SUMMARY
In mammals, hearing loss is irreversible due to the lack of regenerative potential of non-sensory cochlear cells. Neonatal cochlear cells, however, can grow into organoids that harbor sensory epithelial cells, including hair cells and supporting cells. Here, we purify different cochlear cell types from neonatal mice, validate the composition of the different groups with single-cell RNA sequencing (RNA-seq), and assess the various groups’ potential to grow into inner ear organoids. We find that the greater epithelial ridge (GER), a transient cell population that disappears during post-natal cochlear maturation, harbors the most potent organoid-forming cells. We identified three distinct GER cell groups that correlate with a specific spatial distribution of marker genes. Organoid formation was synergistically enhanced when the cells were cultured at increasing density. This effect is not due to diffusible signals but requires direct cell-to-cell contact. Our findings improve the development of cell-based assays to study culture-generated inner ear cell types.
Graphical Abstract
In Brief
Kubota et al. compare the organoid-formation potential of cochlear cell types and show that greater epithelial ridge cells, adjacent to the neonatal organ of Corti, have the strongest potential. Single-cell transcriptomic analysis confirms the identities of all cochlear cell groups. Their findings advance cell-based assays for development of inner ear therapies.
INTRODUCTION
The ability to hear critically depends on the function of a few thousand sensory hair cells inside the cochlea. Noise exposure, certain drugs, aging, and various genetic predispositions contribute to the demise of hair cells, which is the leading cause of sensorineural hearing loss (Brigande and Heller, 2009; Wu et al., 2020). Hair cells are associated with non-sensory cells, such as the anatomically distinct supporting cells of the organ of Corti and the less characterized cell types that make up neighboring tissues. Whereas the non-sensory cochlear cells of non-mammalian vertebrates display robust regenerative potential (reviewed in Janesick and Heller, 2019), their mammalian counterparts are quiescent, which is the principal reason why hearing loss is an irreversible disorder.
Although cochlear hair cell regeneration does not occur in adult mammals, the neonatal mouse cochlea shows limited and transient regenerative potential that can be attributed to non-sensory cells. Neonatal cochlear supporting cells are mitotically quiescent unless cochlear cell death activates their limited regenerative ability (Bramhall et al., 2014; Cox et al., 2014; Hu et al., 2016; Shi et al., 2013). This ability becomes apparent when the neonatal cochlear duct is dissociated and the individual cells are cultured in non-adherent conditions. Dissociated cochlear duct cells grow into spherical organoids that contain proliferating otic progenitors, which can be expanded and differentiated into inner ear supporting and hair cell-like cells (Malgrange et al., 2002; Oshima et al., 2007; Sinkkonen et al., 2011). The organoid-generating potential of cochlear duct cells is limited to the first 2 neonatal weeks and does not exist in the adult cochlea (Oshima et al., 2007; White et al., 2006). Additionally, the potential of some distinct non-sensory cochlear cell types to form organoids has previously been assessed, revealing that different cell groups differ in proliferative capacity and ability to give rise to hair cell marker-expressing cells (Chai et al., 2012; Doetzlhofer et al., 2006; Shi et al., 2012; Sinkkonen et al., 2011; White et al., 2006; Zhang et al., 2018).
We used fluorescence-activated cell sorting (FACS) to isolate different non-sensory cell groups, and we validated the composition of these groups with single-cell RNA sequencing (RNA-seq). We found that single-pass FACS is reasonably accurate but varies with different transgenic marker combinations and gating strategies. Here, we provide as a resource the single-cell RNA-seq profiles of all major cell types of the post-natal day 2 (P2) mouse organ of Corti and neighboring tissues, as well as metadata on their transgenic origin. Comparison of the organoid-formation capacity of different non-sensory cell groups using various media supplements showed that a recently identified combination of growth factors, signaling pathway modulators, and epigenetic modifiers (McLean et al., 2017) has the most profound organoid formation effect on non-sensory cochlear duct cells. We found that the organoid-formation capacity of cochlear duct cells prominently depends on the density of the cultured cells. At higher density, the cells of the greater epithelial ridge (GER), a transient neonatal cell group located medial to the inner hair cells and their surrounding supporting cells, displayed a synergistic organoid-formation ability. We determined that the mechanism for a robust expansion of GER cells requires cell-cell contact and does not depend on diffusible factors. We surmise that the capacity of GER cells can be further exploited for the future development of cell-based bioassays (Roccio and Edge, 2019).
RESULTS
Effective culture conditions for the generation of inner ear organoids
To identify the most efficient cell culture condition for organoid formation from cochlear non-sensory cells, we compared combinations of growth factors and small molecules based on previously reported media supplements that promote three-dimensional colony formation (Figure 1A) (Li et al., 2003a; McLean et al., 2017; Oshima et al., 2007; Sinkkonen et al., 2011). Earlier reports noted that the morphology of organoids that form after 1-week cultures varies from solid round to hollow configurations (Diensthuber et al., 2009; Oshima et al., 2007). Most organoids initially are of the solid type and eventually become single-layered hollow spheres of differentiated flat cells via intermediate stages with increasing vacuoles devoid of cells. Solid-type organoids harbor significantly more cycling cells and contain more cells that express otic progenitor markers compared with hollow and transitional types, indicating that progenitor cells with stem cell capabilities are associated with the solid morphology (Diensthuber et al., 2009). Therefore, our initial goal was to determine the most effective way to generate large solid-type organoids from neonatal cochlear cells while preventing their transition to the hollow configuration.
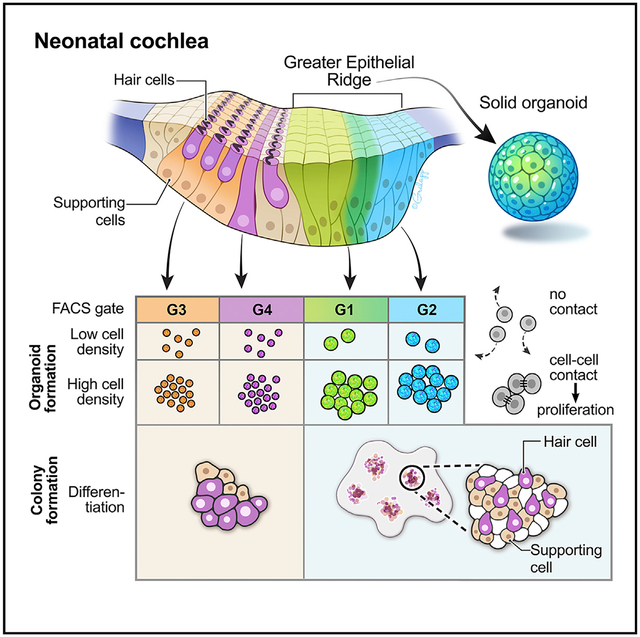
Figure 1. Optimization of organoid culture conditions.
(A) Process of P2 mouse cochlear cell dissociation and suspension culture. Dissociated cells are cultured for 7 days to generate organoids, followed by 14 days of adherent culture. For comparing different media supplements, dissociated cells were plated at 50 cells/μL density in 24-well dishes (25,000 cells/well). Conditions 1–7 are listed in the table.
(B) Representative organoids that formed after 7 days in culture. Conditions 1–7 are shown. Scale bars: 200 μm.
(C and D) The number of organoids (C) and the ratio of the three organoid morphologies (D) after 7 days in culture. Shown are the means ± standard deviations (SD) of triplicate experiments.
(E) Colonies generated from organoids after 21 days in cultures supplemented with EFI_CVPM (condition 5). Myosin 7a-expressing cells were not detected in colonies grown from hollow-type organoids (a, b1, and b2). The supporting cell/otic progenitor cell marker Sox2 was detected. Myosin 7a-expressing cells were detected in colonies grown from solid-type organoids (c, d1, and d2). Asterisks in (a) and (c) highlight the regions shown magnified in (b1), (b2), (d1), and (d2). Scale bars: 500 μm (a); 20 μm (b1 and b2); 500 μm (c); and 20 μm (d1 and d2).
Without adding growth factors and small molecules, we quantified 64 ± 6.5 organoids that formed from 25,000 dissociated P2 organ of Corti cells after 7 days in non-adherent culture conditions (Figures 1A–1C). Addition of epidermal growth factor (EGF), fibroblast growth factor 2 (FGF2), and insulin growth factor 1 (IGF1) (EFI) (Li et al., 2003a) doubled the number of organoids, and adding heparan sulfate (Oshima et al., 2007) further increased organoid numbers. The histone deacetylase (HDAC) inhibitor valproic acid (V) in combination with a glycogen synthase kinase 3 (GSK-3) inhibitor (CHIR99021 = C) (McLean et al., 2017) was equally effective in enhancing organoid formation (Figures 1B and 1C). Supplementing a stable form of vitamin C (2-phospho-l-ascorbic acid = P) and an inhibitor of transforming growth factor β (TGF-β) receptor 1 (2-(3-(6-methylpyridin-2-yl)-1H-pyrazol-4-yl)-1,5-naphthyridine = M), which are both parts of a previously optimized concoction to enhance cochlear colony formation (McLean et al., 2017), did not further increase the number of organoids generated after 7 days in non-adherent culture (Figure 1C).
After classifying organoid morphologies, we found that the combination of EFI with C, V, P, and M resulted in the most robust formation of solid-type organoids and almost complete suppression of hollow-type organoids (EFI_CVPM, condition 5; Figures 1B and 1D). A more extensive screen for additional supplements did not identify any single compound that would achieve similar or increased organoid formation capabilities as CVPM plus growth factors (Figures S1A–S1D). EFI_CVPM has previously been described for expansion of neonatal cochlear supporting cells (McLean et al., 2017), and we conclude that this condition is the most effective of the tested conditions in promoting and preserving a progenitor state in organ of Corti cell-derived colonies because it leads to the formation of large solid organoids and suppresses their transition into a hollow morphology. To support this conclusion, we tested whether the organoids grown in EFI_CVPM are capable of differentiating into hair cell marker-expressing cells by transferring individual organoids onto an adherent culture substrate and maintaining the adherent colonies for 14 days in the same media used for organoid formation (Figure 1A). We found that myosin 7a-positive cells, reminiscent of hair cells, differentiated only in colonies grown from solid EFI_CVPM-derived organoids when compared with hollow-type organoids grown in the same conditions (Figure 1E).
Organoid formation capacity differs among supporting cell subtypes and depends on cell density
We next compared the organoid formation capacities of different neonatal cochlear non-sensory cell subgroups in EFI_CVPM-supplemented suspension cultures. Sox2-GFP mice were utilized to FACS-enrich all supporting cells and GER cells (Figures 2A and 2D; Figure S2A). A more delimited group of supporting cells consisting of Deiters’, inner border, inner phalangeal, and outer pillar cells was isolated from Lfng-GFP mice (Maass et al., 2016; Figures 2B and 2E; Figure S2A). Combining Sox2-GFP, Fgfr3-CreERT2, and Ai14-tdTomato alleles (Waldhaus et al., 2015) enabled us to isolate four distinct groups of cells with different intensities of GFP and tdTomato expression, gated as G1–G4 (Figures 2C and 2F; Figure S2A).
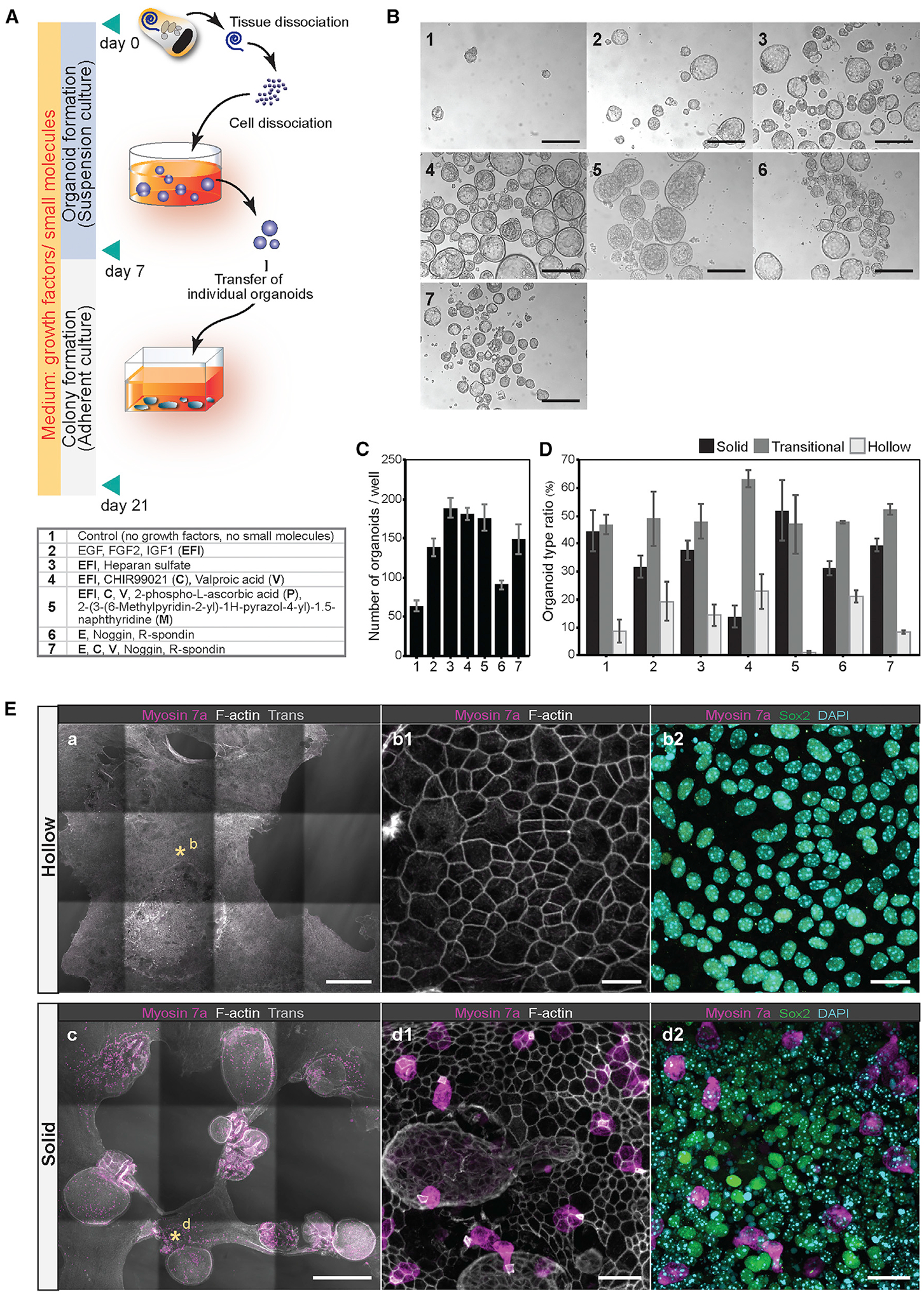
Figure 2. Organoid formation by cochlear cell groups at different cell densities.
(A–C) Fluorescent reporter gene expression in P2 organ of Corti vibratome sections of Sox2-GFP (A), lunatic fringe (Lfng)-GFP (B), and Fgfr3-iCreERT2/Ai14tdTomato/Sox2-GFP (C) mouse strains. Scale bars: 50 μm. Schematic drawings illustrate the different cell groups for each mouse line.
(D–F) FACS plots (left: dot plots; right: zebra plots) and gates applied for cell isolation: Sox2-positive gate (G_Sox2) (D), Lfng-positive gate (G_Lfng) (E), and gates 1–4 (G1–G4) defined by different levels of GFP and tdTomato expression using Fgfr3-iCreERT2, Ai14-tdTomato, and Sox2-GFP triple-transgenic mice (F). We expected that G_Sox2 represents all supporting cells and some GER cells. G_Lfng should consist of DCs, OPCs, IPHs, and IBCs. G1–G4 were expected to discriminate among all Sox2-GFP-positive supporting cells, with G1 and G2 representing enriched GER populations and G3 and G4 representing DCs and pillar cells.
(G) Boxplot presenting the number of organoids generated after 7 days in suspension culture at low cell density (2.5 cells/μL media) from G_Sox2 and G_Lfng cells. The mean ± SD is shown next to each boxplot. ***p < 0.001, two-tailed unpaired Student’s t test; n = 7 for each group.
(H and I) Boxplots presenting the number of organoids from the cells in G1–G4 generated after 7 days in low cell density culture (2.5 cells/μL media) (H) and higher cell density culture (10 cells/μL media) (I). The mean ± SD is shown next to each boxplot. ***p < 0.001, **p < 0.01, *p < 0.05, two-tailed unpaired Student’s t test; n = 11 (G1), n = 11 (G2), n = 11 (G3), and n = 10 (G4) in low cell density plots (H). n = 7 (G1), n = 6 (G2), n = 6 (G3), and n = 4 (G4) in higher cell density plots (I).
(J and K) Colorimetric cell viability assays of G1–G4 cells after 7 days of culture at low density (2.5 cells/μL media) (J) and higher density (10 cells/μL media) (K). Shown are the means ± SD of four experiments. **p < 0.01, *p < 0.05, two-tailed unpaired Student’s t test.
(L) Colorimetric cell viability assays performed after 7-day culture of cells from G1. The absorbance at 490 nm was compared between wells that received at the onset of the experiment 2,000 cells at higher cell density (10 cells/μL media) and the combined cells harvested from four individual wells that received 500 cells each (also a total of 2,000 cells), but at low cell density (2.5 cells/μL media). Shown are the means ± SD of triplicate experiments. The data are representative of three independently performed experiments. **p < 0.01, two-tailed unpaired Student’s t test.
DC, Deiters’ cell; GER, greater epithelial ridge; HeC, Hensen’s cells; IBC, inner border cell; IPC, inner pillar cell; IPH, inner phalangeal cell; ns, not significant; OPC, outer pillar cell.
GFP-expressing cells isolated from Sox2-GFP cochlear ducts generated 4.1 ± 2.1 organoids when cultured in a volume of 200 μL at a density of 2.5 cells/μL media (Figure 2G; Table S1). Lfng-GFP-positive cells gave rise to three times more organoids, indicating an increased proliferative capacity of supporting cells when compared with surrounding epithelial cells, which is consistent with previous reports when cells were cultured at low densities (Oshima et al., 2007; Sinkkonen et al., 2011). GFP-positive and tdTomato-negative cells from gate G1, representing mainly lateral and intermediate GER cells, gave rise to 4.0 ± 3.2 organoids when grown at low density (Figure 2H). Less intensely GFP-expressing G2 cells, likely representing the intermediate and medial population of GER cells and Hensen’s cells, did not differ significantly from G1 cells in their organoid formation capacity. G3 and G4 cells displayed 5- to 6-fold increased organoid formation capacity, which indicates the distinct potential of Deiters’ and pillar cells for colony formation (Sinkkonen et al., 2011; White et al., 2006). Of note, we observed that cells from gates G1 and G2 mostly degenerated and eventually disappeared during the 7-day culture period, whereas the cells from G3 and G4 were viable even without observable proliferation. Organoids derived from G1 and G2 were consistently larger than the organoids derived from G3 and G4 (Table S1). These observations explain the finding that cell proliferation at low density did not differ among the G1–G4 cell groups when directly compared using a colorimetric cell viability assay (Tada et al., 1986; Figure 2J).
We noticed a discrepancy in organoid numbers when the cells were cultured at 4× higher density (10 cells/μL media). All cultures displayed synergistic behavior, with G1 and G2 cells prominently producing ≈15× more organoids than lower density cultures (Figure 2I). It is conceivable that cell aggregation is occurring more frequently at higher culture densities, but we assumed this would not necessarily explain the observed increase of organoid numbers. Compared with low cell density (Figure 2J), we found that G1 cultures showed significantly increased cell numbers over G2, G3, and G4 cells when cultured at a higher density, which indicates higher proliferation rates (Figure 2K). When we directly compared the proliferation of G1-derived cells at high and low densities, we further confirmed the increase of cell numbers in higher density cultures (Figure 2L). These findings support the hypothesis that the proliferation of G1 cells, presumably originating from the GER, is enhanced at higher density. The increase of organoid numbers is not the result of cell aggregation.
Single-cell RNA-seq reveals composition of FACS-gated cochlear cell groups
FACS can be highly precise, and accurate selection of the desired cell type can be achieved, particularly when the gating strategy is carefully authenticated (Hertzano et al., 2020; Waldhaus et al., 2015). In contrast, differences between flow cytometers and gating strategies, as well as transgenic reporter gene expression, can be affected by the genetic backgrounds of mouse strains; this ambiguity requires a careful validation of different FACS-isolated cell groups. In parallel to culturing cells, we harvested individual FACS-sorted G1–G4 and gate Lfng-GFP (G_Lfng) cells and performed single-cell RNA-seq (Figure S3; Picelli et al., 2014). We used CellTrails (Ellwanger et al., 2018) for spectral embedding and unsupervised clustering of the data resulting in 11 distinct cell groups (states S1–S11; Figure 3A). Based on the expression of distinct markers (Tables S2 and S3; Figure S4), we identified supporting cell subtypes, GER cells, and hair cells (Figure 3A). Cluster S11 consists of inner and outer hair cells based on the expression of Fgf8, Slc17a8, Slc26a5, and Insm1 (Seal et al., 2008; Shim et al., 2005; Tateya et al., 2013; Wiwatpanit et al., 2018; Zheng et al., 2000). Pillar cells and Deiters’ cells were identified in clusters S6–S10 by the expression of Fgfr3 and Prox1 (Bermingham-McDonogh et al., 2006; Hayashi et al., 2010). Pillar cells were also found in S6, S9, and S10 by expression of Etv4, Etv5, and Ngfr (Hayashi et al., 2008; Mansour et al., 2013; Shim et al., 2005). Third-row Deiters’ cells were found in S7 through the high expression of Lgr5 (Chai et al., 2011; McLean et al., 2017; Shi et al., 2012). Inner border cells and inner phalangeal cells were identified in S5 based on the expression of Lfng and Otol1 (Deans et al., 2010; Zhang et al., 2000). High expression of Dkk3 indicated that clusters S2–S4 represent GER cells (Huang et al., 2011). Finally, cluster S1 consists of uncharacterized cells, likely of glial character (Table S2).
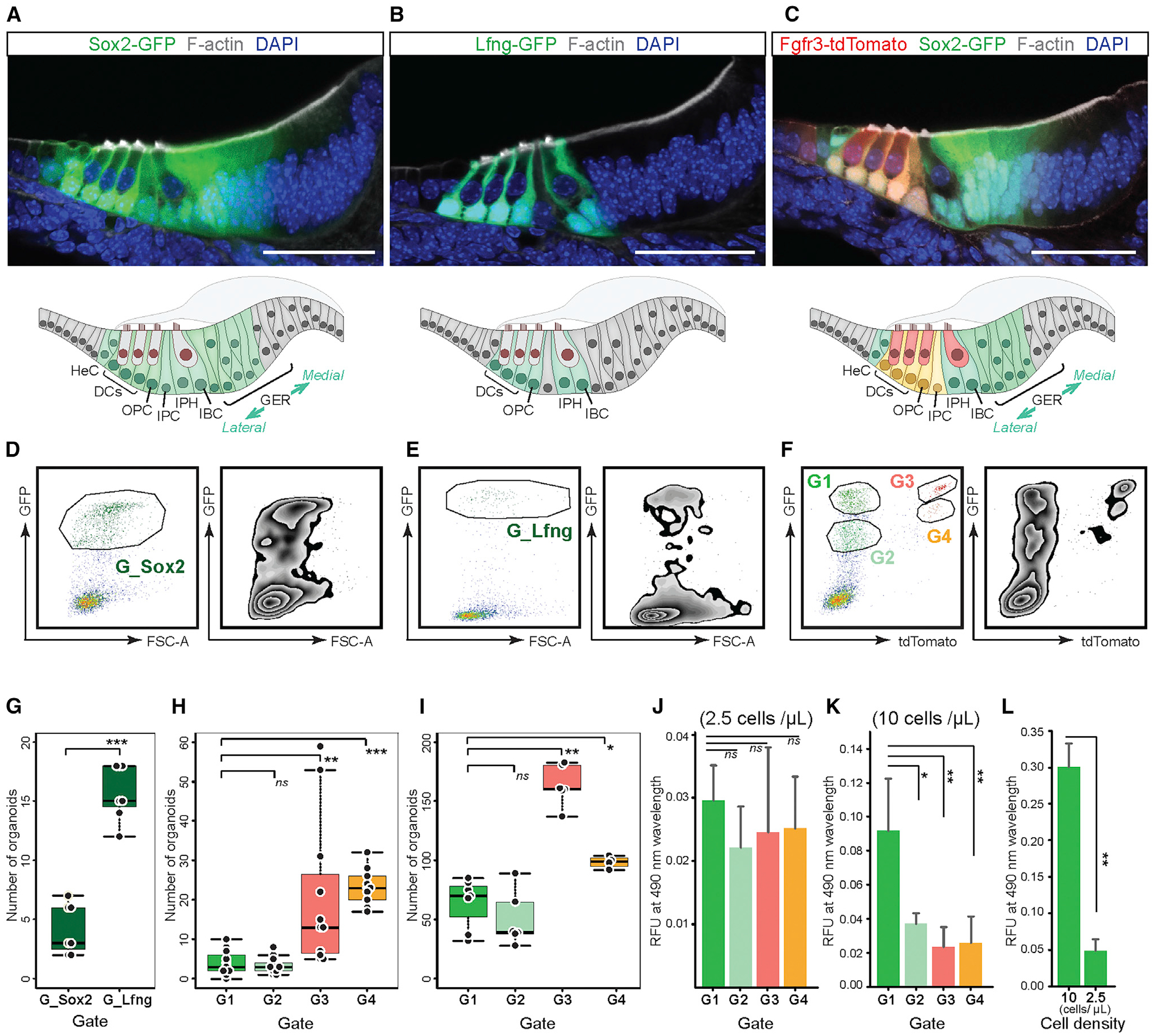
Figure 3. Single-cell RNA-seq confirms distinct FACS-gated cochlear cell subtypes.
(A) CellTrails identified 11 distinct clusters referred to as states (S1–S11). Shown are 409 FACS-isolated cochlear duct single cells projected into two-dimensional space via t-distributed stochastic neighbor embedding (tSNE). Single cells were colored by state affiliation.
(B) Violin plots for differentially expressed genes.
(C) Immunohistochemistry on P2 organ of Corti vibratome sections for Npy, Gsn, Sparcl1, and Crabp1. Schematic drawings highlight protein-positive cells. Npy mRNA expression is high in S10 (B), and the protein is specifically expressed by inner pillar cells. Gsn and Sparcl1 mRNA is enriched in S2–S5 (B), and the corresponding proteins are detected in GER, inner border, and inner phalangeal cells. Crabp1 mRNA is found in S2 (B), and the protein is expressed in the lateral GER. Scale bars: 50 μm.
(D) tSNE for the FACS gates used for cell isolation.
(E) Top (state): CellTrails states are labeled with the identified cochlear cell subtypes. Bottom (G1–G4, and G_Lfng): representation of the different clusters in G1–G4 and G_Lfng groups. State affiliations of the cells in G1–G4 and G_Lfng are shown by proportion indicated on the x axis.
(F) CellTrails trajectory reconstruction results in a furcating trajectory encompassing S2–S11. CellTrails identified S1 as an independent component distinct from S2–S11.
DCr1,2, first and second rows of DCs; DCr3, the third row of DCs; IHC, inner hair cell; OHC, outer hair cell; PC, pillar cell.
To further investigate and validate predicted cell subtypes, we analyzed the expression of markers that were differentially expressed in various clusters. High Npy gene expression was associated with S10 (Figure 3B; Tables S2 and S3). Immunohistochemistry performed on P2 cochlear sections demonstrated specific expression of Npy protein in inner pillar cells, confirming that S10 represents inner pillar cells (Figure 3C; Kolla et al., 2020). Previous studies described the expression of Gsn and Sparcl1 in the neonatal cochlear sensory epithelium (Burns et al., 2015; Mburu et al., 2010). Both genes were expressed in all three GER cell clusters (S2–S4) (Figure 3B; Tables S2 and S3) and were not differentially expressed across the three GER cell populations (Table S4). In accordance with this, the Gsn and Sparcl1 proteins were detected in all P2 GER cells (Figure 3C). Gsn and Sparcl1 immunoreactivity was also found in inner border and inner phalangeal cells, which agrees with the single-cell RNA-seq results that indicate expression also in cluster S5 (Figure 3B). We identified Crabp1 expression in GER cells, as has been previously reported (Kim et al., 2016). Crabp1 protein was localized in medial GER cells at P2 (Figure 3C). Given that Crabp1 expression is distinctively higher in cluster S2 versus S3 or S4 (Figure 3B; Table S4), we conclude that S2 contains medial GER cells.
To further authenticate the identified clusters, we used as an alternative feature selection method, M3Drop (Andrews and Hemberg, 2019), that utilizes a different mathematical approach, and performed graph-based clustering, in which seven main clusters were identified (Figure S5A). GER cells were identified in two clusters (clusters 1 and 3) and further sub-clustered into lateral, intermediate, and medial GER cells (Figure S5C; Tables S2 and S4). Outer pillar cells and Deiters’ cells, identified in cluster 4, were further sub-clustered into three populations that represent the first/second-row Deiters’ cells, third-row Deiters’ cells, and outer pillar cells (Figure S5E; Table S2). The defining genes for each cluster identified with graph-based clustering showed more than 90% overlap with the equivalent cluster identified with the CellTrails clustering method for most of the cell populations. Inner pillar cells, outer pillar cells, and the third-row Deiters’ cells overlapped by 84.9%, 76.0%, and 56.0%, respectively (Table S2). Overall, the results obtained with two independent cluster analyses were compatible and provided confidence that the computationally assigned cell subtypes are reasonably accurate.
We anticipated that FACS gates G1–G4 would contain different subsets of cochlear cell types. When we projected the different gates onto the t-distributed stochastic neighbor embedding (tSNE) representation of the single cells, we were able to directly visualize the accuracy of cell isolation based on fluorescent marker gene expression (Figures 3D and 3E). Following our prediction based on reporter gene expression, GER cells (S2–S4) accounted for 93% and 52% of the G1 and G2 populations, respectively. G1 and G2 differed by the abundance of cluster S3 and S4 cells in G1, which represent the most lateral group of GER cells adjacent to the organ of Corti. Similarly, in graph-based clustering, GER cells accounted for 83% and 55% of G1 and G2 populations, respectively; the G2 population represented the medial group (subcluster 4) of GER cells exclusively, whereas the G1 population represented lateral and intermediate GER cell groups (Figures S5C, S5D, and S5G). The G3 population was represented as predicted: pillar cells and Deiters’ cells, defined by markers present in S6–S10, accounted for 98% of the population (Figures 3D and 3E). In contrast, G4 harbored only 24% pillar cells (S6, S9, and S10), despite them being the primary target. Hair cells (S11) accounted for 46% of the G4 population. G_Lfng covered a wide range of cochlear cell subtypes. Nevertheless, the expected target population of G_Lfng, outer pillar, Deiters’, inner border, and inner phalangeal cells (S5 and S7–S9) accounted for 67% of this population (Figures 3D and 3E).
Spatiotemporal relations among the different organ of Corti cell subtypes were further analyzed with the trajectory inference algorithm provided by CellTrails (Figure 3F). We interpret the trajectory map of cells along a branching trajectory as dominated by latent spatial features. Interestingly, cluster S6, which contains cells expressing some, but not the full complement of, pillar cell markers from S9 and S10 (Figure 3A; Table S3), was placed at a branching point between pillar cells, Deiters’ cells, and hair cells (Figure 3E). This cluster might represent a latent transitional status reflecting the notion that neonatal supporting cells are not fully mature and display limited plasticity to differentiate into hair cells. Bmp4, known to be involved in inner ear sensory organ morphogenesis (Koehler and Hashino, 2014; Li et al., 2005; Munnamalai and Fekete, 2016; Ohyama et al., 2010), was expressed at comparatively high levels in S6 (Table S2). Graph-based clustering did not identify S6 cells as a distinct cluster and distributed the cells among several cell groups (Figures S5H). It is therefore possible that S6 cells either (1) represent multiple cell subtypes, (2) represent cells that respond to cell dissociation, or (3) represent a more plastic cell group. Similarly, the S4 cluster represents a branch point that reflects a possible dynamic state between GER cells and inner border/inner phalangeal cells based on overlapping marker gene expression (Table S2). The transcriptomic concordances of these cell groups along the trajectory support the reported potential of GER cells to migrate and replace lost supporting cells located medially to the tunnel of Corti (Mellado Lagarde et al., 2014). We utilized partition-based graph abstraction (Wolf et al., 2019) to provide an interpretable map of cluster relations of the cell groups shown in Figure S5A and found robust links between medial GER cells (cluster 1), lateral GER cells (cluster 3), and inner border cells and inner phalangeal cells (cluster 2) (Figure S5I). The analysis also showed a link between hair cells (cluster 6) and inner pillar cells (cluster 5) (Figure S5I).
GER cells can be expanded and differentiated into Myosin7a-positive cells
Activation of canonical Wnt signaling, combined with Notch signaling inhibition, has been effective in promoting new hair cell generation in the neonatal mammalian cochlea (Li et al., 2015; Samarajeewa et al., 2019; Shi et al., 2013). Various growth factors have been used to stimulate cell proliferation, particularly in cultures of otic progenitor cells, and growth factor removal was hypothesized to drive cell differentiation, such as into hair cell-like cells from embryonic stem cell-derived otic progenitors (Li et al., 2003b; Oshima et al., 2010). When we initiated cell differentiation in organoids grown in EFI_CVPM in the presence of LY411575 to activate Wnt signaling and C to inhibit Notch signaling, we found after 14 days in culture fewer hair cell marker-positive cells compared with continuous supplementation of the cultures with EFI_CVPM. Conversely, we found that hair cells, as well as supporting cells, were differentiating when the organoids were grown as colonies attached to a substrate in the sustained presence of all factors and compounds used. We consequently did not change the composition of the growth medium after transferring the organoids onto substrate, which resulted in continuous expansion of the resulting colonies. To evaluate the extent of organoid expansion and hair cell differentiation, we first compared organoids from the FACS gate G1, representing lateral and intermediately located GER cells, with G2-gated medial GER cells (Figures 3D and 3E; Figures S5A–S5D and S5G). The organoids from the FACS gate G3, representing mainly pillar and Deiters’ cells, were also compared (Figures 3D and 3E; Figures S5A, S5B, and S5G). Large colonies grew from G1-derived organoids. These colonies robustly harbored myosin7a-expressing cells (Figure 4A; Figure S6A). Organoids grown from G2 also gave rise to myosin7a-expressing cells similar to G1 (Figure 4B; Figure S6B). In contrast, G3-derived organoids showed substantially less expansion and generated only small colonies consisting of a few to tens of cells, yet many of those cells expressed myosin7a (Figure 4C). These observations suggest that medial and lateral GER cells display equally strong proliferative potential and robust ability to generate hair cell marker-positive cells. This is despite medial and lateral GER cell groups being transcriptionally distinct, with 139 and 322 differentially expressed genes detected, respectively (Figure 5; Table S4), which includes Lgr5 in lateral GER cells, a gene previously suggested to be indicative of neonatal cochlear stemness (McLean et al., 2017; Shi et al., 2012). We conclude that GER cells are the primary activated cell type in EFI_CVPM-supplemented media. Pillar and Deiters’ cells, which accounted for 98% of the G3 population, lack a robust proliferative ability in EFI_CVPM-supplemented media, but they showed competence to differentiate into myosin7a-expressing cells. Finally, we generated organoids from the G4 cell population for comparison, which also generated small colonies that efficiently gave rise to myosin7a-expressing cells. These colonies would mainly have pillar cell origin (Figure S6C).
Figure 4. Organoid colonies generated from different cochlear cell subtypes.
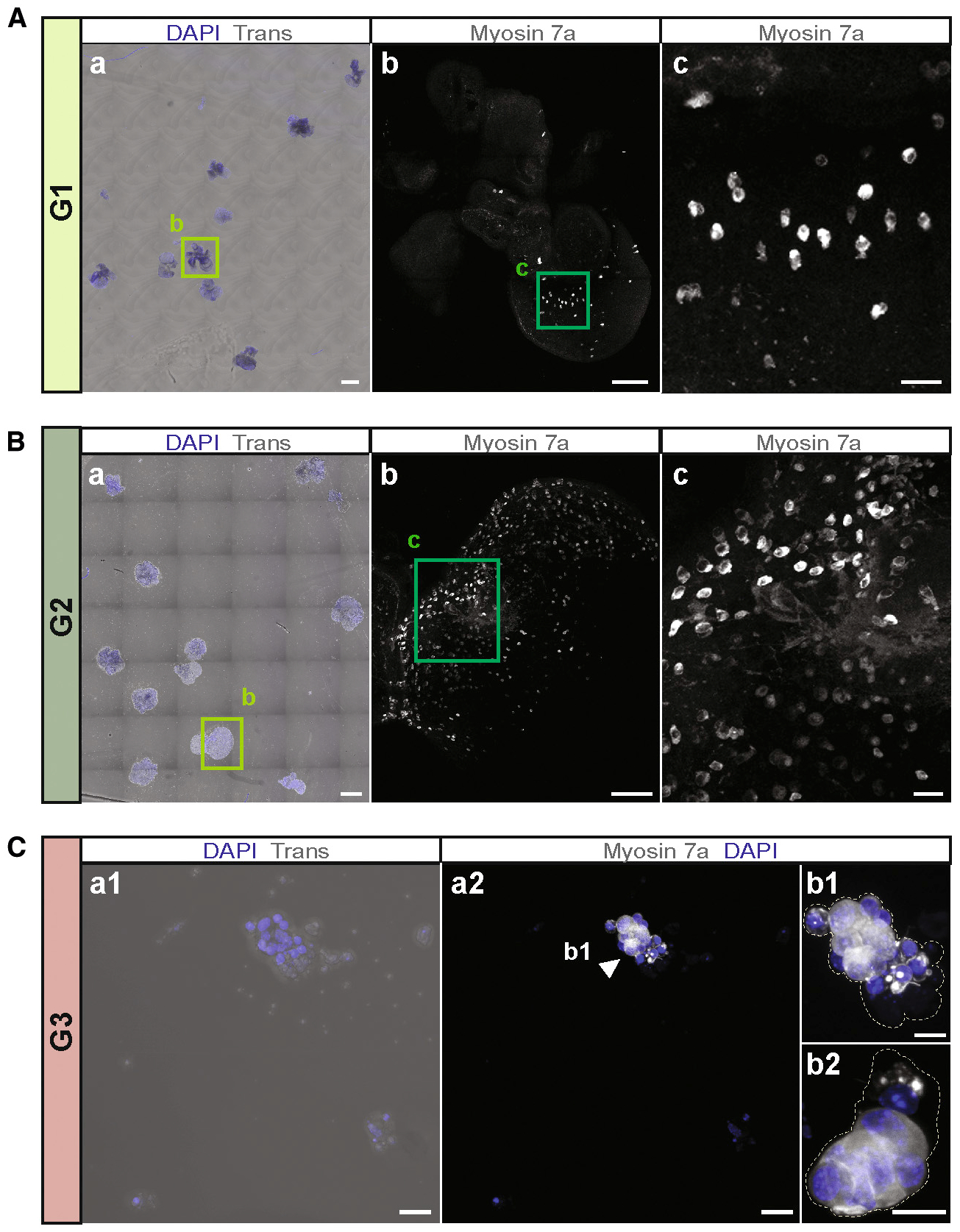
(A) Colonies generated from G1-derived organoids after 14 days of substrate-attached culture in media continuously supplemented with EFI_CVPM. (a) Overview image of the whole chamber illustrates the size and distribution of colonies. Scale bar: 500 μm. (b) Higher magnification of the colony surrounded by the square in (a). Myosin7a-expressing cells are visible. Scale bar: 100 μm. (c) Higher magnification view of the area surrounded by the square in (b). Scale bar: 10 μm.
(B) Colonies generated from G2-derived organoids, using the same experimental conditions as (A). (a) Overview image of the whole chamber shows large colonies. Scale bar: 500 μm. (b) Higher magnification of the colony surrounded by the square in (a). Myosin7a-expressing cells are visible. Scale bar: 100 μm. (c) Higher magnification view of the area surrounded by the square in (b). Scale bar: 20 μm.
(C) Colonies generated from G3-derived organoids, using the same experimental conditions as (A). (a1) Small colonies, consisting of only a few cells formed from G3-derived organoids. (a2) Myosin7a-expressing cells are visible. Scale bars: 20 μm (a1 and a2). (b1) Higher magnification of the colony marked with the arrowhead in (a2). (b2) Another representative colony. Dotted lines outline the colonies. Scale bars: 10 μm (b1 and b2).
Figure 5. Differential gene expression among the three GER clusters.
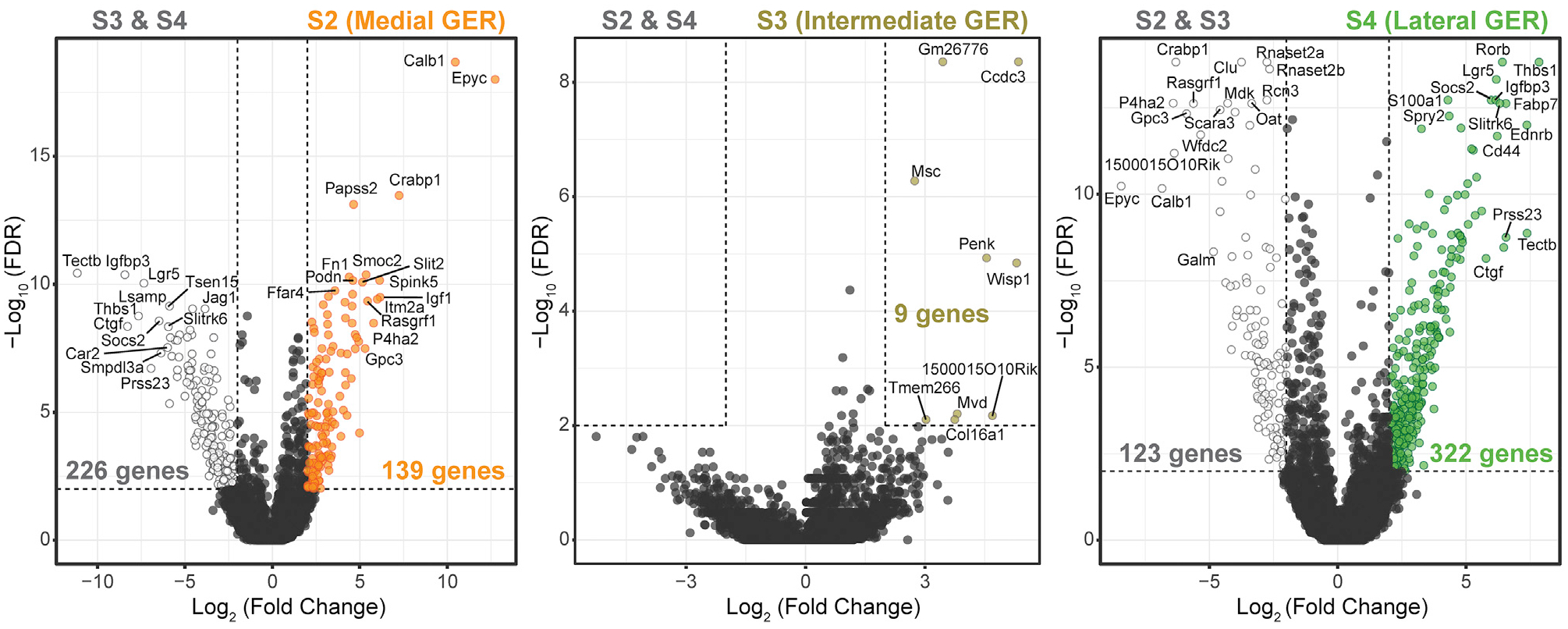
Volcano plots showing comparisons between each GER cell cluster (S2, S3, and S4) with the remaining GER cells. Thresholds are indicated with dotted lines: log2-fold expression difference of >2 or <−2 and −log10(FDR) > 2 (FDR < 1%). Labeled are top differentially expressed genes in medial (S2: left), intermediate (S3: middle), and lateral (S4: right) GER cells. The full list of differentially expressed genes is shown in Table S4. Wilcoxon rank-sum test was used for all comparisons shown here.
Expanded GER cells generate hair cells and supporting cells
As an alternative source for GER cells, we utilized Lfng-GFP mice and FACS-sorted GFP-negative cells (G_Lfng (−)) from dissociated cochlear duct cells at P2 (Figure S7A). We hypothesized that the G_Lfng (−) cell population would consist mainly of GER cells, although with some contribution of other cells (Figure 2B). G_Lfng (−) cells robustly generated organoids when cultured at a density of 10 cells/μL in media supplemented with EFI_CVPM. When further grown for 2 weeks, the organoids attached to substrate and formed large colonies (Figure 6A). The colonies harbored myosin7a-expressing cells, which were closely associated with GFP-expressing cells (Figures 6B and 6C; Figure S7B). Although we cannot exclude that some Lfng-negative hair cells survived sorting and organoid formation, we consider it as not very likely that the many myosin 7a-expressing cells that differentiated in the colonies are surviving hair cells. We conclude that G_Lfng (−) GER cells were capable of growing into large organoids in which myosin7a-positive cells differentiated in close association with GFP-positive cells, indicative of upregulation of the supporting cell marker Lfng. Hair cell-dependent induction of supporting cell markers in GER cells has been previously reported in the native tissue (Woods et al., 2004), as well as in embryonic stem cell-derived organoids (Koehler et al., 2013; Oshima et al., 2010). We note that this orchestration of sensory epithelium differentiation also happens in GER-derived organoids. We conclude that GER cells can be substantially expanded in EFI_CVPM-supplemented media, and these organoids harbor otic progenitors capable of differentiation into nascent otic sensory epithelial cell types.
Figure 6. Hair cells and supporting cells differentiate in GER organoid-derived colonies.
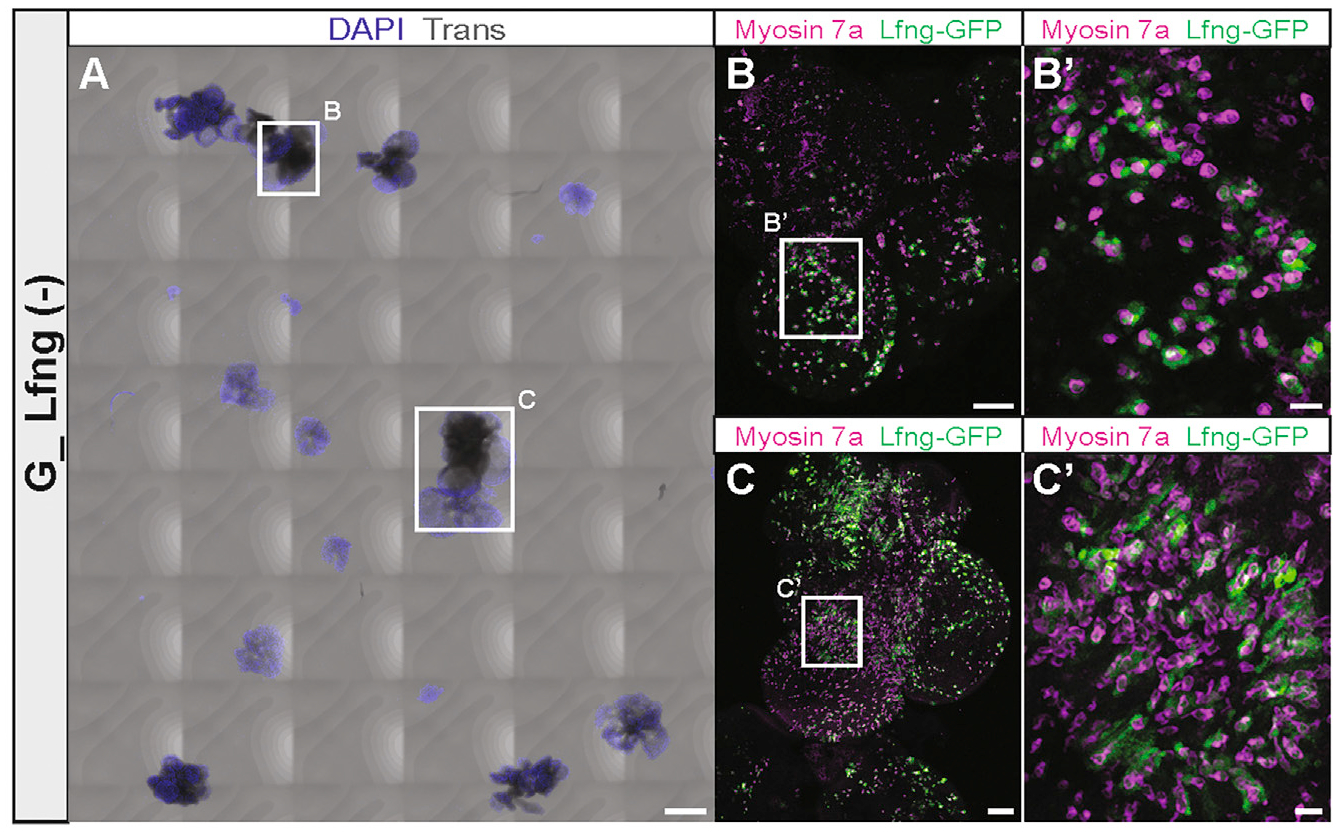
(A) Colonies generated from G1-derived organoids after 14 days of substrate-attached culture in media continuously supplemented with EFI_CVPM. The whole chamber with all colonies is shown. Scale bar: 500 μm.
(B and C) Higher magnification of the colonies surrounded by the squares in (A). Myosin7a-expressing cells and closely associated Lfng-GFP-expressing cells are visible and further magnified in (B′) and (C′). Scale bars: 100 μm (B and C); 20 μm (B′and C′).
GER cell proliferation depends on direct contact
The synergistic growth potential of GER cells at higher density (Figures 2H–2L) suggested that the increased proliferation is enhanced by paracrine signaling. The robust initial growth of GER cell-derived organoids is followed by sustained expansion that ultimately results in effective generation of a large number of new epithelial cells (Figures 4A, 4B, and 6A). To identify whether GER cell proliferation is driven by direct cell-cell contact or by soluble factors secreted and transmitted among the neighboring cells, we performed organoid formation assays in a medium containing 1.27% methylcellulose supplemented with EFI_CVPM. Direct cell-cell contact in this condition is minimized as individual cells are being physically separated due to the viscosity of the gel, whereas released soluble factors can diffuse freely (Kaufman et al., 2001; Le Joncour et al., 2019). Dissociated cochlear duct cells seeded at 2.5 cells/μL media gave rise to 82 ± 13 organoids in this condition. When we increased the cell density 2-fold and 4-fold, we found that the number of organoids grown in the medium increased proportionally, but not synergistically (Figure 7A). This result suggests that the synergistic enhancement of organoid formation in EFI_CVPM-supplemented media depends on direct cell-cell contact and not on soluble/diffusible factors. To further buttress this conclusion, we compared GER cell-derived organoid growth in a transwell co-culture system. We found that organoid growth was not affected by factors released from co-cultured GER cells seeded in excess in an adjacent compartment separated by a membrane permeable to diffusible factors (Figure 7B). These observations indicate that the enhanced proliferation of GER cells at higher densities is dependent on direct cell-cell contact rather than the diffusion of soluble molecules in the medium.
Figure 7. Direct cell-cell contact synergistically increases incident of organoid formation.
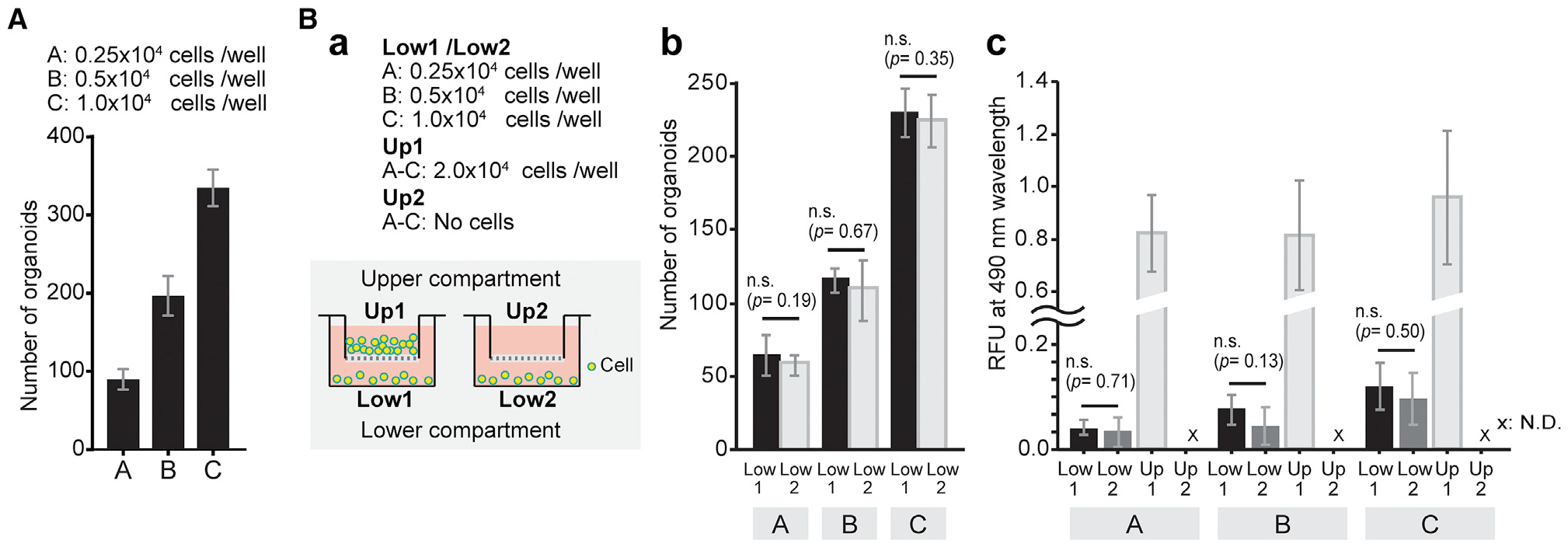
(A) Quantification of the number of organoids generated from dissociated cochlear duct cells after 7 days in EFI_CVPM culture in 1.27% methylcellulose gel. The sequential doubling of seeded cells results in a linear increase of organoid numbers. Presented are mean ± SD of triplicate experiments.
(B) Organoids were grown in a transwell co-culture system. (a) Schematic illustration and experimental details. The cells were seeded at three different densities into the bottom compartments as indicated, and for each cell density, the top compartment harbored either 2.0 × 104 additional cells or no cells. (b) Bar plots show the number of organoids grown in the bottom compartments after 7 days. Presented are mean ± SD of triplicate experiments. (c) Colorimetric cell viability assay of the cells in the bottom compartments after 7 days of culture in transwell co-culture system shown in (b). Shown are the means ± SD of triplicate experiments. The data are representative of three independently performed experiments.
DISCUSSION
Culture conditions to elicit proliferation of non-sensory cochlear cells were initially based on methods for isolation and propagation of neural stem cells (Gritti et al., 1996). These conditions were composed of serum-free media supplemented with growth factors previously reported to have mitogenic effects on the developing inner ear (Li et al., 2003a). The organoids grown from neonatal mouse cochlear duct cells in these conditions were rare, their propagation potential was limited, and the number of hair cell-like cells that differentiated in such cultures was low (Diensthuber et al., 2009; Oshima et al., 2007; Sinkkonen et al., 2011). It was noted that two main types of organoids formed from cochlear duct cells: hollow organoids consisting of epithelial cells and smaller solid ones (Diensthuber et al., 2009). It was further hypothesized, based on the potential of solid organoids for expansion and the ability to generate hair cell-like cells, that solid organoids harbor otic progenitor cells.
A major improvement for unlocking the proliferative capacity of non-sensory cochlear duct cells came when the effects on colony formation of small compounds, other additives, and pathway modulators were systematically assessed. A combination of factors, including V, a GSK-3 inhibitor, vitamin C, and a TGF-β receptor 1 inhibitor, in addition to growth factors EFI, significantly increased the numbers of organoids formed from dissociated cochlear duct cells (McLean et al., 2017). Many organoid cells displayed canonical WNT pathway activation, which was revealed by a transgenic reporter for Lgr5 expression. In the neonatal cochlea, Lgr5 reporter expression is restricted to inner pillar cells, inner border cells, lateral GER, and the third row of Deiters’ cells (Chai et al., 2011, 2012; Shi et al., 2012). This raises the question of whether organoid formation is a feature of Lgr5-expressing neonatal non-sensory cells or whether other Lgr5-negative non-sensory cells can give rise to organoids and Lgr5 expression is upregulated during organoid formation. It has been previously reported that the various non-sensory cochlear cell types have differing organoid formation potential (McLean et al., 2016; Sinkkonen et al., 2011; White et al., 2006). We hypothesized that the organoid formation potential of various defined non-sensory cochlear cell types is substantially higher than previously reported when using the most efficient known culture conditions.
Our experiments confirmed that media supplementation with EFI_CVPM is highly efficient in inducing organoid formation of dissociated cells from the neonatal organ of Corti and adjacent tissues. We cannot dismiss the potential of other supplements to promote organoid formation, but we found that EFI_CVPM not only promoted the formation of solid organoids, which we previously regarded as an indication of increased proliferative potential (Diensthuber et al., 2009), but also suppressed transition to hollow organoids, which is also an indication of maintaining a proliferative state. In contrast with our previous findings, the organoids grown in EFI_CVPM-supplemented media were much larger (up to 1 mm), which indicates that the addition of CVPM elicited substantial cell proliferation.
FACS-based cell sorting of inner ear cells is reasonably accurate, but not perfect, which is shown by our single-cell RNA-seq analysis of sorted cell groups. This is partially owed to the paucity of cochlear cells, which precludes multiple rounds of sorting for increased purity and also an indication that gating strategies for various cell groups can be further improved. Multiple reporters and highly stringent gating strategies can most certainly increase accuracy (Waldhaus et al., 2015). Nevertheless, single-cell RNA-seq allowed us to unequivocally determine the starting composition of cells used for organoid formation. Based on these assessments, we found that the formation of large solid organoids in EFI_CVPM-supplemented media is a distinct feature of GER cells. Moreover, we found that the number of organoids, but not their size, synergistically increased when we quadrupled the density of cells. Finally, we showed that cell-cell contact is essential for the observed synergistic effect. Direct contact and a variety of potential interactions have been previously put forward as a potent stimulator of proliferation, as well as an inhibitor of cell death in various cell types (Nelson and Chen, 2002; Osswald et al., 2015; Venkatesh et al., 2019).
GER cells are a transient cell group, also referred to as Kölliker’s organ (Dayaratne et al., 2014), that are replaced with inner sulcus cells during post-natal maturation of the cochlear duct. We found that GER cells are the most potent cochlear cell group for organoid formation when cultured at a higher density in the presence of EFI_CVPM. In the neonatal cochlea, only the most lateral GER cells express Lgr5, albeit at lower levels than inner pillar, inner border, and the third row of Deiters’ cells (Chai et al., 2011; Shi et al., 2012). Our observations are compatible with the notion that Lgr5-positive lateral GER cells can give rise to the observed large solid organoids. Nevertheless, we found that Lgr5-negative, more medially located GER cells are equally capable of generating large organoids. Therefore, Lgr5-expression in GER cells is not per se an indicator of stemness or proliferative potential.
Single-cell RNA-seq transcriptomes of neonatal organ of Corti and other cochlear duct cell types have recently been reported (Burns et al., 2015; Kolla et al., 2020). We contribute a set of RNA-seq data at high sequencing depth, which serves as an additional resource for interested researchers and is available at the gene Expression Analysis Resource (gEAR) data depository (Orvis et al., 2020) (https://umgear.org/p?l=afd2eb77), as well as from the Gene Expression Omnibus (GEO: GSE162308). Cluster analysis of GER cells revealed heterogeneity, which confirms the results of a recent study (Kolla et al., 2020). We found distinct GER groups, uniquely characterized by marker gene expression combinations in clusters S2, S3, and S4 (Figures 3A and 5; Table S4). These distinctive GER groups were additionally confirmed with a different clustering method and independently performed analyses (Figures S5A and S5C). We noticed that the gene expression profile of the most lateral GER group that borders the region around the inner hair cells has the highest similarity with neonatal inner border and inner phalangeal cells (clusters S4/S5, Figure 3A; cluster 3/cluster 2, Figure S5A). This raises the open question of whether P2 inner border and inner phalangeal cells have a similar organoid-forming potential as GER cells; the answer to this question will require innovative FACS gating strategies. The GER lies outside of the zone of non-proliferating cells that signifies the developing organ of Corti from middle embryonic ages onward (Chen et al., 2002), which suggests more fundamental differences between organ of Corti supporting cells and GER cells.
The competence of GER cells to robustly proliferate and generate large organoids was observed only when we cultured the cells at a higher density. At lower cell densities, GER cells displayed reduced viability, and the generation of organoids occurred only rarely. Interestingly, the few organoids that grew in low-density GER cultures were large, which suggests that individual GER cells have the potential for robust proliferation, but that higher density is required to unlock this potential in more cells. Our experiments suggest that cell-cell contact is essential for robust activation of this feature, which could occur through increased GER cell survival, direct stimulation of cell proliferation, or a combination of both. GER cell-derived large organoids harbored hair cell marker-positive cells that were associated with supporting cell marker-expressing cells. This is a strong indication that GER cells, after mitotic expansion, have otic pro-sensory cell characteristics. This may be an inherent feature of this neonatal cell group, because GER cells can also generate hair cell-like cells following Atoh1 expression (Gubbels et al., 2008; Shou et al., 2003; Woods et al., 2004). These observations and the evidence provided by our study show that GER cells are a distinctive cochlear cell group characterized by responsiveness to various reprogramming strategies, including phenotypic conversion (via Atoh1 expression) and proliferation/de-differentiation (via EFI_CVPM).
GER cell-derived organoids display several unique features that differ from organoids generated from other cochlear non-sensory cell origin. Most prominent is the observation that removal of growth factors impairs the efficacy of hair cell formation in GER organoids. We noticed this when GER organoids were cultured without EFI, with the intention to induce hair cell differentiation, and we found that hair cell marker-expressing cells did not occur in these conditions. This is in contrast with experiments where organoids were grown from non-GER cells, in the presence of growth factors but without CVPM (Li et al., 2003a; Oshima et al., 2007; White et al., 2006). Growth factor-dependent sustained proliferation and massive expansion, therefore, are distinct features of GER organoids that appear to be required for the robust generation of hair cells and supporting cells. This GER organoid feature also varies from organoids generated from pluripotent stem cells that require growth factor withdrawal and self-guided differentiation for efficient hair cell differentiation (Koehler et al., 2013, 2017; Li et al., 2003b; Oshima et al., 2010).
One additional conclusion that we draw from our experiments is that the in vitro proliferative capacity of different non-sensory cochlear cell types depends, not surprisingly, on the composition of media supplements. For example, various supporting cell groups, including pillar and Deiters’ cells, show organoid formation potential when grown at low density in media supplemented with growth factors (Oshima et al., 2007; Sinkkonen et al., 2011; White et al., 2006), but proliferation and ability of these supporting cells to grow into organoids larger than a few cells are severely reduced when grown in EFI_CVPM. Specifically, pillar cells and Deiters’ cells did not proliferate robustly in these conditions, but the cells were not only viable in EFI_CVPM-supplemented media but upregulated hair cell markers. Comparison of single-cell transcriptomic profiles revealed that of all supporting cell subtypes, inner pillar cells were closest in gene expression to hair cells, which might explain this observation.
Limitations of study
Our experiments revealed a distinct feature of GER cells to generate sizeable inner ear organoids. The strong potential of the EFI_CVPM supplement has been previously reported (McLean et al., 2017), and our findings imply that the principal target population of this treatment is GER cells. We show that other cochlear supporting cell types differ in their organoid formation ability. Expansion of the FACS strategy with additional reporter lines and markers would allow a quantitative assessment of each supporting cell subtype, which remains a desirable goal. Moreover, single-cell RNA-seq of organoids derived from GER cells and organoids obtained with the other cochlear supporting cell groups would allow for in-depth characterization and comparison of the different progenitor cell groups’ composition. We envision to pursue this exciting question in the near future, which could provide additional insight into the activation of GER cells’ proliferative potential.
Stringent FACS gating, deep RNA-seq, robust clustering, and immunohistochemical validation resulted in the identification of cochlear cell populations that were in close agreement with results obtained with shallow sequencing but many more cells (Kolla et al., 2020). Of course, the differences between technologies used in the different studies come with distinct limitations, such as the numbers of genes associated with individual cells that could affect clustering outcomes. Integration into the gEAR data depository (Orvis et al., 2020) provides an instrument for directly comparing the datasets. We found that cell density is an important variable for activating GER cell growth, and that direct cell contact is essential. The nature of the signal that mediates this effect remains to be identified. The transcriptomic analysis revealed that the GER consists of at least three distinct cell groups, and that all three groups are receptive to EFI_CVPM-based activation. In summary, our study reveals that GER cells are a potent cochlear cell type that can be utilized for cell-based in vitro assays for inner ear research. We provide mechanistic insight for culture conditions that result in efficient organoid formation and robust generation of cells expressing hair cell and supporting cell markers.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to the Lead Contact, Stefan Heller (hellers@stanford.edu).
Materials availability
The study did not generate new unique reagents.
Data and code availability
The RNA sequencing data generated in this paper is available from GEO with accession number GSE162308. The data are also available at gEAR, a gene Expression Analysis Resource (Orvis et al., 2020), via PERMA-LINK https://umgear.org//index.html?layout_id=afd2eb77&gene_symbol_exact_match=1. In gEAR, it is possible to project each gene’s expression value into the tSNE plot shown in Figure 3A, into a cochlear illustration with all cell types identified (based on Figures 2A–2C), as well as violin plots concordant with Table S3.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
FVB/NJ mice (The Jackson Laboratory: 001800) were used for organoid formation assays in Figures 1 and 7; Figure S1). Sox2-GFP (The Jackson Laboratory: 017592), Lfng-GFP (MMRRC_015881-UCD), Fgfr3-CreERT2 (Young et al., 2010), and Ai14-tdTomato (The Jackson Laboratory: 007908) mice were used for the flow cytometric isolation of cochlear cell subtypes (Figures 2A–2F; Figures S2 and S7A). Recombination of the Stop-tdTomato allele in Fgfr3-tdTomato/Sox2-GFP mice was done with intraperitoneal tamoxifen injection (0.2 mg/g) (Sigma) at P0. We used two-days old mice of both sexes for all experiments. Procedures involving animals were approved by Stanford University’s Institutional Animal Care and Use Committee. Animal experiments were done in conformity to the NIH criteria described in ‘The Guide for the Care and Use of Laboratory Animals’ and observed the Cartagena Protocol.
METHOD DETAILS
Cochlear cell isolation
Temporal bones were removed from P2 mice and cochlear ducts were dissected. The spiral ligament and stria vascularis were removed from each cochlear duct. Cochlear ducts were washed twice with potassium and magnesium-free PBS and incubated in 0.25% Trypsin-EDTA (GIBCO) at 37°C and 5% CO2 for 10 min. After adding soybean trypsin inhibitor (10 mg/mL) (GIBCO) and DNaseI (1 mg/mL) (Stemcell technologies), the cochlear duct tissue was mechanically dissociated and passed through a 70 μm strainer to remove aggregates.
Organoid culture
An overview of the organoid culture procedure is shown in Figure 1A. Cochlear duct cells derived from P2 mice were seeded at 50 cells per μL density into suspension culture dishes and maintained in DMEM/F-12 (GIBCO) supplemented with N-2 Supplement (GIBCO), B-27 Supplement (GIBCO), and 100 μg/mL ampicillin. Growth factors, small molecules, and recombinant proteins were supplemented into the medium throughout the 21 days culture period at 37°C and 5% CO2. Half of the medium was changed every other day. At day 7, the organoids were evaluated for number, size, and morphology. For adherent culture, the organoids were harvested on day 7 and transferred to Lab-Tek Chamber Slides (Nunc), pre-coated with Growth Factor Reduced Basement Membrane Matrix (Matrigel, Corning) at 1:10 dilution. The attached organoids were maintained for another 14 days at 37°C and 5% CO2. The colonies were evaluated for differentiation on day 21. In the procedures involving FACS-sorting, the cochlear duct cells were sorted into a 96-well suspension culture dish at either 2.5 cells/μL or 10 cells/μL densities and maintained in DMEM/F-12 supplemented with N-2 Supplement, B-27 Supplement, and 100 μg/mL ampicillin. In addition, growth factors (EGF, FGF2, IGF1) and small molecules (CHIR99021, valproic acid, 2-phospho-L-ascorbic acid, and TGFß receptor inhibitor 616452) (= EFI_CVPM) (McLean et al., 2017) were added to the medium. The sorted cells were incubated for 7 days at 37°C and 5% CO2 and evaluated for organoid formation and proliferation (Figures 2G–2L). The organoids were then transferred into Lab-Tek Chamber Slides pre-coated with Matrigel, followed by incubation for another 14 days at 37°C and 5% CO2. On day 21, the colonies were evaluated for differentiation (Figures 4 and 6; Figures S6 and S7). Quantifications shown are derived from 3–5 experiments; each series of experiments was conducted at least three times independently.
Growth factors, small molecules, and recombinant proteins were added to the medium at the following working concentrations: EGF (20 ng/mL), FGF2 (10 ng/mL), IGF1 (50 ng/mL), CHIR99021 (3 μM), valproic acid (500 μM), 2-phospho-L-ascorbic acid (100 μg/mL), TGFß receptor inhibitor 616452 (2 μM), heparan sulfate (50ng/ml), Noggin (100ng/ml), R-spondin-1 (500ng/ml), Trichostatin A (0.03 μM), Reversine (0.03 μM), NXT1219 (3 μM), Paullone (3 μM), DAPT (1 μM), Smoothened Agonist (0.3 μM), Vismodegib (3 μM).
Flow cytometry
Dissociated cochlear duct cells were collected in round-bottom tubes and washed twice with phenol red-free DMEM/F-12 medium (GIBCO). Each time, the cells were pelleted at 300 × g for 5 min and gently resuspended in medium. Debris was excluded based on scatter parameters (forward-scatter area [FSC-A] versus side-scatter area [SSC-A]). Singlets (individual cells) were discriminated based on physical parameters via two gating steps: FSC-A versus forward-scatter height [FSC-H] and side-scatter height [SSC-H] versus side-scatter width [SSC-W]. CYTOX Red cell viability marker (Invitrogen) was used for dead cell elimination (Figure S2B). Sox2-GFP expressing cells (G_Sox2) (Figure 2D), and Lfng-GFP expressing and Lfng-GFP-negative cells (G_Lfng and G_Lfng (−)) (Figure 2E; Figure S7A) were sorted individually from Sox2-GFP and Lfng-GFP mice-derived cochlear duct cells. G1-G4 cell populations were sorted from Fgfr3-tdTomato/Sox2-GFP mice-derived cochlear duct cells based on different intensities of GFP and tdTomato fluorescence (Figure 2F) (Waldhaus et al., 2015). FVB/NJ mice-derived cochlear duct cells were used as negative controls to validate GFP fluorescent gate settings for sorts involving Sox2-GFP and Lfng-GFP cells. Non-recombinant Fgfr3-CreERT2/Ai14-tdTomato mice-derived cochlear duct cells were used to adjust the gate settings for G1-G4 (Figure S2C). Sorting was performed with a FACS Aria II (BD Biosciences). The cells were sorted into 96-well suspension culture dishes for cell cultures or 96-well lysis buffer plates for single-cell RNA-seq.
Immunohistochemistry (IHC)
Colonies grown in Lab-Tek Chamber Slides (Figures 1E, 4, and 6; Figures S6 and S7B) were rinsed twice with PBS, fixed with 4% paraformaldehyde for 30 min, and washed three times for 15 min with PBS. 0.5% Triton X-100 in PBS for 2 h was used to permeabilize cells, and blocking was done with 10% fetal bovine serum, 0.5% Triton X-100, 0.05% NaN3 in PBS for 1 h, followed by incubation with the primary antibodies in blocking buffer overnight at 4°C. The colonies were then washed three times for 15 min with PBS and incubated with the secondary antibodies, with or without Alexa Fluor 647-conjugated phalloidin (Invitrogen) and DAPI in blocking buffer at room temperature for 1 h. After washing three times for 15 min with PBS, the media chambers were removed from the Chamber Slides, and the glass slides were sealed with ProLong Diamond Antifade mounting media (Invitrogen) and coverslipped.
For validation of marker gene expression (Figure 3B), whole inner ears were dissected from P2 FVB/NJ mice and fixed with 4% paraformaldehyde for 2 h. After washing three times for 15 min with PBS, the tissues were embedded in 65°C 4% Low Melt Agarose (Bio-Rad) in dH2O. The tissue blocks were cooled until the agarose solidified, and 100 μm sections were cut using a vibratome (VT100S, Leica). The sections were permeabilized for 30 min with 0.5% Triton X-100 in PBS and blocked with 1% BSA, 0.2% Triton X-100 in PBS for 1 h. Incubation with the primary antibodies in blocking buffer was done overnight at 4°C. The sections were then washed three times for 15 min with PBS and incubated with the secondary antibodies, Alexa Fluor 647-conjugated phalloidin, and DAPI in blocking buffer at room temperature for 1 h. After washing three times for 15 min with PBS, the sections were mounted on glass slides using ProLong Diamond Antifade mounting media, Secure-Seal Spacer (Invitrogen), and sealed with coverslips.
Confocal microscopy
Cochlear sections and colonies were imaged with an LSM880 confocal laser scanning microscope (ZEISS) and processed with ZEN (ZEISS) software. Whole chamber images were obtained by using the automated tiling tool of the LSM880.
Cell proliferation assay
CellTiter 96 Aqueous One Solution Cell Proliferation Assay System (Promega) was used to comparatively assess cell mass increases in organoid cultures. 20 μL CellTiter 96 Aqueous One Solution reagent was added per 100 μL culture media and incubated at 37°C and 5% CO2 for 4 h. Absorbance at 490 nm was measured using a plate reader (Infinite M1000, Tecan). The average of six replicate wells per condition was used as one data point; at least three independent experiments were conducted.
Single-cell RNA-seq library preparation and sequencing
Single cells were collected from P2 Fgfr3-tdTomato/Sox2-GFP and Lfng-GFP mice’ cochlear ducts and sorted into 96-well plates pre-filled with lysis buffer (2U/μL RNase inhibitor, 0.2% (vol/vol) Triton X-100, 2.5 μM modified Oligo-dT primer (5′- AAG CAG TGG TAT CAA CGC AGA GTA CTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TVN - 3′). Reverse transcription was performed with SMARTScribe Reverse Transcriptase (Clontech) followed by cDNA synthesis according to the SmartSeq2 protocol (Picelli et al., 2014). Synthesized cDNA was purified with AMPure XP beads (Beckman Coulter) and then quantified and assessed for size distribution with the HS NGS Fragment Kit (1–6000bp) (Agilent) and a fragment analyzer (Advanced Analytical Technologies, Inc.). Samples with more than 0.05 ng/μL of cDNA were selected using PACTAN software (Sinha et al., 2017). 98.7% of the samples passed the criteria and were concentration-normalized and tagmented with the Nextra XT (Illumina) sample preparation system, adding cell-specific barcodes to the fragments. The samples were amplified by PCR and purified with AMPure XP beads. Purified cDNA library was quality assessed with a Bioanalyzer 2100 (Agilent). Sequencing was performed on an Illumina NextSeq 500 high output flow cell configuration with paired-end sequencing of 150 bp.
Sequencing data processing
Sequenced reads were aligned to the mouse reference genome release GECm38 mm10, GENCODE annotation vM17, using custom scripts on the Sherlock Supercomputer Cluster (Stanford). STAR aligner was used to map the sequencing raw reads. RSEM was used to quantify the transcriptome Bam files. Count matrices were loaded into R and processed with the scater package version 1.14.6. (McCarthy et al., 2017). 537 cells were sequenced, yielding 52,732 features (genes). Per-cell library size, number of detected genes, mitochondrial genes, and spike-in control (ERCCs) percentages were identified using the perCellQCMetrics function (Lun et al., 2016) and 92 low quality cells were excluded. Batch effects from metadata - gate and plate ID - were computed, and an additional set of 36 non-conforming cells were excluded. Spike-in ERCCs and genes expressed in fewer than 3 cells were excluded, resulting in a total 409 cells and 22,000 features (Figure S3). The data were normalized with the SCnorm package version 1.8.2 (Bacher et al., 2017). CellTrails package version 1.4.0 (Ellwanger et al., 2018) was used for feature selection, and 11 states (clusters) were identified and visualized with tSNE plots (Figure 3A). Trajectory maps were created using the spatiotemporal trajectory inference function of CellTrails (Figure 3F). In parallel, we used the M3Drop package version 1.12.0 (Andrews and Hemberg, 2019) for variable feature selection and identified 2,457 differentially expressed genes with a False Discovery Rate (FDR) of less than 1%. These genes were used for performing principal component analysis. Eight principal components were chosen by identifying an elbow point indicating a gap invariance along successively plotted variance values. Graph-based clustering analysis using scater identified seven clusters at k = 10 that were visualized with tSNE plots (Figure S5A). The cells representing Deiters’ and outer pillar cells (cluster 4) and GER cells (clusters 1 and 3) were individually extracted and analyzed for differentially expressed genes. M3Drop identified 1,123 genes and 551 differentially expressed genes, respectively, at 5% FDR. Cluster 4 and Cluster 1 and 3 were both divided into three sub-clusters at k = 5 and k = 10, respectively (Figures S5C and S5E). The relationship between clusters was visualized in a force-based layout based on the log-ratio of high-weight paths between clusters (Wolf et al., 2019) (Figure S5I). Wilcoxon rank-sum test was used for the identification of differentially expressed genes or marker genes among clusters with an FDR of less than 1% (Figure 5; Tables S2, S3, and S4).
Organoid culture in methylcellulose
Dissociated cochlear duct cells derived from P2 FVB/NJ mice were serially plated in a 12-well dish at 0.5 × 104 cells (5 cells/μL), 1.0 × 104 cells (10 cells/μL), and 2.0 × 104 cells (20 cells/μL) per a well and maintained in DMEM/F-12 containing 1.27% methylcellulose supplemented with N-2 Supplement, B-27 Supplement, 100 μg/mL ampicillin, and EFI_CVPM. Methylcellulose Stock Solution (R&D Systems) was added to each cell suspension to a final concentration of 1.27%. The cells were incubated at 37°C and 5% CO2 for 7 days, and the number of organoids was quantified (Figure 7A).
Transwell co-culture
Individual wells of a 24-well dish were separated into upper and lower compartments with Millicell Cell Culture Inserts of 40 μm pore size (Millipore). Dissociated cochlear duct cells derived from P2 FVB/NJ mice were passed through 40 μm strainers twice and serially plated into lower compartments at 0.25 × 104 cells (3.3 cells/μL), 0.5 × 104 cells (6.7 cells/μL), and 1.0 × 104 cells (13.3 cells/μL) per well into 750 μL medium. For each cell density, the upper compartment either received 2.0 × 104 cells or no cells. The cells were incubated at 37°C and 5% CO2 for 7 days in DMEM/F-12 supplemented with N-2 Supplement, B-27 Supplement, 100 μg/mL ampicillin, and EFI_CVPM. On day 7, the number of organoids grown in the lower compartments was determined. Relative increases in cell numbers, a measure of proliferation, were determined in the lower compartments using CellTiter 96 Aqueous One Solution Cell Proliferation Assay system (Figure 7B).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed in R (version 3.6.1). Data are presented as mean ± standard deviation. Statistical details of experiments are provided in the figure legends. Statistical significance was performed using two-tailed unpaired Student’s t test. Differences were considered statistically significant when p < 0.05. Single-cell RNA-seq data analysis utilized the Mann-Whitney U test, also known as the Wilcoxon rank-sum test in R (wilcox.test), for ranking differential gene expression.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti Myosin-Vlla (1:1000) | Proteus | Cat# 25-6790; RRID:AB_10015251 |
| Goat anti SOX2 (1:200) | R&D Systems | Cat# AF2018; RRID:AB_355110 |
| Rabbit anti Neuropeptide Y (NPY) (1:50) | proteintech | Cat# 12833-1-AP; RRID:AB_10791890 |
| Rabbit anti Gelsolin (GSN) (1:200) | proteintech | Cat# 11644-2-AP; RRID:AB_2295090 |
| Goat anti Sparc-like 1 (SPARCL1) (1:500) | R&D Systems | Cat# AF2836; RRID:AB_2195097 |
| Rabbit anti CRABP1 (1:100) | proteintech | Cat# 12588-1-AP; RRID:AB_2292271 |
| Donkey anti-Rabbit IgG, Alexa Fluor 594 | Invitrogen | Cat# A-21207; RRID:AB_141637 |
| Donkey anti-Goat IgG, Alexa Fluor 488 | Invitrogen | Cat# A-11055; RRID:AB_2534102 |
| Donkey anti-Rabbit IgG, Alexa Fluor 488 | Invitrogen | Cat# A-21206; RRID:AB_2535792 |
| Donkey anti-Rabbit IgG, Alexa Fluor 647 | Invitrogen | Cat# A-31573; RRID:AB_2536183 |
| Alexa Fluor 647 Phalloidin | Invitrogen | Cat# A22287; RRID:AB_2620155 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat# D1306; RRID:AB_2629482 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse EGF | R&D systems | Cat# 2028-EG |
| Recombinant Mouse FGF basic (FGF2) | R&D systems | Cat# 3139-FB |
| Recombinant mouse IGF-1 | R&D systems | Cat# 791-MG |
| CHIR99021 | LC laboratories | Cat# C-6556 |
| Valproic Acid Sodium Salt | Sigma-Aldrich | Cat# P4543 |
| 2-Phospho-L-ascorbic acid trisodium salt | Sigma-Aldrich | Cat# 49752 |
| TGF-b RI Kinase Inhibitor II | Millipore | Cat# 616452 |
| Heparan sulfate sodium salt from bovine kidney | Sigma-Aldrich | Cat# H7640 |
| Recombinant Murine Noggin | Peprotech | Cat# 250-38 |
| Recombinant Murine R-spondin-1 | Peprotech | Cat# 315-32 |
| Trichostatin A | SelleckChem | Cat# S1045 |
| Reversine | Sigma-Aldrich | Cat# R3904 |
| NXT1219 | Inception Sciences | N/A |
| Paullone | Santa Cruz Biotechnology | Cat# sc-208152 |
| DAPT | Sigma-Aldrich | Cat# D5942 |
| Smoothened Agonist (SAG) | Sigma-Aldrich | Cat# 566660 |
| Vismodegib | SeleckChem | Cat# GDC-0449 |
| N-2 Supplement (100X) | GIBCO | Cat# 17502048 |
| B-27 Supplement (50X) | GIBCO | Cat# 17504044 |
| Matrigel | Corning | Cat# 356230 |
| Trypsin-EDTA (0.25%) | GIBCO | Cat# 25200-056 |
| Soybean Trypsin Inhibitor | GIBCO | Cat# 17075029 |
| DNaseI | Stemcell technologies | Cat# 07469 |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| SYTOX® Red dead cell stain | Invitrogen | Cat# S34859 |
| SMARTScribe Reverse Transcriptase | Clontech | Cat# 639538 |
| ERCC | Thermo Fisher Scientific | Cat# 4456653 |
| Recombinant ribonuclease inhibitor | Clontech | Cat# 2313A |
| 5x First Strand Buffer | Takara Bio | Cat# ST0062 |
| (Betaine solution) | Sigma-Aldrich | Cat# B0300 |
| Kapa HiFi HotStart ReadyMix | KAPA Biosystems | Cat# KK2602 |
| AMPure XP beads | Beckman Coulter | Cat# A63881 |
| ProLong Diamond Antifade mountant | Invitrogen | Cat# P36970 |
| Methylcellulose Stock Solution | R&D Systems | Cat# HSC001 |
| Critical commercial assays | ||
| Smart-seq2 | Picelli et al., 2014 | https://www.illumina.com/science/sequencing-method-explorer/kits-and-arrays/smart-seq2.html |
| HS NGS Fragment kit | Agilent | DNF-474 Fragment analyzer method: DNF-474-33 - HS NGS Fragment 1-6000bp.mthds |
| Nextra XT DNA Library Prep Kit | illumina | Cat# FC-131-1096 |
| High Sensitivity DNA kit | Agilent | Cat# 5067-4626 |
| CellTiter96 | Promega | Cat# G3582 |
| Deposited data | ||
| Single cell RNA-seq | NextSeq 500 (illumina) | GEO accession: GSE162308, gEAR: https://umgear.org/p?l=afd2eb77 |
| Experimental models: organisms/strains | ||
| FVB/NJ Mice | The Jackson Laboratory | Stock# 001800; MGI:2163709; RRID:IMSR_JAX:001800 |
| Sox2-GFP Mice | The Jackson Laboratory | Stock# 017592;MGI:5298206; RRID:IMSR_JAX:017592 |
| Lfng-GFP Mice | MMRRC | Stock# 015881-UCD; MGI:5003715; RRID:MMRRC_015881-UCD |
| Fgfr3-CreERT2 Mice | The Jackson Laboratory | Stock# 025809; MGI:5603119; RRID:IMSR_JAX:025809 |
| Ai14-tdTomato Mice | The Jackson Laboratory | Stock# 007908; MGI:3817869; RRID:IMSR_JAX:007908 |
| Oligonucleotides | ||
| TSO 5’-AAGCAGTGGTATCAACGCAG AGTACATrGrG | N/A | N/A |
| Oligo-dT 5’- AAG CAG TGG TAT CAA CGC AGA GTA CTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TVN - 3’ | N/A | N/A |
| ISPCR primer 5’-AAGCAGTGGTATCA ACGCAGAGT-3’ | N/A | N/A |
| Software and algorithms | ||
| Fiji/ImageJ | Fiji | https://fiji.sc; RRID:SCR_002285 |
| Zen Black | Zeiss | https://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html; RRID:SCR_018163 |
| Zen Blue | Zeiss | https://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html; RRID:SCR_013672 |
| PACTAN | Sinha et al., 2017 | N/A |
| FlowJo | BD | https://www.flowjo.com |
| AxioImager | Zeiss | https://www.zeiss.com/microscopy/us/products/light-microscopes/axio-imager-2-for-biology.html; RRID:SCR_018876 |
| R | R Project for Statistical Computing | https://www.r-project.org; RRID:SCR_001905 |
| SCnorm R package | Bioconductor Bacher et al., 2017 | https://bioconductor.org/packages/release/bioc/html/SCnorm.html |
| CellTrails R package | Bioconductor Ellwanger et al., 2018 | https://www.bioconductor.org/packages/devel/bioc/html/CellTrails.html |
| Scater R package | Bioconductor McCarthy et al., 2017 | https://bioconductor.org/packages/release/bioc/html/scater.html; RRID:SCR_015954 |
| M3Drop R package | Bioconductor Andrews and Hemberg, 2019 | https://www.bioconductor.org/packages/release/bioc/html/M3Drop.html |
| ggplot2 R package | (Wickham, 2016) | https://cran.r-project.org/web/packages/ggplot2/index.html; RRID:SCR_014601 |
Highlights.
Efficient culture conditions for the generation of inner ear organoids
Single-cell RNA-seq resource of all principal neonatal cochlear cell groups
Greater epithelial ridge cells display robust potential to generate otic organoids
Organoid formation is synergistically enhanced by direct cell-cell contact
ACKNOWLEDGMENTS
We thank P.K. Lee and A. Ling for their technical help, S.P. Monti for inspiring illustrations, C. Carswell-Crumpton for providing training and assistance, S. Sim and M.C. Yee for single-cell library preparation and sequencing support, Y. Song and R. Hertzano for expert assistance with gEAR, and the members of the Heller laboratory and R. Hertzano for comments on the manuscript. We also thank T. Nakagawa for support and advice. We acknowledge the Institute for Stem Cell Biology and Regenerative Medicine’s FACS Core facility, the Stanford Shared FACS Facility, the Stanford Functional Genomics Facility, the Stanford Research Computing Center, the Stanford Animal Care Facility, and the Otolaryngology Imaging Core. This work was supported by National Institutes of Health Grants R01DC015201 (to S.H.) and T32DC015209 (to T.A.J.), by the Hearing Health Foundation’s Hearing Restoration Project, and through the Stanford Initiative to Cure Hearing Loss by generous gifts from the Evslin Family Foundation and the Bill and Susan Oberndorf Foundation. M.K. was supported in part by the Japan Society for the Promotion of Science, the Uehara Memorial Foundation, and the Soda Toyoji Memorial Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108646.
DECLARATION OF INTERESTS
S.H. is a paid consultant of Pipeline Therapeutics.
REFERENCES
- Andrews TS, and Hemberg M (2019). M3Drop: dropout-based feature selection for scRNASeq. Bioinformatics 35, 2865–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Chu LF, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, and Kendziorski C (2017). SCnorm: robust normalization of single-cell RNA-seq data. Nat. Methods 14, 584–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, and Hayashi T (2006). Expression of Prox1 during mouse cochlear development. J. Comp. Neurol 496, 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhall NF, Shi F, Arnold K, Hochedlinger K, and Edge AS (2014). Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Reports 2, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigande JV, and Heller S (2009). Quo vadis, hair cell regeneration? Nat. Neurosci 12, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Kelly MC, Hoa M, Morell RJ, and Kelley MW (2015). Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun 6, 8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Xia A, Wang T, Jan TA, Hayashi T, Bermingham-McDonogh O, and Cheng AG (2011). Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J. Assoc. Res. Otolaryngol 12, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, Liu Z, Taketo MM, Oghalai JS, Nusse R, et al. (2012). Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. USA 109, 8167–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, and Segil N (2002). The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505. [DOI] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, Chalasani K, Steigelman KA, Fang J, Rubel EW, et al. (2014). Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaratne MW, Vlajkovic SM, Lipski J, and Thorne PR (2014). Kölliker’s organ and the development of spontaneous activity in the auditory system: implications for hearing dysfunction. BioMed Res. Int 2014, 367939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR, Peterson JM, and Wong GW (2010). Mammalian Otolin: a multimeric glycoprotein specific to the inner ear that interacts with otoconial matrix protein Otoconin-90 and Cerebellin-1. PLoS ONE 5, e12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber M, Oshima K, and Heller S (2009). Stem/progenitor cells derived from the cochlear sensory epithelium give rise to spheres with distinct morphologies and features. J. Assoc. Res. Otolaryngol 10, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, White P, Lee YS, Groves A, and Segil N (2006). Prospective identification and purification of hair cell and supporting cell progenitors from the embryonic cochlea. Brain Res. 1091, 282–288. [DOI] [PubMed] [Google Scholar]
- Ellwanger DC, Scheibinger M, Dumont RA, Barr-Gillespie PG, and Heller S (2018). Transcriptional Dynamics of Hair-Bundle Morphogenesis Revealed with CellTrails. Cell Rep. 23, 2901–2914.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritti A, Parati EA, Cova L, Frolichsthal P, Galli R, Wanke E, Faravelli L, Morassutti DJ, Roisen F, Nickel DD, and Vescovi AL (1996). Multipotential stem cells from the adult mouse brain proliferate and self-renew in response to basic fibroblast growth factor. J. Neurosci 16, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, and Brigande JV (2008). Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 455, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, and Bermingham-McDonogh O (2008). Fgf20 is required for sensory epithelial specification in the developing cochlea. J. Neurosci 28, 5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Younkins C, and Bermingham-McDonogh O (2010). Expression patterns of FGF receptors in the developing mammalian cochlea. Dev. Dyn 239, 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Gwilliam K, Rose K, Milon B, and Matern MS (2020). Cell Type-Specific Expression Analysis of the Inner Ear: A Technical Report. Laryngoscope, Published online June 24, 2020. 10.1002/lary.28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Lu J, Chiang H, Wu H, Edge AS, and Shi F (2016). Diphtheria Toxin-Induced Cell Death Triggers Wnt-Dependent Hair Cell Regeneration in Neonatal Mice. J. Neurosci 36, 9479–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Sage C, Tang Y, Lee SG, Petrillo M, Hinds PW, and Chen ZY (2011). Overlapping and distinct pRb pathways in the mammalian auditory and vestibular organs. Cell Cycle 10, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick AS, and Heller S (2019). Stem Cells and the Bird Cochlea-Where Is Everybody? Cold Spring Harb. Perspect. Med 9, a033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman DS, Hanson ET, Lewis RL, Auerbach R, and Thomson JA (2001). Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 98, 10716–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Ibrahim LA, Wang SZ, Yuan W, Evgrafov OV, Knowles JA, Wang K, Tao HW, and Zhang LI (2016). EphA7 regulates spiral ganglion innervation of cochlear hair cells. Dev. Neurobiol 76, 452–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, and Hashino E (2014). 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat. Protoc 9, 1229–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Mikosz AM, Molosh AI, Patel D, and Hashino E (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Nie J, Longworth-Mills E, Liu XP, Lee J, Holt JR, and Hashino E (2017). Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol 35, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, Palermo AT, So KS, Mays JC, Orvis J, et al. (2020). Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun 11, 2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Joncour V, Filppu P, Hyvönen M, Holopainen M, Turunen SP, Sihto H, Burghardt I, Joensuu H, Tynninen O, Jääskeläinen J, et al. (2019). Vulnerability of invasive glioblastoma cells to lysosomal membrane destabilization. EMBO Mol. Med 11, e9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu H, and Heller S (2003a). Pluripotent stem cells from the adult mouse inner ear. Nat. Med 9, 1293–1299. [DOI] [PubMed] [Google Scholar]
- Li H, Roblin G, Liu H, and Heller S (2003b). Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. USA 100, 13495–13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Corrales CE, Wang Z, Zhao Y, Wang Y, Liu H, and Heller S (2005). BMP4 signaling is involved in the generation of inner ear sensory epithelia. BMC Dev. Biol 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, and Li H (2015). Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. USA 112, 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass JC, Gu R, Cai T, Wan YW, Cantellano SC, Asprer JS, Zhang H, Jen HI, Edlund RK, Liu Z, and Groves AK (2016). Transcriptomic Analysis of Mouse Cochlear Supporting Cell Maturation Reveals Large-Scale Changes in Notch Responsiveness Prior to the Onset of Hearing. PLoS ONE 11, e0167286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgrange B, Belachew S, Thiry M, Nguyen L, Rogister B, Alvarez ML, Rigo JM, Van De Water TR, Moonen G, and Lefebvre PP (2002). Proliferative generation of mammalian auditory hair cells in culture. Mech. Dev 112, 79–88. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Li C, and Urness LD (2013). Genetic rescue of Muenke syndrome model hearing loss reveals prolonged FGF-dependent plasticity in cochlear supporting cell fates. Genes Dev. 27, 2320–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu P, Romero MR, Hilton H, Parker A, Townsend S, Kikkawa Y, and Brown SD (2010). Gelsolin plays a role in the actin polymerization complex of hair cell stereocilia. PLoS ONE 5, e11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun AT, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean WJ, McLean DT, Eatock RA, and Edge AS (2016). Distinct capacity for differentiation to inner ear cell types by progenitor cells of the cochlea and vestibular organs. Development 143, 4381–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean WJ, Yin X, Lu L, Lenz DR, McLean D, Langer R, Karp JM, and Edge ASB (2017). Clonal Expansion of Lgr5-Positive Cells from Mammalian Cochlea and High-Purity Generation of Sensory Hair Cells. Cell Rep. 18, 1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado Lagarde MM, Wan G, Zhang L, Gigliello AR, McInnis JJ, Zhang Y, Bergles D, Zuo J, and Corfas G (2014). Spontaneous regeneration of cochlear supporting cells after neonatal ablation ensures hearing in the adult mouse. Proc. Natl. Acad. Sci. USA 111, 16919–16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, and Fekete DM (2016). Notch-Wnt-Bmp crosstalk regulates radial patterning in the mouse cochlea in a spatiotemporal manner. Development 143, 4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, and Chen CS (2002). Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 514, 238–242. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, and Groves AK (2010). BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J. Neurosci 30, 15044–15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J, Gottfried B, Kancherla J, Adkins RS, Song Y, Dror AA, Olley D, Rose K, Chrysostomou E, Kelly MC, et al. (2020). gEAR: gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. bioRxiv. 10.1101/2020.08.28.272039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, Géléoc GS, Edge A, Holt JR, and Heller S (2007). Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J. Assoc. Res. Otolaryngol 8, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, and Heller S (2010). Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 141, 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, et al. (2015). Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Roccio M, and Edge ASB (2019). Inner ear organoids: new tools to understand neurosensory cell development, degeneration and regeneration. Development 146, dev177188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarajeewa A, Jacques BE, and Dabdoub A (2019). Therapeutic Potential of Wnt and Notch Signaling and Epigenetic Regulation in Mammalian Sensory Hair Cell Regeneration. Mol. Ther 27, 904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal RP, Akil O, Yi E, Weber CM, Grant L, Yoo J, Clause A, Kandler K, Noebels JL, Glowatzki E, et al. (2008). Sensorineural deafness and seizures in mice lacking vesicular glutamate transporter 3. Neuron 57, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Kempfle JS, and Edge AS (2012). Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J. Neurosci 32, 9639–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, and Edge AS (2013). Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. USA 110, 13851–13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, and Martin GR (2005). Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell 8, 553–564. [DOI] [PubMed] [Google Scholar]
- Shou J, Zheng JL, and Gao WQ (2003). Robust generation of new hair cells in the mature mammalian inner ear by adenoviral expression of Hath1. Mol. Cell. Neurosci 23, 169–179. [DOI] [PubMed] [Google Scholar]
- Sinha R, Stanley G, Gulati GS, Ezran C, Travaglini KJ, Wei E, Chan CKF, Nabhan AN, Su T, Morganti RM, et al. (2017). Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv. 10.1101/125724. [DOI] [Google Scholar]
- Sinkkonen ST, Chai R, Jan TA, Hartman BH, Laske RD, Gahlen F, Sinkkonen W, Cheng AG, Oshima K, and Heller S (2011). Intrinsic regenerative potential of murine cochlear supporting cells. Sci. Rep 1, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada H, Shiho O, Kuroshima K, Koyama M, and Tsukamoto K (1986). An improved colorimetric assay for interleukin 2. J. Immunol. Methods 93, 157–165. [DOI] [PubMed] [Google Scholar]
- Tateya T, Imayoshi I, Tateya I, Hamaguchi K, Torii H, Ito J, and Kageyama R (2013). Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development 140, 3848–3857. [DOI] [PubMed] [Google Scholar]
- Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, Tam LT, Espenel C, Ponnuswami A, Ni L, et al. (2019). Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhaus J, Durruthy-Durruthy R, and Heller S (2015). Quantitative High-Resolution Cellular Map of the Organ of Corti. Cell Rep. 11, 1385–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, and Segil N (2006). Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag; ). https://ggplot2.tidyverse.org, ISBN 978-3-319-24277-4. [Google Scholar]
- Wiwatpanit T, Lorenzen SM, Cantú JA, Foo CZ, Hogan AK, Márquez F, Clancy JC, Schipma MJ, Cheatham MA, Duggan A, and García-Añoveros J (2018). Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, and Theis FJ (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, and Kelley MW (2004). Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci 7, 1310–1318. [DOI] [PubMed] [Google Scholar]
- Wu PZ, O’Malley JT, de Gruttola V, and Liberman MC (2020). Age-related hearing loss is dominated by damage to inner ear sensory cells, not the cellular battery that powers them. J. Neurosci 40, 6357–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, and Richardson WD (2010). An Fgfr3-iCreER(T2) transgenic mouse line for studies of neural stem cells and astrocytes. Glia 58, 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Martin GV, Kelley MW, and Gridley T (2000). A mutation in the Lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr. Biol 10, 659–662. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo L, Lu X, Cheng C, Sun S, Li W, Zhao L, Lai C, Zhang S, Yu C, et al. (2018). Characterization of Lgr6+ Cells as an Enriched Population of Hair Cell Progenitors Compared to Lgr5+ Cells for Hair Cell Generation in the Neonatal Mouse Cochlea. Front. Mol. Neurosci 11, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Shen W, He DZ, Long KB, Madison LD, and Dallos P (2000). Prestin is the motor protein of cochlear outer hair cells. Nature 405, 149–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated in this paper is available from GEO with accession number GSE162308. The data are also available at gEAR, a gene Expression Analysis Resource (Orvis et al., 2020), via PERMA-LINK https://umgear.org//index.html?layout_id=afd2eb77&gene_symbol_exact_match=1. In gEAR, it is possible to project each gene’s expression value into the tSNE plot shown in Figure 3A, into a cochlear illustration with all cell types identified (based on Figures 2A–2C), as well as violin plots concordant with Table S3.


