalian sarcoma retroviruses. The products encoded by these genes function in signal transduction pathways. There is almost a perfect overlap between the WT and mutated structures. Still, there is a noticeable change in the local structure in which the secondary structure at position 13 changes dramatically.
Figure 12.
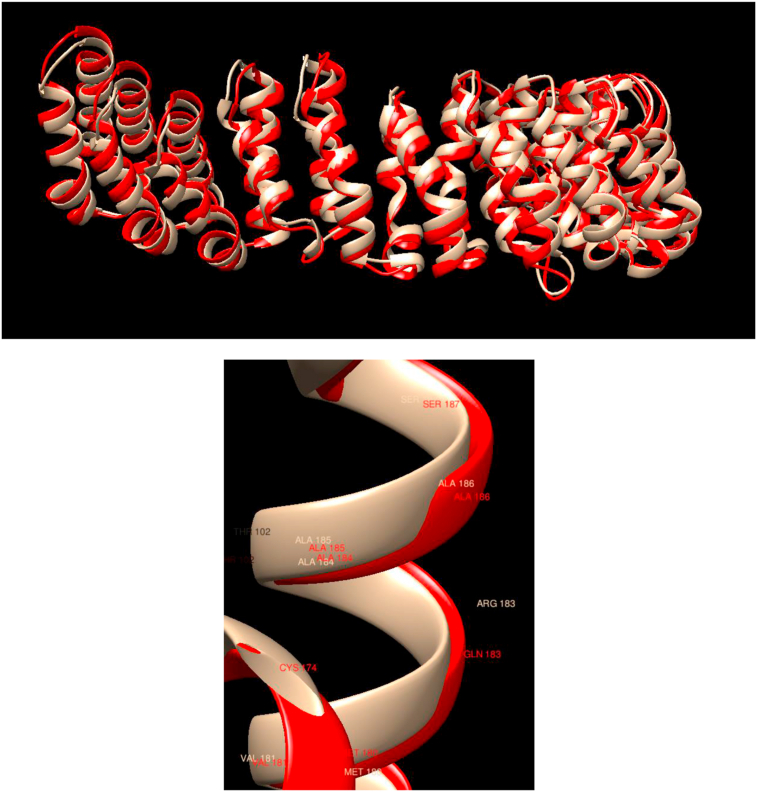
Comparison of WT and Mutated Structures for PPP2R1A: WT: Gold; Mutant 1: Red (Arg183Gln). This gene encodes a constant regulatory subunit of protein phosphatase 2. Protein phosphatase 2 is one of the four major Ser/Thr phosphatases, and it is implicated in the negative control of cell growth and division. In this case, it is observed that the mutation does not affect either the overall structure or the local one, with both showing almost perfect overlap.
Figure 13.
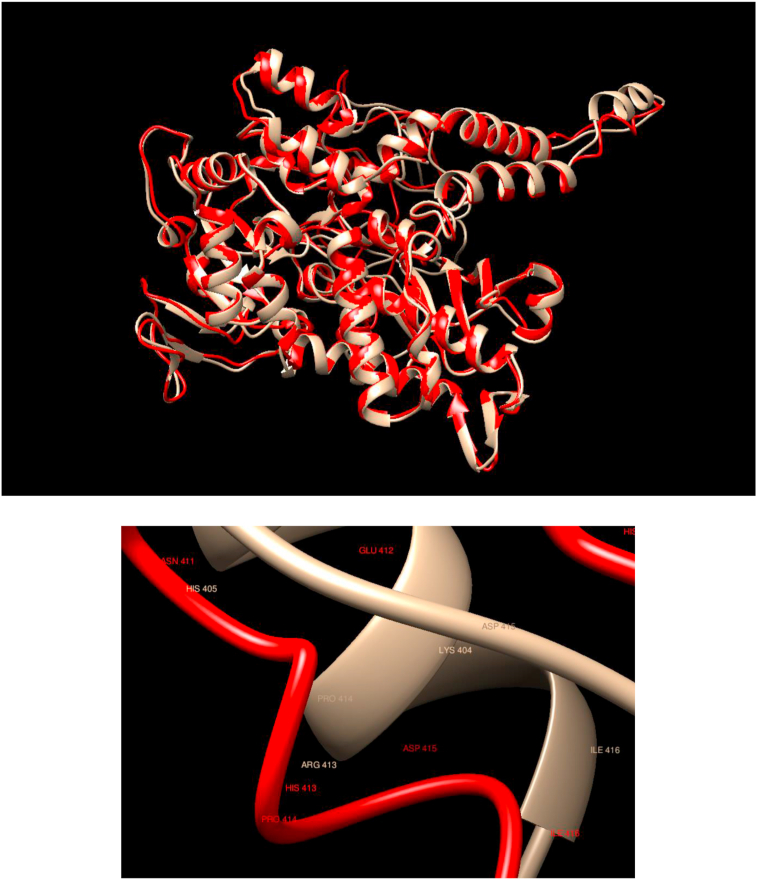
Comparison of WT and Mutated Structures for NT5C2: WT: Gold; Mutant 1: Red (Arg413His). This gene encodes a hydrolase that serves as an important role in cellular purine metabolism by acting primarily on inosine 5′-monophosphate and other purine nucleotides. In this case, there is an excellent overlap between the WT and mutated structures, but the secondary structure at the position of the substitution changes dramatically.
Figure 14.
Comparison of WT and Mutated Structures for FH: WT: Gold; Mutant 1: Red (Asp179Asn). The protein encoded by this gene is an enzymatic component of the tricarboxylic acid (TCA) cycle, or the Krebs cycle, and catalyzes the formation of L-malate from fumarate. For this mutation, it is observed that both the overall and local structures for the WT and mutated proteins show almost perfect overlap.
Figure 15.
Comparison of WT and Mutated Structures for MAP2K1: WT: Gold; Mutant 1: Red (Lys57Asn). The protein encoded by this gene is a member of the dual-specificity protein kinase family, which acts as a mitogen-activated protein (MAP) kinase. MAP kinases, also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals. In this case, it is observed that there is a substantial overlap of the overall structure as well as the local one in the neighborhood of the mutation.
Figure 16.
Comparison of WT and Mutated Structures for PIK3CA: WT: Gold; Mutant 1: Red (Met1Val). This protein encoded by this gene represents the catalytic subunit, which uses ATP to phosphorylate PtdIns, PtdIns4P, and PtdIns(4,5)P2. The overall structures of the WT and mutated proteins show substantial overlap, except for the terminal coil of the WT. Still, the local region of the mutation does not show any noticeable changes.
Figure 17.
Comparison of WT and Mutated Structures for MAP2K4: WT: Gold; Mutant 1: Red (Arg134Trp). This gene encodes a member of the mitogen-activated protein kinase (MAPK) family. Members of this family act as an integration point for multiple biochemical signals and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development. In this case, we observe that the mutation produces quite noticeable changes in the overall structure. Still, there is almost a perfect overlap between the WT and mutated structures in the region of the mutation.
In all cases, the figures depict the structures showing the maximum overlap among them. The discussion of the most remarkable features observed is provided for convenience in the Figure captions. All the gene descriptions were taken from GeneCards [31]. It is important to realize that the observations reported here are quite generic and do not attempt to provide detailed insight on the structural changes observed for each protein, but to extract overall observations leading towards identifying key (if any) structural factors that may determine pathogenicity. All structures depicted in the figures were obtained using I-TASSER and they are available at: http://home.chpc.utah.edu/∼u0033399/Protein%20Structural%20Changes%20for%20Oncogenic%20Missense%20Variants/.
From the discussions in Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17, it is difficult to extract any significant trends that associate changes in the 3D structures with pathogenicity. We observe cases in which there are no changes in either the overall or the local structures (TP53, CALR, FANCF, MUTYH, PPP2R1A, FH, MAP2K1, MAP2K4), cases in which there are noticeable changes in the overall structure, but not in the local one (CADH1, PIK3CA), cases in which these are no noticeable changes in the overall structure but relevant ones in the local one (RB1, HRAS, NT5C2) and finally examples in which both the local and overall structures are changed by the mutation (CBL, RET). Even for the case in which no noticeable changes have been noticed in this analysis, we can't conclude that these minor changes in geometry and/or electrostatics may not play a critical role in defining the pathogenicity of the variant [11, 12].
Table 3 presents the results of comparing the distributions of the structural features in Table 2 for the WT and mutated 3D predicted structures. The Mann-Whitney-Wilcoxon [28] test examines whether two samples are likely to derive from the same population taking as the null hypothesis that the distributions are equal. Therefore, a low p-value after corrections, here p = 0.05, is interpreted as that the feature may be used to distinguish between WT and mutated protein structures. It is apparent that this includes most of the structural properties in Table 2 except for the root mean square deviation of the atomic position between WT and mutated and the Template Modeling (TM) score. Thus the remaining features that were tested for statistical significance lead to the possibility that they could be used to classify the structures using, for instance, clustering analysis.
Table 3.
Probability that the distributions of Table 2 features for WT and mutated structures are the same. The Mann-Whitney-Wilcoxon [28] test examines whether two samples are likely to derive from the same population taking as the null hypothesis that the distributions are equal. Therefore, a low p-value, here p = 0.05, after corrections is interpreted as that the feature may be used to distinguish between WT and mutated protein structures. The p values below 0.05 are presented in bold.
| Predicted Structural Feature | P-value After Correction (BH) |
|---|---|
| Root mean square deviation of atomic position | 5.815e-01 |
| Confidence score | 2.032e-02 |
| Template modeling Score | 5.6e-01 |
| Solvent accessible surface area | 1.6e-05 |
| Total Gibbs free energy | 2.03e-02 |
| Pairwise distance-dependent energy | 2.56e-02 |
| Sidechain orientation-dependent energy | 2.56e-02 |
| Distance-related energy | 2.56e-02 |
| Angle-related energy | 4.78e-02 |
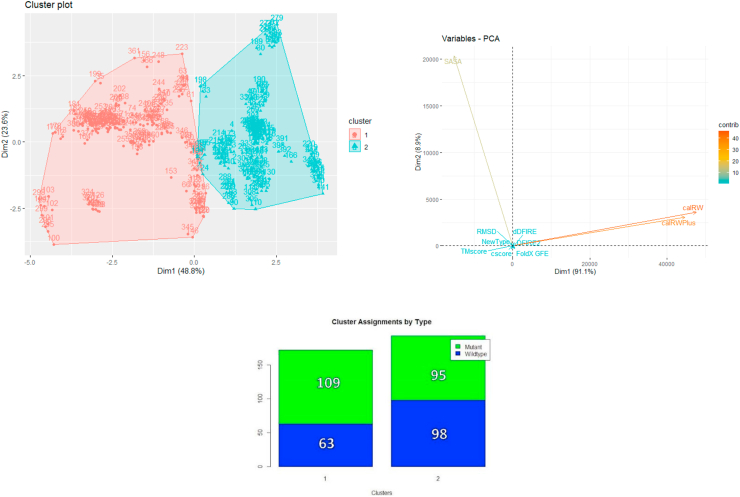
A cluster density calculation (within-cluster sum of squares) was performed on the dataset, and it was found that k = 2 clusters were the optimum number to use in the clustering analysis. K-means (k = 2) clustering was performed on the dataset, which yielded a visible separation when visualized (Figure 18). However, a purity analysis of the clusters showed that the clustering assignments of the WT and mutant proteins were not separated into the 2 clusters. Analysis of the principal components (Figure 18) indicates that the first dimension by which the clusters are divided by the solvent accessible surface area (SASA). This suggests that the proteins studied here can be classified into those that are relatively extended and those that are more compact, but that this classification is not changed by the oncogenic mutations considered here. This is consistent with the observations derived from Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17, which do not show any consistent changes in the overall protein structures upon mutation. Furthermore, we performed density cluster calculations consecutively eliminating the solvent accessible surface area of protein structure (SASA), pair-wise distance-dependent energy (calRW), side-chain orientation-dependent energy (calRWPlus) and total stability energy (FoldX). This order of elimination was dictated by choosing to eliminate the most distinctive component of the most significant contribution to the first principal component (see Figures S1-S4). In all cases, we observe good clustering according to the first principal component, but the clustering also does not discriminate (purity analysis) between the WT and the mutated structures. No differences were noticed between the clustering properties of structures calculated with full or partial template inclusions. It was again showing consistency with the lack of clear trends defining the structural changes upon mutation for this set of oncogenic proteins. The RMSD values that were calculated by comparing our wild type and mutant structures to their respective reference structures. Here they are used as an internal control of the quality of the structure predicted by I-TASSER because small RMSD between reference structure and the other four predicted structures is considered as an indication of a reliable prediction, but they do not differentiate WT from mutated structures.
Figure 18.
Cluster analysis performed using all features from Table 2.
4. Conclusions
Based on the above findings, there were several significant features that could be useful for distinguishing predicted WT and mutated protein structures, but the clustering analyses showed that there were no discernible differences in structural properties that could be found using an unsupervised method, i.e., k-means clustering. This is consistent with the observations from Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17, which do not show any consistent changes in the overall protein structures upon mutation, precluding any further statistical analysis. While we did not find overall principles that can be used to classify WT and oncogenic variables in this dataset, the judicious use of 3D structure prediction methods remains a valuable tool to understand oncogenic mutations further, as demonstrated previously [11, 12, 32, 33, 34, 35]. We recognize that comparing these results with those that could be obtained from neutral non-pathogenic variants could provide more details on the problem and perhaps could help in differentiating the structural effects in different types of variants. Additionally, we also seek to highlight that due to the limited availability of experimental structures that still should be considered the gold standard, the use of 3D structure prediction to understand molecular mechanisms of pathogenicity, in general, is a necessary albeit difficult task due to difficulties in experimental determination of mutated structures and the large volume of reported protein-coding sequences. In our study, we have had to understand that although we found statistically-significant differences in the predicted features of our dataset between wild type structures and those affected by oncogenic mutations, these changes were not conclusively detected in our other analyses. This could be due to the variety in size and function of proteins in our dataset, which could have contributed to inconclusive clustering analyses despite finding these statistically-significant predicted structural features. Therefore, in future work, the possibility of finding higher-order structural features that can be used to classify WT and oncogenic protein structures could be explored using a much larger dataset, which includes nonpathogenic variants, more structural features, and supervised machine learning algorithms, e.g. Random Forest, for binary classification, i.e., WT/mutant classification.
Declarations
Author contribution statement
Rolando Hernandez: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Julio C. Facelli: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the Utah Center for Clinical and Translational Science funded by NCATS award 1ULTR002538 and the NLM Training grant T15 LM00712418.
Data availability statement
No data was used for the research described in the article.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Computer resources were provided by the University of Utah Center for High-Performance Computing, which has been partially funded by the NIH Shared Instrumentation Grant 1S10OD02164401A1.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Material
References
- 1.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34(Database issue):D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arodź T., Płonka P.M. Effects of point mutations on protein structure are nonexponentially distributed. Proteins: Struct. Funct. Bioinform. 2012;80(7):1780–1790. doi: 10.1002/prot.24073. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C. Iowa State University; 2016. Protein Wild-type and Mutant Ensemble Database. [Google Scholar]
- 7.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J. Critical assessment of methods of protein structure prediction (CASP)—round XIII. Proteins: Struct. Funct. Bioinform. 2019;87(12):1011–1020. doi: 10.1002/prot.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pejaver V., Urresti J., Lugo-Martinez J., Pagel K.A., Lin G.N., Nam H.-J. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv. 2017:134981. doi: 10.1038/s41467-020-19669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2017;34(3):511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan L., Lv Q., Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32(19):2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teerlink C.C., Huff C., Stevens J., Yu Y., Holmen S.L., Silvis M.R. A nonsynonymous variant in the GOLM1 gene in cutaneous malignant melanoma. JNCI: J. Natl. Cancer Inst. 2018;110(12):1380–1385. doi: 10.1093/jnci/djy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C., Liu T., Liu B., Hernandez R., Facelli J.C., Grossman D. A novel CDKN2A variant (p16L117P) in a patient with familial and multiple primary melanomas. Pigm. Cell Melanoma Res. 2019;32(5):734–738. doi: 10.1111/pcmr.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bamford S., Dawson E., Forbes S., Clements J., Pettett R., Dogan A. The COSMIC (Catalogue of somatic mutations in cancer) database and website. Br. J. Canc. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium T.U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen J., Scoles D.R., Facelli J.C. Structure prediction of polyglutamine disease proteins: comparison of methods. BMC Bioinf. 2014;15 doi: 10.1186/1471-2105-15-S7-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A., Kucukural A., Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(SUPPL. 2):W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Zhou Y. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions. Protein Sci. 2008;17(7):1212–1219. doi: 10.1110/ps.033480.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Zhou Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins: Struct. Funct. Bioinform. 2008;72(2):793–803. doi: 10.1002/prot.21968. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PloS One. 2010;5(10) doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan L., Lv Q., Zhang Y.J.B. STRUM: structure-based prediction of protein stability changes upon single-point mutation. 2016;32(19):2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;18(1):50–60. [Google Scholar]
- 29.Haynes W. Benjamini–hochberg method. In: Dubitzky W., Wolkenhauer O., Cho K.-H., Yokota H., editors. Encyclopedia of Systems Biology. Springer New York; New York, NY: 2013. p. 78. [Google Scholar]
- 30.Allaire J. RStudio: integrated development environment for R. Boston, MA. 2012;770:394. [Google Scholar]
- 31.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020–baq. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerasimavicius L., Liu X., Marsh J.A. Identification of pathogenic missense mutations using protein stability predictors. Sci. Rep. 2020;10(1):15387. doi: 10.1038/s41598-020-72404-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casadio R., Vassura M., Tiwari S., Fariselli P., Luigi Martelli P. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum. Mutat. 2011;32(10):1161–1170. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen S.V., Stein A., Dinitzen A.B., Papaleo E., Tatham M.H., Poulsen E.G. Predicting the impact of Lynch syndrome-causing missense mutations from structural calculations. PLoS Genet. 2017;13(4) doi: 10.1371/journal.pgen.1006739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pey A.L., Stricher F., Serrano L., Martinez A. Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am. J. Hum. Genet. 2007;81(5):1006–1024. doi: 10.1086/521879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
No data was used for the research described in the article.