ABSTRACT
The ability of zebrafish to heal their heart after injury makes them an attractive model for investigating the mechanisms governing the regenerative process. In this study, we show that the gene cellular communication network factor 2a (ccn2a), previously known as ctgfa, is induced in endocardial cells in the injured tissue and regulates CM proliferation and repopulation of the damaged tissue. We find that, whereas in wild-type animals, CMs track along the newly formed blood vessels that revascularize the injured tissue, in ccn2a mutants CM proliferation and repopulation are disrupted, despite apparently unaffected revascularization. In addition, we find that ccn2a overexpression enhances CM proliferation and improves the resolution of transient collagen deposition. Through loss- and gain-of-function as well as pharmacological approaches, we provide evidence that Ccn2a is necessary for and promotes heart regeneration by enhancing the expression of pro-regenerative extracellular matrix genes, and by inhibiting the chemokine receptor gene cxcr3.1 through a mechanism involving Tgfβ/pSmad3 signaling. Thus, Ccn2a positively modulates the innate regenerative response of the adult zebrafish heart.
KEY WORDS: Ccn2a, Ctgf, Heart regeneration, Zebrafish, Extracellular matrix, TGFβ
Highlighted Article: Loss- and gain-of-function approaches and pharmacological studies reveal that Ccn2a promotes heart regeneration by enhancing pro-regenerative extracellular matrix gene expression and by inhibiting the chemokine receptor gene cxcr3.1.
INTRODUCTION
Heart disease is the leading cause of death in industrialized nations [https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)]. Adult mammalian hearts fail to regenerate after myocardial damage, typically leading to persistent scarring and reduced heart function (Engel et al., 2006; Doppler et al., 2017). In contrast, zebrafish can regenerate their injured myocardium through cardiomyocyte (CM) proliferation (Poss et al., 2002) aided by a specific immune response (Hui et al., 2017; Lai et al., 2017) and the ability to clear transient collagen deposition (Poss et al., 2002; González-Rosa et al., 2011) (Fig. 1A). Like zebrafish, neonatal mice exhibit a robust capacity for heart regeneration for a few days after birth (Porrello et al., 2011). A low level of CM proliferation can also be detected in adult humans (Bergmann et al., 2009) and mice (Senyo et al., 2013). Another study has provided evidence that, in humans, a high level of CM cytokinesis can be detected in infants, which was not visible after 20 years of age (Mollova et al., 2013). Importantly, Mohamed et al. (2018) have reported that overexpression of a combination of cell cycle regulators can induce cell division in post-mitotic human cardiomyocytes. These observations have led to the recent hypothesis that adult mammals, including humans, which have a limited heart regeneration capacity, could be induced to regenerate cardiac tissue by identifying and activating regenerative genes.
Fig. 1.
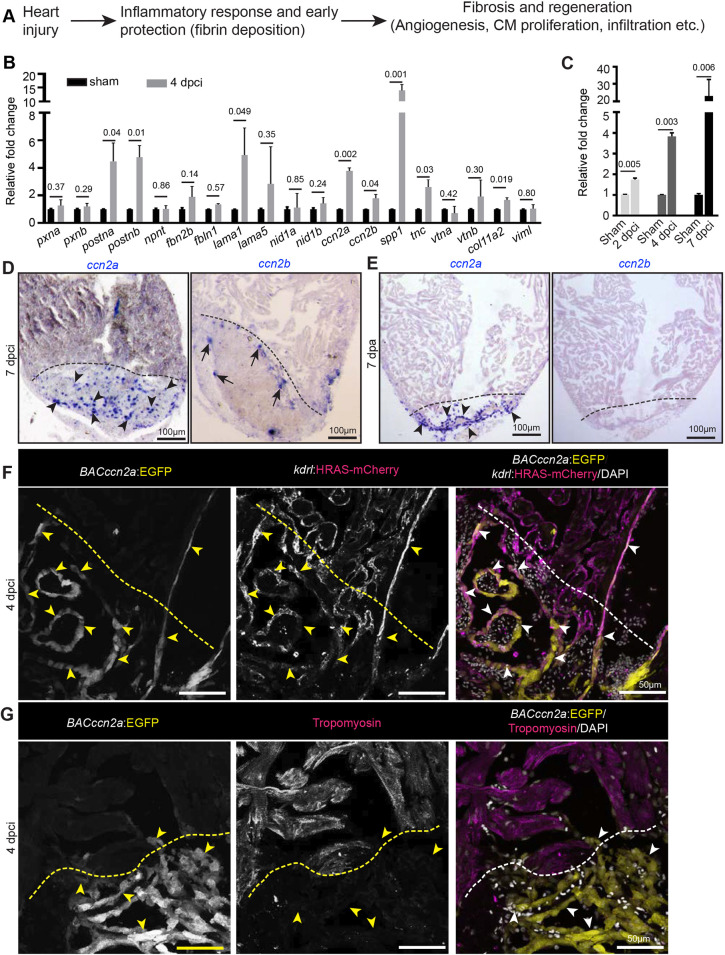
ccn2a is expressed by endocardial cells in injured zebrafish hearts. (A) Schematic depiction of the known processes involved in heart regeneration. (B,C) qPCR-based small-scale screen to identify dynamically expressed extracellular matrix genes in 4 dpci adult zebrafish hearts (B), and qPCR analysis of ccn2a expression during the early stages of heart regeneration (C) (n=3, each sample represents a pool of 6 hearts). Data are mean±s.d. Values are normalized to the mean of the sham control. The statistical significance of differences was evaluated using a two-tailed Student's t-test (GraphPad Prism). Mean Ct values are provided in Table S4. (D,E) Representative images of ccn2a and ccn2b expression on sagittal sections of 7 dpci (D) and 7 dpa (E) adult zebrafish hearts. Arrowheads and arrows indicate ccn2a- and ccn2b-expressing cells in the injured tissue, respectively. (F) BACccn2a:EGFP and kdrl:HRAS-mCherry expression in a sagittal section of a 4 dpci heart. Arrowheads indicate EGFP expression overlapping with mCherry positive-cells in the injured tissue. (G) BACccn2a:EGFP expression and tropomyosin (magenta; marks CMs) immunostaining on a sagittal section of a 4 dpci heart. Arrowheads indicate EGFP-expressing cells. Dashed lines indicate the wound border. dpci, days post cryoinjury; dpa, days post amputation.
Historically, the extracellular matrix (ECM) was thought to serve as an inert scaffold, providing mechanical support to tissues. However, work in recent decades suggests that the ECM plays vital roles in regulating signaling cascades involved in organ development (O'Shea et al., 1990; Fogerty et al., 1994; Linton et al., 2007; Li et al., 2014), maintenance (Fang et al., 2010), disease (Sasaki et al., 2001; Jacobetz et al., 2013; Saupe et al., 2013) and regeneration (González-Rosa et al., 2011; Wang et al., 2013; Bassat et al., 2017; Wehner et al., 2017). To identify potential pro-regenerative ECM genes, we screened various extracellular genes that are induced in the zebrafish heart at 4 days post cryoinjury (dpci). This small-scale candidate gene screen identified nine ECM genes whose expression was significantly increased at 4 dpci (Fig. 1B and Table S1). A few of these genes, including periostin a (postna) have been reported to induce cardiac regeneration in mammalian models (Kuhn et al., 2007; Ladage et al., 2013). Other hits from our screen, such as osteopontin (spp1) and tenascin C (tnc), have previously been reported to be expressed in dilated cardiomyopathy patients (Satoh et al., 2005; Yokokawa et al., 2016). Recently, Pfefferli and Jaźwińska showed that cellular communication network factor 2a (ccn2a) [previously known as connective tissue growth factor a (ctgfa)] is expressed in injured zebrafish cardiac tissue (Pfefferli and Jaźwińska, 2017).
In our injury-induced ECM screen, the expression of both paralogs of cellular communication network factor 2 (CCN2; ccn2a and ccn2b) were increased in the regenerating heart, suggesting a function during cardiac regeneration. Many studies suggest that, in mammals, CCN2 expression is related to the progression of hepatic (Tamatani et al., 1998), renal (Jaffa et al., 2008), skin (Mori et al., 1999), lung (Ponticos et al., 2009) and cardiac (Dean et al., 2005; Angelini et al., 2015) fibrosis. Recent reports indicate that CCN2 promotes ligament regeneration in rabbits (Zhang et al., 2017), β-cell proliferation in mice (Riley et al., 2015) and spinal cord regeneration in zebrafish (Mokalled et al., 2016). Gravning et al. showed that transgenic overexpression of CCN2 in the permanent coronary artery ligation mouse model partially protected the heart from scarring and cardiac function deterioration, suggesting a beneficial role for CCN2 after cardiac injury (Gravning et al., 2012). In contrast, a recent report suggests that perturbation of CCN2 levels by deletion or overexpression does not affect cardiac fibrosis or remodeling in the adult mouse heart subjected to multiple acute stresses (Accornero et al., 2015). Taken together, the reported functions of CCN2 in organ/cardiac regeneration appear contradictory. Here, we have sought to explore the role of Ccn2a in heart regeneration in adult zebrafish.
We show that, upon cardiac injury in adult zebrafish, ccn2a expression is induced in endocardial cells in the injured tissue. In wild-type animals CMs track along new coronary vessels into the wound, as previously shown (Marín-Juez et al., 2019). Upon loss of ccn2a, there is a decrease in CM proliferation and diminished CM tracking along the new coronary vessels, thereby disrupting cardiac muscle regeneration. Consistent with this observation, ccn2a overexpression enhances CM proliferation and expedites the resolution of collagenous tissue. Molecular analysis shows that Ccn2a regulates heart regeneration by positively regulating pSmad3 signaling and a subset of its downstream targets, including ECM genes and the cxcr3.1 chemokine receptor gene. Thus, our study finds that Ccn2a is necessary for heart regeneration in zebrafish, and that it can accelerate this process by promoting CM proliferation and repopulation of the injured tissue via the upregulation of pSmad3 signaling.
RESULTS
ccn2 expression is induced in the zebrafish heart upon cardiac injury
We first analyzed mRNA levels of ECM genes using total RNA isolated from sham or 4 dpci whole cardiac ventricles. The expression levels of selected genes were normalized to ef1α. Our qPCR-based screen found that, in adult zebrafish cardiac ventricles, the mRNA levels of both paralogs of the CCN2 gene (ccn2a and ccn2b) were upregulated upon cardiac injury (Fig. 1B), although the mRNA levels of ccn2b were much lower compared with those of ccn2a (Fig. S1A). On a temporal scale, ccn2a expression in cardiac ventricles was found to be upregulated starting at 2 dpci onwards during the early stages of heart regeneration (Fig. 1C). Spatial RNA expression analysis by in situ hybridization showed that ccn2a was expressed in the primordial layer and cortical myocardium of the uninjured adult cardiac ventricle. However, in situ hybridization failed to detect ccn2b transcripts in the uninjured cardiac ventricle (Fig. S1B).
We studied the spatiotemporal expression of ccn2a and ccn2b in injured heart tissue in two cardiac regeneration models: the cryoinjury model, where a large amount of transient collagen accumulates in the injured tissue (González-Rosa et al., 2011); and the ventricular resection model, where limited amounts of transient collagen deposits along the resection plane (Poss et al., 2002). Upon cryoinjury, at 7 dpci, ccn2a transcripts were predominantly induced in the injured tissue (Fig. 1D). The expression pattern of ccn2a in the uninjured part of the ventricle (>200 µm away from the wound edge) did not appear to be affected (Fig. S1C). Similarly, in the amputation model, ccn2a transcripts were found predominantly in the wound region from 3 to 14 days post-amputation (dpa) (Fig. 1E and Fig. S1D). Corroborating the qPCR data, in both injury models, ccn2b was detected only in the border zone with relatively lower expression than its paralog (Fig. 1D,E and Fig. S1E).
To further investigate the expression pattern of ccn2a, we generated a TgBAC(ccn2a:EGFP) reporter line (BACccn2app01) using a bacterial artificial chromosome (BAC), where EGFP was placed at the start codon of ccn2a and was flanked by a ∼53-kb upstream regulatory genomic sequence and a ∼110-kb downstream genomic sequence (Fig. S2A,B). The EGFP expression pattern in uninjured and injured TgBAC(ccn2a:EGFP) hearts resembled the endogenous ccn2a mRNA expression observed by in situ hybridization (Fig. 1F,G and Fig. S2C,D). Relatively weak TgBAC(ccn2a:EGFP) expression was observed along the primordial layer and in a few cells in the cortical myocardium of the uninjured heart (Fig. S2C). Interestingly, upon injury TgBAC(ccn2a:EGFP)-positive cells were prominently detected in the injured tissue (Fig. S2D). At 4 dpci, TgBAC(ccn2a:EGFP)-expressing cells largely overlapped with kdrl+ endocardial cells at the injury site (Fig. 1F). However, at this stage, the CMs at the border zone remained devoid of TgBAC(ccn2a:EGFP) expression (Fig. 1G). Overall, ccn2a expression appears to be induced upon injury and is observed mainly in endocardial cells in the injured cardiac tissue.
ccn2a is necessary for zebrafish heart regeneration after cardiac injury
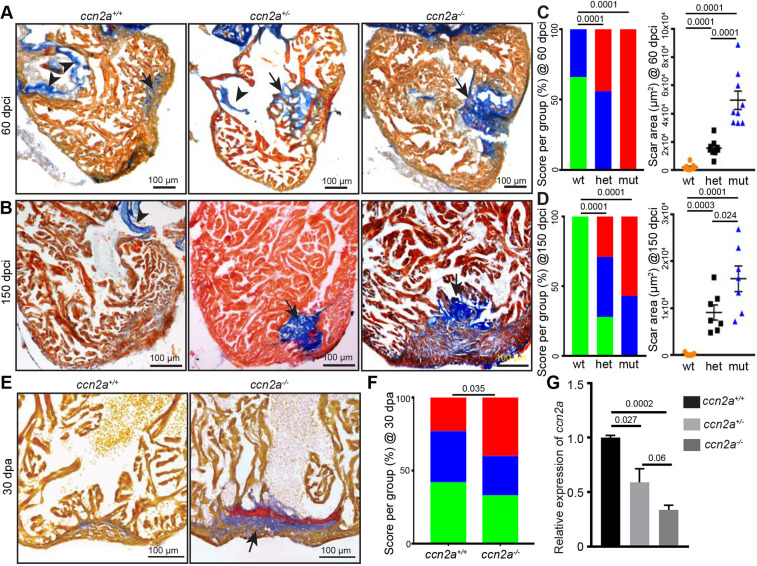
To explore the role of Ccn2a in heart regeneration, we employed a loss-of-function ccn2a mutant allele (the ctgfabns50 allele referred to as ccn2a−) (Mokalled et al., 2016). ccn2a−/− animals develop into viable adults with no visible structural abnormalities in the heart. However, upon cryoinjury, ccn2a+/− and ccn2a−/− animals showed impaired heart regeneration. We used a semi-quantitative method to measure regeneration by scoring for the presence of the collagenous scar in heart sections taken at 60 and 150 dpci. This analysis revealed that, although ∼60% of wild-type injured hearts show vigorous regeneration at 60 dpci, all ccn2a−/− and ∼50% of ccn2a+/− injured hearts displayed minimal regeneration at this stage, evident by the presence of a large collagenous scar at the site of injury (Fig. 2A,C). Interestingly, a persistent collagenous scar was observed in ∼14% ccn2a+/− and 60% ccn2a−/− hearts, even at 150 dpci (Fig. 2B,D), indicating that these phenotypes are not merely delays in regeneration but are indicative of poor repair. Similarly, in the ventricular resection model, ccn2a−/− animals showed a reduction in the efficiency of heart regeneration at 30 dpa (Fig. 2E,F). As heterozygous animals also showed poor heart regeneration, we measured expression levels of ccn2a transcripts in ccn2a+/+, ccn2a+/− and ccn2a−/− embryos at 48 hpf. Our qPCR analysis showed ∼60% and 34% transcripts are present in ccn2a+/− and ccn2a−/− animals, respectively, relative to the wild-type embryos (Fig. 2G). Given the nature of the mutation, we cannot formally rule out the possibility that the mutant allele gives rise to a truncated protein. Nevertheless, more severe cardiac phenotype in ccn2a−/− animals compared with ccn2a+/− siblings and the decrease in transcript level in the heterozygous and mutants suggest that the phenotype is likely to be associated with loss of gene function. Together, these results thus support a necessary role for Ccn2a in heart regeneration.
Fig. 2.
ccn2a mutant hearts exhibit increased scarring after injury. (A,B) Representative bright-field images of 12 μm sagittal paraffin sections of 60 dpci (A) and 150 dpci (B) hearts stained with acid fuchsin-orange G (AFOG; muscle, yellowish red; fibrin, brick red; collagen, blue). Arrows and arrowheads indicate the collagenous scar and atrioventricular valves, respectively. (C,D) Semi-quantitative analysis of scarring in ccn2a+/+, ccn2a+/− and ccn2a−/− hearts (n=9 each) at 60 dpci (C), and ccn2a+/+, ccn2a+/− and ccn2a−/− hearts (n=7 each) at 150 dpci (D). Color-coded bar chart indicates the degree of regeneration: green, complete; blue, moderate; red, very poor. Data indicate the percentage of total hearts represented by each score. The statistical significance of differences was evaluated by a wound-recovery χ2 test. Dot plots show highest area covered by collagenous scar on a tissue section from each heart. Data are mean±s.e.m. (E) Sagittal sections of 30 dpa ventricles stained with AFOG. Example of a vigorously regenerating ccn2a+/+ heart and a poorly regenerating ccn2a−/− heart section. Arrow indicates the collagenous scar. (F) Semi-quantitative analysis of wound recovery in 25 ccn2a+/+ and 30 ccn2a−/− animals. Average of area covered by collagen on three histological sections in each heart was considered for scar quantification. Color-coded bar chart indicates degree of regeneration: green, complete; blue, moderate; red, very poor. Data indicate the percentage of total hearts represented by each score. The statistical significance of differences was evaluated by a wound-recovery χ2 test. (G) qPCR analysis of ccn2a transcripts in 48 hpf embryos from three genotypes. Values are normalized to the mean of the wild type. Data are mean±s.d. dpci, days post cryoinjury; dpa, days post amputation; hpf, hours post fertilization.
Cardiomyocytes fail to track along new coronary vessels in ccn2a mutants post-cardiac injury
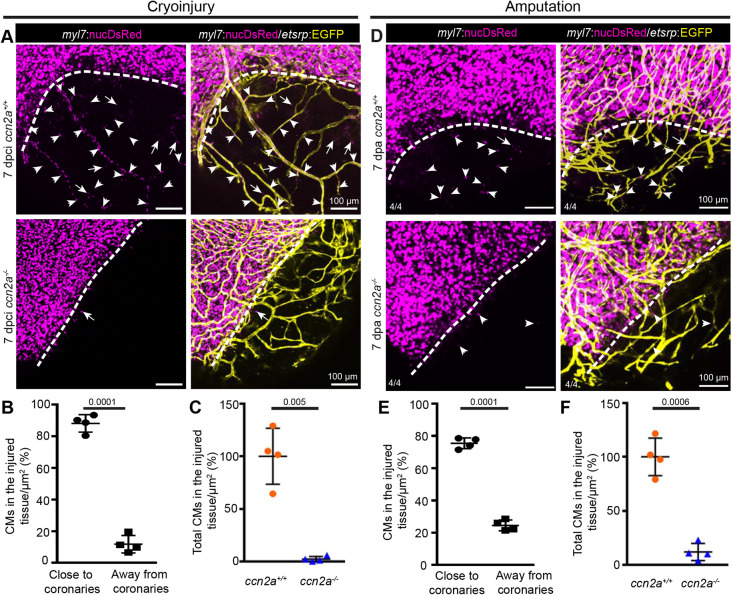
Much remains to be learned about how CMs repopulate a cardiac wound. An earlier study by Itou et al. used a photoconvertible protein, Kaede, to reveal evidence for CM migration after resection injury, and suggested that this process is required for heart regeneration (Itou et al., 2012). Published evidence also indicates that new coronary vessel formation starts as early as 15 h post cryoinjury (hpci) (Marín-Juez et al., 2016) and is crucial for heart regeneration (Harrison et al., 2015). As ccn2a transcripts are detected during the early stages post-injury, we sought to investigate the role of Ccn2a in coronary angiogenesis and CM colonization at the site of injury. To explore the relationship between coronary angiogenesis and CM infiltration into the wound, we used double-transgenic [Tg(etv2:EGFP)/Tg(myl7:nucDsRed)] (Mably et al., 2003; Proulx et al., 2010) reporter animals that expressed EGFP in endothelial cells and DsRed in CM nuclei. In wild-type cryoinjured hearts, even though new coronary vessels begin to sprout at ∼15 hpci (Marín-Juez et al., 2016), we did not observe myl7:nucDsRed+ CMs in the injured tissue of whole-mount heart at this stage (Fig. S3A). Interestingly, at 7 dpci, myl7:nucDsRed+ CMs were visible in the damaged tissue of wild-type hearts with the majority of these cells (∼88%) aligned with the new coronary vessels (Marín-Juez et al., 2019), suggesting that CMs track along the new coronary vessels (Fig. 3A,B). In contrast, in ccn2a−/− animals, even though coronary angiogenesis after injury appears unaffected, CM repopulation of the cardiac wound was impaired, as evaluated using both whole-mount imaging (Fig. 3A,C and Fig. S3B) and histology (Fig. S3C,D). CM infiltration was also impaired in ccn2a+/− animals (Fig. S3B). These data suggest that Ccn2a is required for CM infiltration into the cardiac wound during regeneration.
Fig. 3.
Cardiomyocyte tracking along new coronary vessels into injured tissue fails in ccn2a mutants. (A) Maximum intensity projections of confocal images of freshly isolated whole-mount heart evaluated for coronary angiogenesis and CM tracking along the new coronary vessels into the injured tissue. DsRed expression marks CM nuclei (magenta); EGFP expression marks coronary vessels (yellow). Arrowheads and arrows indicate CMs along and distant from new coronary vessels in the injured tissue, respectively. Dotted lines indicate the injury border. (B) Analysis of the percentage of the total infiltrated CMs observed along and distant from coronary vessels in the injured tissue quantified in wild-type whole-mount hearts (n=4). The total number of infiltrated CMs in the injured tissue of each sample was considered as 100%. (C) A comparison of the total infiltrated CMs in wild-type and ccn2a mutant whole-mount hearts at 7 dpci (n=4). The average for wild type was considered to be 100%. Data are mean±s.d. (D) Representative maximum intensity projections of confocal images of whole-mount heart freshly isolated at 7 dpa. DsRed marks CM nuclei; EGFP marks coronary vessels. Arrowheads and arrows indicate CMs residing along and distant from, respectively, new coronary vessels in the wound. Dotted lines indicate the wound border. (E) Analysis of the percentage of the total infiltrated CMs seen along and distant from coronary vessels in the injured tissue quantified in whole-mount wild-type hearts (n=4). Total number of CMs in the injured tissue of each sample was considered to be 100%. (F) A comparison of the total number of CMs in the injured tissue of wild-type and ccn2a mutant whole-mount hearts at 7 dpa. The average for wild type (n=4) was considered to be 100%. Data are mean±s.d. The statistical significance of differences was evaluated by a two-tailed Student's t-test (GraphPad Prism). Thickness of maximum intensity projections: 30 to 40 µm.
In the cryoinjury model, dead tissue persists at the site of injury and is eventually replaced by new tissue. Therefore, it was essential to determine that the appearance of CMs in the injured tissue is due to CMs infiltration into the wound and not because of the proliferation of residual viable CMs. To address this, we examined the process of regeneration using the ventricular resection model, in which heart regeneration occurs through repopulation of the apical wound with CMs (Poss et al., 2002). Our analysis, using whole-mount confocal imaging, showed that new coronary vessels covered the entire apical wound at 4 dpa in wild-type and ccn2a−/− hearts (Fig. S4A). However, the apical wound remained devoid of myl7:nucDsRed+ CMs at this stage in both genotypes (Fig. S4A,B). Corroborating the cryoinjury model, at 7 dpa, myl7:nucDsRed+ CMs were visible in the apical wound of wild-type hearts (Fig. 3D,E), as previously shown (Lepilina et al., 2006), and most of these cells were positioned along the new coronary vessels (Fig. 3D,E), suggesting that coronary angiogenesis precedes the appearance of CMs in the injured tissue and that these cells track along the new coronary vessels. Moreover, similar to the cryoinjury model, CM infiltration into the injured tissue was impaired in ccn2a−/− hearts (Fig. 3D-F and Fig. S5A,B). Taken together, our data suggest that CMs track along new coronary vessels into damaged tissue and that Ccn2a is required for these events but not for coronary angiogenesis during heart regeneration.
ccn2a is required for CM proliferation in injured hearts
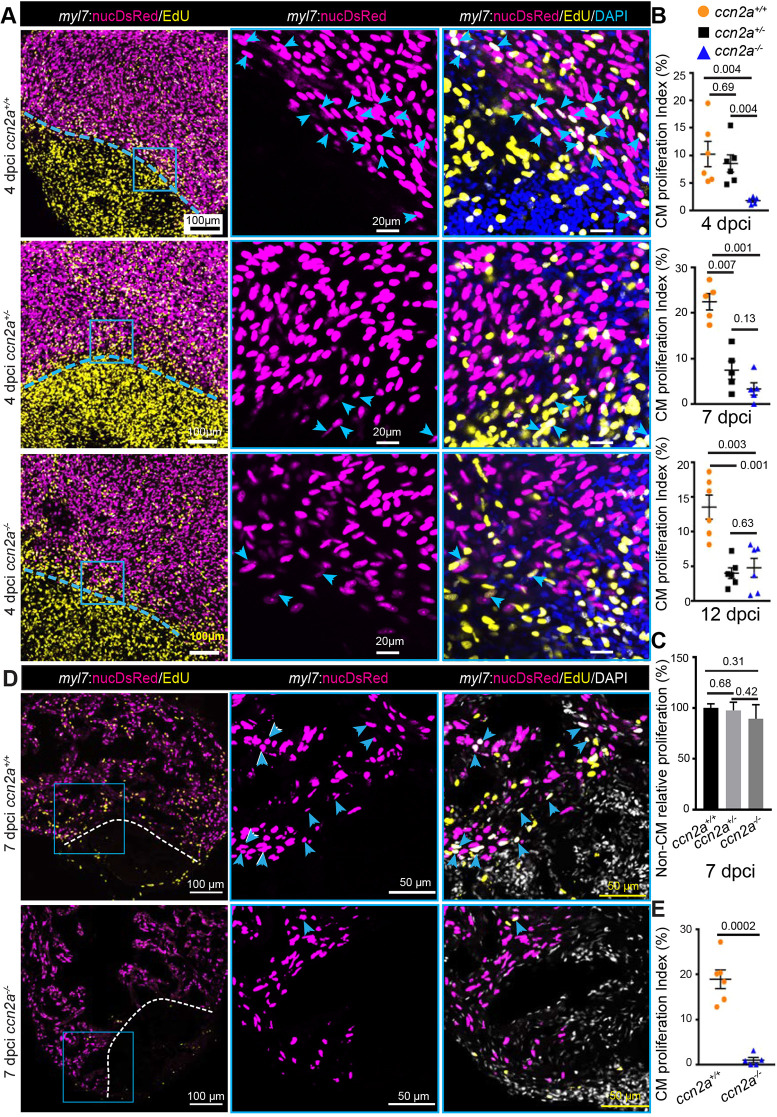
One of the factors affecting regeneration in injured ccn2a−/− hearts could be inefficient CM proliferation. We therefore performed a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay with the cryoinjury model to quantify CM proliferation indices. EdU incorporation by CM nuclei (myl7:nucDsRed+) was estimated in whole-mount hearts and sagittal sections. Whole-mount assessments revealed ∼75%, 90% and 60% reductions in CM proliferation in ccn2a−/− hearts relative to wild-type hearts at 4, 7 and 12 dpci, respectively (Fig. 4A,B and Fig. S6). Although CM proliferation was not affected in 4 dci ccn2a+/− hearts, an ∼65% reduction in CM proliferation was detected in ccn2a+/− hearts relative to wild-type hearts at 7 and 12 dpci (Fig. 4A,B and Fig. S6). Proliferation of non-CMs in 7 dpci hearts remains indistinguishable among the three genotypes (Fig. 4C). As laser scanning confocal microscopy can detect signals from a maximum tissue depth of ∼20 µm, the whole-mount data thus represents CM proliferation of the cortical myocardium. To determine overall CM cell cycle entry, we analyzed EdU incorporation in sagittal sections of the ventricle at 7 dpci and found an ∼90% reduction in the border zone of ccn2a−/− hearts relative to wild type (Fig. 4C,D). Both ccn2a+/− and ccn2a−/− animals displayed abnormal heart regeneration. However, the impairment in heart regeneration capacity was more prominent in ccn2a−/− animals than in ccn2a+/− siblings, suggesting that the influence of Ccn2a on heart regeneration is dependent on the amount of Ccn2a. To further test the effects of Ccn2a on CM proliferation, we carried out Ccn2a gain-of-function experiments using hsp70:ctgfa-FL-2a-EGFP (hereafter hsp70:ctgfa) transgenic fish (Mokalled et al., 2016), which express Ccn2a ubiquitously upon heat-shock. Upon cryoinjury, the fish were subjected to daily heat shocks from 1 to 6 dpci and then assessed for CM proliferation (Fig. 5A). EdU incorporation assay revealed a ∼45% increase in CM proliferation in the ccn2a-overexpressing fish compared with controls (Fig. 5B,C). Continued overexpression of Ccn2a for 1 month diminished scarring in ccn2a-overexpressing fish relative to controls (Fig. 5A,D-F). These results indicate that overexpression of Ccn2a enhances CM proliferation and heart regeneration.
Fig. 4.
ccn2a mutants display reduced cardiomyocyte proliferation post-cryoinjury. (A) Panels in the left column show maximum intensity projections (25-35 µm) of confocal images of 4 dpci whole-mount hearts stained for EdU (yellow; marks proliferating cells) and stained with DAPI (blue; marks all nuclei). DsRed marks CM nuclei. The corresponding high-magnification single-plane optical sections are shown in the middle and right columns. Arrowheads indicate EdU+/DsRed+ cells. Dotted lines mark the injury border. (B) The percentage of proliferating CMs in the border zone (up to 100 µm away from the injury border) was quantified in whole-mount ccn2a+/+, ccn2a+/− and ccn2a−/− hearts at 4 dpci (n=6 each), 7 dpci (n=5 each) and 12 dpci (n=6 each). Graphs represent the percentage ratio of DsRed+/EdU+ CMs to the total number of DsRed+ CMs. (C) Percentage of non-CM proliferation quantified in whole-mount ccn2a+/+, ccn2a+/− and ccn2a−/− hearts at 7 dpci (n=3). The graph represents the ratio of EdU+/DAPI+ nuclei to the total number DAPI+ nuclei in the injured tissue. The mean wild-type control value was set to 100%. (D) Maximum intensity projections of confocal images of a 10 µm ventricular sagittal cryosection stained for EdU (yellow) and with DAPI (white; marks all nuclei). DsRed marks CM nuclei; arrowheads indicate EdU+/DsRed+ cells. Dotted lines indicate the injury border. (E) The percentage of proliferating CMs in the border zone (up to 100 µm away from the injury border) was quantified in ccn2a+/+ and ccn2a−/− hearts at 7 dpci (n=6). At least two sections from each heart were analyzed. Data are mean±s.e.m. in B and E, and mean±s.d. in C. The statistical significance of differences was evaluated using Mann–Whitney nonparametric tests (GraphPad Prism).
Fig. 5.
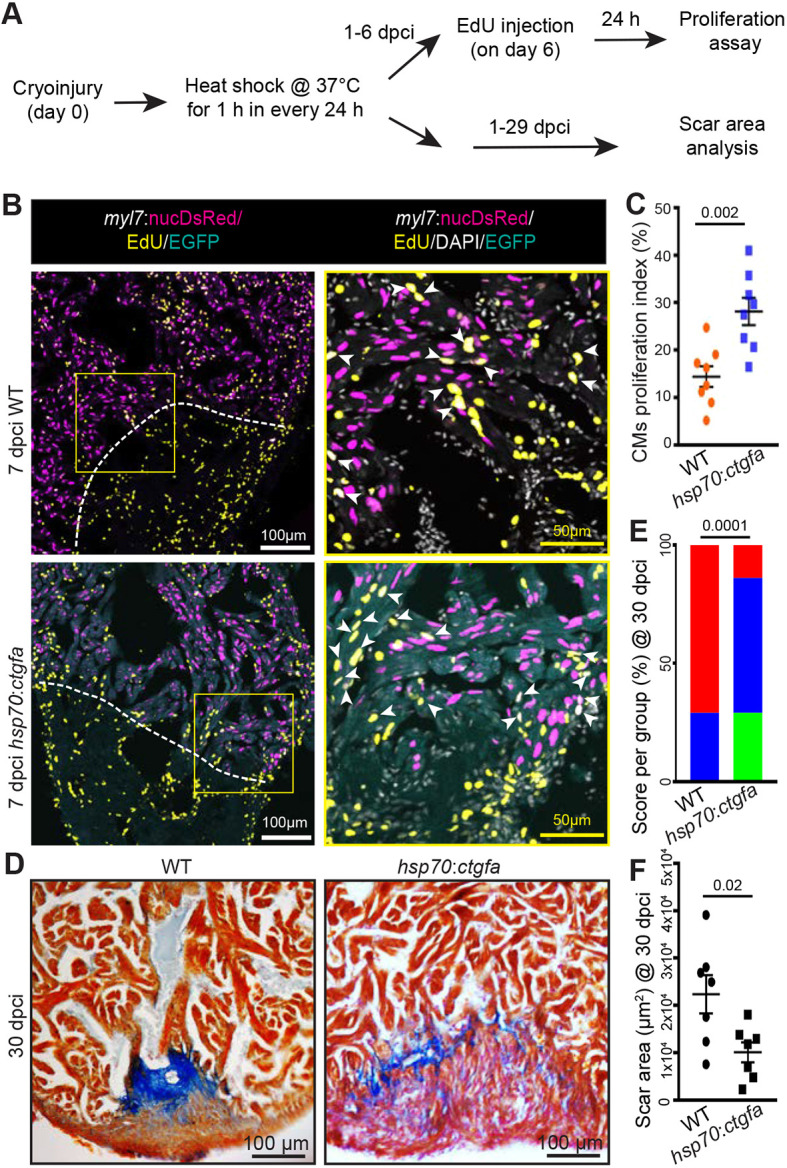
Ccn2a promotes cardiomyocyte proliferation and heart regeneration post-injury. (A) Schematic of the experimental procedures. Post-cryoinjury, wild-type and ccn2a-overexpressing transgenic animals were subjected to daily 1 h heat shock. CM proliferation was assessed at 7 dpci and scarring at 30 dpci. (B) Representative maximum intensity projections of confocal images of 10 μm sagittal cryosections of 7 dpci heart expressing DsRed in CM nuclei, immunostained for EGFP (cyan; marks cells expressing ectopic Ccn2a), stained for EdU (yellow; marks proliferating cells) and stained with DAPI (white; marks all nuclei). Arrowheads indicate EdU+/DsRed+ cells. (C) CM proliferation quantified in wild-type and ccn2a-overexpressing (hsp70:ctgfa) hearts at 7 dpci (n=8 each). At least two sagittal sections of each heart were analyzed for quantification. (D) Representative bright-field images of 12 μm sagittal paraffin sections of 30 dpci wild-type and hsp70:ctgfa hearts stained with AFOG (muscle, yellowish red; fibrin, brick red; collagen, blue). (E) Semi-quantitative analysis of scarring in wild-type and ccn2a-overexpressing (hsp70:ctgfa) hearts at 30 dpci (n=7 each). The histological section with the largest area covered by collagen in each heart was considered for scar quantification. Color-coded bar chart indicates degree of regeneration: green, complete; blue, moderate; red, very poor. Data indicate the percentage of total hearts represented by each score and the statistical significance of differences was evaluated by a wound-recovery χ2 test. (F) Dot plot shows highest area covered by collagenous scar on a tissue section from each heart. Data are mean±s.e.m. in C,F.
By contrast, when we analyzed the effect of ccn2a loss of function on CM proliferation in the ventricular resection model, no significant difference in CM proliferation was detected between wild-type and mutants (Fig. S7). Taken together, these results suggest that, although Ccn2a is a positive regulator of CM infiltration independent of the type of injury, its effect on CM proliferation depends on the type of injury. Nonetheless, the fact that mutant animals display defects in heart muscle regeneration in both injury models indicates the importance of this gene in regeneration.
Ccn2a regulates ECM gene expression in injured hearts
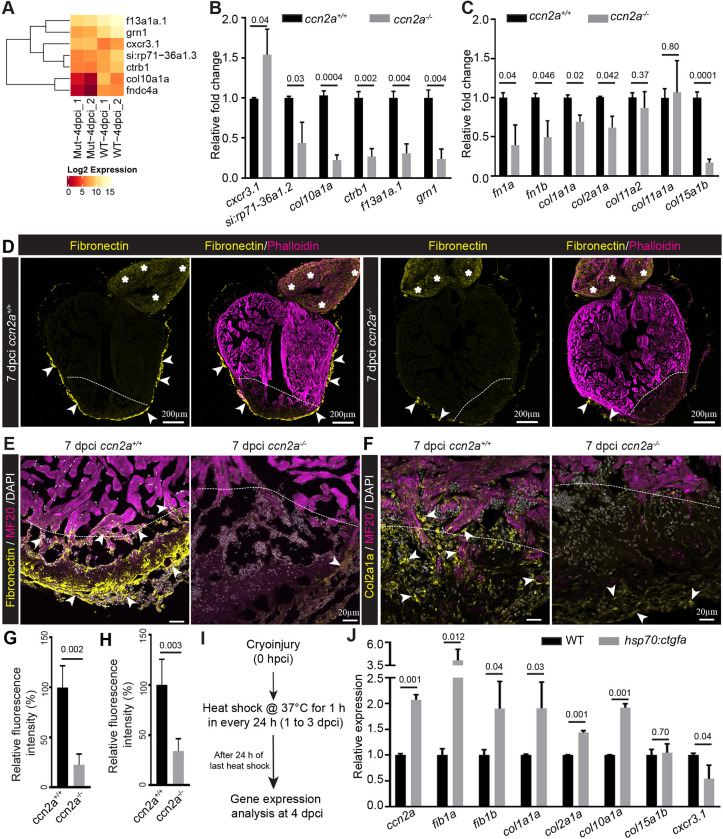
To identify the underlying molecular mechanisms involving Ccn2a in heart regeneration, we used the cardiac cryoinjury model, as it produced a more severe defect in ccn2a mutants. Comparative transcriptomic analysis was performed at 4 dpci to identify potentially affected downstream signaling cascades in injured ccn2a−/− hearts. Unexpectedly, only seven genes were found to be differentially expressed in ccn2a−/− hearts compared with wild type (Fig. 6A and Table S2). The expression of these genes was analyzed by qPCR. In line with the RNA-Seq data, the qPCR data showed that the expression levels of the chemokine receptor gene cxcr3.1 were increased by ∼50%; those of col10a1, an unannotated gene (si:rp71-36a1.2), ctrb1, f13a1a.1 and grn1 were decreased by ∼50 to 70% in mutant hearts at 4 dpci (Fig. 6B). As collagens and fibronectins are known to be important regulators of heart regeneration (Wang et al., 2013) and our transcriptome analysis showed that col10a1a and fibronectin type III domain containing 4a gene expression levels were altered in injured ccn2a−/− hearts, we analyzed the expression of other collagen and fibronectin genes. A decrease in expression of fn1a, fn1b, col1a1a, col2a1a, col10a1a and col15a1b was observed in ccn2a−/− hearts relative to wild type (Fig. 6C), whereas the expression of other collagens, i.e. col11a2 or col11a1a, remained unchanged (Fig. 6C).
Fig. 6.
Ccn2a regulates secreted protein gene expression in injured heart. (A) Heat map with color key showing changes in gene expression between ccn2a mutant (Mut) and wild-type (WT) hearts at 4 dpci based on RNA-sequencing analysis. (B) qPCR analysis to validate the RNA sequencing data (A) at 4 dpci (n=3, each sample is a pool of 6 hearts). (C) Quantification of fibronectin and collagen gene expression at 4 dpci (n=3, each sample is a pool of 6 hearts). (D) Sagittal cryosections of 7 dpci heart immunostained for fibronectin (yellow) and stained for F-actin (phalloidin; marks all cells). Arrows and asterisks indicate fibronectin localization in the ventricle and bulbus arteriosus, respectively. (E) Fibronectin and MF20 immunohistochemical staining of ventricular sagittal cryosection of 7 dpci heart. Arrowheads indicate fibronectin localization in the injured tissue. (F) Col2a1a and MF20 immunohistochemical staining of a ventricular sagittal cryosection of 7 dpci heart. Arrowheads indicate Col2a1a localization in the injured tissue. Dotted lines mark the injury border. (G,H) Quantification of the percent relative fluorescence intensity of fibronectin (G) and Col2a1a (H) in wild-type and ccn2a−/− heart sections (n=4). The heart section from each heart showing the highest fluorescence intensity for fibronectin or Col2a1a was considered for analysis, and the mean of the wild-type control value was set to 100%. (I) Schematic of the experimental protocol used to measure gene expression post-cryoinjury in wild-type and hsp70:ctgfa transgenic animals. After cryoinjury, animals were subjected to daily 1 h heat shocks for 3 days. RNA was isolated 24 h after the last heat shock. (J) Quantitative analysis of the expression of fib1a, fib1b, col1a1a, col2a1a, col10, col15a1b and cxcr3.1 in injured wild-type and ccn2a overexpressing (hsp70:ctgfa) hearts at 4 dpci (n=3, each sample represents a pool of six hearts). Data are mean±s.d. Values in B,C and J are normalized to the mean of the control. The statistical significance of differences was evaluated using a two-tailed Student's t-test (GraphPad Prism). The thickness of each maximum projection is 10-12 µm. Mean Ct values for this figure are provided in Table S4.
Corroborating the RNA expression data, protein localization of fibronectin and Col2a1a was also affected in injured ccn2a−/− ventricles (Fig. 6D-H). In wild-type hearts, fibronectin accumulated in the damaged cardiac tissue and bulbus arteriosus (BA) (Fig. 6D). In ccn2a−/− hearts, fibronectin expression was substantially reduced in the injured ventricle, with BA-localized expression unaffected (Fig. 6D,E,G). Similarly, Col2a1a expression was also reduced in the damaged ccn2a−/− cardiac tissue (Fig. 6F,H).
To further test whether these target genes are regulated by Ccn2a, we examined their expression levels upon ectopic expression of ccn2a during cardiac regeneration. Heat-shock treatments (Fig. 6I) resulted in an approximately two-fold increase in ccn2a expression in hsp70:ctgfa hearts relative to wild-type hearts (Fig. 6J). Supporting the loss-of-function data, expression of genes such as fn1a, fn1b, col1a1a, col2a1a and col10a1a were significantly increased upon overexpression of ccn2a in wild-type injured hearts (Fig. 6J). Moreover, cxcr3.1, the expression of which was increased in injured ccn2a−/− hearts, decreased upon ccn2a overexpression in wild-type injured hearts (Fig. 6J).
During heart regeneration, fibronectin is predominantly synthesized by epicardial cells, which migrate to cover the wound after injury (Wang et al., 2013). Because fibronectin expression is reduced in injured ccn2a−/− hearts, and Ccn2a is necessary for CM infiltration into the wound, we examined whether epicardial cell migration is also Ccn2a dependent. We performed immunostaining on sagittal ventricular sections for Cav1, an epicardial marker (Cao et al., 2016). However, no evidence of epicardial migration defects was observed in ccn2a−/− (Fig. S8). Taken together, our experiments suggest that Ccn2a negatively regulates the chemokine receptor gene cxcr3.1 and positively regulates the expression of the ECM genes fn1a, fn1b, col1a1a, col2a1a, col10a1a and col15a1b without affecting col11a2 or col11a1a expression, or epicardial cell migration into the injured cardiac tissue.
Ccn2a positively regulates nuclear pSmad3 localization in injured hearts
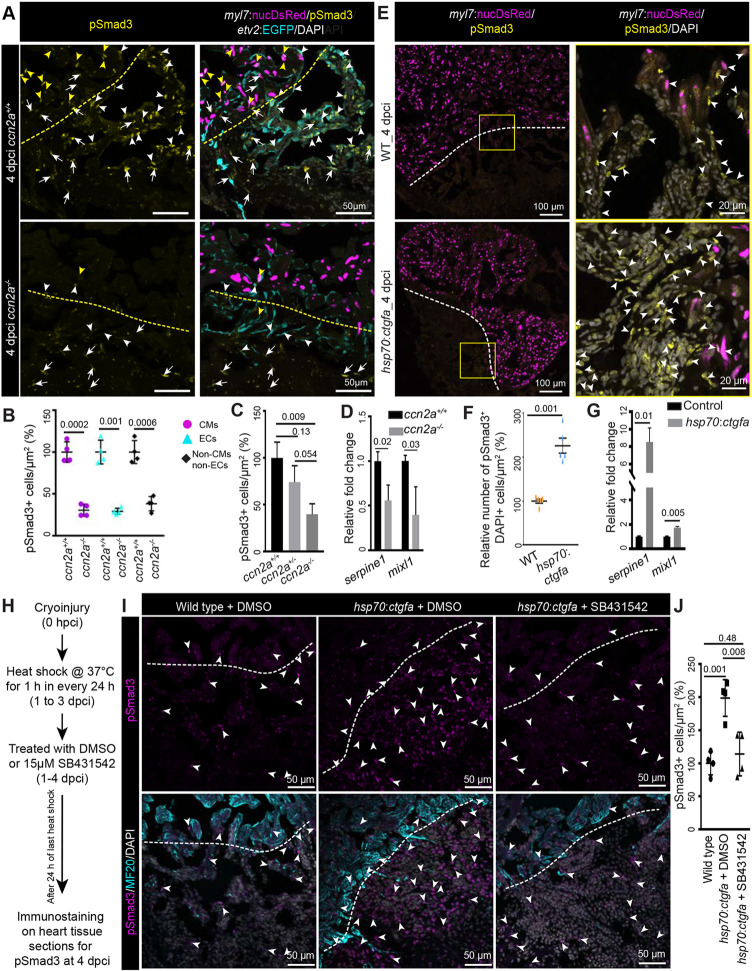
Next, we sought to understand how Ccn2a regulates ECM or cxcr3.1 chemokine receptor gene expression in damaged heart muscle. It has been described that CCN2 regulates TGFβ1-induced expression of fibronectin in Graves' orbital fibroblasts (Tsai et al., 2018). Another in vitro study reported a direct role for TGFβ signaling in the regulation of fibronectin and collagen expression (Ignotz and Massague, 1986), which we find are also regulated by Ccn2a in regenerating hearts. Thus, we investigated whether Ccn2a modulates Tgfβ signaling in injured cardiac tissue by analyzing the levels of phosphorylated nuclear-translocated Smad3, a transducer of the Tgfβ signal. Histological analysis showed that, in injured tissue, pSmad3-positive CM, endocardial cells and other cell nuclei are reduced by ∼69%, 71% and 62%, respectively, in ccn2a−/− hearts relative to wild-type hearts (Fig. 7A,B). Moreover, statistical analysis indicated a trend toward a decreasing number of nuclear pSmad3-positive cells in injured ccn2a heterozygous hearts compared with wild-type controls (Fig. 7C). Corroborating the histological data, expression of serpine1 and mixl1, which are reported to be direct Smad3 target genes (Mizutani et al., 2011; Xi et al., 2011; Zhang et al., 2011), was decreased by ∼50% in ccn2a−/− hearts relative to wild-type hearts at 4 dpci (Fig. 7D). Supporting the loss-of-function data, the nuclear pSmad3 level was significantly increased and expression of serpine1 and mixl1, was increased by ∼8- and 1.6-fold, respectively, upon ccn2a overexpression at 4 dpci (Fig. 7E-G), suggesting that Ccn2a regulates Tgfβ/pSmad3 signaling cascade in injured heart tissue. Next, we investigated whether Ccn2a-mediated activation of nuclear pSmad3 localization acts through the Tgfβ/pSmad3 pathway. We found that pharmacological inhibition of Tgfβ signaling cascade suppresses the Ccn2a gain-of-function-mediated activation of pSmad3 nuclear localization (Fig. 7H-J), consistent with a mechanism in which Ccn2a positively regulates nuclear pSmad3 localization through the Tgfβ/pSmad3 signaling cascade.
Fig. 7.
Ccn2a levels positively modulate nuclear pSmad3 localization in injured hearts. (A) Maximum intensity projections of confocal images of sagittal cryosections of 4 dpci hearts expressing DsRed in CM nuclei (magenta) and EGFP in endocardial/endothelial cells (cyan), immunostained for pSmad3 (yellow) and stained with DAPI (white; marks all nuclei). Yellow and white arrowheads indicate DsRed+/pSmad3+ and EGFP+/pSmad3+ cells, respectively. White arrows indicate cells that are DAPI+/pSmad3+ but DsRed− and EGFP−. Dotted lines indicate the wound edge. (B) Percentage of Tgfβ activity quantified in ccn2a+/+ and ccn2a−/− hearts (n=4). The graph represents the ratio of nuclear DsRed+/pSmad3+ CMs to the total number of nuclear DsRed+ CMs in the border zone (CMs); the ratio of EGFP+/pSmad3+ cells to the total number of EGFP+ endocardial/endothelial cells in the injured tissue (ECs); and the ratio of pSmad3+/DAPI+ cells to the total number of EGFP− and DsRed− cells per unit area in the injured tissue (non-CMs and non-ECs). The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. (C) Percentage of Tgfβ activity quantified in ccn2a+/+, ccn2a+/− and ccn2a−/− hearts at 4 dpci (n=3). The graph represents the ratio of pSmad3+/DAPI+ nuclei to the total number DAPI+ nuclei in the injured tissue. The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. (D) qPCR analysis of serpine1 and mixl1 expression at 4 dpci (n=3, each sample is a pool of six hearts). (E) Maximum intensity projection of confocal images of sagittal cryosection through 4 dpci wild-type and ccn2a-overexpressing (hsp70:ctgfa) heart expressing DsRed in CM nuclei (magenta), immunostained for pSmad3 (yellow) and stained with DAPI (white; marks all nuclei). Arrowheads indicate pSmad3+/DAPI+ cells. Dotted lines indicate the wound edge. (F) Nuclear pSmad3+ cells in the injured tissue quantified in wild-type and ccn2a-overexpressing (hsp70:ctgfa) heart (n=5 each). Two sagittal heart sections from each heart were considered for quantification. The mean wild-type control value was set to 100%. (G) qPCR analysis of serpine1 and mixl1 expression at 4 dpci (n=3, each sample is a pool of four hearts). (H) Schematic of the experimental procedures used in I and J. (I) Maximum intensity projection of optical sections of sagittal cryosections through 4 dpci wild-type heart and DMSO or SB431542 treated ccn2a-overexpressing (hsp70:ctgfa) heart, immunostained for pSmad3 (magenta) and with MF20 (cyan), and stained with DAPI (white; marks all nuclei). Arrowheads indicate pSmad3+/DAPI+ cells. Dotted lines indicate the wound edge. (J) Percentage of Tgfβ activity quantified in wild-type, and in DMSO or SB431542-treated ccn2a-overexpressing hearts at 4 dpci (n=4). The graph represents the ratio of pSmad3+/DAPI+ nuclei to the total number DAPI+ nuclei in the injured tissue. The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. Data are mean±s.d. A two-tailed Student's t-test was used to evaluate the statistical significance of the differences (GraphPad Prism). Thickness of the each maximum projection is 10-12 µm. Mean Ct values are provided in Table S4.
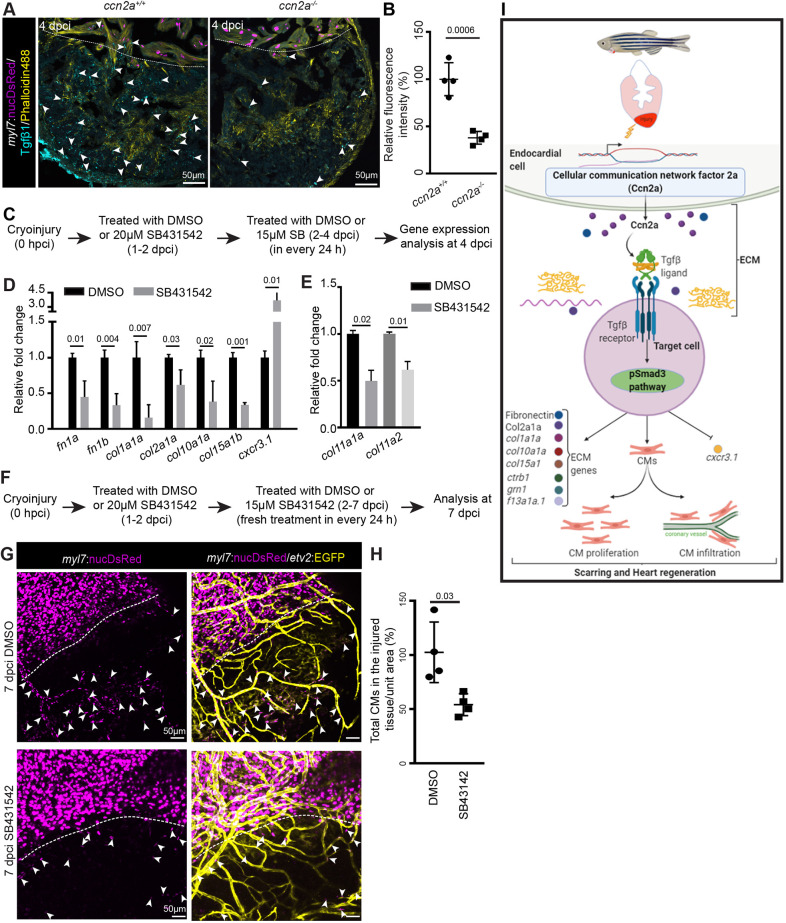
To test the level of Tgfβ ligands, we performed immunohistochemistry to detect Tgfβ1, one of the known ligands of this pathway. The data revealed a ∼60% reduction in Tgfβ1 expression in ccn2a−/− hearts relative to wild type (Fig. 8A,B), suggesting that Ccn2a positively regulates the level of Tgfβ1. To understand whether Cc2a regulates Tgfβ ligand expression at the transcriptional level, we performed qPCR analysis and found that Ccn2a does not regulate the expression of tgfb1, tgfb2 and tgfb3, as their transcript levels remain indistinguishable between ccn2a+/+ and ccn2a−/− hearts (Fig. S9A). Their expression was also indistinguishable between injured wild-type and ccn2a-overexpressing hearts (Fig. S9B). This result suggests Ccn2a regulates Tgfβ/pSmad3 signaling likely by modulating stability or bioavailability of Tgfβ ligand in regenerating heart.
Fig. 8.
Decreased Tgfβ1 expression is observed in injured ccn2a mutant heart and Tgfβ/pSmad3 inhibition recapitulates the genetic regulation by ccn2a in injured heart. (A) Maximum intensity projection of confocal images of 10 μm sagittal cryosection of wild-type and ccn2a−/− heart at 4 dpci, expressing DsRed in CM nuclei (magenta), immunostained for Tgfβ1 (cyan) and stained for F-actin (yellow; marks all cells). Arrowheads indicate Tgfβ1 expression. Dotted lines mark the injury edge. (B) Percentage of relative fluorescence intensity of Tgfβ1 (n=4 each). The heart section from each heart showing the highest fluorescence intensity for Tgfβ1 was considered for quantification, and the mean of the wild-type control value was set to 100%. (C) Schematic of the experimental procedures used in D,E. (D,E) qPCR-based quantification of gene expression from DMSO- or SB431542 (TGFβ type I receptor inhibitor)-treated animals at 4 dpci (n=3, each sample is a pool of six hearts). The mean value of the DMSO-treated control for each gene was set to 1. (F) Schematic of the experimental procedures used in G,H. (G) Maximum intensity projections of confocal images of 7 dpci whole-mount heart expressing DsRed in CM nuclei and EGFP in endothelial/endocardial cells. Arrowheads indicate infiltrated CMs in the injured tissue. Dotted lines indicate the injury edge. (H) Analysis of infiltrated CMs in injured cardiac tissue quantified four DMSO-treated and four SB431542-treated wild-type animals from two independent experiments. The mean of the DMSO-treated control value was set to 100%. Data are mean±s.d. (I) Model of the regulatory role of Ccn2a in heart regeneration. CM, cardiomyocyte; ECM, extracellular matrix. The statistical significance of differences was evaluated by a two-tailed Student's t-test (GraphPad Prism). Mean Ct values are provided in Table S4.
Ccn2a regulates ECM gene expression and CM infiltration through Tgfβ/pSmad3 signaling in injured hearts
Our results implicated Ccn2a in mediating the downregulation of fn1a, fn1b, col1a1a, col2a1a, col10a1a and col15a1b, and upregulation of cxcr3.1. To test whether this regulation is dependent on the Tgfβ pathway, we used the pharmacological inhibitor SB431542 to disrupt Tgfβ signaling from 1 to 4 dpci (Fig. 8C). We found a significant decrease in expression of fn1a, fn1b, col1a1a, col2a1a, col10a1a and col15a1b (Fig. 8D). Additionally, cxcr3.1 expression increased approximately fourfold upon inhibition of the Tgfβ pathway, as also observed in ccn2a−/− animals (Fig. 8D). However, the expression of col11a1a or col11a2, genes that were unaltered in ccn2a−/− injured hearts, was decreased upon Tgfβ signaling inhibition (Fig. 8E).
Next, we explored whether global pharmacological perturbation of Tgfβ signaling had an impact on the Tgfβ-mediated functions in other tissues. We analyzed the expression of col1a1a, col11a1, col15a1b, fn1b and fn1b in non-cardiac tissues upon pharmacological inhibition of Tgfβ pathway by SB431542. Decreased expression of col11a1 and col1a1a was observed in the healthy vertebral tissue and eyes, respectively, upon inhibition of the Tgfβ pathway (Fig. S10). However, the expression of other collagen and fibronectin genes remained unaltered in these tissues. Moreover, in the brain, inhibition of Tgfβ pathway had no detectable effect on the expression of these genes (Fig. S10). Taken together, these results indicate that pharmacological inhibition of the Tgfβ pathway has context-specific targets.
Tgfβ signaling is essential for CM proliferation during zebrafish heart regeneration (Chablais and Jazwinska, 2012). To investigate whether the deficiency in CM repopulation of the injured heart tissue in ccn2a−/− animals was due to the suppression of the Tgfβ pathway, we assessed the effects of pharmacological Tgfβ inhibition (Fig. 8F). SB431542-mediated inhibition of the Tgfβ pathway from 1 to 7 dpci resulted in diminished CM repopulation of the injured tissue at 7 dpci (Fig. 8G,H). The observation that nuclear pSmad3 levels were reduced in injured ccn2a mutant hearts, and increased upon transgenic ectopic overexpression of Ccn2a in injured hearts, suggests that Tgfβ/pSmad3 signaling is likely to function downstream of Ccn2a.
DISCUSSION
Based on our findings, we propose the following model. Upon injury, Ccn2a is synthesized and secreted into the ECM from endocardial cells of the wound. Here, it functions to modulate Tgfβ/pSmad3 signaling to promote CM proliferation and infiltration, and expression of pro-regenerative ECM genes. The regulation of Tgfβ signaling by Ccn2a could be either direct, through interaction with the Tgfβ ligands, or indirect, by influencing the mechanical properties of the ECM (Fig. 8I).
It is interesting to note that the role of Ccn2a in CM proliferation is mode-of-injury dependent. One of the crucial differences between the cryoinjury and resection models is the amount of fibrotic tissue produced. After cryoinjury, massive transient collagen appears in place of the dead tissue mass (González-Rosa et al., 2011); in contrast, in resection-injured hearts, a smaller amount of collagen localizes mostly superficially on the cut surface of the ventricle (Poss et al., 2002). Thus, the influence of Ccn2a on CM proliferation may be dependent on the amount or composition of collagen. This finding also points to the possibility that although both cardiac injury models are well established and many cellular and molecular responses are shared, fine-tuning of the signaling cascades involved in these heart regeneration models may differ. Further exploration of these differences would be helpful to gain insights into the mode of repair.
Genetic analysis indicated that Ccn2a positively regulates nuclear pSmad3 localization and ECM genes such as fibronectin and collagen gene expression, and negatively regulates chemokine receptor cxcr3.1 expression, during heart regeneration. It is known that some collagens and fibronectin are downstream targets of the TGFβ/pSMAD3 pathway (Ignotz and Massague, 1986), and CCN2 regulates TGFβ1-induced expression of fibronectin in vitro (Tsai et al., 2018). Chemical inhibition of Tgfβ pathway in wild-type animals shows that, although all the regulated genes in injured ccn2a mutant hearts are pSmad3 targets, some of the pSmad3 target ECM genes uch as col11a2 and col11a1a are not regulated by Ccn2a in injured hearts. This indicates that, in regenerating cardiac tissue, Ccn2a does not work through global activation of pSmad3 signaling. Heterogeneity in molecular signaling and behavior exists even within the same cell type, due to them being present in different microenvironments (desJardins-Park et al., 2018; Rognoni and Watt, 2018). Thus, regulation of Tgfβ/pSmad3 targets by Ccn2a may be dependent on the microenvironment of the target cells in the damaged cardiac tissue.
All TGFβ ligands are synthesized as inactive precursor molecules containing a propeptide region and secreted into the ECM as a large latent complex. Physical interactions between the latent TGFβ and ECM molecules are essential for the transformation into an active form of TGFβ ligand (Dabovic et al., 2002; Yoshinaga et al., 2008; Shi et al., 2011). Another study using human fibroblasts suggested that CCN2 regulates TGFβ-induced phosphorylation of Smad1 through the integrin αvβ3 receptor (Nakerakanti et al., 2011). Our finding that Ccn2a positively regulates Tgfβ1 localization in injured cardiac tissue without apparent effects on expression of its mRNA suggests a role for Ccn2a in regulating the processing or stability of Tgfβ1. Abreu et al. (2002) reported that CCN2 can enhance TGFβ1 binding to its receptor and signaling in vitro. In addition to the direct interaction of ECM molecules with latent TGFβ, the mechanical state of the ECM also regulates TGFβ activity (Wipff et al., 2007; Giacomini et al., 2012; Henderson et al., 2013). These pieces of evidence suggest that a plausible mechanism underlying the regulation of Tgfβ1 processing or stability in injured zebrafish hearts could involve either a physical interaction of Ccn2a with latent Tgfβ or other complex mechanical properties of the wound, but these possibilities need to be further elucidated.
The immune response plays a crucial role in regeneration (Hui et al., 2017; Lai et al., 2017). In mammals, immune cells express CXCR3 and preferentially help in the migration of immune cells into inflamed tissue (Torraca et al., 2015). In teleosts, including zebrafish, there are two paralogs: cxcr3.1 and cxcr3.2. Although the expression of cxcr3.1 and cxcr3.2 in relation to infection or inflammation in zebrafish has not been determined, in other teleosts, such as ayu (Plecoglossus altivelis), cxcr3.1- and cxcr3.2-positive macrophages exhibit M1 and M2 polarization, respectively, upon bacterial infection (Lu et al., 2017). M1 macrophages are pro-inflammatory; in contrast, M2 macrophages are associated with anti-inflammatory states (Liu et al., 2014). In ccn2a mutants and animals treated with chemical inhibitors of Tgfβ signaling, cxcr3.1 expression was increased in injured heart without affecting the expression of its paralog cxcr3.2 (data not shown), indicating that Ccn2a likely suppresses inflammatory signals by inhibiting M1 polarization of macrophages through pSmad3 signaling. In the future, it will be interesting to explore the functional role of cxcr3.1 in heart regeneration or fibrosis.
Taken together, our study highlights the importance of Ccn2a function in heart regeneration and shows that secreted signaling cues in early injury sites play an important role in determining the extent of scarring; specifically, whether a transient collagen can be efficiently replaced with functional tissue. Thus, it would be worth exploring whether targeted manipulation of ECM production can serve as a strategy to induce mammalian heart regeneration and improve cardiac function.
MATERIALS AND METHODS
Zebrafish maintenance
Wild-type AB and transgenic Tg(–5.1myl7:nDsRed2)f2 (Mably et al., 2003) [hereafter Tg(myl7:nucDsRed)], TgBAC(etv2:EGFP)ci1 (Proulx et al., 2010) [hereafter Tg(etv2:EGFP)], Tg(kdrl:HRAS-mCherry)s896 (Chi et al., 2008) and Tg(hsp70:ctgfa-FL-2a-EGFP) (Mokalled et al., 2016) zebrafish were maintained in a state-of-the-art zebrafish aquarium as described previously (Kimmel et al., 1995). Four- to 10-month-old male and female animals of ∼2.5 to 3 cm in length were used.
Ethics statement
For zebrafish maintenance and experimentation, the guidelines recommended by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India, were followed. The Institute Animal Ethics Committee (IAEC) approved the animal care procedures and protocols used in this study.
qPCR and gene expression analysis
For qPCR analysis, total RNA was isolated from sham, cryoinjured cardiac ventricles and non-cardiac tissues (eyes, brain and vertebral tissue) at desired time points using an RNeasy Micro kit (Qiagen) according to the manufacturer's instructions. In brief, five hearts were pooled per biological replicate, and 1 µg of total RNA was reverse transcribed into cDNA with a SuperScriptIII cDNA synthesis kit (Invitrogen). For ccn2a transcript stability analysis, total RNA was isolated from 48 hpf ccn2a+/+, ccn2a+/− and ccn2a−/− embryos using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Forty embryos were pooled per biological replicate, and 2 µg of total RNA was reverse transcribed into cDNA with MMLV reverse transcriptase (Invitrogen). The prepared cDNA was used to perform qPCR with a PCR Max Real-time PCR detection system (Cole Parmer). The primer sequences used in this study are provided in Table S3. mRNA expression levels relative to ef1α or rpl13 expression levels were calculated using the ΔCt method. Mean Ct values are listed in Table S4.
In situ hybridization
For RNA probe synthesis, a partial cDNA template of ccn2a or ccn2b was amplified from an embryonic cDNA library using primers shown in Table S1 and cloned into a pGEMT easy vector. Digoxigenin (DIG)-labeled ccn2a and ccn2b riboprobes were synthesized using linearized vectors and SP6 and T7 RNA polymerase, respectively.
For in situ hybridization on sections, tissue samples were processed for wax sectioning and in situ hybridization was performed as previously described (Lepilina et al., 2006). Briefly, 15 µm zebrafish heart tissue sections were rehydrated, permeabilized, washed in acid, blocked and hybridized with a DIG-labeled riboprobe (500 ng/ml in hybridization buffer) at 65°C overnight. Subsequently, the tissue sections were incubated with an alkaline phosphatase-conjugated anti-DIG antibody (Roche). Finally, after washing, the signal was detected with an NBT/BCIP staining solution (Roche), and the sections were mounted in Kaiser's glycerol gelatin (Merck) for imaging.
Generation of a BAC recombineering-based TgBAC(ccn2a:EGFP) line
Using the UCSC genome browser (Zv6 assembly, https://genome-euro.ucsc.edu), a BAC clone encompassing both the upstream and downstream sequences around the ATG start site of ccn2a was identified. BAC clone CH211-156I24 (ID: 23907380), which covers ∼53 kb of regulatory sequences upstream from the start ATG and ∼55 kb downstream from the last exon of ccn2a, was purchased from the BACPAC Resources Center, Children's Hospital Oakland (https://bacpacresources.org/). BAC recombineering was performed as described previously (Bussmann and Schulte-Merker, 2011). Briefly, in the first step, the BAC clone was transformed with the pRedET plasmid (Gene Bridges), which enabled arabinose-inducible homologous recombination. In the next step, PCR-amplified Tol2 arms in opposing directions flanking an ampicillin resistance cassette were inserted into the BAC backbone. Finally, a combination of EGFP and a kanamycin-resistance cassette was inserted at the start ATG of ccn2a. The final BAC DNA was purified using a NucleoBond XTra BAC kit (Machery Nagel), and correct insertion of the EGFP-kanamycin-resistance cassette was verified by sequencing. In total, 10-15 pg of BAC DNA was co-injected with 50 pg of Tol2 transposase mRNA into a one-cell wild-type embryo. Founders were outcrossed, and two F1 animals were isolated and maintained. Outcrossing of both GFP-positive F1 transgenic lines (and the following generations) revealed segregation of GFP expression at Mendelian ratios, suggesting that each line carried a single insertion. The reporter line was named TgBAC(ccn2a:EGFP)pp01. Hemizygous TgBAC(ccn2a:EGFP) animals were analyzed in this study.
Cryoinjury and heat-shock experiments
Cardiac ventricular cryoinjuries were made as described previously (González-Rosa and Mercader, 2012). In summary, animals were anesthetized by immersion in 0.02% tricaine and immobilized into a wet foam holder. The heart ventricle was exposed by making a small thoracic incision and gently squeezing the abdomen. To injure the heart, a 0.4 mm diameter copper filament linked to a polypropylene insulation tube was cooled by dipping in liquid nitrogen and placed on the ventricular apex until thawing was observed (a few seconds). After the operation, the fish were transferred to a tank with fresh system water and, after recovery, the fish were placed in the aquarium. For heat-shock experiments, wild-type (as control) and clutch mate Tg(hsp70:ctgfa:2a:EGFP) fish received heat shock by incubation in preheated system water at 37°C for 1 h (maximum six fish per liter) in 24 h intervals for the desired period.
Coronary growth and CM infiltration in a damaged tissue assay
Cryoinjured or amputated Tg(etv2:EGFP)::Tg(myl7:nucDsRed) animals were kept in the aquarium until 4, 7 or 12 dpci or dpa. For whole-mount analysis, freshly isolated hearts collected at the desired time points were immediately embedded in 1% low-melting point agarose/1×PBS containing 0.001% tricaine in a glass-bottomed dish. Confocal sections were imaged with a Leica SP8 confocal microscope and processed to obtain maximal intensity projections. DsRed+ nuclei in the injured/scar tissue were considered infiltrated CMs.
Sagittal cryosections (10 µm) through Tg(myl7:nucDsRed)+ hearts were stained with Alexa-488-conjugated phalloidin and 4′,6′-diamidino-2-phenylindole (DAPI) to study CM infiltration into the damaged tissue in histological sections. Confocal sections were imaged with a Leica SP8 confocal microscope and processed to obtain maximal intensity projection.
EdU incorporation assay
For EdU incorporation analysis, injured fish were anesthetized with 0.02% tricaine, and 10 µl of 10 mM EdU (Click-iT EdU, Invitrogen) was injected intraperitoneally into each fish at 24 h before heart isolation. Harvested hearts were fixed in 4% PFA for 20 min at room temperature and processed for whole-mount EdU labeling or cryosectioning. EdU labeling was performed on the whole-mount hearts or cardiac tissue sections according to the manufacturer's instructions. For co-immunostaining with EdU detection, immunostaining was performed before EdU staining. DAPI (0.5 mg/ml water, Sigma) was used to detect DNA. For imaging, the whole-mount stained hearts were embedded in 1% low-melting point agarose in a glass-bottomed dish. Optical sections were captured using a Leica SP8 confocal microscope and quantified using LAS X software.
Histological and immunohistochemical analyses
Cardiac tissue was embedded in paraffin following standard protocols, and sagittal sections of the heart were prepared using a microtome (Bright Instruments). For histological analysis, acid fuchsin-orange G (AFOG) staining was performed using an AFOG staining kit (BioGnost) following the manufacturer's instructions. The slides were mounted with Entellan (Merck) and bright-field images were captured using a Leica stereoscope. The degree of heart regeneration was analyzed based on the presence of collagenous scar on the 60 dpci or 150 dpci heart tissue sections. For each heart, the section showing the highest area covered by scar tissue was considered for analysis. If the highest area covered by the collagenous scar without any visible invading muscle on the tissue section was <2000 µm2, then that heart was considered as ‘vigorously regenerating’ and if the scar area was >15,000 µm2, then that heart was considered as ‘poorly regenerating’ or having a persistent collagenous scar at that time point. To group the hearts based on degree of regeneration, we considered complete regeneration when highest scar area was <2000 µm2, moderate regeneration when highest scar area was in between 2000 and 15,000 µm2, and very poor regeneration when highest scar area was >15,000 µm2.
For immunohistochemical analysis, cardiac tissue was embedded in tissue-freezing medium following standard protocols, and the heart was sagittally sectioned (10 µm, cryotome, Leica). Immunohistochemical analysis was performed on the tissue sections as described previously (Patra et al., 2017). Briefly, PBS-washed tissue sections were re-fixed, permeabilized, blocked and incubated with primary antibodies [mouse MF20, 1:40 (DSHB); rabbit anti-GFP, 1:400 (Novus Biologicals, NB600-308); rabbit anti-TGFβ1, 1:100 (Cloud-Clone, PAA124Mu01); rabbit anti-tropomyosin, 1:50 (Cloud-Clone, PAD449Mu01); rabbit anti-pSMAD3, 1:200 (Abcam, ab52903); rabbit anti-fibronectin, 1:200 (Sigma, F3648); and rabbit anti-Col2a1a, 1:100 (Genetex, GTX127988)] overnight at 4°C after blocking for 1 h in a blocking solution [5% goat serum (MP Biomedicals)/0.2% Tween 20/PBS]. Primary immune complexes were detected by AlexaFluor488-, AlexaFluor 555- or AlexaFluor 647-conjugated antibodies (1:400; Molecular Probes, A11034, A21428 and A21244). DAPI (0.5 mg/ml water, Sigma) was used to detect DNA. Optical sections were captured using a Leica SP8 or Zeiss LSM 710 confocal microscope.
RNA-sequencing
RNA was isolated from 4 dpci wild-type and ccn2a mutant cardiac ventricles (bulbus arteriosus and atrium were dissected out from each heart) using a RNeasy micro Kit (Qiagen). Five cardiac ventricles were pooled per biological replicate and two samples each from wild-type and ccn2a−/− animals were analyzed. To avoid genomic DNA contamination, samples were treated by on-column DNase digestion (DNase-Free DNase Set, Qiagen) during isolation. Total RNA and library integrity were verified with a LabChip Gx Touch 24 (Perkin Elmer) instrument. 2.2 µg of total RNA was used as input for Truseq Stranded mRNA Library preparation following the low sample protocol (Illumina). Sequencing was performed with the NextSeq500 instrument (Illumina) using v2 chemistry, resulting in an average of 23 M reads per library with 1×75 bp single end setup. The raw reads were assessed for quality, adapter content and duplication rates with FastQC. To trim reads after a quality drop below a mean of Q20 in a window of 10 nucleotides, Reaper version 13-100 was employed (Davis et al., 2013). Reads between 30 and 150 nucleotides were selected for further analyses. Selected reads were aligned versus the Ensembl Zebrafish genome version DanRer10 (GRCz10.87) using STAR 2.4.0a with the parameter ‘--outFilterMismatchNoverLmax 0.1’ to increase the maximum ratio of mismatches to mapped length to 10% (Dobin et al., 2013). The FeatureCounts 1.4.5-p1 tool from the Subread package was used to count the number of reads aligning to genes (Liao et al., 2014). Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. Differentially expressed genes were identified using DESeq2 version 1.62 (Love et al., 2014). Only genes with a minimum fold change of ±1.5 (log2±0.59), a maximum Benjamini-Hochberg corrected P-value of 0.05 and a minimum combined mean of five reads was deemed to be significantly differentially expressed. The Ensemble annotation was enriched with UniProt data (release 06.06.2014) based on Ensembl gene identifiers.
Drug treatment
The TGFβ type I receptor inhibitor SB431542 (Sigma) was dissolved at a stock concentration of 30mM in DMSO and used for treatment. Zebrafish were treated with a 20 µM or 15 µM SB431542 solution in system water from 1 to 2 dpci and from 2 to q4 or 7 dpci, respectively. Control animals were kept in system water containing 63 µl of DMSO/100 ml of system water. During the experiment, all animals were maintained at a density of four fish per 125 ml system water. At 4 or 7 dpci, animals were anesthetized, and hearts were isolated and processed for total RNA isolation and histology.
Microscopy and quantification
For whole-mount bright-field and confocal microscopy, hearts were embedded in 1% low–melting-point agarose in PBS and imaged with a Leica SP8 or M205FA (Leica). Immunostained slides were imaged using a Leica SP8 confocal microscope. After imaging, the acquired confocal z-stacks were processed, and cell counting was performed with LAS×(Leica) or ImageJ/Fiji software. Quantification of CM proliferation was performed in an area 100 μm from the border zone. Bright-field images were obtained with stereomicroscopes (M205FA, Leica; Stereodiscovery V8, Zeiss). Quantifications of CM proliferation, appearance in the wound, and pSmad3+ cells were performed using LAS X software. Fluorescence intensity analysis was performed using ImageJ/Fiji software. For fluorescence intensity measurement, images were converted to 8-bit and the scale was ensured to be equal for all images. Mean intensity and area was measured using the ROI manager tool in ImageJ/Fiji software. Degree of heart regeneration was considered based on the highest area covered by a collagenous scar on the heart section in each heart and a semi-quantitative scoring-based analysis was performed.
Statistical analysis
Statistical differences in qPCR expression, CM tracking along new coronary vessels into the injured tissue, nuclear pSmad3 quantification, and fibronectin, Col2a1a and Tgfβ1 immunofluorescent intensity were analyzed using a two-tailed Student's t-test. CM proliferation was analyzed using Mann–Whitney nonparametric tests. For scar analysis, statistical significance of differences was evaluated by a wound-recovery χ2 test. In all analysis, changes were considered to be statistically significant when P<0.05. Data were processed with the GraphPad Prism7 software. Values are represented as mean±s.d. or mean±s.e.m.
Supplementary Material
Acknowledgements
C.P. is grateful to Dr Ratnaparkhi, and Dr M. Housley for revising the manuscript critically. We thank S. Bhujbal and M. Müller-Boche for excellent fish care at Agharkar Research Institutee and the Max Planck Institute, respectively, and H. M. Maischein at the Max Planck Institute for microinjections.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.M., C.P.; Methodology: D.M., G.W., K.D.P., D.Y.R.S., C.P.; Validation: D.M., G.W., A.R.; Formal analysis: D.M., G.W., M.H.M., Z.K., A.L.D., A.R., S.G.; Investigation: D.M., G.W., M.H.M., A.L.D., C.P.; Resources: K.D.P., C.P.; Data curation: D.M.; Writing - original draft: D.M., C.P.; Writing - review & editing: D.M., Z.K., K.D.P., D.Y.R.S., C.P.; Supervision: C.P.; Project administration: D.M., C.P.; Funding acquisition: K.D.P., C.P.
Funding
This work was supported in part by the Max-Planck-Gesellschaft (to C.P. and D.Y.R.S.) and Department of Science and Technology India (Max-Planck-partner group award to C.P.), by an Agharkar Research Institute internal grant, and by the National Institutes of Health (R01 HL081674 and R01 HL131319 to K.D.P.). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in GEO under accession number GSE164491.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193219.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193219.reviewer-comments.pdf
References
- Abreu J. G., Ketpura N. I., Reversade B. and De Robertis E. M. (2002). Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat. Cell Biol. 4, 599-604. 10.1038/ncb826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accornero F., van Berlo J. H., Correll R. N., Elrod J. W., Sargent M. A., York A., Rabinowitz J. E., Leask A. and Molkentin J. D. (2015). Genetic analysis of connective tissue growth factor as an effector of transforming growth factor β signaling and cardiac remodeling. Mol. Cell. Biol. 35, 2154-2164. 10.1128/MCB.00199-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini A., Li Z., Mericskay M. and Decaux J. F. (2015). Regulation of connective tissue growth factor and cardiac fibrosis by an SRF/MicroRNA-133a axis. PLoS ONE 10, e0139858 10.1371/journal.pone.0139858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassat E., Mutlak Y. E., Genzelinakh A., Shadrin I. Y., Baruch Umansky K., Yifa O., Kain D., Rajchman D., Leach J., Riabov Bassat D. et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179-184. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Bhardwaj R. D., Bernard S., Zdunek S., Barnabe-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B. A., Druid H. et al. (2009). Evidence for cardiomyocyte renewal in humans. Science 324, 98-102. 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J. and Schulte-Merker S. (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development138, 4327-4332. 10.1242/dev.068080 [DOI]
- Cao J., Navis A., Cox B. D., Dickson A. L., Gemberling M., Karra R., Bagnat M. and Poss K. D. (2016). Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development 143, 232-243. 10.1242/dev.130534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F. and Jazwinska A. (2012). The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 139, 1921-1930. 10.1242/dev.078543 [DOI] [PubMed] [Google Scholar]
- Chi N. C., Shaw R. M., De Val S., Kang G., Jan L. Y., Black B. L. and Stainier D. Y. (2008). Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734-739. 10.1101/gad.1629408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B., Chen Y., Colarossi C., Obata H., Zambuto L., Perle M. A. and Rifkin D. B. (2002). Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J. Cell Biol. 156, 227-232. 10.1083/jcb.200111080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. P., van Dongen S., Abreu-Goodger C., Bartonicek N. and Enright A. J. (2013). Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63, 41-49. 10.1016/j.ymeth.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. G., Balding L. C., Candido R., Burns W. C., Cao Z., Twigg S. M. and Burrell L. M. (2005). Connective tissue growth factor and cardiac fibrosis after myocardial infarction. J. Histochem. Cytochem. 53, 1245-1256. 10.1369/jhc.4A6560.2005 [DOI] [PubMed] [Google Scholar]
- desJardins-Park H. E., Foster D. S. and Longaker M. T. (2018). ‘Fibroblasts and wound healing: an update’, Regen. Med. 13: 491-495. 10.2217/rme-2018-0073 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler S. A., Deutsch M. A., Serpooshan V., Li G., Dzilic E., Lange R., Krane M. and Wu S. M. (2017). Mammalian heart regeneration: the race to the finish line. Circ. Res. 120, 630-632. 10.1161/CIRCRESAHA.116.310051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel F. B., Hsieh P. C., Lee R. T. and Keating M. T. (2006). FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. USA 103, 15546-15551. 10.1073/pnas.0607382103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Kahai S., Yang W., He C., Seth A., Peng C. and Yang B. B. (2010). Transforming growth factor-beta inhibits nephronectin-induced osteoblast differentiation. FEBS Lett. 584, 2877-2882. 10.1016/j.febslet.2010.04.074 [DOI] [PubMed] [Google Scholar]
- Fogerty F. J., Fessler L. I., Bunch T. A., Yaron Y., Parker C. G., Nelson R. E., Brower D. L., Gullberg D. and Fessler J. H. (1994). Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for Drosophila alpha PS2 beta PS integrins. Development 120, 1747-1758. [DOI] [PubMed] [Google Scholar]
- Giacomini M. M., Travis M. A., Kudo M. and Sheppard D. (2012). Epithelial cells utilize cortical actin/myosin to activate latent TGF-beta through integrin alpha(v)beta(6)-dependent physical force. Exp. Cell Res. 318, 716-722. 10.1016/j.yexcr.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rosa J. M. and Mercader N. (2012). Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat. Protoc. 7, 782-788. 10.1038/nprot.2012.025 [DOI] [PubMed] [Google Scholar]
- González-Rosa J. M., Martin V., Peralta M., Torres M. and Mercader N. (2011). Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138, 1663-1674. 10.1242/dev.060897 [DOI] [PubMed] [Google Scholar]
- Gravning J., Ørn S., Kaasbøll O. J., Martinov V. N., Manhenke C., Dickstein K., Edvardsen T., Attramadal H. and Ahmed M. S. (2012). Myocardial connective tissue growth factor (CCN2/CTGF) attenuates left ventricular remodeling after myocardial infarction. PLoS ONE 7, e52120 10.1371/journal.pone.0052120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. R. M., Bussmann J., Huang Y., Zhao L., Osorio A., Burns C. G., Burns C. E., Sucov H. M., Siekmann A. F. and Lien C.-L. (2015). Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell 33, 442-454. 10.1016/j.devcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson N. C., Arnold T. D., Katamura Y., Giacomini M. M., Rodriguez J. D., McCarty J. H., Pellicoro A., Raschperger E., Betsholtz C., Ruminski P. G. et al. (2013). Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617-1624. 10.1038/nm.3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S. P., Sheng D. Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D. and Kikuchi K. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659-672.e5. 10.1016/j.devcel.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Ignotz R. A. and Massague J. (1986). Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 261, 4337-4345. [PubMed] [Google Scholar]
- Itou J., Oishi I., Kawakami H., Glass T. J., Richter J., Johnson A., Lund T. C. and Kawakami Y. (2012). Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 139, 4133-4142. 10.1242/dev.079756 [DOI] [PubMed] [Google Scholar]
- Jacobetz M. A., Chan D. S., Neesse A., Bapiro T. E., Cook N., Frese K. K., Feig C., Nakagawa T., Caldwell M. E., Zecchini H. I. et al. (2013). Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 62, 112-120. 10.1136/gutjnl-2012-302529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffa A. A., Usinger W. R., McHenry M. B., Jaffa M. A., Lipstiz S. R., Lackland D., Lopes-Virella M., Luttrell L. M. and Wilson P. W. (2008). Connective tissue growth factor and susceptibility to renal and vascular disease risk in type 1 diabetes. J. Clin. Endocrinol. Metab. 93, 1893-1900. 10.1210/jc.2007-2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kuhn B., del Monte F., Hajjar R. J., Chang Y. S., Lebeche D., Arab S. and Keating M. T. (2007). Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med. 13: 962-969. 10.1038/nm1619 [DOI] [PubMed] [Google Scholar]
- Ladage D., Yaniz-Galende E., Rapti K., Ishikawa K., Tilemann L., Shapiro S., Takewa Y., Muller-Ehmsen J., Schwarz M., Garcia M. J. et al. (2013). Stimulating myocardial regeneration with periostin Peptide in large mammals improves function post-myocardial infarction but increases myocardial fibrosis. PLoS ONE 8, e59656 10.1371/journal.pone.0059656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S. L., Marín-Juez R., Moura P. L., Kuenne C., Lai J. K. H., Tsedeke A. T., Guenther S., Looso M. and Stainier D. Y. (2017). Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife 6, e25605 10.7554/eLife.25605.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G. and Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell127, 607-619. 10.1016/j.cell.2006.08.052 [DOI] [PubMed]
- Li R., Luo M., Ren M., Chen N., Xia J., Deng X., Zeng M., Yan K., Luo T. and Wu J. (2014). Vitronectin regulation of vascular endothelial growth factor-mediated angiogenesis. J. Vasc. Res. 51, 110-117. 10.1159/000360085 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Linton J. M., Martin G. R. and Reichardt L. F. (2007). The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development 134, 2501-2509. 10.1242/dev.005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C., Zou X.-B., Chai Y.-F. and Yao Y.-M. (2014). Macrophage polarization in inflammatory diseases. Int. J. Biol. Sci. 10, 520-529. 10.7150/ijbs.8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.-J., Chen Q., Rong Y.-J., Chen F. and Chen J. (2017). CXCR3.1 and CXCR3.2 Differentially Contribute to Macrophage Polarization in Teleost Fish. J. Immunol. 198, 4692-4706. 10.4049/jimmunol.1700101 [DOI] [PubMed] [Google Scholar]
- Mably J. D., Mohideen M. A., Burns C. G., Chen J.-N. and Fishman M. C. (2003). Heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138-2147. 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- Marín-Juez R., Marass M., Gauvrit S., Rossi A., Lai S.-L., Materna S. C., Black B. L. and Stainier D. Y. R. (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 113, 11237-11242. 10.1073/pnas.1605431113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Juez R., El-Sammak H., Helker C. S. M., Kamezaki A., Mullapuli S. T., Bibli S. I., Foglia M. J., Fleming I., Poss K. D. and Stainier D. Y. R. (2019). Coronary revascularization during heart regeneration is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation. Dev. Cell 51, 503-515.e4. 10.1016/j.devcel.2019.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A., Koinuma D., Tsutsumi S., Kamimura N., Morikawa M., Suzuki H. I., Imamura T., Miyazono K. and Aburatani H. (2011). Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4alpha in HepG2 cells. J. Biol. Chem. 286, 29848-29860. 10.1074/jbc.M110.217745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed T. M. A., Ang Y. S., Radzinsky E., Zhou P., Huang Y., Elfenbein A., Foley A., Magnitsky S. and Srivastava D. (2018). Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104-116.e12. 10.1016/j.cell.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled M. H., Patra C., Dickson A. L., Endo T., Stainier D. Y. and Poss K. D. (2016). Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630-634. 10.1126/science.aaf2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M., Bersell K., Walsh S., Savla J., Das L. T., Park S.-Y., Silberstein L. E., dos Remedios C. G., Graham D., Colan S. et al. (2013). Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 110, 1446-1451. 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Kawara S., Shinozaki M., Hayashi N., Kakinuma T., Igarashi A., Takigawa M., Nakanishi T. and Takehara K. (1999). Role and interaction of connective tissue growth factor with transforming growth factor-beta in persistent fibrosis: a mouse fibrosis model. J. Cell. Physiol. 181, 153-159. [DOI] [PubMed] [Google Scholar]
- Nakerakanti S. S., Bujor A. M. and Trojanowska M. (2011). CCN2 is required for the TGF-beta induced activation of Smad1-Erk1/2 signaling network. PLoS ONE 6, e21911 10.1371/journal.pone.0021911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea K. S., Liu L. H., Kinnunen L. H. and Dixit V. M. (1990). Role of the extracellular matrix protein thrombospondin in the early development of the mouse embryo. J. Cell Biol. 111(6 Pt 1), 2713-2723. 10.1083/jcb.111.6.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra C., Kontarakis Z., Kaur H., Rayrikar A., Mukherjee D. and Stainier D. Y. R. (2017). The zebrafish ventricle: A hub of cardiac endothelial cells for in vitro cell behavior studies. Sci. Rep. 7, 2687. 10.1038/s41598-017-02461-1 [DOI] [PMC free article] [PubMed]
- Pfefferli C. and Jaźwińska A. (2017). The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat. Commun. 8, 15151 10.1038/ncomms15151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticos M., Holmes A. M., Shi-wen X., Leoni P., Khan K., Rajkumar V. S., Hoyles R. K., Bou-Gharios G., Black C. M., Denton C. P. et al. (2009). Pivotal role of connective tissue growth factor in lung fibrosis: MAPK-dependent transcriptional activation of type I collagen. Arthritis. Rheum. 60, 2142-2155. 10.1002/art.24620 [DOI] [PubMed] [Google Scholar]
- Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N. and Sadek H. A. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078-1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Proulx K., Lu A. and Sumanas S. (2010). Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Dev. Biol. 348, 34-46. 10.1016/j.ydbio.2010.08.036 [DOI] [PubMed] [Google Scholar]
- Riley K. G., Pasek R. C., Maulis M. F., Peek J., Thorel F., Brigstock D. R., Herrera P. L. and Gannon M. (2015). Connective tissue growth factor modulates adult beta-cell maturity and proliferation to promote beta-cell regeneration in mice. Diabetes 64, 1284-1298. 10.2337/db14-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni E. and Watt F. M. (2018). Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 28, 709-722. 10.1016/j.tcb.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Dai M., Auclair D., Fukai I., Kiriyama M., Yamakawa Y., Fujii Y. and Chen L. B. (2001). Serum level of the periostin, a homologue of an insect cell adhesion molecule, as a prognostic marker in nonsmall cell lung carcinomas. Cancer 92, 843-848. [DOI] [PubMed] [Google Scholar]
- Satoh M., Nakamura M., Akatsu T., Shimoda Y., Segawa I. and Hiramori K. (2005). Myocardial osteopontin expression is associated with collagen fibrillogenesis in human dilated cardiomyopathy. Eur. J. Heart Fail 7, 755-762. 10.1016/j.ejheart.2004.10.019 [DOI] [PubMed] [Google Scholar]
- Saupe F., Schwenzer A., Jia Y., Gasser I., Spenlé C., Langlois B., Kammerer M., Lefebvre O., Hlushchuk R., Rupp T. et al. (2013). Tenascin-C downregulates wnt inhibitor dickkopf-1, promoting tumorigenesis in a neuroendocrine tumor model. Cell Rep 5, 482-492. 10.1016/j.celrep.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Senyo S. E., Steinhauser M. L., Pizzimenti C. L., Yang V. K., Cai L., Wang M., Wu T.-D., Guerquin-Kern J.-L., Lechene C. P. and Lee R. T. (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433-436. 10.1038/nature11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhu J., Wang R., Chen X., Mi L., Walz T. and Springer T. A. (2011). Latent TGF-beta structure and activation. Nature 474, 343-349. 10.1038/nature10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamatani T., Kobayashi H., Tezuka K., Sakamoto S., Suzuki K., Nakanishi T., Takigawa M. and Miyano T. (1998). Establishment of the enzyme-linked immunosorbent assay for connective tissue growth factor (CTGF) and its detection in the sera of biliary atresia. Biochem. Biophys. Res. Commun. 251, 748-752. 10.1006/bbrc.1998.9543 [DOI] [PubMed] [Google Scholar]
- Torraca V., Cui C., Boland R., Bebelman J.-P., van der Sar A. M., Smit M. J., Siderius M., Spaink H. P. and Meijer A. H. (2015). The CXCR3-CXCL11 signaling axis mediates macrophage recruitment and dissemination of mycobacterial infection. Dis. Model. Mech. 8, 253-269. 10.1242/dmm.017756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-C., Wu S.-B., Kau H.-C. and Wei Y.-H. (2018). Essential role of connective tissue growth factor (CTGF) in transforming growth factor-beta1 (TGF-beta1)-induced myofibroblast transdifferentiation from Graves’ orbital fibroblasts. Sci. Rep. 8, 7276 10.1038/s41598-018-25370-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Karra R., Dickson A. L. and Poss K. D. (2013). Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 382, 427-435. 10.1016/j.ydbio.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner D., Tsarouchas T. M., Michael A., Haase C., Weidinger G., Reimer M. M., Becker T. and Becker C. G. (2017). Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat. Commun. 8, 126 10.1038/s41467-017-00143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff P.-J., Rifkin D. B., Meister J.-J. and Hinz B. (2007). Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J. Cell Biol. 179, 1311-1323. 10.1083/jcb.200704042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q., Wang Z., Zaromytidou A.-I., Zhang X. H.-F., Chow-Tsang L.-F., Liu J. X., Kim H., Barlas A., Manova-Todorova K., Kaartinen V. et al. (2011). A poised chromatin platform for TGF-β access to master regulators. Cell 147, 1511-1524. 10.1016/j.cell.2011.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa T., Sugano Y., Nakayama T., Nagai T., Matsuyama T. A., Ohta-Ogo K., Ikeda Y., Ishibashi-Ueda H., Nakatani T., Yasuda S. et al. (2016). Significance of myocardial tenascin-C expression in left ventricular remodelling and long-term outcome in patients with dilated cardiomyopathy. Eur. J. Heart Fail 18, 375-385. 10.1002/ejhf.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K., Obata H., Jurukovski V., Mazzieri R., Chen Y., Zilberberg L., Huso D., Melamed J., Prijatelj P., Todorovic V. et al. (2008). Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors. Proc. Natl. Acad. Sci. USA 105, 18758-18763. 10.1073/pnas.0805411105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Handley D., Kaplan T., Yu H., Bais A. S., Richards T., Pandit K. V., Zeng Q., Benos P. V., Friedman N. et al. (2011). High throughput determination of TGFβ1/SMAD3 targets in A549 lung epithelial cells. PLoS ONE 6, e20319 10.1371/journal.pone.0020319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zheng J., Chen J. and Huang L. (2017). The influence of connective tissue growth factor on rabbit ligament injury repair. Saudi Pharm. J. 25, 498-503. 10.1016/j.jsps.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.