ABSTRACT
Human lifespan is now longer than ever and, as a result, modern society is getting older. Despite that, the detailed mechanisms behind the ageing process and its impact on various tissues and organs remain obscure. In general, changes in DNA, RNA and protein structure throughout life impair their function. Haematopoietic ageing refers to the age-related changes affecting a haematopoietic system. Aged blood cells display different functional aberrations depending on their cell type, which might lead to the development of haematologic disorders, including leukaemias, anaemia or declining immunity. In contrast to traditional bulk assays, which are not suitable to dissect cell-to-cell variation, single-cell-level analysis provides unprecedented insight into the dynamics of age-associated changes in blood. In this Review, we summarise recent studies that dissect haematopoietic ageing at the single-cell level. We discuss what cellular changes occur during haematopoietic ageing at the genomic, transcriptomic, epigenomic and metabolomic level, and provide an overview of the benefits of investigating those changes with single-cell precision. We conclude by considering the potential clinical applications of single-cell techniques in geriatric haematology, focusing on the impact on haematopoietic stem cell transplantation in the elderly and infection studies, including recent COVID-19 research.
KEY WORDS: Haematopoietic ageing, Single-cell multiomics, Single-cell epigenomics, Single-cell transcriptomics
Summary: We discuss the cellular changes that occur during haematopoietic ageing at the different molecular levels, and provide an overview of the benefits of investigating those changes with single-cell precision.
Introduction
Since 2009, access to single-cell techniques has broadened from a few expert laboratories to research groups around the world (Lim et al., 2020). This shift was possible due to the commercialisation of these technologies (e.g. 10× Genomics, Pacific Biosciences, Illumina) and resulted in an increased application of single-cell techniques to study biological phenomena, from basic research (Canu et al., 2020; Farnsworth et al., 2020; Pijuan-Sala et al., 2020) through understanding intercellular interactions in the human body during homeostasis (James et al., 2020; Litviň uková et al., 2020; Park et al., 2020) to tracking disease-related changes (Martin et al., 2019; Nam et al., 2019; Vieira Braga et al., 2019). Haematology as a clinical discipline has been known as an early adopter of technological developments (Laurenti and Göttgens, 2018); therefore, it comes as little surprise that this branch of science quickly added single-cell techniques to its portfolio. Single-cell approaches provide the highest-resolution insight into the cellular makeup of complex systems possible, thus constituting a great tool for dissecting the dynamics of haematopoietic ageing. On a phenotypic level, haematopoietic ageing manifests through the loss of adaptive immunity, an increased rate of cytopaenias (see Glossary, Box 1) and an increased risk of developing myeloid malignancies (Gazit et al., 2008). Behind this phenotype lay molecular changes in the genome, transcriptome and epigenome. Single-cell technologies hold promise for a better understanding of ageing mechanisms by uncovering the changes happening in the body in a cell-dependent manner. This is important from a clinical pathology point of view, as connecting altered pathways with molecular changes in the context of cell types opens new avenues for possible pharmaceutical manipulation to reverse the ageing phenotype. In this Review, we focus on new insights gained from the application of single-cell technologies to study haematopoietic ageing over the past 5 years. The technical aspects of single-cell approaches are not covered here, as several excellent reviews on this topic are already available (Clark et al., 2016; Gawad et al., 2016; Mincarelli et al., 2018; Svensson et al., 2018). However, for a short overview of each technique please refer to Box 2.
Box 1. Glossary.
Clonal repertoire of adaptive lymphocytes: adaptive lymphocytes (T and B cells) are characterised by the presence of antigen-specific receptors, T cell receptors (TCRs) and B cell receptors (BCRs), respectively. Binding of an antigen to the receptor triggers an immune response. Cells from the same clone share the sequence of the receptor. Different clones have different antigen specificity. Therefore, higher variability of the clonal repertoire (T and B cells with distinct TCR/BCR sequences) provides protection against more pathogens.
CpG islands: parts of the genome characterised by the presence of large numbers of CG dinucleotide repeats.
Cytopaenia: deficiency in the number of mature blood cells.
Engraftment: ability of transplanted haematopoietic stem cells (HSCs) to contribute to the stable blood production in a new host upon transplantation.
Homing: ability of HSCs to migrate to bone marrow (BM), where they can contribute to the production of new blood cells.
Immunosenescence: changes in the immune system caused by ageing and resulting in the deterioration of the immune response.
Inflammaging: chronic low-grade inflammation typical of ageing.
Lineage bias: increased production of one or more blood lineages at the expense of the other lineages (e.g. aged HSCs were shown to produce more myeloid cells and fewer lymphoid cells compared to their young counterparts).
Long-term HSCs (LT-HSCs): subset of HSCs responsible for the long-term production of blood cells.
Peripheral blood mononuclear cells (PBMCs): population of blood cells with single nuclei (monocytes, lymphocytes, dendritic cells).
Poly(dT) primer: single-stranded sequence of deoxythymidine (dT), which anneals to the poly(A) tail of mRNA molecules and is used during the reverse transcription reaction.
Pseudotime analysis: method of single-cell trajectory analysis, which measures the progression of a cell through a biological process, e.g. differentiation.
Quiescence: ability of HSCs to stay dormant (not to divide).
Self-renewal: ability of HSCs to give rise to new HSCs without differentiation.
Box 2. Overview of single-cell techniques.
Single-cell DNA sequencing (scDNAseq): provides information about the DNA content of single cells. The most common parameters investigated using this approach include single-nucleotide variants (Lodato et al., 2015) and copy number variations (Knouse et al., 2016).
Single-cell RNA sequencing (scRNAseq): provides information about the transcriptome profile of a single cell. Polyadenylated mRNA is separated using oligo-dT primers and converted into complementary DNA (cDNA), followed by amplification, library construction and sequencing. Currently available scRNAseq methods can be roughly divided into plate-based and droplet-based techniques. One of the main differences between those techniques is throughput: the plate-based approach allows the analysis of up to hundreds of cells, whereas droplet-based assays interrogate up to tens of thousands of cells (Haque et al., 2017; Knouse et al., 2016).
Single-cell transposase-accessible chromatin sequencing (scATACseq): a method to study genome-wide chromatin accessibility in single cells. It allows the identification of regulatory regions of open chromatin throughout the genome. The method uses Tn5 transposase and inserts sequencing primers into regions of open chromatin (Chen et al., 2018).
Single-cell chromatin immunoprecipitation sequencing (scChIPseq): analyses DNA-protein interactions. Combination of chromatin immunoprecipitation with DNA sequencing allows the identification of binding sites of DNA-associated proteins. This method can capture repressive and active chromatin states (Grosselin et al., 2019; Rotem et al., 2015).
Single-cell bisulphite sequencing (scBSseq): provides information about the methylation profile of single cells. It measures DNA methylation at cytosine residues (5mC), an epigenetic mark critical in the regulation and maintenance of cell-type-specific transcriptional programs (Smallwood et al., 2014).
Single-cell mass cytometry (CyTOF): this method allows the analysis of the protein content of single cells using a combination of flow cytometry and mass spectrometry. CyTOF uses antibodies conjugated to rare heavy metal isotopes and measures its time of flight (TOF), a parameter characteristic to the atom's mass. The use of metal isotope-conjugated antibodies in lieu of fluorophore-conjugated ones overcomes the problem of spectral overlap in multicolour flow cytometry and allows for the measurements of over 40 parameters simultaneously (Gadalla et al., 2019; Kay et al., 2016).
Single-cell multiomics: the combination of usually two single-cell techniques (for example, scRNAseq and scATACseq) (Granja et al., 2019) to investigate multiple molecular features in the same cell (Hu et al., 2018).
Haematopoietic ageing
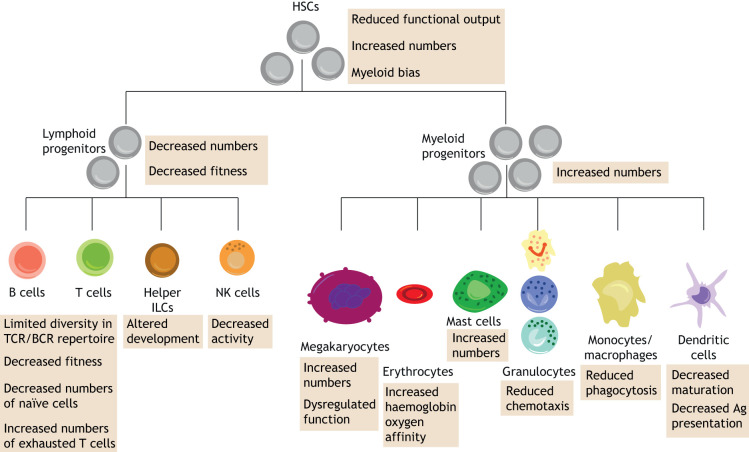
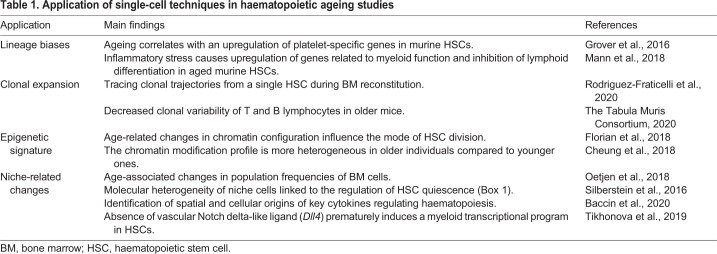
Ageing leads to numerous changes that affect multiple components of the body. The haematopoietic system consists of blood cells and their niche, the bone marrow (BM). The alterations in BM and haematopoietic stem cells (HSCs) caused by ageing disturb tissue homeostasis. A key discovery in HSC ageing research revealed that HSCs from old mice had only 25% of the efficiency of young HSCs in terms of homing (Box 1) and engraftment (Box 1) into the BM of a transplant recipient (Morrison et al., 1996). Since then, it has been shown that aged HSCs have reduced functional output, which is compensated for by their increased numbers (de Haan et al., 1997). Aged HSCs also have reduced lymphoid potential, whereas their myeloid potential (particularly granulopoiesis) is increased (Rossi et al., 2005; Sudo et al., 2000) (Fig. 1). Similarly, age-related changes also affect the BM. Some factors from the BM microenvironment (such as cytokines and enzymes) are crucial contributors to the ageing process (Frisch et al., 2019). Aged BM has a reduced capability of retaining HSCs in the niche, and increased mobilisation of HSCs was observed in older mice (Xing et al., 2006). Thus, changes related to ageing can be roughly classified as intrinsic (occurring within the blood cells themselves) and extrinsic (affecting the BM microenvironment). Single-cell technologies are increasingly contributing to a better understanding of the molecular mechanisms underlying haematopoietic ageing. Early studies within the field focused on an HSC-centric view of ageing (Grover et al., 2016; Kowalczyk et al., 2015) and are currently shifting towards a new emerging concept, which looks at haematopoietic ageing from the microenvironmental angle (Dorshkind et al., 2019; Ho et al., 2019; Maryanovich et al., 2018; Pioli et al., 2019). In general, haematopoietic studies applying single-cell technologies in the context of haematopoietic ageing can roughly be divided into four main areas: (1) lineage biases (Box 1) (Grover et al., 2016), (2) clonal expansion (Rodriguez-Fraticelli et al., 2020), (3) epigenetic signature (Cheung et al., 2018; Florian et al., 2018) and (4) niche-related changes (Baccin et al., 2020; Oetjen et al., 2018; Silberstein et al., 2016; Tikhonova et al., 2019) (Table 1).
Fig. 1.
Age-related changes affecting haematopoietic hierarchy. Age-related changes have different effects on haematopoietic cells depending on their lineage identity. Aged HSCs are characterised by decreased functional output, which is compensated for by their increased numbers. They also display myeloid bias (generating reduced numbers of lymphoid progeny). Haematopoietic ageing affects mature blood cells in a cell-type-specific manner. Depending on the cell type, it can impact cell numbers (B and T lymphocytes, megakaryocytes, mast cells), functional activity (B and T lymphocytes, NK cells, megakaryocytes, erythrocytes, granulocytes, monocytes/macrophages, dendritic cells) or development (helper ILCs, dendritic cells). Ag, antigen; BCR, B cell receptor; HSC, haematopoietic stem cell; ILC, innate lymphoid cell; NK, natural killer; TCR, T cell receptor.
Table 1.
Application of single-cell techniques in haematopoietic ageing studies
Single-cell techniques in geriatric haematology
Geriatric haematology is a great example of a clinical research field that can benefit from dissecting the changes that occur during haematopoietic ageing with single-cell precision. In this context, advances in single-cell techniques can shed new light on the dynamics of blood cell reconstitution upon transplantation in elderly recipients (Frick et al., 2019; Rodriguez-Fraticelli et al., 2020), provide information on how to improve immune system function and decrease the risk of developing age-associated myeloid malignancies (Giladi and Amit, 2018; Paguirigan et al., 2015), identify biomarkers of ageing (Ma et al., 2020) or help to develop rejuvenation strategies (Li et al., 2020).
Age-related changes in DNA
Accumulation of DNA damage is considered a hallmark of ageing. However, establishing the causal relationship between ageing and somatic mutations remains challenging. This is a field in which single-cell DNA sequencing can be successfully applied.
In general, most somatic mutations are characterised by their random nature and low abundance; therefore, population-level studies would predominantly highlight the germline genotype instead of de novo somatic mutations. Numerous somatic mutations accumulate during ageing. As blood is one of the most proliferative systems in the human body (Gondek and DeZern, 2020), it is also prone to acquiring somatic mutations at a higher rate. The presence of certain somatic mutations in HSCs adds to their fitness advantage and results in the clonal expansion of mutation-positive HSCs, which contributes to clonal haematopoiesis (Jaiswal et al., 2014). In healthy young individuals, all HSCs are equally capable of producing all of the mature blood cells, which maintains polyclonal haematopoiesis (Doulatov et al., 2012). By contrast, during age-related clonal haematopoiesis, a substantial proportion of mature blood cells is derived from a dominant HSC clone. It is estimated that at least 30% of all elderly people are affected by clonal haematopoiesis (Jaiswal et al., 2014; McKerrell et al., 2015; Young et al., 2016). The presence of clonal haematopoiesis is linked to an elevated risk of haematological malignancies and cardiovascular disease, contributing to an increased mortality rate overall (Steensma, 2018). Therefore, an in-depth understanding of somatic mutations that lead to clonal haematopoiesis is crucial (Arends et al., 2018; Mylonas et al., 2020; Ortmann et al., 2015), especially since it has been shown that the order in which mutations are acquired can influence the phenotypic manifestation of the pathology, including the response to targeted therapy and the biology of stem and progenitor cells (Ortmann et al., 2015).
Somatic mutations at single-cell resolution
Several methods that allow investigation of somatic mutations at single-cell level are now available (Albertí-Servera et al., 2020; Lareau et al., 2020; Nam et al., 2019; Rodriguez-Meira et al., 2019; Zhang et al., 2019). TARGET-seq combines single-cell whole transcriptome analysis with single-cell targeted locus genotyping, thus enabling researchers to link mutation status with a gene expression profile in heterogeneous tumour cell populations (Rodriguez-Meira et al., 2019). Similarly, application of single-cell DNA sequencing helped to reveal clonal heterogeneity of acute leukaemias (Albertí-Servera et al., 2020; Miles et al., 2020; Morita et al., 2020). Finally, single-cell methods that use mutations within mitochondrial DNA to infer clonal relationships between cells have also been reported (Lareau et al., 2020; Ludwig et al., 2019). Single-cell studies investigating somatic mutations in blood cells focus on the characterisation of mutational signatures and their impact on cellular functions (Nam et al., 2019; Zhang et al., 2019). Application of single-cell whole-genome sequencing has already provided detailed information about somatic mutations that affect B cells during the human lifespan (Zhang et al., 2019). This approach has allowed researchers to distinguish mutational signatures specific to development from age-related ones. It revealed that an age-related mutational signature correlates with the signature of B cell leukaemia, highlighting the role of ageing as a cancer risk factor (Zhang et al., 2019). Single-cell techniques can be also applied to investigate the functional consequences of somatic mutations. This is possible when the presence of a mutation affects a protein-coding region of the mRNA sequence (Nam et al., 2019; van Galen et al., 2019). The Landau group developed a novel Genotyping of Transcriptomes (GoT) method, which allows the distinguishing of mutation-positive from mutation-negative cells within the same cell type (Nam et al., 2019). Application of this method enables the direct comparison of transcriptional profiles between cells of the same identity (e.g. monocytes). It thus gives a unique opportunity to pinpoint the alterations in gene expression stemming from the presence of the mutation. Recent studies implicate an altered inflammation profile of monocyte-derived macrophages as one of the factors contributing to cardiovascular disease in elderly people with clonal haematopoiesis (Jaiswal and Ebert, 2019). Application of GoT in clonal haematopoiesis studies will provide an in-depth understanding of how the presence of such mutations affects inflammatory pathways. This knowledge could later be translated into new pharmaceutical strategies to alleviate the risk of cardiovascular disease in individuals affected by clonal haematopoiesis.
The transcriptome during ageing
Transcriptome analysis of different cell types provides valuable insight into their functional properties linked to their particular role in the body. Additionally, changes in a cell's microenvironment (such as inflammation) can also be detected as specific changes within the transcriptome (e.g. increased production of anti-inflammatory cytokines) (Butler et al., 2018; Hernández et al., 2018). Thus, the readout of the transcriptome is a good reflection of the cell state. Comparison of the transcriptomes between ‘young’ and ‘old’ cells within the same cell type coupled with functional assays therefore constitutes an excellent tool to dissect age-related changes in a cell's behaviour.
Age-related gene expression changes in HSCs
Among single-cell studies, large-scale single-cell transcriptomic projects are leading the way in providing valuable insight into haematopoietic ageing (Grover et al., 2016; Kowalczyk et al., 2015; Mann et al., 2018). One of the advantages offered by single-cell-level analysis in those settings is the power to determine whether alterations occurring during ageing are homogeneously distributed across each cell population or rather specific to a subset of cells within the population. Even before the single-cell era, the decline of HSC function during ageing was well documented (de Haan et al., 1997; Rossi et al., 2005; Sudo et al., 2000); however, the underlying molecular mechanisms were not yet fully understood. Single-cell RNA sequencing (scRNAseq) deciphers the gene expression changes in individual HSCs associated with the ageing phenotype, such as lineage bias, imbalance between self-renewal (Box 1) and differentiation, and declined engraftment in transplantation assays (Dykstra et al., 2011). The landmark studies in the field are the studies of Kowalczyk et al. (2015) and Grover et al. (2016), which were among the first ones investigating the differences between young (2- to 3-month-old) and old (>22-month-old) murine HSCs at single-cell resolution. These studies provided information about the transcriptional mechanisms behind the imbalance in blood cell production observed in ageing. This insight into gene expression showed that young and old HSCs are transcriptionally different enough to form separate clusters during the analysis. The most prominent differences were observed in the expression of cell cycle-related genes and genes characteristic for the megakaryocyte/platelet lineage (Grover et al., 2016; Kowalczyk et al., 2015). Interestingly, old long-term HSCs (LT-HSCs) (Box 1) were significantly under-represented among cells in the G1/S cell cycle phase, suggesting a more rapid transition through this phase (Kowalczyk et al., 2015). It has been established that self-renewing cells have a shorter G1 phase (Coronado et al., 2013; Li et al., 2012). Therefore, this observation provides a possible mechanistic explanation behind the increased numbers of HSCs in aged BM (de Haan et al., 1997). ScRNAseq has also explained changes underlying the observed myeloid bias and decreased lymphoid potential of old HSCs (Fig. 1) (Rossi et al., 2005; Sudo et al., 2000). Comparison of individual HSC transcriptomes isolated from old and young mice showed that ageing correlated with an upregulation of platelet-specific gene expression to the point that the majority of aged HSCs almost exclusively produced platelets (Grover et al., 2016). The lymphoid potential of aged HSCs was successfully restored by knocking out the zinc finger protein, multitype 1 (Zfpm1) gene, which is involved in erythroid and megakaryocytic cell differentiation. Those findings illustrate the power of single-cell transcriptome analysis in highlighting possible intervention strategies for reversing an aged phenotype.
Age-dependent inflammatory response of HSCs
Apart from investigating age-related changes in HSCs during homeostasis, scRNAseq has also provided insight into the age-dependent inflammatory response of HSCs (Mann et al., 2018). Old LT-HSCs display unique biases upon immunological stimulation. Under inflammatory stress, HSCs isolated from elderly mice switch to a myeloid-biased expression programme (upregulation of genes related to myeloid function and inhibition of lymphoid differentiation). By contrast, stimulated young HSCs upregulate genes related to lymphocyte development (adaptive immune response) and acute inflammatory response. Aged mice demonstrate a strong acute increase in myeloid output (innate immune response) following the inflammatory challenge, which was not observed in young mice (Mann et al., 2018). Those results are in line with the decreased competency of the adaptive immune system observed in the elderly (Fuentes et al., 2017), which relies mainly on B and T lymphocytes.
Age-related gene expression changes in immune cells
Age-related changes in the transcriptome of single mature blood cells can also explain their altered functional properties observed at later stages of life. This is especially valuable in the study of the ageing immune system. Elderly people have a declining immune system, which manifests as an increased vulnerability to infectious diseases, diminished responses to vaccination and susceptibility to age-related inflammatory diseases (Aiello et al., 2019). Whole-blood and peripheral blood mononuclear cells (PBMCs) (Box 1), collected from healthy donors in a wide range of age groups, have been used to investigate the age-associated decline in immunity (Ademokun et al., 2010; Dunn-Walters and Ademokun, 2010; Sansoni et al., 1993). In line with that goal, scRNAseq is helping to better understand the mechanisms of immunosenescence and inflammaging (Box 1) in blood cells. A recent study from the Tabula Muris Consortium applied scRNAseq to create a comprehensive atlas of age-associated changes in different murine organs and tissues (The Tabula Muris Consortium, 2020). Their findings revealed that old leukocytes have increased expression of pro-inflammatory markers (Cd14, Lgals3, Tnfrsf12a) and decreased expression of anti-inflammatory markers (Cd9, Cd81), an observation consistent with the chronic low-grade inflammation typical of ageing. Additionally, analysis of the clonal repertoire of adaptive (T and B) lymphocytes (Box 1) from single-cell data pointed at a decreased clonal variability in older mice (Fig. 1) (The Tabula Muris Consortium, 2020). This explains, in part, the higher vulnerability of the elderly to infections [e.g. influenza, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] (Mueller et al., 2020) and lower benefits from vaccination reported in older individuals (Goronzy and Weyand, 2013). Another interesting study applied scRNAseq to investigate the population of peripheral blood cells from supercentenarians (people who reached at least 110 years of age) to understand the pathways and mechanisms that promote healthy ageing (Hashimoto et al., 2019). Supercentenarians display a delayed onset of age-related diseases (Andersen et al., 2012); therefore, they are considered a model of successful ageing. The analysis revealed the existence of a specific hybrid population of T cells, combining the features of helper and cytotoxic T cells. This discovery would not be possible in experiments on a population level. The existence of such hybrid T cells might be considered a specific adaptation that results in exceptional longevity (Hashimoto et al., 2019). Further single-cell studies focusing on generating an in-depth understanding of the mechanisms sustaining the development of these cells can benefit the modern ageing society.
The epigenome during ageing
The ageing process is also responsible for the alteration of the chromatin structure and the epigenetic signature within HSCs. Epigenetics refers to all mechanisms regulating gene expression independent of the DNA sequence (Pagiatakis et al., 2019). Epigenetic changes associated with ageing include changes in DNA methylation, reorganisation of chromatin (Moskowitz et al., 2017; Ucar et al., 2017) and post-translational modifications of histones (Pal and Tyler, 2016) (Fig. 2). Over the past decade, several studies strongly implicated epigenetic mechanisms in the dysregulation of gene expression that is observed during the ageing process (Pagiatakis et al., 2019). Ageing in somatic tissues is linked to global hypomethylation of DNA (Gonzalo, 2010). Regulators of DNA methylation include DNA methyltransferases, which drive methylation of CpG islands (Box 1), and the ten-eleven translocation (Tet) enzymes, which regulate demethylation (Pan et al., 2017; Rodríguez-Ubreva et al., 2017). Expression of both types of enzymes differs between young and old HSCs (Beerman et al., 2013; Sun et al., 2014). Post-translational histone modifications can also disturb gene expression patterns in aged HSCs. Aged HSCs display broad peaks of H3K4me3 signal around self-renewal genes (Beerman et al., 2013; Sun et al., 2014), which are associated with higher gene expression (Dhar et al., 2018). Studying the epigenome plays a crucial role in age-related research, because it has been shown that DNA methylation can be used as a reliable biomarker of biological age, thus constituting a so-called ‘epigenetic clock’ (Horvath and Raj, 2018). For epigenomic profiling at the single-cell level, researchers can choose from single-cell assay for transposase accessible chromatin sequencing (scATACseq) (Chen et al., 2018; Satpathy et al., 2019), single-cell chromatin immunoprecipitation sequencing (scChIPseq) (Grosselin et al., 2019), single-cell bisulphite sequencing (scBSseq) (Smallwood et al., 2014) or single-cell reduced-representation bisulphite sequencing (scRRBS) (Guo et al., 2013). Examining epigenome-related changes in the context of haematopoietic ageing is appealing, as an investigation of potential biological ageing estimators showed the superiority of epigenetic clock estimations (Jylhävä et al., 2017). As epigenetic ageing manifests via alterations that affect a minute number of cells, it is perfect for the application of single-cell techniques (Horvath and Raj, 2018).
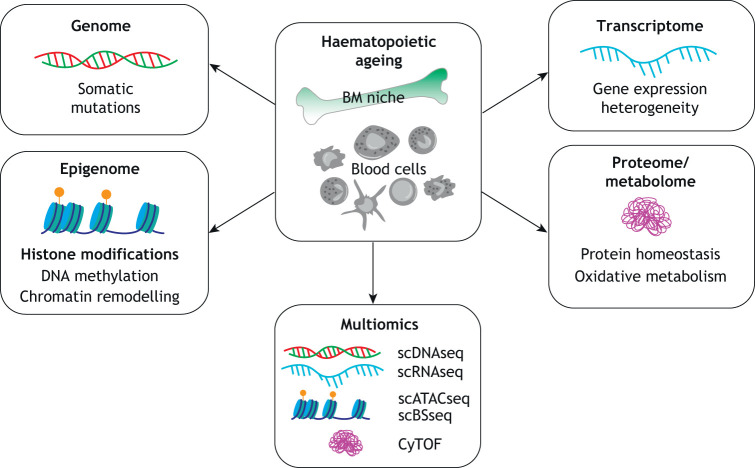
Fig. 2.
Molecular changes related to haematopoietic ageing at different molecular levels. Single-cell technologies provide insight into cell-to-cell variation in the context of haematopoietic ageing at various molecular levels. Current single-cell techniques enable the investigation of somatic mutations, epigenetic changes, gene expression heterogeneity and changes in the proteome and metabolome related to the process of ageing. The combination of different single-cell techniques (multiomics) allows for the investigation of multiple molecular features from the same cell, thus providing more data to understand the process of haematopoietic ageing. Yellow circles indicate methylation. BM, bone marrow; CyTOF, single-cell mass cytometry; scATACseq, single-cell assay for transposase accessible chromatin sequencing; scBSseq, single-cell bisulphite sequencing; scDNAseq, single-cell DNA sequencing; scRNAseq, single-cell RNA sequencing.
Influence of chromatin structure on HSC division
With recent developments in scATACseq scalability allowing it to be used in both droplet-based mode (interrogates up to tens of thousands of cells) (Satpathy et al., 2019) and plate-based mode (allows the analysis of up to hundreds of cells) (Chen et al., 2018), its use to decipher specific cellular mechanisms increases (Popescu et al., 2019; Ranzoni et al., 2020). Recently, scATACseq analysis revealed the involvement of chromatin structure in executing different modes of HSC division (Florian et al., 2018). Old HSCs were shown to divide symmetrically, supporting self-renewal. By contrast, young HSCs preferentially underwent asymmetric division, generating progenitor cells (Florian et al., 2018). This yet again provided a direct link to the increased number of HSCs in the BM upon ageing (de Haan et al., 1997). Solely based on transcriptome profiles obtained from scRNAseq experiments, it was not possible to predict the mode of the division. Only insight into the open chromatin configuration obtained from the analysis of scATACseq peaks showed the correlation between the mode of division and HSC age, and allowed the prediction of the fate of the daughter cell (retaining stem cell potential or differentiating) (Florian et al., 2018). This supports the importance of multimodal measurements on single cells to decipher the mechanisms underlying the altered phenotype in ageing.
Chromatin profile of aged immune cells
Another study employed a single-cell method based on mass cytometry, epigenetic landscape profiling using cytometry by time-of-flight [EpiTOF; a variation of single-cell mass cytometry (CyTOF; Box 2)], to look at the chromatin modification profiles across immune cells during ageing (Cheung et al., 2018). This method allows the measurement of diverse chromatin marks in single cells and reveals an increased cell-to-cell variability in chromatin marks as a signature of immune cell ageing. Chromatin modification profiles in younger individuals (<25 years old) are homogeneous, whereas those of older individuals (>65 years old) are heterogeneous. Interestingly, an analysis of cells from monozygotic twins pointed out that these variations are largely caused by non-heritable factors (Cheung et al., 2018). This single-cell approach contributed to the creation of a comprehensive immune cell epigenetic atlas of age-associated changes based on chromatin modification profiles. Thus, the above examples illustrate how application of single-cell epigenomics provides a deeper understanding of the mechanisms behind the regulation and maintenance of the aberrant gene expression that is observed in ageing.
Ageing and metabolomics research
Metabolomics is one of the novel approaches in systems biology with a high potential to provide important information about the ageing process. Metabolites, the end products of a biological process, provide a quick readout of the function-related phenotype. The metabolic composition of the cell is strictly dependent on physiological conditions (Onjiko et al., 2015). Studies in animal models show that calorie restriction, which directly influences metabolism, is an effective method against age-related diseases (Balasubramanian et al., 2017). Additionally, glucose metabolism has been shown to influence chromatin structure and the transcription process (Heiden et al., 2009), and to interfere with stem cell fate decisions in terms of proliferation, differentiation and dormancy (Shyh-Chang et al., 2013). Therefore, applying metabolomics to study haematopoietic ageing can provide an additional layer of understanding of this process.
The influence of fasting on the ageing process
Recently, a new study provided an atlas of single-cell transcriptomic changes that are associated with the influence of fasting on the ageing process. It demonstrated how metabolic intervention can rewire ageing programmes in multiple cell types, including blood cells and the BM (Ma et al., 2020). Calorie restriction successfully replenishes the number of certain blood cells in the BM (including precursors of B cells and erythrocytes, T cells and dendritic cells) that are reduced during ageing (Ma et al., 2020). Additional pseudotime analysis (Box 1) also revealed that, during ageing, neutrophils acquire a migratory phenotype and infiltrate peripheral tissue, thus explaining the inflammaging phenotype. This migration was reversed by a calorie restriction strategy (Ma et al., 2020).
Glycolysis in aged haematopoietic progenitor cells (HPCs)
The combination of scRNAseq with metabolomic and proteomic approaches has provided interesting insights into the behaviour of aged HPCs (Hennrich et al., 2018; Poisa-Beiro et al., 2020). Aged HPCs with myeloid bias potential displayed elevated glycolysis (Hennrich et al., 2018). The increase in glycolytic enzyme levels was attributed to the expansion of HPC subpopulations that become more glycolytic upon ageing (Poisa-Beiro et al., 2020). Even though these findings derive from the metabolomics analysis at the population level, single-cell metabolomics holds promise to further advance haematopoietic ageing research. Single-cell metabolomics is still under development, but already constitutes an exciting research area. This approach is particularly challenging, as mass spectrometry methods struggle with the identification of short-lived biomolecules that are not chemically stable in vitro. Nevertheless, there are reports of the successful detection of small amounts of metabolites from single cells (Comi et al., 2017; Duncan et al., 2019; Zhang and Vertes, 2018). In the future, it is likely that the research field of haematopoietic ageing will benefit from the development and application of single-cell metabolomics.
Functional single-cell methods
Apart from purely sequencing-based methods, single-cell functional assays such as single-cell in vitro assays (Chapman et al., 2020; Lee-Six et al., 2018) or single-cell transplantations (Carrelha et al., 2018; Yamamoto et al., 2018) are also improving our understanding of haematopoietic ageing. In the landmark study by Lee-Six et al. (2018), single HSCs and HPCs from BM aspirate obtained from an adult man were expanded in vitro in single-cell liquid cultures. Subsequently, whole-genome sequencing of DNA isolated from single cell-derived colonies was performed to identify somatic mutations. In this setting, somatic mutations served as clonal markers to quantify the number, activity and longevity of human HSCs during normal haematopoiesis (Lee-Six et al., 2018). The same approach has been applied to study blood cells isolated from human foetuses, which shed light on the dynamics of foetal haematopoiesis (Chapman et al., 2020). Age-related functional changes in HSCs can also be successfully evaluated via single-cell transplantation assays (Yamamoto et al., 2018). Yamamoto et al. (2018) transplanted single HSCs isolated from young and old mice into lethally irradiated recipients and discovered a population of ‘latent HSCs’ exclusively in the aged BM. Latent HSCs showed a restricted myeloid potential in primary recipients but regained lymphoid potential when re-transplanted into secondary recipients. A better understanding of the development of latent HSCs could help in designing strategies to rebalance haematopoiesis in the elderly (Yamamoto et al., 2018).
HSC transplantation at single-cell resolution
Advanced age is one of the risk factors for myeloid malignancies. HSC transplantation is a part of the standard treatment protocol for many haematological malignancies. Initially, this treatment strategy was restricted to younger patients, but its application in older adults has been increasing recently (Wildes et al., 2014). An interplay between the aged BM niche and blood reconstitution dynamics upon transplantation has been demonstrated (Ergen et al., 2012; Ho et al., 2019). Aged HSCs transplanted into young mice show improved capacity to generate lymphoid progeny, suggesting that the old BM microenvironment is involved in myeloid skewing (Ergen et al., 2012). Indeed, the absence of Notch delta-like ligand (Dll4) produced by BM vascular cells causes the premature upregulation of the myeloid transcriptional program in HSCs (Tikhonova et al., 2019). Single-cell-level analysis offers the opportunity to investigate context-dependent crosstalk between transplanted HSCs and BM cells. Tools like CellPhone DB (Efremova et al., 2020) enable the inference of cell-to-cell communication networks from scRNAseq data and can thus help to dissect the role of the microenvironment in myeloid skewing. Transplantation involves the subsampling of a genetically heterogeneous population of blood cells and then repopulation of the recently emptied BM niche. This subsampling of a genetically heterogeneous population creates an interesting genetic bottleneck that influences the dynamics of blood cell reconstitution. The increased occurrence of clonal haematopoiesis is one of the signs of haematopoietic ageing (Arends et al., 2018; Jaiswal et al., 2014). Several studies have investigated the impact of clonal haematopoiesis on the outcome of HSC transplantation therapy (Frick et al., 2019; Gibson et al., 2017; Mouhieddine et al., 2020; Ortmann et al., 2019). However, the implications of clonal haematopoiesis for stem cell transplantation are difficult to identify owing to the high degree of mutational heterogeneity present within the genetically distinct subclones (Park et al., 2019). Therefore, studying it at single-cell resolution can significantly improve our understanding of the process. The transplanted graft contains a heterogeneous population of cells, with only some of the cells harbouring clonal haematopoiesis-associated mutations. Advances in scRNAseq have revealed the transcriptional diversity among HSCs, thus providing a possible explanation behind their functional heterogeneity (Buenrostro et al., 2018; Giladi et al., 2018; Velten et al., 2017). However, the destructive nature of sequencing assays, which require cell lysis, prevents the simultaneous assessment of the transcriptome and functional output of the same cell, which is an important aspect from a transplantation point of view. This challenge was addressed by the application of lentiviral barcoding, which enables the simultaneous analysis of lineage outputs and transcriptomes from single HSCs during long-term bone marrow reconstitution in mice (Rodriguez-Fraticelli et al., 2020). Lentiviral barcoding has contributed to the identification of a mechanism for the maintenance of the self-renewing HSC state, which depends on transcription factor 15 (Tcf15) (Rodriguez-Fraticelli et al., 2020). This example highlights how the development of single-cell lineage-tracing methods, together with further application of single-cell techniques to study graft cells and blood cell reconstitution dynamics in patients, can bring us closer to better understanding the exact mechanisms active in the human body upon HSC transplantation. This knowledge could be translated into the optimisation of future transplantation strategies for elderly patients.
Single-cell view on infections in elderly
Age-associated changes affecting the immune system are under extensive investigation in the clinic (Esme et al., 2019). The myeloid skewing of the blood compartment and immunosenescence occurring with advancing age can be associated with a decline in the adaptive immune response (Choudry and Frontini, 2016). With advancing age, infections become more frequent, are generally more severe and can present different symptoms than those in younger adults (Htwe et al., 2007). Infections are one of the leading causes of morbidity and mortality in people with advanced age (Esme et al., 2019). Seniors have an elevated risk of respiratory infections, such as severe influenza illness, which often requires hospitalisation (Nguyen et al., 2017). Advanced age is also considered a risk factor for severe outcome of Clostridium difficile infection (Shin et al., 2016). Finally, skin and soft tissue infections, e.g. with Staphylococcus aureus, in the elderly are frequently encountered both in hospitals and in the community (Laube and Farrell, 2002). Single-cell omics techniques allow the dissection of important cell-to-cell variation in the immune response that contributes to the infection outcome (Strzelecka et al., 2018). Accordingly, single-cell-level analysis of T cell receptors (TCRs) in cytotoxic T cells revealed that ageing was associated with reduced numbers of influenza-specific cytotoxic T cells (Nguyen et al., 2017). Recently, several groups applied a single-cell approach to understand the dynamics of SARS-CoV-2 (Schulte-Schrepping et al., 2020; Wen et al., 2020; Wilk et al., 2020; Zhang et al., 2020; Zheng et al., 2020). An interesting study deciphered the landscape of human circulating immune cells in young and old individuals and in COVID-19 patients (Zheng et al., 2020), applying scRNAseq, scATACseq and CyTOF to capture age- and COVID-19-associated changes at the transcriptomic, chromatin and protein level. The results revealed that SARS-CoV-2 infection amplifies the age-induced upregulation of inflammatory genes in immune cells, providing a possible explanation for the increased number of severe and fatal COVID-19 cases in elderly patients (Zheng et al., 2020). Single-cell approaches offer an in-depth understanding of the molecular changes underlying age-related immune dysfunction and can thus provide crucial insight into the development of elderly-targeted infection care.
A glimpse into the future: concluding remarks
One of the biggest advantages of single-cell techniques is the broad readout of cellular composition from a relatively small amount of tissue. Genomic, transcriptomic and epigenomic technologies combined with proteome and metabolome analysis will generate multidimensional data with unprecedented insight into the ageing process. The ability to simultaneously detect changes at various molecular levels (epigenomic, transcriptomic, proteomic and metabolomic) is providing even more data to understand haematopoietic ageing. Each of these omics data can reveal useful markers of the process and provide insights into perturbed biological pathways. However, when analysed individually, omics data are just a snapshot of the processes happening within the cell and thus not sufficient to reveal the causal relationship between molecular signatures and the manifestation of a pathology. Integration of the different omics data types has the potential to uncover the causative changes underlying the age-associated phenotypes of blood cells.
Single-cell techniques advancing haematopoietic ageing studies
There are several areas in which different single-cell techniques can make a unique contribution to haematopoietic ageing (Fig. 2). Currently, single-cell transcriptomics is the most widely used method (Ranzoni et al., 2019), followed closely by single-cell epigenomics (Kelsey et al., 2017). ScRNAseq has the power to detect ageing-related genes with limited expression level or expression restricted to a specific cell type. When combined with somatic mutation tracking (i.e. in clonal haematopoiesis studies), it can highlight changes that are directly linked to the mutation status of the cell (Nam et al., 2019). Analysis of population dynamics with scRNAseq precision can also uncover unique features such as the exhaustion of stem cells or cell population shifts (Fischer et al., 2019). Similarly, single-cell epigenetics approaches can reveal cell-type-specific epigenetic alterations emerging during ageing and its influence on gene expression patterns (Cheung et al., 2018). In a haematological context, the application of the multiomics approaches is already contributing to the better understanding of the human HSC compartment and the changes affecting immune cells (Granja et al., 2019). Further single-cell studies on the ageing blood system will help to answer the question whether altered signatures are indeed drivers or consequences of ageing. The global initiative of the Human Cell Atlas (HCA) (www.humancellatlas.org) is gathering multimodal single-cell data from various human tissues at an unprecedented scale. Comparison of the collected data between individuals of different ages will help to develop a geriatric atlas of the human body similar to Tabula Muris Sensis (The Tabula Muris Consortium, 2020). This will further advance our understanding of haematopoietic ageing.
Limitations of single-cell techniques
Some limitations need to be taken into consideration when applying single-cell techniques in causative analyses to understand haematopoietic ageing. The majority of currently available scRNAseq platforms use poly(dT) primers (Box 1) to capture the poly(A) fraction of a cell's total RNA. This approach excludes non-coding RNA, which was shown to regulate the ageing processes (Sousa-Franco et al., 2019). Another drawback includes the identification of apparent changes in gene expression that are in fact caused by sample processing (van den Brink et al., 2017). To address this problem, new experimental protocols are being developed (Lambrechts et al., 2018), aiming to minimise gene expression alterations that are a consequence of experimental processing and that affect the downstream analysis of the sequencing data. There is also a specific problem related to the granulocytic population in droplet-based scRNAseq approaches. Granulocytes are characterised by relatively low RNA content and relatively high levels of RNases (Zilionis et al., 2019), which could degrade RNA during sample processing and thus interfere with the reliability of the resulting data. Therefore, supplementing buffers with RNase inhibitor might be beneficial when handling granulocyte samples. Additionally, during the data analysis step, a low filtering threshold for transcript count is recommended because granulocytes (especially neutrophils) can be inadvertently excluded when using the data filters commonly used in scRNAseq studies (Zilionis et al., 2019). Finally, analysing single-cell data from different platforms is a challenging task. Therefore, there is a constant need for the development of new computational methods capable of integrative data analysis. Currently available methods for single-cell multiomics data analysis have recently been reviewed in detail (Efremova and Teichmann, 2020). The development of single-cell multimodal omics tools will bring us closer to successfully dissecting the operational principles of biological systems.
Bringing single-cell techniques to the clinical field
The future applicability of single-cell techniques in the clinical field will largely depend on the successful development of robust sample preprocessing techniques. Single-cell analysis in large clinical cohorts still remains expensive and faces technical difficulties. Several groups are putting an effort into benchmarking available protocols and optimizing the workflows for clinical settings to ensure the generation of valid high-quality results (Ding et al., 2020; Hanamsagar et al., 2020; Mereu et al., 2020). The development of compact single-cell work stations suitable for clinical laboratories will facilitate the application of single-cell approaches to clinical settings. Further advances in single-cell techniques will enable their transition from the bench to the bedside.
In conclusion, single-cell approaches keep refining our perception of haematopoietic ageing. In the future, integrative analyses of single-cell multiomics data will contribute to the creation of a detailed atlas highlighting age-associated alterations. This knowledge will have a direct potential to be translated into new geroprotective interventions benefiting the elderly population.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
P.M.S. received a Postdoctoral Research Fellowship from Alexander von Humboldt-Stiftung. F.D. receives research funding from Deutsche Krebshilfe (70113643).
References
- Ademokun A., Wu Y.-C. and Dunn-Walters D. (2010). The ageing B cell population: composition and function. Biogerontology 11, 125-137. 10.1007/s10522-009-9256-9 [DOI] [PubMed] [Google Scholar]
- Aiello A., Farzaneh F., Candore G., Caruso C., Davinelli S., Gambino C. M., Ligotti M. E., Zareian N. and Accardi G. (2019). Immunosenescence and its hallmarks: how to oppose aging strategically? a review of potential options for therapeutic intervention. Front. Immunol. 10, 2247 10.3389/fimmu.2019.02247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertí-Servera L., Demeyer S., Govaerts I., Swings T., De Bie J., Gielen O., Brociner M., Michaux L. M., Maertens J., Uyttebroeck A. et al. (2020). Single-cell DNA amplicon sequencing reveals clonal heterogeneity and evolution in T-cell acute lymphoblastic leukemia. Blood blood.2020006996 10.1182/blood.2020006996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Sebastiani P., Dworkis D. A., Feldman L. and Perls T. T. (2012). Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J. Gerontol. A Biol. Sci. Med. Sci 67, 395-405. 10.1093/gerona/glr223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends C. M., Galan-Sousa J., Hoyer K., Chan W., Jäger M., Yoshida K., Seemann R., Noerenberg D., Waldhueter N., Fleischer-Notter H. et al. (2018). Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 32, 1908-1919. 10.1038/s41375-018-0047-7 [DOI] [PubMed] [Google Scholar]
- Baccin C., Al-Sabah J., Velten L., Helbling P. M., Grünschläger F., Hernández-Malmierca P., Nombela-Arrieta C., Steinmetz L. M., Trumpp A. and Haas S. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol 22, 38-48. 10.1038/s41556-019-0439-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian P., Howell P. R. and Anderson R. M. (2017). Aging and caloric restriction research: a biological perspective with translational potential. EBioMedicine 21, 37-44. 10.1016/j.ebiom.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Bock C., Garrison B. S., Smith Z. D., Gu H., Meissner A. and Rossi D. J. (2013). Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 12, 413-425. 10.1016/j.stem.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Buenrostro J. D., Ryan Corces M., Lareau C. A., Wu B., Schep A. N., Aryee M. J., Majeti R., Chang H. Y. and Greenleaf W. J. (2018). Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535-1548.e16. 10.1016/j.cell.2018.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E. and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canu G., Athanasiadis E., Grandy R. A., Garcia-Bernardo J., Strzelecka P. M., Vallier L., Ortmann D. and Cvejic A. (2020). Analysis of endothelial-to-haematopoietic transition at the single cell level identifies cell cycle regulation as a driver of differentiation. Genome Biol. 21, 157 10.1186/s13059-020-02058-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrelha J., Meng Y., Kettyle L. M., Luis T. C., Norfo R., Alcolea V., Boukarabila H., Grasso F., Gambardella A., Grover A. et al. (2018). Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106-111. 10.1038/nature25455 [DOI] [PubMed] [Google Scholar]
- Chapman M. S., Ranzoni A. M., Myers B., Williams N., Coorens T., Mitchell E., Butler T., Dawson K. J., Hooks Y., Moore L. et al. (2020). Lineage tracing of human embryonic development and foetal haematopoiesis through somatic mutations.
- Chen X., Miragaia R. J., Natarajan K. N. and Teichmann S. A. (2018). A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 9, 5345 10.1038/s41467-018-07771-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Vallania F., Warsinske H. C., Donato M., Schaffert S., Chang S. E., Dvorak M., Dekker C. L., Davis M. M., Utz P. J. et al. (2018). Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 173, 1385-1397.e14. 10.1016/j.cell.2018.03.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudry F. A. and Frontini M. (2016). Epigenetic control of haematopoietic stem cell aging and its clinical implications. Stem Cells Int. 2016, 5797521 10.1155/2016/5797521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. J., Lee H. J., Smallwood S. A., Kelsey G. and Reik W. (2016). Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 17, 72 10.1186/s13059-016-0944-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi T. J., Do T. D., Rubakhin S. S. and Sweedler J. V. (2017). Categorizing cells on the basis of their chemical profiles: progress in single-cell mass spectrometry. J. Am. Chem. Soc. 139, 3920-3929. 10.1021/jacs.6b12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tabula Muris Consortium (2020). A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590-595. 10.1038/s41586-020-2496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.-Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R. et al. (2013). A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Research 10, 118-131. 10.1016/j.scr.2012.10.004 [DOI] [PubMed] [Google Scholar]
- de Haan G., Nijhof W. and Van Zant G. (1997). Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood 89, 1543-1550. 10.1182/blood.V89.5.1543 [DOI] [PubMed] [Google Scholar]
- Dhar S. S., Zhao D., Lin T., Gu B., Pal K., Wu S. J., Alam H., Lv J., Yun K., Gopalakrishnan V. et al. (2018). MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol. Cell 70, 825-841.e6. 10.1016/j.molcel.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Adiconis X., Simmons S. K., Kowalczyk M. S., Hession C. C., Marjanovic N. D., Hughes T. K., Wadsworth M. H., Burks T., Nguyen L. T. et al. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737-746. 10.1038/s41587-020-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshkind K., Höfer T., Montecino-Rodriguez E., Pioli P. D. and Rodewald H.-R. (2019). Do haematopoietic stem cells age? Nat. Rev. Immunol. 20, 196-202. 10.1038/s41577-019-0236-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Laurenti E. and Dick J. E. (2012). Hematopoiesis: a human perspective. Cell Stem Cell 10, 120-136. 10.1016/j.stem.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Duncan K. D., Fyrestam J. and Lanekoff I. (2019). Advances in mass spectrometry based single-cell metabolomics. Analyst 144, 782-793. 10.1039/C8AN01581C [DOI] [PubMed] [Google Scholar]
- Dunn-Walters D. K. and Ademokun A. A. (2010). B cell repertoire and ageing. Curr. Opin. Immunol. 22, 514-520. 10.1016/j.coi.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Dykstra B., Olthof S., Schreuder J., Ritsema M. and de Haan G. (2011). Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208, 2691-2703. 10.1084/jem.20111490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M. and Teichmann S. A. (2020). Computational methods for single-cell omics across modalities. Nat. Methods 17, 14-17. 10.1038/s41592-019-0692-4 [DOI] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S. A. and Vento-Tormo R. (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc 15, 1484-1506. 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- Ergen A. V., Boles N. C. and Goodell M. A. (2012). Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 119, 2500-2509. 10.1182/blood-2011-11-391730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esme M., Topeli A., Yavuz B. B. and Akova M. (2019). Infections in the elderly critically-Ill patients. Front. Med. 6, 118 10.3389/fmed.2019.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth D. R., Saunders L. M. and Miller A. C. (2020). A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100-108. 10.1016/j.ydbio.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D. S., Fiedler A. K., Kernfeld E. M., Genga R. M. J., Bastidas-Ponce A., Bakhti M., Lickert H., Hasenauer J., Maehr R. and Theis F. J. (2019). Inferring population dynamics from single-cell RNA-sequencing time series data. Nat. Biotechnol. 37, 461-468. 10.1038/s41587-019-0088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian M. C., Klose M., Sacma M., Jablanovic J., Knudson L., Nattamai K. J., Marka G., Vollmer A., Soller K., Sakk V. et al. (2018). Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 16, e2003389 10.1371/journal.pbio.2003389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M., Chan W., Arends C. M., Hablesreiter R., Halik A., Heuser M., Michonneau D., Blau O., Hoyer K., Christen F. et al. (2019). Role of donor clonal hematopoiesis in allogeneic hematopoietic stem-cell transplantation. J. Clin. Oncol. 37, 375-385. 10.1200/JCO.2018.79.2184 [DOI] [PubMed] [Google Scholar]
- Frisch B. J., Hoffman C. M., Latchney S. E., LaMere M. W., Myers J., Ashton J., Li A. J., Saunders J II, Palis J., Perkins A. S. et al. (2019). Aged marrow macrophages expand platelet-biased hematopoietic stem cells via Interleukin1B. JCI Insight 5, e124213 10.1172/jci.insight.124213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes E., Fuentes M., Alarcón M. and Palomo I. (2017). Immune system dysfunction in the elderly. An. Acad. Bras. Cienc. 89, 285-299. 10.1590/0001-3765201720160487 [DOI] [PubMed] [Google Scholar]
- Gadalla R., Noamani B., MacLeod B. L., Dickson R. J., Guo M., Xu W., Lukhele S., Elsaesser H. J., Razak A. R. A., Hirano N. et al. (2019). Validation of CyTOF against flow cytometry for immunological studies and monitoring of human cancer clinical trials. Front. Oncol. 9, 415 10.3389/fonc.2019.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C., Koh W. and Quake S. R. (2016). Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175-188. 10.1038/nrg.2015.16 [DOI] [PubMed] [Google Scholar]
- Gazit R., Weissman I. L. and Rossi D. J. (2008). Hematopoietic stem cells and the aging hematopoietic system. Semin. Hematol. 45, 218-224. 10.1053/j.seminhematol.2008.07.010 [DOI] [PubMed] [Google Scholar]
- Gibson C. J., Coleman Lindsley R., Tchekmedyian V., Mar B. G., Shi J., Jaiswal S., Bosworth A., Francisco L., He J., Bansal A. et al. (2017). Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J. Clin. Oncol. 35, 1598-1605. 10.1200/JCO.2016.71.6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi A. and Amit I. (2018). Single-cell genomics: a stepping stone for future immunology discoveries. Cell 172, 14-21. 10.1016/j.cell.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Giladi A., Paul F., Herzog Y., Lubling Y., Weiner A., Yofe I., Jaitin D., Cabezas-Wallscheid N., Dress R., Ginhoux F. et al. (2018). Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836-846. 10.1038/s41556-018-0121-4 [DOI] [PubMed] [Google Scholar]
- Gondek L. P. and DeZern A. E. (2020). Assessing clonal haematopoiesis: clinical burdens and benefits of diagnosing myelodysplastic syndrome precursor states. Lancet Haematol 7, e73-e81. 10.1016/S2352-3026(19)30211-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S. (2010). Epigenetic alterations in aging. J. Appl. Physiol. 109, 586-597. 10.1152/japplphysiol.00238.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy J. J. and Weyand C. M. (2013). Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 14, 428-436. 10.1038/ni.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja J. M., Klemm S., McGinnis L. M., Kathiria A. S., Mezger A., Corces M. R., Parks B., Gars E., Liedtke M., Zheng G. X. Y. et al. (2019). Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458-1465. 10.1038/s41587-019-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., Dahmani A., Lameiras S., Reyal F., Frenoy O. et al. (2019). High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060-1066. 10.1038/s41588-019-0424-9 [DOI] [PubMed] [Google Scholar]
- Grover A., Sanjuan-Pla A., Thongjuea S., Carrelha J., Giustacchini A., Gambardella A., Macaulay I., Mancini E., Luis T. C., Mead A. et al. (2016). Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7, 11075 10.1038/ncomms11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu P., Wu X., Li X., Wen L. and Tang F. (2013). Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126-2135. 10.1101/gr.161679.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R., Reizis T., Chamberlain M., Marcus R., Nestle F. O., de Rinaldis E. and Savova V. (2020). An optimized workflow for single-cell transcriptomics and repertoire profiling of purified lymphocytes from clinical samples. Sci. Rep. 10, 2219 10.1038/s41598-020-58939-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A., Engel J., Teichmann S. A. and Lönnberg T. (2017). A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 10.1186/s13073-017-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Kouno T., Ikawa T., Hayatsu N., Miyajima Y., Yabukami H., Terooatea T., Sasaki T., Suzuki T., Valentine M. et al. (2019). Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA 116, 24242-24251. 10.1073/pnas.1907883116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden M. G. V., Vander Heiden M. G., Cantley L. C. and Thompson C. B. (2009). Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029-1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennrich M. L., Romanov N., Horn P., Jaeger S., Eckstein V., Steeples V., Ye F., Ding X., Poisa-Beiro L., Lai M. C. et al. (2018). Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. 9, 4004 10.1038/s41467-018-06353-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P. P., Strzelecka P. M., Athanasiadis E. I., Hall D., Robalo A. F., Collins C. M., Boudinot P., Levraud J.-P. and Cvejic A. (2018). Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 3, eaau5265 10.1126/sciimmunol.aau5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.-H., del Toro R., Rivera-Torres J., Rak J., Korn C., García-García A., Macías D., González-Gómez C., del Monte A., Wittner M. et al. (2019). Remodeling of bone marrow hematopoietic stem cell niches promotes myeloid cell expansion during premature or physiological aging. Cell Stem Cell 25, 407-418.e6. 10.1016/j.stem.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. and Raj K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371-384. 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- Htwe T. H., Mushtaq A., Robinson S. B., Rosher R. B. and Khardori N. (2007). Infection in the elderly. Infect. Dis. Clin. North Am. 21, 711-743. 10.1016/j.idc.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Hu Y., An Q., Sheu K., Trejo B., Fan S. and Guo Y. (2018). Single cell multi-omics technology: methodology and application. Front. Cell Dev. Biol. 6, 28 10.3389/fcell.2018.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S. and Ebert B. L. (2019). Clonal hematopoiesis in human aging and disease. Science 366, eaan4673 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P. V., Mar B. G., Lindsley R. C., Mermel C. H., Burtt N., Chavez A. et al. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med 371, 2488-2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K. R., Gomes T., Elmentaite R., Kumar N., Gulliver E. L., King H. W., Stares M. D., Bareham B. R., Ferdinand J. R., Petrova V. N. et al. (2020). Distinct microbial and immune niches of the human colon. Nat. Immunol 21, 343-353. 10.1038/s41590-020-0602-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jylhävä J., Pedersen N. L. and Hägg S. (2017). Biological age predictors. EBioMedicine 21, 29-36. 10.1016/j.ebiom.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay A. W., Strauss-Albee D. M. and Blish C. A. (2016). Application of mass cytometry (CyTOF) for functional and phenotypic analysis of natural killer cells. Methods Mol. Biol. 1441, 13-26. 10.1007/978-1-4939-3684-7_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G., Stegle O. and Reik W. (2017). Single-cell epigenomics: recording the past and predicting the future. Science 358, 69-75. 10.1126/science.aan6826 [DOI] [PubMed] [Google Scholar]
- Knouse K. A., Wu J. and Amon A. (2016). Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 26, 376-384. 10.1101/gr.198937.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M. S., Tirosh I., Heckl D., Rao T. N., Dixit A., Haas B. J., Schneider R. K., Wagers A. J., Ebert B. L. and Regev A. (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860-1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Eynde V. d. (2018). Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277-1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lareau C. A., Ludwig L. S., Muus C., Gohil S. H., Zhao T., Chiang Z., Pelka K., Verboon J. M., Luo W., Christian E. et al. (2020). Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 10.1038/s41587-020-0645-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube S. and Farrell A. M. (2002). Bacterial skin infections in the elderly. Drugs Aging 19, 331-342. 10.2165/00002512-200219050-00002 [DOI] [PubMed] [Google Scholar]
- Laurenti E. and Göttgens B. (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418-426. 10.1038/nature25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Six H., Øbro N. F., Shepherd M. S., Grossmann S., Dawson K., Belmonte M., Osborne R. J., Huntly B. J. P., Martincorena I., Anderson E. et al. (2018). Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473-478. 10.1038/s41586-018-0497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V. C., Ballabeni A. and Kirschner M. W. (2012). Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc. Natl. Acad. Sci. USA 109, 12550-12555. 10.1073/pnas.1206740109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zeng X., Xu Y., Wang B., Zhao Y., Lai X., Qian P. and Huang H. (2020). Mechanisms and rejuvenation strategies for aged hematopoietic stem cells. J. Hematol. Oncol. 13, 31 10.1186/s13045-020-00864-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B., Lin Y. and Navin N. (2020). Advancing cancer research and medicine with single-cell genomics. Cancer Cell 37, 456-470. 10.1016/j.ccell.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litviň uková M., Talavera-López C., Maatz H., Reichart D., Worth C. L., Lindberg E. L., Kanda M., Polanski K., Heinig M., Lee M. et al. (2020). Cells of the adult human heart. Nature 588, 466-472. 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato M. A., Woodworth M. B., Lee S., Evrony G. D., Mehta B. K., Karger A., Lee S., Chittenden T. W., D'Gama A. M., Cai X. et al. (2015). Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94-98. 10.1126/science.aab1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig L. S., Lareau C. A., Ulirsch J. C., Christian E., Muus C., Li L. H., Pelka K., Ge W., Oren Y., Brack A. et al. (2019). Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325-1339.e22. 10.1016/j.cell.2019.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Sun S., Geng L., Song M., Wang W., Ye Y., Ji Q., Zou Z., Wang S., He X. et al. (2020). Caloric restriction reprograms the single-cell transcriptional landscape of rattus norvegicus aging. Cell 180, 984-1001.e22. 10.1016/j.cell.2020.02.008 [DOI] [PubMed] [Google Scholar]
- Mann M., Mehta A., de Boer C. G., Kowalczyk M. S., Lee K., Haldeman P., Rogel N., Knecht A. R., Farouq D., Regev A. et al. (2018). Heterogeneous responses of hematopoietic stem cells to inflammatory stimuli are altered with age. Cell Rep 25, 2992-3005.e5. 10.1016/j.celrep.2018.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J. A., Gettler K., Chuang L. S., Nayar S G. A. J., Dubinsky M. et al. (2019). Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493-1508.e20. 10.1016/j.cell.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanovich M., Zahalka A., Pierce H., Pinho S., Nakahara F., Asada N., Wei Q., Wang X. and Frenette P. (2018). Adrenergic nerve degeneration in the bone marrow drives aging of the hematopoietic stem cell niche. Exp. Hematol. 64, S83-S84. 10.1016/j.exphem.2018.06.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrell T., Park N., Moreno T., Grove C. S., Ponstingl H., Stephens J., Crawley C., Craig J., Scott M. A., Hodkinson C. et al. (2015). Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 10, 1239-1245. 10.1016/j.celrep.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereu E., Lafzi A., Moutinho C., Ziegenhain C., McCarthy D. J., Álvarez-Varela A., Batlle E., Sagar G., Lau D., Boutet S. C. et al. (2020). Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747-755. 10.1038/s41587-020-0469-4 [DOI] [PubMed] [Google Scholar]
- Miles L. A., Bowman R. L., Merlinsky T. R., Csete I. S., Ooi A. T., Durruthy-Durruthy R., Bowman M., Famulare C., Patel M. A., Mendez P. et al. (2020). Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587, 477-482. 10.1038/s41586-020-2864-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincarelli L., Lister A., Lipscombe J. and Macaulay I. C. (2018). Defining cell identity with single-cell omics. Proteomics 18, e1700312 10.1002/pmic.201700312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Wang F., Jahn K., Hu T., Tanaka T., Sasaki Y., Kuipers J., Loghavi S., Wang S. A., Yan Y. et al. (2020). Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat. Commun. 11, 5327 10.1038/s41467-020-19119-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. J., Wandycz A. M., Akashi K., Globerson A. and Weissman I. L. (1996). The aging of hematopoietic stem cells. Nat. Med. 2, 1011-1016. 10.1038/nm0996-1011 [DOI] [PubMed] [Google Scholar]
- Moskowitz D. M., Zhang D. W., Hu B., Le Saux S., Yanes R. E., Ye Z., Buenrostro J. D., Weyand C. M., Greenleaf W. J. and Goronzy J. J. (2017). Epigenomics of human CD8 T cell differentiation and aging. Sci. Immunol. 2, eaag0192 10.1126/sciimmunol.aag0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhieddine T. H., Sperling A. S., Redd R., Park J., Leventhal M., Gibson C. J., Manier S., Nassar A. H., Capelletti M., Huynh D. et al. (2020). Clonal hematopoiesis is associated with adverse outcomes in multiple myeloma patients undergoing transplant. Nat. Commun. 11, 2996 10.1038/s41467-020-16805-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A. L., McNamara M. S. and Sinclair D. A. (2020). Why does COVID-19 disproportionately affect older people? Aging 12, 9959-9981. 10.18632/aging.103344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas E., Yoshida K., Frick M., Hoyer K., Christen F., Kaeda J., Obenaus M., Noerenberg D., Hennch C., Chan W. et al. (2020). Single-cell analysis based dissection of clonality in myelofibrosis. Nat. Commun. 11, 73 10.1038/s41467-019-13892-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam A. S., Kim K.-T., Chaligne R., Izzo F., Ang C., Taylor J., Myers R. M., Abu-Zeinah G., Brand R., Omans N. D. et al. (2019). Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature 571, 355-360. 10.1038/s41586-019-1367-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. H. O., Sant S., Bird N. L., Grant E. J., Bridie Clemens E., Koutsakos M., Valkenburg S. A., Gras S., Lappas M., Jaworowski A. et al. (2017). Perturbed CD8 T cell immunity across universal influenza epitopes in the elderly. J. Leukoc. Biol. 103, 321-339. 10.1189/jlb.5MA0517-207R [DOI] [PubMed] [Google Scholar]
- Oetjen K. A., Lindblad K. E., Goswami M., Gui G., Dagur P. K., Lai C., Dillon L. W., McCoy J. P. and Hourigan C. S. (2018). Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 3, e124928 10.1172/jci.insight.124928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onjiko R. M., Moody S. A. and Nemes P. (2015). Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc. Natl. Acad. Sci. USA 112, 6545-6550. 10.1073/pnas.1423682112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann C. A., Kent D. G., Nangalia J., Silber Y., Wedge D. C., Grinfeld J., Baxter E. J., Massie C. E., Papaemmanuil E., Menon S. et al. (2015). Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 372, 601-612. 10.1056/NEJMoa1412098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann C. A., Dorsheimer L., Abou-El-Ardat K., Hoffrichter J., Assmus B., Bonig H., Scholz A., Pfeifer H., Martin H., Schmid T. et al. (2019). Functional dominance of CHIP-mutated hematopoietic stem cells in patients undergoing autologous transplantation. Cell Reports 27, 2022-2028.e3. 10.1016/j.celrep.2019.04.064 [DOI] [PubMed] [Google Scholar]
- Pagiatakis C., Musolino E., Gornati R., Bernardini G. and Papait R. (2019). Epigenetics of aging and disease: a brief overview. Aging Clin. Exp. Res. 10.1007/s40520-019-01430-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paguirigan A. L., Smith J., Meshinchi S., Carroll M., Maley C. and Radich J. P. (2015). Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci. Transl. Med. 7, 281re2 10.1126/scitranslmed.aaa0763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. and Tyler J. K. (2016). Epigenetics and aging. Sci. Adv. 2, e1600584 10.1126/sciadv.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Zhu S., Qu K., Meeth K., Cheng J., He K., Ma H., Liao Y., Wen X., Roden C. et al. (2017). The DNA methylcytosine dioxygenase Tet2 sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression. Immunity 47, 284-297.e5. 10.1016/j.immuni.2017.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. S., Akuffo A. A., Muench D. E., Grimes H. L., Epling-Burnette P. K., Maini P. K., Anderson A. R. A. and Bonsall M. B. (2019). Clonal hematopoiesis of indeterminate potential and its impact on patient trajectories after stem cell transplantation. PLoS Comput. Biol. 15, e1006913 10.1371/journal.pcbi.1006913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Botting R. A., Domínguez Conde C., Popescu D.-M., Lavaert M., Kunz D. J., Goh I., Stephenson E., Ragazzini R., Tuck E. et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 10.1126/science.aay3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B., Wilson N. K., Xia J., Hou X., Hannah R. L., Kinston S., Calero-Nieto F. J., Poirion O., Preissl S., Liu F. et al. (2020). Single-cell chromatin accessibility maps reveal regulatory programs driving early mouse organogenesis. Nat. Cell Biol. 22, 487-497. 10.1038/s41556-020-0489-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli P. D., Casero D., Montecino-Rodriguez E., Morrison S. L. and Dorshkind K. (2019). Plasma cells are obligate effectors of enhanced myelopoiesis in aging bone marrow. Immunity 51, 351-366.e6. 10.1016/j.immuni.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisa-Beiro L., Thoma J., Landry J., Sauer S., Yamamoto A., Eckstein V., Romanov N., Raffel S., Hoffmann G. F., Bork P. et al. (2020). Glycogen accumulation, central carbon metabolism, and aging of hematopoietic stem and progenitor cells. Sci. Rep. 10, 11597 10.1038/s41598-020-68396-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D.-M., Botting R. A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E. F., Polanski K., Goh I., Efremova M. et al. (2019). Decoding human fetal liver haematopoiesis. Nature 574, 365-371. 10.1038/s41586-019-1652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzoni A. M., Strzelecka P. M. and Cvejic A. (2019). Application of single-cell RNA sequencing methodologies in understanding haematopoiesis and immunology. Essays Biochem. 63, 217-225. 10.1042/EBC20180072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzoni A. M., Tangherloni A., Berest I., Riva S. G., Myers B., Strzelecka P. M., Xu J., Panada E., Mohorianu I., Zaugg J. B. et al. (2020). Integrative single-cell RNA-Seq and ATAC-Seq analysis of human foetal liver and bone marrow haematopoiesis. Cell Stem Cell S1934-5909(20)30553-1 10.1016/j.stem.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A. E., Weinreb C., Wang S.-W., Migueles R. P., Jankovic M., Usart M., Klein A. M., Lowell S. and Camargo F. D. (2020). Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature 583, 585-589. 10.1038/s41586-020-2503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Meira A., Buck G., Clark S.-A., Povinelli B. J., Alcolea V., Louka E., McGowan S., Hamblin A., Sousos N., Barkas N. et al. (2019). Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell 73, 1292-1305.e8. 10.1016/j.molcel.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ubreva J., Català-Moll F., Obermajer N., Álvarez-Errico D., Ramirez R. N., Carlos Company Vento-Tormo, R., Moreno-Bueno G., Edwards R. P., Mortazavi A. et al. (2017). Prostaglandin E2 leads to the acquisition of DNMT3A-dependent tolerogenic functions in human myeloid-derived suppressor cells. Cell Reports 21, 154-167. 10.1016/j.celrep.2017.09.018 [DOI] [PubMed] [Google Scholar]
- Rossi D. J., Bryder D., Zahn J. M., Ahlenius H., Sonu R., Wagers A. J. and Weissman I. L. (2005). Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 102, 9194-9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A., Ram O., Shoresh N., Sperling R. A., Goren A., Weitz D. A. and Bernstein B. E. (2015). Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165-1172. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansoni P., Cossarizza A., Brianti V., Fagnoni F., Snelli G., Monti D., Marcato A., Passeri G., Ortolani C. and Forti E. (1993). Lymphocyte subsets and natural killer cell activity in healthy old people and centenarians. Blood 82, 2767-2773. 10.1182/blood.V82.9.2767.2767 [DOI] [PubMed] [Google Scholar]
- Satpathy A. T., Granja J. M., Yost K. E., Qi Y., Meschi F., McDermott G. P., Olsen B. N., Mumbach M. R., Pierce S. E., Corces M. R. et al. (2019). Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925-936. 10.1038/s41587-019-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. et al. (2020). Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419-1440.e23. 10.1016/j.cell.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H., High K. P. and Warren C. A. (2016). Older is not wiser, immunologically speaking: effect of aging on host response to Clostridium difficile Infections. J. Gerontol. A Biol. Sci. Med. Sci. 71, 916-922. 10.1093/gerona/glv229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G. Q. and Cantley L. C. (2013). Stem cell metabolism in tissue development and aging. Development 140, 2535-2547. 10.1242/dev.091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein L., Goncalves K. A., Kharchenko P. V., Turcotte R., Kfoury Y., Mercier F., Baryawno N., Severe N., Bachand J., Spencer J. A. et al. (2016). Proximity-based differential single-cell analysis of the niche to identify stem/progenitor cell regulators. Cell Stem Cell 19, 530-543. 10.1016/j.stem.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S. A., Lee H. J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S. R., Stegle O., Reik W. and Kelsey G. (2014). Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817-820. 10.1038/nmeth.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Franco A., Rebelo K., da Rocha S. T. and Bernardes de Jesus B. (2019). LncRNAs regulating stemness in aging. Aging Cell 18, e12870 10.1111/acel.12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma D. P. (2018). Clinical consequences of clonal hematopoiesis of indeterminate potential. Hematol. Am. Soc. Hematol. Educ. Program 2018, 264-269. 10.1182/asheducation-2018.1.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka P. M., Ranzoni A. M. and Cvejic A. (2018). Dissecting human disease with single-cell omics: application in model systems and in the clinic. Dis. Model. Mech. 11, dmm036525 10.1242/dmm.036525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y. and Nakauchi H. (2000). Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192, 1273-1280. 10.1084/jem.192.9.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K. F., Chen R. et al. (2014). Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673-688. 10.1016/j.stem.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V., Vento-Tormo R. and Teichmann S. A. (2018). Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599-604. 10.1038/nprot.2017.149 [DOI] [PubMed] [Google Scholar]
- Tikhonova A. N., Dolgalev I., Hu H., Sivaraj K. K., Hoxha E., Cuesta-Domínguez Á., Pinho S., Akhmetzyanova I., Gao J., Witkowski M. et al. (2019). The bone marrow microenvironment at single-cell resolution. Nature 569, 222-228. 10.1038/s41586-019-1104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar D., Márquez E. J., Chung C.-H., Marches R., Rossi R. J., Uyar A., Wu T.-C., George J., Stitzel M. L., Palucka A. K. et al. (2017). The chromatin accessibility signature of human immune aging stems from CD8 T cells. J. Exp. Med. 214, 3123-3144. 10.1084/jem.20170416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink S. C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C. S., Robin C. and van Oudenaarden A. (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935-936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- van Galen P., Hovestadt V., Wadsworth M. H., Ii H., Griffin T. K., Battaglia G. K., Verga S., Stephansky J. A., Pastika J., Lombardi Story T. J. (2019). Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265-1281.e24. 10.1016/j.cell.2019.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten L., Haas S. F., Raffel S., Blaszkiewicz S., Islam S., Hennig B. P., Hirche C., Lutz C., Buss E. C., Nowak D. et al. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271-281. 10.1038/ncb3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Braga F. A., Kar F. A., Berg G., Carpaij M., Polanski O. A., Simon K., Brouwer L. M., Gomes S., Hesse T., Jiang L. (2019). A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153-1163. 10.1038/s41591-019-0468-5 [DOI] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J. et al. (2020). Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov 6, 31 10.1038/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildes T. M., Stirewalt D. L., Medeiros B. and Hurria A. (2014). Hematopoietic stem cell transplantation for hematologic malignancies in older adults: geriatric principles in the transplant clinic. J. Natl. Compr. Canc. Netw. 12, 128-136. 10.6004/jnccn.2014.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A. J., Rustagi A., Zhao N. Q., Roque J., Martínez-Colón G. J., McKechnie J. L., Ivison G. T., Ranganath T., Vergara R., Hollis T. et al. (2020). A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070-1076. 10.1038/s41591-020-0944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z., Ryan M. A., Daria D., Nattamai K. J., Van Zant G., Wang L., Zheng Y. and Geiger H. (2006). Increased hematopoietic stem cell mobilization in aged mice. Blood 108, 2190-2197. 10.1182/blood-2005-12-010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R., Wilkinson A. C., Ooehara J., Lan X., Lai C.-Y., Nakauchi Y., Pritchard J. K. and Nakauchi H. (2018). Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 22, 600-607.e4. 10.1016/j.stem.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. L., Challen G. A., Birmann B. M. and Druley T. E. (2016). Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun 7, 12484 10.1038/ncomms12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. and Vertes A. (2018). Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angewandte Chemie International Edition 57, 4466-4477. 10.1002/anie.201709719 [DOI] [PubMed] [Google Scholar]
- Zhang L., Dong X., Lee M., Maslov A. Y., Wang T. and Vijg J. (2019). Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc. Natl Acad. Sci. USA 116, 9014-9019. 10.1073/pnas.1902510116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y. et al. (2020). Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 21, 1107-1118. 10.1038/s41590-020-0762-x [DOI] [PubMed] [Google Scholar]
- Zheng Y., Liu X., Le W., Xie L., Li H., Wen W., Wang S., Ma S., Huang Z., Ye J. et al. (2020). A human circulating immune cell landscape in aging and COVID-19. Protein Cell 11, 740-770. 10.1007/s13238-020-00762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H. D., Krishnan I., Maroni G., Meyerovitz C. V., Kerwin C. M. et al. (2019). Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317-1334.e10. 10.1016/j.immuni.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]