Abstract
Comprehensive and spatially mapped molecular atlases of organs at a cellular level are a critical resource to gain insights into pathogenic mechanisms and personalized therapies for diseases. The Kidney Precision Medicine Project (KPMP) is an endeavor to generate three-dimensional (3-D) molecular atlases of healthy and diseased kidney biopsies by using multiple state-of-the-art omics and imaging technologies across several institutions. Obtaining rigorous and reproducible results from disparate methods and at different sites to interrogate biomolecules at a single-cell level or in 3-D space is a significant challenge that can be a futile exercise if not well controlled. We describe a “follow the tissue” pipeline for generating a reliable and authentic single-cell/region 3-D molecular atlas of human adult kidney. Our approach emphasizes quality assurance, quality control, validation, and harmonization across different omics and imaging technologies from sample procurement, processing, storage, shipping to data generation, analysis, and sharing. We established benchmarks for quality control, rigor, reproducibility, and feasibility across multiple technologies through a pilot experiment using common source tissue that was processed and analyzed at different institutions and different technologies. A peer review system was established to critically review quality control measures and the reproducibility of data generated by each technology before their being approved to interrogate clinical biopsy specimens. The process established economizes the use of valuable biopsy tissue for multiomics and imaging analysis with stringent quality control to ensure rigor and reproducibility of results and serves as a model for precision medicine projects across laboratories, institutions and consortia.
Keywords: imaging, kidney disease, metabolomics, proteomics, transcriptomics
INTRODUCTION
Recent advances in biotechnology allow capturing the state of a tissue in health and disease at an unprecedented structural and molecular resolution (9). Application of these technologies at the level of the genome, transcriptome, proteome, and metabolome have enabled identification of regulatory cascades and their mapping into tissue compartments at a single-cell resolution (12, 17, 18, 22, 23, 25, 27). There is an urgent need to apply these technologies to clinical samples from patients with the two most devastating categories of kidney diseases, chronic kidney disease (CKD) and acute kidney injury (AKI). With a prevalence as high as 14% (37 million people) for CKD and high mortality in AKI patients, deciphering the underlying molecular and architectural complexity can result in better treatment in these conditions (20). Investigators have begun to apply single-cell omics and high-resolution imaging technologies to healthy or diseased kidney biopsy tissue to provide important information on the cell type composition and spatial relationships in diseases like lupus nephritis, diabetes, and in healthy tissue (21, 24). Integrating multimodal information derived from existing datasets and emerging technologies is a major challenge, because protocols, biological terms, and experimental standards are not uniform. In addition, applying multiple cutting-edge technologies frequently involve the collaboration of several laboratories with specialized practices and protocols.
Recognizing these limitations, one of the goals of the Kidney Precision Medicine Project (KPMP) is to establish rigorous preanalytical and analytical protocols with highly standardized and controlled workflows to interrogate biopsies of AKI and CKD patients by using cutting-edge omics and imaging technologies. Here, we describe a quality-controlled tissue interrogation pipeline component of the KPMP for multimodal analysis of kidney biopsies. This pipeline realizes the power of combined analyses of well-vetted and -curated data from different technologies to ensure rigor, reproducibility, and complementarity to generate molecular atlases of healthy and diseased kidneys that can impact patient care while extracting maximum data from a limited amount of tissue. The framework established serves as a paradigm for similar atlas efforts and precision medicine projects of other organs systems and diseases and a guide to investigators to generate high quality and reproducible data from limited tissue (1, 9, 10, 17).
RESULTS AND DISCUSSION
Guiding Principles for Multimodal Quality-Controlled Tissue Interrogation
The KPMP encompasses a diverse set of technologies to generate a robust molecular, spatial, and structural atlas (Figs 1 and 2 and Supplemental File S1; all supplemental materials are available available at https://github.com/tashkar/Q_C_KPMP). A major strength of multimodal interrogation (Table 1 lists specialized terms used) of various biomolecules including RNA, proteins, and metabolites using different technologies is to ensure a comprehensive coverage of these biomolecules in case a single technology does not capture (“blind spots”) expression of a particular gene, protein or metabolite. Redundancy among the different technologies further provides orthogonal validation that lends confidence to the discovered genes/proteins/metabolites/cell types and cell states. To enhance data quality, reproducibility and identify weaknesses and strengths of each technology (Fig. 2) in this multimodal approach our guiding principle was to harmonize (Table 1) tissue collection, processing, preservation, and analytical steps.
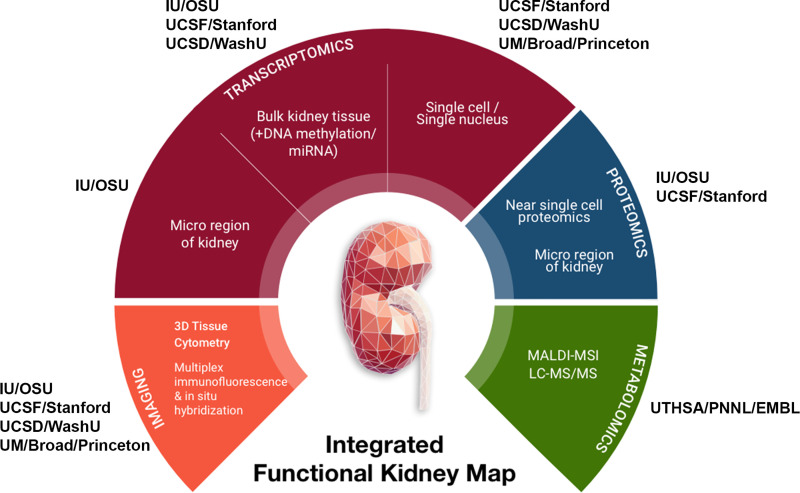
Figure. 1.
Overview of tissue interrogation sites and technologies in the Kidney Precision Medicine Project (KPMP). IU, Indiana University; OSU, Ohio State University; UCSD, University of California, San Diego; WU, Washington University in St. Louis; UM, University of Michigan; UCSF, University of California, San Francisco; UTHSA, University of Texas Health Sciences, San Antonio; PNNL, Pacific Northwest National Laboratory; EMBL, European Molecular Biology Laboratory.
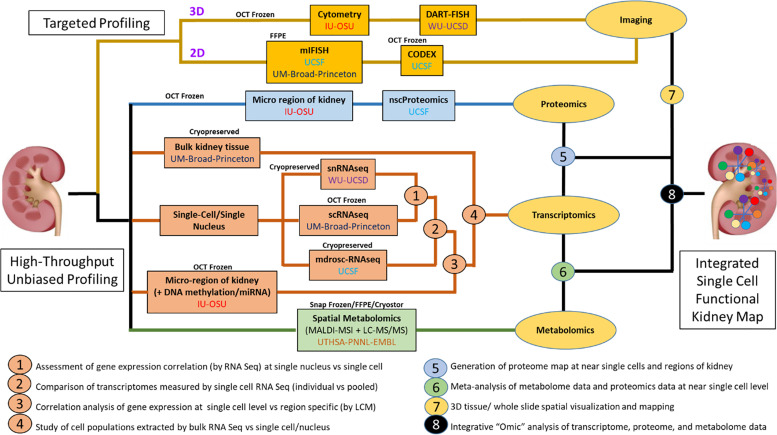
Figure 2.
Blueprint for interconnectivity for multimodal molecular assessment of kidney tissue at the initiation of the Kidney Precision Medicine Project (KPMP). IU, Indiana University; OSU, Ohio State University; UCSD, University of California, San Diego; WU, Washington University in St. Louis; UCSF, University of California, San Francisco.
Table 1.
Description of selected quality control terms as they relate to the Kidney Precision Medicine Project
| Term | Description |
|---|---|
| Complementarity | Different types of information gained from different technologies to describe the tissue composition and the relationships among its constituents |
| Follow the tissue | The different steps that the tissue undergoes during its interrogation from collection to preservation, processing, storage, shipping, assay, analysis, and data dissemination |
| Harmonization | Efforts to combine data from different sources including file formats, nomenclature into a cohesive and comparable format for analysis, and interpretation |
| Metadata | Information or data associated with each aspect of the process of tissue collection, its interrogation, and analysis |
| Metadata modeling | Development of rules and framework to define how to build relationships between concepts of a defined domain |
| Multimodal | Different ways (technologies) to interrogate the tissue in KPMP |
| Ontologize | The act of converting into ontological terms or entities |
| Ontology | A human- and computer-interpretable set of terms and relations that represent entities in a specific domain and how they relate to each other |
| Orthogonal validation | Using different technologies or assays to verify a given observation |
| Pipeline | The process through which each aspect of tissue interrogation is systematically done (example, tissue preservation pipeline, tissue processing pipeline) |
| Standardization | The process by which critical elements of quality control parameters in “follow the tissue” are established and implemented to minimize sources of errors and technical variations and ensure consistent results are produced for a particular technology |
| Technology drift | Changes in a technology or its components over time that can impact the reproducibility of results |
| Tissue interrogation | The act of using different techniques and tools to identify, analyze and determine the 2-D/3-D relationships between cells, their extracellular environment, and their molecular components including gene, protein, and metabolite expressions |
However, applying multiscalar technologies on a limited amount of tissue in a collaborative manner among the various KPMP tissue interrogation sites (TISs) posits unprecedented challenges. Errors are compounded as the data are dependent on multiple processes and steps that begin from specimen procurement to data generation and analysis. There are several sources of random and technical variations that confound the outcomes and impact biological reproducibility and interpretation of data. An additional challenge is maximizing the application of these sensitive, big-data technologies to clinical biopsies with limited tissue available for research. We developed a process to standardize and harmonize (Table 1), where possible, the entire pipeline from tissue procurement to analysis (termed “follow the tissue”) to overcome these challenges. Key factors considered were economizing tissue usage, maximizing preservation and use for multiple technologies, and documenting quality of intermediate steps with clearly defined quality assurance and control criteria. We implemented rigorous procedures for transparency, technical and biological reproducibility across multiple sources of tissue procurement, analysis, and interpretation and standards for quality control (QC).
Overview of the strategy: follow the tissue.
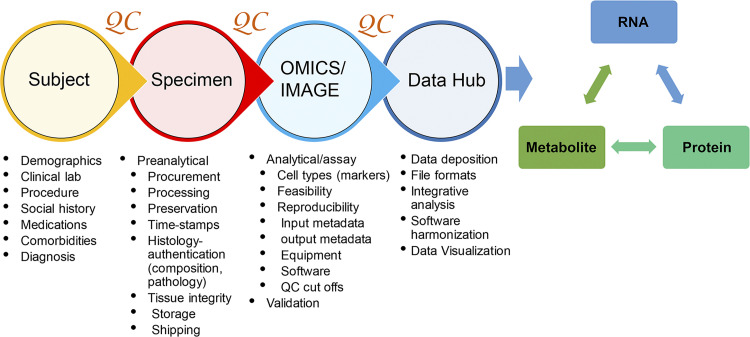
Since we anticipated that the tissue would come from different recruitment sites, our approach was to develop a tissue-processing pipeline that could be easily implemented at the bedside and applicable to multiple state-of-the-art interrogation technologies. We required from each TIS to demonstrate feasibility and reproducibility, apply rigorous analytics to their technology, develop processing methods that economizes tissue use, and foster integrated analysis and quality control measures in collaboration with the other TISs. The challenges in the “follow the tissue” process included variable standards in collecting metadata, procurement, processing, sample preparation, analytical parameters, analysis, and data sharing and deposition in a repository for public access. To tackle these challenges, we formed working groups for tissue processing that included expertise in omics, imaging, and pathology. These groups addressed quality control measures across four main categories: 1) participants, 2) tissue, 3) assays and analysis, and 4) data hub (Fig. 3). The objective was to identify critical parameters and data to be collected in each of the four categories that are important for standardization and QC. The outcomes of the meetings and decision process were documented at the KPMP management website in “Basecamp” (www.basecamp.com) to enable easy access to archived documents and meeting minutes for future reference. A critical aspect was formulating and capturing well-vetted relevant metadata in each of these categories to enable the interpretation of molecular discoveries among the underlying biological variations related to healthy and disease phenotypes (Table 2). We established benchmarks for the entire process and a clear approach for data visualization and sharing for various types of users. Once the overall vision or “blueprint” was established, the pipeline was pressure-tested with a pilot project using adult human kidney tissue (see the section discussing the consortium-wide pilot experiment). We describe below the individual steps of this workflow.
Figure 3.
Overview of “follow the tissue” pipeline with essential quality assurance/quality control (QA/QC) parameters.
Table 2.
Preanalytical parameters for rigor, reproducibility, quality assessment, and control
| Subject | Specimen Procurement | Specimen Morphology | Specimen Storage/Shipping |
|---|---|---|---|
| Age | Procedure data | Gross assessment | Storage time |
| Race | Procedure type (U/S, IR, Surgical) | Integrity (intact, fragmented) | Storage temperature |
| Sex | Location/site | %Cortex | Storage medium |
| Height | Procurement time | %Medulla | Storage container |
| Weight | Specimen collection media | Microscopic assessment | Shipping time |
| BMI | Tissue transport time to lab | # Glomeruli | Shipping container |
| Clinical labs (blood, urine) | Transport temperature | %Global glomerulosclerosis (age corrected) | Shipping image |
| eGFR | Preservation media/condition | Necrosis | Receiving time |
| Clinical diagnosis | Supplies/reagent details | Hemorrhage | Receiving image |
| Pathology diagnosis | Processing time | Autolysis | Storage time receiving |
| Comorbidities | Size | Tubular atrophy | Storage temp receiving |
| Medications | Image | Interstitial fibrosis | |
| Social history | Interstitial inflammation | ||
| Family history | Image (gross, H&E, PAS) | ||
| Freezing artifacts |
Subject metadata.
Detailed relevant parameters of participant-associated metadata were identified through a team effort of all the recruitment sites and TISs and are described elsewhere (Table 2). This was important to interpret variations in data due to contributions of patient attributes. For example, sex differences, age, race, or medications such as diuretics or antihypertensives can contribute to changes in molecular or cellular distribution of transporters in the kidney. The metadata fields are being modeled and represented using the Kidney Tissue Atlas Ontology (KTAO) (7) and Ontology of Precision Medicine and Investigation (OPMI) (6) two open-source biomedical ontologies that are being developed by collaborative efforts between the KPMP and ontology communities.
Tissue preanalytical considerations.
We identified and harmonized several preanalytical parameters for QC related to specimen handling, processing, preservation, orientation, quantity used, shipping, and storage to minimize their impact on technical variations and decreased data quality (Table 2). An infrastructure to track specimen movement from origin to specimen processing sites was developed (SpecTrack). A key feature was the ability to record deidentified specimens and documentation of the time, temperature, and state in which the specimens were procured, shipped, and received from the recruitment sites. All the sites were required to show successful use of this system before being qualified to receive specimens. An important consideration in designing the pipeline was to assess the quality and composition of the tissue being analyzed to best interpret the molecular outcomes. Whereas some technologies depended on complete dissociation of tissue (scRNA-seq), others had the opportunity to register tissue composition before dissociation or analysis. As such, guidelines were developed by the KPMP pathologists and TIS investigators for high-level quality assessment of composition and integrity of tissue sections and included relative proportions of cortex, medulla, glomerulosclerosis, and features compromising integrity (Table 2).
Tissue analytical and assay considerations.
We focused on three main tenets: 1) assay metadata, 2) assay quality assurance (QA) parameters, and 3) assay QC parameters. We emphasized from the beginning to define and record key metadata associated with assay performance to ensure transparency and reproducibility. QA is linked to understanding and applying the best practices recommended in the field for that particular technology, using a specific instrument set(s), protocols, or platform. The reliance on data from the manufacturer or postmarketing analysis when available is essential. We ensured that all platforms used in data production are optimized to produce the best possible results or operating under bone fide core facilities. For the QC component, we expected to meet and exceed a set of criteria guaranteeing that the assay works properly. We harmonized metadata collections, assays, instrumentations, and post hoc analyses for similar biomolecules and used standard terms where possible. QC parameters and minimum attributes that were relevant for the performance of the assay were identified, and common terms were used for similar types of technologies (RNA or protein or imaging). Each TIS was expected to come up with concrete criteria for QC of each technology that could be tracked throughout and demonstrate pass-fail rates and reproducibility in pilot experiments (see consortium-wide pilot experiment). Furthermore, within each technology, implementation of measures that allow detection and control of batch effects and assay drift were also incorporated.
These criteria also set a benchmark to give reproducible data for building the kidney atlas (Supplemental Tables S1–S3).
Identifying and annotating cell types.
We followed the concept of building an iterative marker list derived from published data and data generated from the KPMP. This served several purposes: 1) to qualify the identity and composition of the tissue being interrogated, 2) to validate and optimize tissue processing pipelines, and 3) to identify regions or cell types for integrative quality check and analysis to build the kidney atlas. Our initial list (made in 2018) of a subset of cell type markers relied mainly on rodent studies and bulk RNAseq data, with corresponding evidence from the human protein atlas (Supplemental File S2) (2–5, 13, 14, 19, 26, 28, 30). Later iterations of the potential cell types/states were heavily dependent on the data generated from the KPMP pilot project. Similarly, for imaging studies, a number of parameters were established to best standardize the formats of image acquisition, analysis, and deposition (Supplemental Table S4).
Data quality check, visualization, and sharing.
After passing the local TIS QC, the data were required to be deposited in the “data hub” that is managed by non-TIS members. The roles of the data hub team are 1) examination of the associated metadata for completeness, 2) independent analysis of the data for passing QC thresholds, 3) enhancement of data availability to other sites of the KPMP for integrated analysis and quality check, and 4) planning for public sharing. An essential component of data output is making it accessible to the public. In this regard, the KPMP data hub is tasked with a team dedicated for building tools for summary analysis and visualization of the integrated results generated by the various technologies.
A Consortium-Wide Pilot Experiment to Test the “Follow the Tissue” Pipeline
Rationale.
The objective of a consortium-wide pilot experiment was to use a same-source kidney specimen to 1) standardize tissue processing/handling, storage, and shipping steps; 2) establish feasibility and validate the QA-QC parameters for all the technologies in the interrogation pipeline; 3) compare, when applicable across sites, the performance of molecular interrogation and identify sources of variabilities and concordance; 4) lay out a blueprint for harmonization and complementarity across technologies (Table 1); and 5) identify gaps and weaknesses in the interrogation pipeline. An important outcome was to define a protocol that is harmonized across technologies and that could ultimately be used for interrogating biopsies from patients in an economical and efficient manner.
Design.
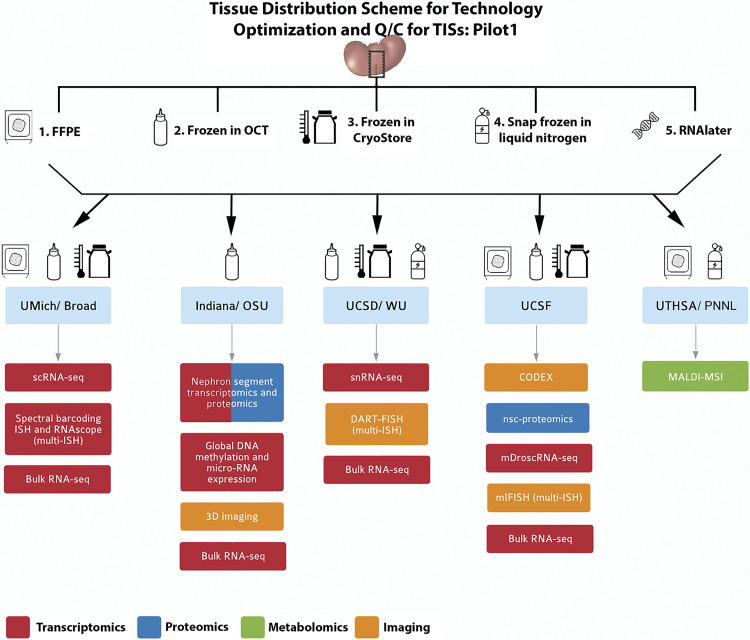
Tumor-free kidney cortical tissues from nephrectomy specimens were procured from the University of Michigan tissue collection center, preserved in different types of media according to the needs of the various TISs, and distributed to each TIS for testing feasibility, validation, and identification of the QC metrics for their respective technologies. Specifically, contiguous serial sections (∼1 cm × 2 mm × 2 mm) in the shape of rectangular cuboids were cut for processing and preservation and shipped to each TIS designated by a code (Fig. 4 and Supplemental Fig. S1 for the preservation methods used). In total, six different nephrectomy specimens were processed as described above and used by all the TISs. Hence, not only did each site have access to the same tissue source, but there were also six biological replicates distributed for the purpose of testing reproducibility, as discussed below.
Figure 4.
Pilot 1 tissue processing and distribution. A single-source kidney tissue from a tumor nephrectomy was sectioned into equal blocks (schema in Supplemental Fig. S1) and processed into 5 tissue processing methods according to the downstream applications listed. Blocks of the tissue were distributed to the sites according to the technology, as indicated by the drawings. Formalin-fixed paraffin embedded (FFPE); in situ hybridization (ISH); single-cell (sc) and single nuclear (sn)RNA sequencing (RNA-seq). Near-single-cell (nsc)-proteomics; matrix-assisted laser desorption-ionization mass spectrometry imaging (MALDI-MSI). OSU, Ohio State University; UCSD, University of California, San Diego; UCSF, University of California, San Francisco; UTHSA, University of Texas Health Sciences, San Antonio; PNNL, Pacific Northwest National Laboratory
Quality Control Outcomes and Observations Based on the Pilot Experiment
- Tissue procurement/preservation: preanalytical parameters. The following outcomes were directly derived from this pilot experience:
-
(A). better definition of the metadata associated with tissue procurement, preservation, integrity, and composition. We identified commonalities among tissue procurement, processing, assessment, and storage that enabled the use of similar conditions for multiple technologies. For example, snRNAseq, 3D tissue cytometry, laser microdissection (LMD) transcriptomics, LMD proteomics, mDroscRNAseq, spatial metabolomics, miFISH, and DART-FISH could all use fresh frozen optimal cutting temperature (OCT) blocks (Supplemental Tables S1–S3 and detailed in TIS manual of operations at www.kpmp.org/resources).
-
(B). ontology-based metadata modeling and representation. The identified metadata elements confirmed the need to implement this ontology-based approach to represent relationships between different metadata types more meaningful and machine interpretable, supporting advanced data analytics and knowledge discovery (Table 1) (8, 15, 29).
-
(C). real-time testing of specimen tracking using SpecTrack software. This live tracking of the tissue revealed weaknesses in the pipeline and allowed improvements including better documentation of tissue and temperature states of shipments and appropriate packaging materials (Supplemental Fig. S2; see pathology protocols at https://kpmp.org/researcher-resources/).
-
(D). effect of shipping and best practices establishment. To determine the effect of shipping on tissue quality, an assessment of RNA integrity was performed on bulk tissue preserved in RNAlater by using independent RNA preparation methods at two different sites. All the bulk RNA samples (total 12; 6 nephrectomy samples in RNAlater shipped to each site) were sequenced at a central site. These results showed strong correlation among adjacent tissue samples from the same subject for all six subjects and established the shipping conditions for frozen tissue that do not adversely affect tissue state as measured by RNA expression and integrity analysis (Fig. 5). We noted several observations during the pilot experiment that could in general impact tissue integrity including insufficient dry ice, the contents not well embedded in dry ice upon receipt due to movement during transit, and frozen slides not secured in secondary box during transit (detailed shipping conditions are in the online pathology and biospecimen protocols at https://kpmp.org/researcher-resources/).
-
(E). initial processing at the TISs. This experiment also provided an opportunity to examine the initial processing steps at each TIS and explore the potential to standardize common procedures. This resulted in the implementation of common procedures at each site, which were incorporated in the KPMP TIS manual of procedures (www.KPMP.org). For example, this pilot experiment identified the need to obtain histology section flanking areas of interrogation within the tissue, to inform on the state, composition, and orientation of the tissue. This process also allowed the same OCT block to be exchanged by two interrogation sites to perform successful molecular interrogation simultaneously with three different techniques (Fig. 5). The pilot experiment was also crucial to verify, validate, and expand the metadata variables that needed to be captured for faithful documentation of the tissue journey from harvesting to interrogation.
-
(A).
Analytical QC parameters for each technology. One of the main goals of this experiment was to test and validate the QC parameters for each technology in different sites. The design of the pilot experiment allowed repeat testing on a single specimen to assess technical reproducibility, and the use of tissue from different donors and tissue from different sources (pilot and local samples) ensured testing the methodologies for rigor and biological reproducibility. The QC parameters adopted by each technology based on this pilot experiment are summarized in Supplemental Tables S1–S4.Cross-validation with existing data/standards or by cross-validating outcomes from various technologies performed on the single-source kidney tissue provided in this pilot provided another level of confirmation to the QC parameters. Examples of this include detection of the same molecules/metabolites in the same samples by using different technologies and concordant “derived” readouts such as pathway analyses. For example, the TIS technologies can detect different genes/molecules/metabolites, but these molecular entities can be part of the same signaling pathway. Examples of orthogonal validation approaches are shown in Fig. 5, and integrated analyses will be presented in a separate manuscript.
Postanalytical outcomes. In addition to the cross-validation benefits discussed above, examining the outputs from various technologies promoted integration efforts and helped determine the extent of complementary information provided by each technology and further metadata harmonization at the various levels of tissue processing, analytics, and analysis. Additionally, parameters for diagnostic features, composition, and integrity of the tissue that are applicable to all the TISs were further refined and led to a protocol for interrogating patient biopsies in the KPMP described in a comprehensive pathology protocol document (https://kpmp.org/researcher-resources/). This ensures that a comprehensive cellular and molecular converge is provided by the consortium to make a robust kidney atlas and a platform for discovery.
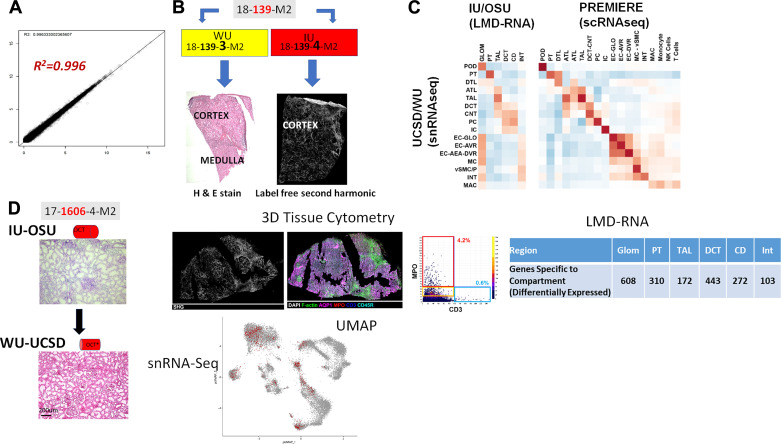
Figure 5.
Testing the “follow the tissue” pipeline with data generated from pilot samples and demonstrating feasibility of multiple technologies on limited tissue sample across sites. A: high correlation of bulk RNA-seq from adjacent samples shipped and processed at different sites. B: adjacent samples from same patient shipped to different tissue investigative sites (TISs) show different types of information from different technologies in Kidney Precision Medicine Project (KPMP) with histological tissue composition verification for snRNAseq at WU-UCSD and label-free second harmonic generation highlighting extent of fibrosis at IU-OSU. C: correlation between cell types and regions among snRNA-seq, laser microdissection (LMD)transcriptomics of the indicated regions by IU/OSU, and between snRNA-seq and scRNA-seq. D: economizing tissue usage by rotating fresh optimal cutting temperature (OCT) frozen kidney tissue between IU and WU/UCSD. Here. frozen sections were processed and analyzed for 3-D tissue cytometry and LMD-RNA analysis, and then the block was shipped to WU/UCSD for histological registration and subsequently generating snRNA-seq. Genes specific to each LMD compartment were determined by their upregulation in that compartment compared with other compartments as well as differential expression at a nominal P < 0.001. The UMAP plot shows the sample (∼900 nuclei) contributing to many of the cortical cell types in the kidney snRNAseq atlas (∼18,000 nuclei) (11). IU, Indiana University; OSU, Ohio State University; UCSD, University of California, San Diego; WU, Washington University in St. Louis; UM, University of Michigan; UCSF.
An additional important outcome was that significant amounts of gene and protein expression data were generated from the pilot samples. These data collected from multiple sites provide an initial view of cellular diversity in the human kidney (Table 3 and Supplemental Fig. S3). The analysis also revealed stress states related to processing of tissue and underlying pathology that could not have been predicted from gross evaluations in presumably healthy tissue. In fact, some novel discoveries have already emerged in the initial version of the kidney atlas from the pilot project (11, 16).
Table 3.
Cell types and associated markers from KPMP Pilot 1 transcriptomic studies
| Structure/Region | Substructure/Subregion | Cell Type | Abbreviation | Subset of Marker Genes | Pertinent negatives/comments |
|---|---|---|---|---|---|
| Renal corpuscle | Bowman’s (glomerular) capsule | Parietal epithelial cell | PEC | CRB2*, CLDN1* | Podocytes are tightly associated with the glomerular tuft |
| Visceral epithelial cell (Podocyte) | POD | NPHS2*, PODXL*, NPHS1* | |||
| Glomerular tuft | Capillary endothelial cell | GC-EC | EHD3*, EMCN*, HECW2*, FLT1*, AQP1* | ||
| Mesangial Cell | MC | POSTN*, PIEZO2*, ROBO1*, ITGA8* | |||
| Tubules | Proximal tubule | Proximal tubule epithelial cell (general) | PT | CUBN*, LRP2*, SLC13A1*, ALDOB*, GATM* | |
| Proximal convoluted tubule epithelial cell segment 1 | PT-S1 | SLC5A2*, SLC5A12* | There is overlap among the segments | ||
| Proximal tubule epithelial cell segment 2 | PT-S2 | SLC22A6* | |||
| Proximal tubule cell epithelial segment 3 | PT-S3 | PDZK1IP1*, MT1G* | |||
| Loop of Henle, thin limb | Descending thin limb cell (general) | DTL | CRYAB*, VCAM1*, AQP1*, SPP1* | CLDN10 low. AQP1 also in PT and a subset of ECs. May have 3 main subtypes | |
| Ascending thin limb cell (general) | ATL | CRYAB*, TACSTD2*, CLDN3* | AQP1 low to none | ||
| Loop of Henle, thick limb | Thick ascending limb cell (general) | TAL | SLC12A1*, UMOD* | SLC12A3 Low to none | |
| Cortex-TAL cell | C-TAL | SLC12A1*, UMOD* | |||
| Medulla-TAL cell | M-TAL | SLC12A1*, UMOD* | |||
| TAL-macula densa cell | TAL-MD | NOS1*, SLC12A1* | |||
| Distal convolution | Distal convoluted tubule cell (general) | DCT | SLC12A3*, TRPM6* | ||
| DCT type 1 cell | DCT-1 | SLC12A3*, TRPM6 | SLC8A1, HSD11B2 (Low to none) | ||
| DCT type 2 cell | DCT-2 | SLC12A3*, SLC8A1*, HSD11B2 | Has CNT and DCT signature | ||
| Connecting tubule | connecting tubule cell (general) | CNT | SLC8A1*, CALB1, TRPV5 | SLC12A3 Low to none. IC or PC without SLC8A1 could be in the CNT structure | |
| CNT-principal cell | CNT-PC | SLC8A1*, AQP2*, SCNN1G* | |||
| CNT-intercalated cell | CNT-IC | SLC8A1*, CA2, ATP6VOD2* | |||
| CNT-IC-A cell | CNT-IC-a | SLC8A1*, SLC4A1*, SLC26A7* | |||
| CNT-IC-B Cell | CNT-IC-B | SLC8A1*, SLC26A4*, SLC4A9* | |||
| Collecting Duct | Collecting Duct (General) Cell | CD | GATA3* | GATA3 May Be in subpopulation of DCT, CNT and vSMC/P. SLC8A1, CALB1, TRPV5 (low to none); Low to No CALCA and KIT in C-CD-IC-A. It may not be possible to assign IC or PC to CNT or CD structures without regional information of their source. | |
| CD-PC (general) | CD-PC | AQP2*, AQP3*, FXYD4*, SCNN1G*, GATA3* | |||
| C-CD-PC | C-CD-PC | ||||
| M-CD-PC | M-CD-PC | ||||
| Outer medulla-CD-PC | OM-CD-PC | ||||
| Inner medulla-CD cell | IM-CD | AQP2*, SLC14A2 | |||
| Transitional PC-IC cell | tPC-IC | FXYD4*, SLC4A9*/SLC26A7* | |||
| CD-IC (general) cell | CD-IC | CA2, ATP6VOD2* | |||
| CD-IC-A (general) cell | CD-IC-a | SLC4A1, SLC26A7*, TMEM213* | |||
| C-CD-IC-A cell | C-CD-IC-a | SLC26A7*, SLC4A1* | |||
| M-CD-IC-A cell | M-CD-IC-a | SLC26A7*, SLC4A1, KIT*, CALCA | |||
| CD-IC-B (general) cell | CD-IC-B | SLC4A9*, SLC26A4* | |||
| C-CD-IC-B cell | C-CD-IC-B | ||||
| M-CD-IC-B cell | M-CD-IC-B | ||||
| Vessels | Endothelial cells (non glomerular) | Endothelial cell (general) | EC | EMCN*, PECAM1*, FLT1* | |
| EC-afferent/efferent arteriole | EC-AEA | SERPINE2*, TM4SF1* | Likely PALMD | ||
| EC-peritubular capillaries | EC-PTC | PLVAP* | |||
| EC-descending vasa recta | EC-DVR | TM4SF1*, PALMD | |||
| EC-ascending vasa recta | EC-AVR | DNASEIL3* | Low to none | ||
| EC-lymphatics | EC-LYM | MMRN1*, PROX1 | |||
| Interstitium | Stroma (nonglomerular) | Vascular smooth muscle/pericyte (general) | vSMC/P | TAGLN*, ACTA2*, MYH11*, NTRK3, MCAM | |
| vSMC/P-renin | vSMC/P-REN | REN | |||
| Fibroblast | FIB | DCN*, ZEB2, C7, LUM | |||
| Immune | Macrophages-resident | MAC-R | CD163*, IL7R* | ||
| Macrophage | MAC | S100A9 | |||
| Natural killer cell | NKC | NKG7 | |||
| Dendritic cell | DC | APOE | |||
| Monocyte | MON | C1QA, HLA-DRA | |||
| T lymphocyte (general) | T | CD3 | |||
| T cytotoxic | T-CYT | GZMA | |||
| B lymphocyte | B | IGJ |
*Genes detected by more than one technology.
Identification of Gaps and Improvement of the Process
An area of priority identified during the integration efforts was the need to establish benchmarks for the nomenclature of cell types, regions, and associated genes, proteins, and metabolites for reference and disease atlas and various injury states. A promising methodology of analysis that could link multiple technologies is a cell-centric approach, whereby the outputs can reflect changes at the cell level in a tissue specimen. This was essential, as several groups are investigating the single-cell transcriptome or proteome of the kidney but there is lack of conformity regarding nomenclature and annotations. However, this analytical process requires an initial definition of cell types based on a set of criteria, such as gene expression (RNA and protein), cell state (baseline, stress, injury), spatial localization, and associations, among others. The pilot studies generated an initial working list delineating the complexity of cell types and a subset of associated marker genes in the adult human kidney, which could serve as a starting point for kidney omics and imaging studies for classification of cell types and states and harmonization with recent renal tubule epithelial cell nomenclature (Table 3) (3). An ontological representation of the cell markers has been initiated to seamlessly link gene, cell type, and spatial tissue location at an integrative semantic level (7).
Implementation of Best Practices to Perform Tissue Interrogation on Biopsies from the KPMP: the TISAC Process
Approval of TISs to receive biopsies for interrogation.
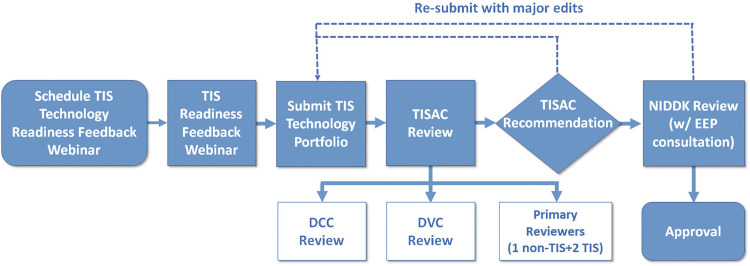
To rigorously evaluate each technology and eliminate self-approved bias by each TIS, we established a Tissue Interrogation Site Approval Committee (TISAC) (Fig. 6). The committee is composed of representation across the KPMP, The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and external ad hoc members as required to provide sufficient expertise to review the technology. This committee evaluates all technologies before approval to perform studies on precious biopsy specimens and ensures that the technology under consideration has presented evidence for robust QC metrics, sample handling, addressing batch effects, and assay drift and is complementary with other technologies. These elements are summarized in Fig. 3 and Supplemental File S3. The TISAC provides constructive feedback to the TIS whose technology is under review to enhance rigor, reproducibility, and complementary aspects of each technology as well as to identify areas that require additional supporting data. Once satisfied with a given technology’s readiness, the TISAC recommends approval and notifies the Steering Committee (Fig. 6). The TISAC provides its report to the NIDDK and the KPMP external expert panel, who ultimately approve the technology and TIS for receipt of patient samples. Each TIS is further expected to report on the state of its technology at least annually, and sooner if there are any modifications in the protocol used.
Figure 6.
Tissue interrogation site approval committee (TISAC) workflow. This workflow includes completion of a recorded webinar and submission of a portfolio that details the quality control metrics and technology outcomes. The TISAC reviews this material, providing recommendations to the tissue interrogation site for improvement before a site is approved to receive biopsies. The TISAC ultimately forwards their recommendations to the The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)and external expert panel (EEP) for final approval. DVC, data visualization center; DCC, data coordination center.
Ongoing Progress, Challenges, and Future Outlook
Evaluating progress and perceiving challenges require a continuous process of self-evaluation at several levels in the KPMP including the expertise of external investigators and interaction with large national and international consortia. The important components for a healthy and sustainable workflow for “follow the tissue” are: a) identifying opportunities to improve the quality control process, b) tackling challenges introduced by evolving or newer technologies, and c) mitigating potential threats or unforeseen errors. Examples of ongoing areas of development or challenges in the immediate future are described in Table 4.
Table 4.
Challenges and opportunities ahead
| Challenges | Explanations |
|---|---|
| Evolving ontology representation of various data and metadata elements | Generation of data and metadata will continue to grow. The need to link different and novel data types, cell markers, assays, and assay components is a dynamic approach and needs constant community engagement. |
| Data visualization, integration, and dissemination of results | Data (raw and processed), metadata and all QC elements will become publicly available for all types of users in a way easy to find, access, interoperate, reuse, and interpret. What is the best way to do this? |
| Incorporation of external data | The external data would need to meet KPMP QC standards for meaningful interpretation and relevance and reach of KPMP and non-KPMP-generated data for discoveries |
| KPMP policies to address changes in technologies | Some technologies and platforms may change. How will data be acquired and archived to be compatible with data from future platforms? This may need a large source of reference tissues that can be interrogated and shared by the TISs. |
| Software changes (analytical or visual software) | New softwares present challenges in compatibility, reliability, and security. |
| Strategies to test batch effect and technology or assay drifts | Current and future technologies are expected to provide an a priori plan to detect batch effect and technology drifts and provide solutions. What reference tissue standards are suitable for this purpose? |
| Validation of emerging technologies and incorporation into KPMP | Will there be a need for a standard tissue used for validating new technologies? What should this tissue be? Is there unlimited supply? |
| Hyperdimensional data management, storage, and sharing | An increasingly problematic issue when big data will be generated from each tissue specimen. |
| Patient protection in the era of artificial intelligence | The risk of linking patients to deidentified raw data may increase as machine learning tools develop further. Steps to mitigate risks in publicly available data will need to be implemented. |
| Justifying the use of limited renal biopsy tissue for research that is unlikely to benefit the patient and could compromise diagnostic yield | The follow-the-tissue pipeline enables multimodal analysis on leftover tissue from diagnostic specimens and would enhance current diagnostic pipeline, as these technologies may lead to new validated clinical tests that can improve diagnosis and management of patients with kidney disease. |
KPMP, Kidney Precision Medicine Project; QC, quality control; TIS, tissue investigative site.
One of the limitations of implementing novel high-throughput technologies on human biopsy samples is the lack of knowledge of assay variances and the biological variability between samples resulting from several factors (discussed above). This is further complicated by the complexity of cell types and 3-D relationships that have never been explored with the resolution of the current scale of technologies. These factors pose a challenge in performing power calculations and estimating sample size for reference or kidney disease atlas. For example, the variance in mean gene expression could be different in distinct cell types depending on demographics or underlying pathology. It is likely that analyzing reference kidney and disease biopsy tissue from the first 20 participants will provide insights into these variations and inform on the sample sizes needed for different cell types under different healthy or disease contexts. In the KPMP, we will analyze results from 20, 50, 100, and 200 biopsies to get a better understanding of the sample size needed for each of the disease categories. In addition, there is already an inherent sampling bias to derive conclusions because a biopsy represents only a fraction of the entire kidney. This limitation already exists in current clinical and pathological evaluations for any organ; however, the scale of multimodal analysis presented above provides a more rigorous analysis of the tissue and when enhanced by increased sample size, will likely overcome this limitation (Table 4).
Conclusions
With the implementation of a standardized multimodal and integrated pipeline for molecular interrogation of kidney biopsy specimens, the goal of the KPMP is to set high standards for quality control, rigor, and reproducibility. Vetted technologies participating in the KPMP will undergo careful scrutiny to comply with these goals of quality control while at the same time allowing a dynamic and iterative approach that promotes improvement and transparency. In doing so, the KPMP could become a model for other national and international efforts that also seek to decipher human disease and build a dynamic tissue atlas. With the QC infrastructure in place, the KPMP will achieve its goal to improve patient care and provide data to develop therapeutics for kidney disease with rigor and reproducibility.
GRANTS
The Kidney Precision Medicine Project is supported by the National Institute of Diabetes and Digestive and Kidney Diseases through the following grants: UH3 DK-114923, UH3 DK-114920, UH3 DK-114933, UH3 DK-114937, UH3 DK-114907, and U2C DK-114886.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.M.E., K.W.D., C.P., M.T.E., E.U.A., S.W., Y.H., V.D.D., R.I., O.G.T., L.B., J.G., K.Z., Z.L., B.R., P.C.D., K.S., M.S., J.B.H., C.A., M.K., S.J., T.K.S., T.A., and S.P. conceived and designed research; T.M.E., K.W.D., E.A.O., C.R.A., J.L., C.P., P.H., A.S., M.T.E., M.J., E.U.A., R.S., S.W., B.S., R.M., J.B.H., B.B.L., S.J., T.K.S., S.P., G.Z., and D.D. performed experiments; T.M.E., K.W.D., E.A.O., C.R.A., J.M.C., J.L., H.H., J.Z., P.H., A.S., M.J., E.U.A., R.S., S.W., B.S., Y.H., R.M., J.G., K.Z., J.B.H., B.B.L., S.J., T.K.S., T.A., S.P., G.Z., and D.D. analyzed data; T.M.E., K.W.D., E.A.O., C.R.A., R.S., S.W., V.D.D., R.I., O.G.T., L.B., J.G., Z.L., B.R., P.C.D., K.S., M.S., J.B.H., C.A., M.K., B.B.L., S.J., T.A., S.P., and G.Z. interpreted results of experiments; T.M.E., J.B.H., and S.J. prepared figures; T.M.E. and S.J. drafted manuscript; T.M.E., K.W.D., C.R.A., Y.H., V.D.D., R.I., O.G.T., L.B., J.G., B.R., P.C.D., K.S., C.A., M.K., B.B.L., S.J., T.K.S., T.A., and S.P. edited and revised manuscript; T.M.E., K.W.D., E.A.O., C.R.A., J.M.C., J.L., C.P., H.H., J.Z., P.H., A.S., M.T.E., M.J., E.U.A., R.S., S.W., B.S., Y.H., V.D.D., R.I., O.G.T., L.B., R.M., J.G., K.Z., Z.L., B.R., P.C.D., K.S., M.S., J.B.H., C.A., M.K., B.B.L., S.J., F.t., T.K.S., T.A., S.P., G.Z., and D.D. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the Kidney Precision Medicine Project (KPMP) patient participants, scientific officers from the National Institute of Diabetes and Digestive and Kidney Diseases, recruitment sites, Central Hub, and all the tissue interrogation sites for many valuable discussions and feedback towards the quality control efforts. We are grateful to the KPMP Publications and Presentation Committee for suggestions and review of this manuscript. A complete list of all KPMP members can be found at kpmp.org.
REFERENCES
- 1.All of Us Research Program I, Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, Jenkins G, The DE. “All of Us”” Research Program. N Engl J Med 381: 668–676, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunskill EW, Potter SS. Gene expression programs of mouse endothelial cells in kidney development and disease. PLoS One 5: e12034, 2010. doi: 10.1371/journal.pone.0012034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Clark JZ, Nelson JW, Kaissling B, Ellison DH, Knepper MA. Renal-tubule epithelial cell nomenclature for single-cell RNA-sequencing studies. JASN 30: 1358–1364, 2019. doi: 10.1681/ASN.2019040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Lee JW, Chou C-L, Nair AV, Battistone MA, Păunescu TG, Merkulova M, Breton S, Verlander JW, Wall SM, Brown D, Burg MB, Knepper MA. Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci USA 114: E9989–E9998, 2017. doi: 10.1073/pnas.1710964114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fetting JL, Guay JA, Karolak MJ, Iozzo RV, Adams DC, Maridas DE, Brown AC, Oxburgh L. FOXD1 promotes nephron progenitor differentiation by repressing decorin in the embryonic kidney. Development 141: 17–27, 2014. doi: 10.1242/dev.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Ong E, Schaub J, Dowd F, O’Toole JF, Siapos A, Reich C, Seager S, Wan L, Yu H, Zheng J, Stoeckert C, Yang X, Yang S, Steck B, Park C, Barisoni L, Kretzler M, Himmelfarb J, Iyengar R, Mooney SD. OPMI: the Ontology of Precision Medicine and Investigation and its support for clinical data and metadata representation and analysis. In: The 10th International Conference on Biomedical Ontology (ICBO-2019). Buffalo, NY: 2019. [Google Scholar]
- 7.He Y, Steck B, Ong E, Mariani L, Lienczewski C, Balis U, Kretzler M, Himmelfarb J, Bertram JF, Azeloglu E, Iyengar R, Hoshizaki D, Mooney SD. KTAO: a kidney tissue atlas ontology to support community-based kidney knowledge base development and data integration. In: International Conference on Biomedical Ontology 2018 (ICBO-2018). Corvallis, OR: 2018. [Google Scholar]
- 8.He Y, Xiang Z, Zheng J, Lin Y, Overton JA, Ong E. The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability. J Biomed Semant 9: 3, 2018. doi: 10.1186/s13326-017-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hood L, Rowen L. The Human Genome Project: big science transforms biology and medicine. Genome Med 5: 79, 2013. doi: 10.1186/gm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubmap Consortium The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574: 187–192, 2019. doi: 10.1038/s41586-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lake BB, Chen S, Hoshi M, Plongthongkum N, Salamon D, Knoten A, Vijayan A, Venkatesh R, Kim EH, Gao D, Gaut J, Zhang K, Jain S. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun 10: 2832–2832, 2019. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ledergor G, Weiner A, Zada M, Wang SY, Cohen YC, Gatt ME, , et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med 24: 1867–1876, 2018. doi: 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- 13.Lee JW, Chou CL, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. JASN 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Krautzberger AM, Sui SH, Hofmann OM, Chen Y, Baetscher M, Grgic I, Kumar S, Humphreys BD, Hide WA, McMahon AP. Cell-specific translational profiling in acute kidney injury. J Clin Invest 124: 1242–1254, 2014. doi: 10.1172/JCI72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Romero M, O'Connor MJ, Shankar RD, Panahiazar M, Willrett D, Egyedi AL, Gevaert O, Graybeal J, Musen MA. Fast and accurate metadata authoring using ontology-based recommendations. AMIA Annu Symp Proc 2017: 1272–1281, 2017. [PMC free article] [PubMed] [Google Scholar]
- 16.Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, Berthier CC, Eichinger F, Subramanian L, Harder J, Ju W, Nair V, Larkina M, Naik AS, Luo J, Jain S, Sealfon R, Troyanskaya O, Hacohen N, Hodgin JB, Kretzler M, (Kpmp) KPMP. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight, 5, 2020. doi: 10.1172/jci.insight.133267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, , et al. The Human Cell Atlas. eLife 6: e27041, 2017. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, , et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 199: 1517–1536, 2019. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers NM, Ferenbach DA, Isenberg JS, Thomson AW, Hughes J. Dendritic cells and macrophages in the kidney: a spectrum of good and evil. Nat Rev Nephrol 10: 625–643, 2014. doi: 10.1038/nrneph.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saran R, Robinson B, Abbott KC, Agodoa LYC, Bragg-Gresham J, Balkrishnan R, , et al. US Renal Data System 2018 Annual Data Report: epidemiology of kidney disease in the United States. Am J Kidney Dis 73: A7–A8, 2019. doi: 10.1053/j.ajkd.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, Pu M, Sharma S, You Y-H, Wang L, Diamond-Stanic M, Lindenmeyer MT, Forsblom C, Wu W, Ix JH, Ideker T, Kopp JB, Nigam SK, Cohen CD, Groop P-H, Barshop BA, Natarajan L, Nyhan WL, Naviaux RK. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. JASN 24: 1901–1912, 2013. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shema E, Bernstein BE, Buenrostro JD. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat Genet 51: 19–25, 2019. doi: 10.1038/s41588-018-0290-x. [DOI] [PubMed] [Google Scholar]
- 23.Shin S-Y, Fauman EB, Petersen A-K, Krumsiek J, Santos R, Huang J, , et al. ; The Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat Genet 46: 543–550, 2014. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart BJ, Clatworthy MR. Applying single-cell technologies to clinical pathology: progress in nephropathology. J Pathol 250: 693–704, 2020. doi: 10.1002/path.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, , et al. Subcellular map of the human proteome. Science 356: eaal3321, 2017. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 26.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, , et al. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419–1260419, 2015. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 27.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, Kriegstein AR. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364: 685–689, 2019. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkert G, Jahn A, Dinkel C, Fahlbusch F, Zurn C, Hilgers KF, Rascher W, Hartner A, Marek I. Contribution of the alpha8 integrin chain to the expression of extracellular matrix components. Cell Commun Adhes 21: 89–98, 2014. doi: 10.3109/15419061.2013.876012. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, , et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3: 160018, 2016. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeisberg M, Kalluri R. Physiology of the renal interstitium. CJASN 10: 1831–1840, 2015. doi: 10.2215/CJN.00640114. [DOI] [PMC free article] [PubMed] [Google Scholar]