Abstract
The coronavirus (CoV) infects a broad range of hosts including humans as well as a variety of animals. It has gained overwhelming concerns since the emergence of deadly human coronaviruses (HCoVs), severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, followed by Middle East respiratory syndrome coronavirus (MERS-CoV) in 2015. Very recently, special attention has been paid to the novel coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 due to its high mobility and mortality. As the COVID-19 pandemic continues, despite vast research efforts, the effective pharmaceutical interventions are still not available for clinical uses. Both expanded knowledge on structure insights and the essential function of viral nucleocapsid (N) protein are key basis for the development of novel, and potentially, a broad-spectrum inhibitor against coronavirus diseases. This review aimed to delineate the current research from the perspective of biochemical and structural study in cell-based assays as well as virtual screen approaches to identify N protein antagonists targeting not only HCoVs but also animal CoVs.
Abbreviations: CoV, coronavirus; HCoVs, human coronaviruses; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; COVID-19, coronavirus disease 2019; SP, spike protein; RNP, ribonucleoproteins; MP, membrane protein; nsp3, the nonstructural protein 3; E, envelope protein; IFN, interferon; NTD, N-terminus RNA-binding domain; CTD, C-terminus dimerization domain; RBD, RNA-binding domain; MHV, mouse hepatitis virus; IBV, infectious bronchitis virus; AMP, UMP, GMP and CMP, ribonucleoside 5′-monophosphates; HIV, human immunodeficiency virus; FIPV, feline infectious peritonitis virus; CCoV, canine coronavirus; BCoV, bovine coronavirus; ECoV, equine coronavirus; TCoV, turkey coronavirus; PEDV, Porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; PDCoV, porcine deltacoronavirus; PRCV, porcine respiratory coronavirus; SeCoV, swine enteric coronavirus; shRNAs, short hairpin RNAs; FECV, feline enteric coronavirus; siRNA, small interfering RNA
Keywords: Coronavirus, COVID-19, SARS-CoV-2, N protein, Antagonists, Inhibitors
1. Introduction
Human coronaviruses (HCoVs), including HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1 typically cause subclinical infections [1]. However, three novel coronaviruses (CoVs) with high mortality rates have emerged since 2003 and possess severe threats to humans. CoVs have drawn great concern since the outbreak of severe acute respiratory syndrome (SARS) that led to approximately 800 deaths with a fatality rate of approximately 40% in aged people [2]. Ten years later, another CoV, Middle East respiratory syndrome (MERS), had 30% mortality rate and caused more than 2,000 cases of infection worldwide [3].
However, neither specific therapeutic agents nor approved vaccines are approved for the treatment or prevention of CoV infections. In January 2020, an outbreak of COVID-19 caused by a novel CoV, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was documented in China [4]. This newly emerging coronavirus has rapidly spread worldwide and been declared a pandemic. Up to January 2021, over 96 million people have been infected by SARS-CoV-2 and the death toll has reached 2 million cases.
Several vaccine candidates are currently undergoing clinical trials, with results indicating that adenovirus-based and mRNA-based SARS-CoV-2 vaccines are clinically effective, as both can induce significant humeral immune responses with minimal side effects [5], [6], [7]. Despite promising progress in vaccine development, total cases of hospitalization and death are still increasing, making development of therapeutics an urgent matter. As the interface between spike protein of CoVs and its host receptor is essential for virus entry, efforts have been notably made in finding molecules against the function of CoV spike [8], [9]. For instance, two research groups have developed in silico platform to screen FDA-approved small molecule libraries and natural compounds that can target the interface between spike and host receptor [10], [11]. However, accumulated evidence indicated SARS-CoV spike protein (SP) had a higher mutation rate and less stable than nucleocapsid protein (NP) [12], [13]. Hence, although most studies have focused on the SP of CoVs, a growing amount of evidence showed that the NP is a potential target for drug development.
The NP structures of many CoVs, including those highly fatal emerging CoVs, reveal similarities in the structure and function of the N- and C- termini, which are responsible for RNA binding and oligomerization, respectively. Identifying such residues yields an opportunity to design novel inhibitors or screen for potentially effective molecules in current drug libraries. Due to these unique features, the NP is a promising target for development of broad-spectrum anti-coronavirus therapeutics [14].
In this mini-review, we emphasized the structure–function analysis of the CoVs’ NPs and summarized current findings on structure-based development of therapeutics for both human and animal CoVs by targeting NPs.
2. The function of NPs in coronavirus
The NP is a versatile protein which has various bio-functions, including oligomerizing NPs, packing viral genome RNA into ribonucleoproteins (RNP) and interacting with other viral proteins. For instance, the NP associates with the membrane (MP) and the nonstructural protein 3 (nsp3) [15]. Throughout viral replication, NP-MP interaction contributes to viral core formation, assembly, budding, and envelope formation. An ionic interaction between the C-terminal region (composed of the residues 237–252) of the MP and the NP leads to genome encapsidation of budding viral particles [16], [17], [18]. Moreover, assembly of coronavirus virions requires dimerization of NPs [19], [20], [21] and association with viral genomic RNA that ultimately forms RNPs [16], [22], [23], [24], [25], [26]. In addition to RNP formation, interactions among the four structural proteins (NP, MP, E, and SP) and acquirement of viral envelopes from the host membrane at budding sites are also critical for virion formation and infectivity. Although NPs are not required for the virion envelope formation [27], [28], [29], overexpression of NP significantly increased virus production [30].
In addition to the regulatory role in viral RNP assembly and genome budding, CoV NPs facilitate viral propagation by modulation of cellular machinery. For instance, the SARS-CoV NP could interrupt the host cell cycle via inhibition of cyclin-CDK activity, which leads to the arrest of S phase progression [31]. Furthermore, CoVs counteract cellular innate immunity, particularly regulation of interferon (IFN) production, to facilitate viral infection. It is noted that the SARS-CoV NP is one of the effectors of this mechanism, antagonizing IFN-β production through the inhibitory effect on IRF-3 and NF-κb activation [32].
3. Structure and function analysis of NPs
NPs are abundant structural proteins in CoVs. The primary function of NPs is to bind the viral RNA genome, form the RNP, and further compress it into a compact virion core. Previous studies [12], [13] and as illustrated in Fig. 1, sequences and structures of NPs are relatively conserved among CoVs. Two functional domains were found in the NPs of CoVs, the N-terminus RNA-binding domain (NTD) and the C-terminus dimerization domain (CTD), which are connected by a central Ser/Arg-rich flexible linker. The NTD and CTD of NPs are responsible for association with viral RNA and formation of NP oligomers, respectively [20], [33]. The central linker region, with its multiple phosphorylation sites [34], has also been demonstrated to be essential in RNA-binding [35]. Initially, the CTD was the first domain found to perform oligomerization; more recent evidence has indicated that NTDs also form homodimers, further leading to NP oligomerization via protein–protein interactions [36].
Fig. 1.
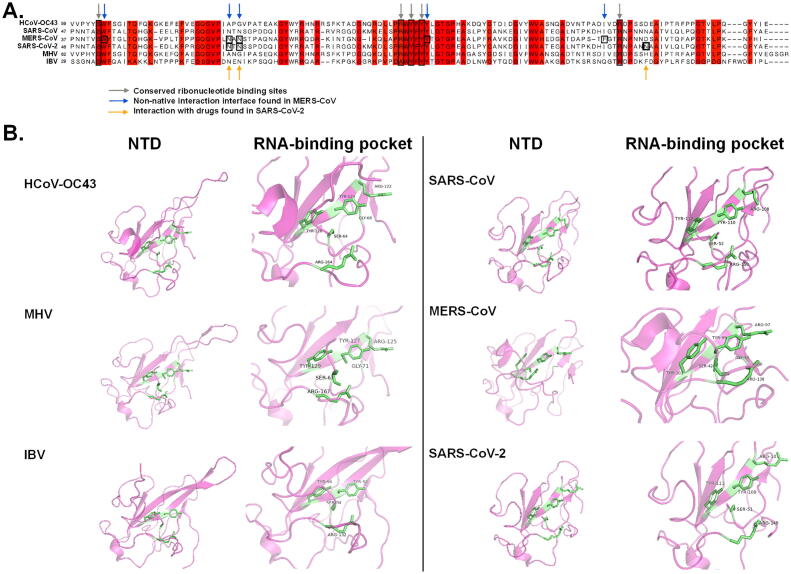
Sequence alignment and structure analyses of NTD of CoV NP. (A) Multiple sequence alignment of HCoV-OC43 (NC005147), SARS-CoV (NC004718), MERS-CoV (NC019843), SARS-CoV-2 (NC045512), MHV (NC001846) and IBV (AY692454). The highly conserved residues were highlighted in red. Grey arrows indicate conserved RNA binding sites identified in previous reports [39], [48], [51], [61]. Blue arrows indicate important residues for non-native oligomerization [48]. Yellow arrows indicate binding sites between potential anti-SARS-CoV-2 compounds and N protein [51]. (B) NTD and RNA binding pockets of HCoV-OC43 (PDB: 4J3K), MHV (PDB: 3HD4), IBV (PDB: 2GEC), SARS-CoV (PDB: 2OG3), MERS-CoV (PDB: 6KL2) and SARS-CoV-2 (PDB: 6WKP) N protein. Green sticks indicate RNA-interaction residues. HCoV-OC43, human coronavirus OC43; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MHV, mouse hepatitis virus; IBV, infectious bronchitis virus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The RNA-binding domain (RBD) has been mapped to the NPs. The minimal region responsible for RNA-binding activity was located from residue 177 to 231 in mouse hepatitis virus (MHV) [22]. Of note, the NP NTD of various CoVs shares conserved secondary structures, i.e. right-handed (loops)-(β-sheet core)-(loops) sandwiched folds [14], which possess a key scaffold for RNA binding. Generally speaking, an NTD is composed of a five-stranded antiparallel β sheet structure with the topology β4-β2-β3-β1-β5 [37]. Several positively charged amino acids — for instance Arg 106, Arg 107, and Arg 117 residues — located at the protruding loop of the NTD in HCoV-OC43, are important for electrostatic interactions with the phosphate backbone of RNA [38]. Moreover, two tyrosine residues in the β5 region of NTDs are conserved in both human and animal CoVs (Fig. 1A, grey arrows), which were proposed to be essential for ribonucleotide binding [14] and correlated with viral replication. Substitution of Tyr 127 and Tyr 129 with alanine resulted in attenuation of MHV replication [39]. Alanine substitution of Tyr 94 at the NTD of infectious bronchitis virus (IBV), corresponding to Tyr 126 of HCoV-OC43 and Tyr 129 of MHV, has consistently led to significantly reduced RNA-binding activity and consequently, decreased infectivity has been demonstrated in Vero cells [40]. In order to understand molecular interaction between NTDs and RNA, the crystal structure of the NTD of HCoV-OC43 in a complex with ribonucleoside 5′-monophosphates (AMP, UMP, GMP and CMP), has been resolved by means of X-ray diffraction [36]. Residues including Ser 64, Gly 68, Arg 122, Tyr 124, Tyr 126, and Arg 164 are located in the center of the RNA-binding pocket in HCoV-OC43 NTD, a composition conserved in other CoV viruses (Fig. 1B). Lin and colleagues, further showed that the interaction between an NP and ribonucleoside 5′-monophosphate mainly relies on hydrogen (H) bonds, ionic bonds, and π-π stacking [36]. The depiction of the crystal structure of HCoV-OC43 NP-NTD complexed with AMP identified a distinct ribonucleotide-binding pocket composed of Arg 122, Tyr 124, Tyr 126, and Arg 164. Tyr 124, located on the surface of the NP, directly interacted with the AMP base through π-π stacking. Moreover, H-bonds were formed between the phenolic hydroxyl group of Tyr 126 and the adenine ring of AMP. This implies that Tyr124 and Tyr126 are essential for recognition and interaction with RNA. Since the two residues are sequentially and structurally conserved in NPs among other CoVs, they could be a potential cross-strain target for drug development.
In addition to the one on the N-terminus, a second RBD has been mapped to the C-terminal region of the NP, between residues 363 to 382 [41]. In addition to RNA-binding, CTD also contributes to the formation of oligomers. As revealed by native gel electrophoresis and size-exclusion chromatography, in the absence of viral genomic RNA, NPs of SARS-CoV form dimers at low concentrations and high-order oligomers at higher concentrations [42]. It has been demonstrated that oligomerization of NPs maintains intact function and is regarded as a crucial step for coronavirus genome assembly [43]. Based on previous reports, the formation of high order NP oligomers is possibly due to self-association of CTDs (residues 283–422) in a concentration-dependent manner [41].
4. Inhibitors of NPs
The emergence of high mortality SARS-CoV in 2003 increased the urgency to develop effective drugs against human CoVs. As the NP is an essential viral protein with multiple functions and shares a relatively high level of sequence similarity with other members of Coronaviridae, it is a suitable target for drug development.
Many of proteins exert their bio-functions by formation of protein: protein interactions (PPI). The interfaces of native PPI are specific and flexible that serve as promising scaffolds for drug design. Of note, interactions of the genomic RNA of CoVs occur through the NTDs of NPs [36], and owing to the conserved native of NP structure, attempts were made to identify small molecules that could bind and further modulate its physiological function [44]. Several signature residues involved in the intermolecular interaction of NPs have been depicted. A ribonucleotide-binding pocket with conserved residues (Arg 122, Tyr 124, Tyr 126, and Arg 164) has been found among different human and animal CoVs (as indicated with grey arrows in Fig. 1A). Based on this structural feature, Chang et al., identified a lead compound (H3) that could inhibit the RNA-binding activity of HCoV-OC43 NPs [44]. As revealed in the structure prediction of HCoV-OC43 NP-NTD in a complex with 2-morpholin-4-yl-ethylamino, Tyr124 directly interacted with quinolone group, and residue Arg164 bonded with the quinoline-5,8-dione ring of 2-morpholin-4-yl-ethylamino with hydrogen bonds, which form a specific binding area with Phe57 and Arg171. Previously, another HCoV-OC43 NP inhibitor, PJ34, has been shown to interact with Phe66 and Tyr124 of NPs through π-π stacking, albeit with a lesser inhibitory effect [43]. Structural superimposition of the HCoV-OC43 NP-NTD with H3 and PJ34 revealed that the branched moiety of PJ34 and morpholin moiety of H3 docked in opposite directions on the HCoV-OC43 NP. Since interaction of H3 molecule highly resembles that of nucleotide AMP, the difference in docking orientation of PJ34 moiety could partially explain its decreased inhibitory effect relative to H3.
Changhyun Roh and colleagues developed a platform for screening inhibitors of the SARS-CoV NP. Candidate compounds were selected based on their ability to compete with quantum dots-conjugated specific RNA oligonucleotides at the RNA-binding site of NP. (−)-catechin gallate and (−)-gallocatechin gallate showed high binding ability with the RBD of SARS-CoV NPs [45]. Both compounds are phytochemicals with antioxidant, anti-inflammatory, and antiviral activities; the former has been reported to inhibit human immunodeficiency virus (HIV) infection via binding with CD4, the receptor for HIV, which ultimately blocks virus entry [46], [47]. As analysis on an RNA oligonucleotide biochip platform, (−)-catechin gallate and (−)-gallocatechin gallate were able to interrupt the binding between NP and RNA; the concentration required for 50% inhibition (IC50 value) was approximately 0.05 μg/ml. Nevertheless, the anti-SARS-CoV activity of these compounds against genuine virus has not yet been investigated in in vitro or in animal models.
More recently, instead of stabilization of native PPI, an alternative strategy for anti-viral drug discovery was established to identify small molecules that could stabilize the non-native PPI interface of NP [48]. Lin et al., revealed that 5-benzyloxygr-amine, in the Acros and ZINC drug databases, can bind and stabilize non-native interfaces between MERS-CoV NP oligomers, ultimately jeopardizing biochemical activity of the target protein [48]. Specific stabilization mediated by 5-benzyloxygr-amine led to abnormal aggregation of the full-length MERS-CoV NPs, inhibiting MERS-CoV replication in Vero E6 cells with EC50 of 32.1 μM. The underlying mechanism was further investigated through structure-based study. It was proposed that Trp43, Asn66, Asn68, Tyr102, and Phe135 on the flexible region of NTDs form a conserved hydrophobic pocket (Fig. 1A, blue arrows), which accommodates the side chain of Met38 of the other NP monomer via hydrophobic contact. The Trp43 residue is important in maintaining the hydrophobic pocket for interaction with the vector-fusion residues of His37 and Met38 from the second monomer of MERS-CoV NP. It was demonstrated that 5-benzyloxygr-amine was able to replace the vector-fusion residues, and that in turn stabilized the NTD of MERS-CoV NPs. Although developing compounds to stabilize the non-native interfaces between proteins is not a conventional means for screening therapeutic compounds, it has been used to identify new anti-viral lead compounds. Two such molecules, nucleozin and S119, have been found to alter the oligomerization state of influenza virus NPs. Abnormal NP aggregates were formed and antagonized the original nuclear localization [49]. As the relatively high similarity of influenza NPs has been documented, the broad-spectrum antiviral effects of these drugs were demonstrated in cell cultures and mouse models.
Structural analysis indicated that the residues (Asn48, Asn49, Thr50, and Ala51) in the N terminal tail of NPs in HCoV-OC43 are more flexible and possibly serve as the binding pocket for viral RNA genomic high order structure [14]. By in-silico screening 56,079 compounds, Sarma and colleagues identified two compounds (ZINC00003118440 and ZINC0000146942, the theophylline and pyrimidone derivatives, respectively) that effectively interact with the NTD of the NPs (i.e. PDB ID: 2OFZ) of SARS-CoV-2 [50]. More specifically, by means of molecular modeling and docking, Yadav et al., conducted a high throughput virtual screening against the RNA-binding potential of NPs (PDB ID: 6YVO) [51]. First, five active sites responsible for RNA-binding were identified, of which three with the highest scores were chosen for docking against the selected ligands: 8722 antiviral drugs from Asinex databases and 265 FDA-approved drugs (from PubChem) for infectious diseases. Ultimately, two antiviral moieties (5817 and 6799) and one FDA drug molecule (Zidovudine) demonstrated great affinity to the three active sites. As evidenced by molecular dynamics simulations, each drug molecule forms 4 to 5 significant hydrogen bond interactions with NPs. Interestingly, most hydrogen bond interactions of the drugs are associated with Asn residues of the NP (Asn75, Asn77, and Asn154 residues, as indicated in yellow arrows in Fig. 1A). Moreover, results from this screening also indicated that many FDA-approved molecules (e.g. anti-HIV drugs and anti-herpes drugs) are active in interactions with the NP. Hence, for urgent use, drug repurposing is cost-effective and crucial for delivering new treatments against COVID-19. The inhibitors against human CoVs described in this review was concisely summarized in Table 1.
Table 1.
Therapeutics against NPs of human CoVs.
| Compounds | Virus | Binding sites | Mechanism | Assay system or simulation modelling | Dose | Reference |
|---|---|---|---|---|---|---|
| H3, 6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5,8-dione | HCoV-OC43 | Phe66, Tyr124, Arg164, Phe57, Ala171 | Decrease of RNA-binding capacity of NP | In vitro assay, surface plasmon resonance (SPR) | 2 mM | [44] |
| PJ34, N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino)acetamide hydrochloride | HCoV-OC43 | Ser 64, Phe 66, Tyr 124, Tyr 126, His 104. | Reduction of the RNA-binding capacity of NP | Cell based infection | 10 μM | [36] |
| (−)-catechin gallate | SARS-CoV | N/A | Attenuation of NP binding affinity to RNA oligonucleotide | In vitro assay, a nanoparticle-based RNA oligonucleotide biochip system. | *IC50 of 0.05 μg/ mL | [45] |
| (−)-gallocatechin gallate. | SARS-CoV | N/A | Attenuation of NP binding affinity to RNA oligonucleotide | In vitro assay, a nanoparticle-based RNA oligonucleotide biochip system. | *IC50 of 0.05 μg/ mL | [45] |
| P3, 5-benzyloxygr- amine | MERS-CoV | Monomer1: W43, N66, N68, S69, T70, N73, F135; Monomer2: V41, G104, T105, G106, A109, T137 | Resulting in abnormal N protein oligomerization | Cell based infection | #EC50 of 32.1 μM | [48] |
| ZINC00003118440, theophylline derivatives | SARS-CoV-2 | Most potent hits: Gln72, Val73, Pro74, Asn76, Thr136, Thr166 | Possible inhibitors of RNA-NP NTD interaction | Molecular dynamics simulation | N/A | [50] |
| ZINC0000146942, pyrimidone derivatives | SARS-CoV-2 | Most potent hits: Gly70, Val73, Gln84 | Possible inhibitors of RNA-NP NTD interaction | Molecular dynamics simulation | N/A | [50] |
| 5817 (from Asinex databases) | SARS-CoV-2 | Most potent hits: Ala55, Arg149, Asn77, Asn153, Asn154 | Interaction with NP NTD | Molecular dynamics simulation | N/A | [51] |
| 6799 (from Asinex databases) | SARS-CoV-2 | Most potent hits: Ala55, Arg107, Asn75, Asn153, Asn154 | Interaction with NP NTD | Molecular dynamics simulation | N/A | [51] |
| Zidovudine (from PubChem database) | SARS-CoV-2 | Most potent hits: Ala55, Asn75, Asn77, Arg 107, Thr148, Asn150, Asn153, Asn154, | Interaction with NP NTD | Molecular dynamics simulation | N/A | [51] |
*IC50, half maximal inhibitory concentration.
# EC50, half maximal effective concentration.
5. Therapeutics against NPs of domestic animal CoVs
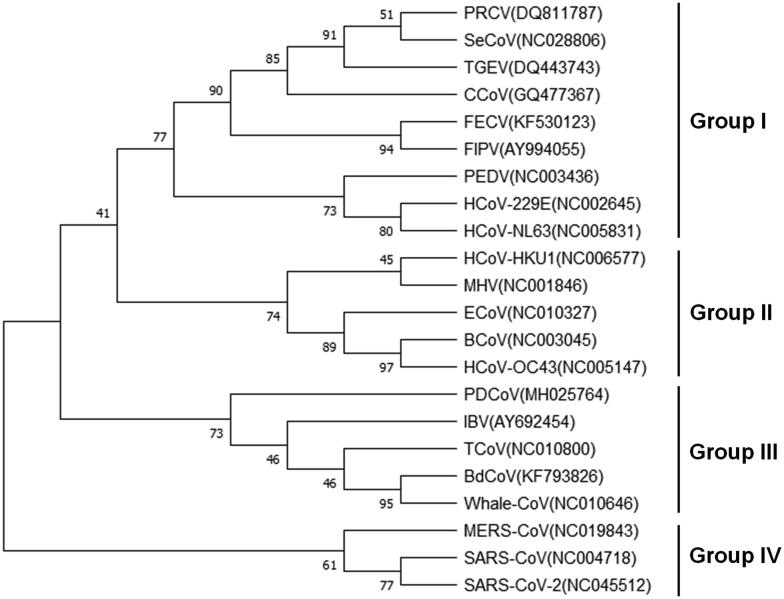
CoVs also infect farm animals and companion animals, causing huge economic impact and loss of nonhuman companions. The total range of pathogenicity within the broad coronavirus family encompasses but is not limited to gastroenteritis in pigs, bronchitis in chickens, and peritonitis in felines. Phylogenetic analysis of NPs within the family of CoVs shows four major groups: feline infectious peritonitis virus (FIPV), canine coronavirus (CCoV) and two porcine coronaviruses in group I; bovine coronavirus (BCoV), equine coronavirus (ECoV), and MHV in group II; turkey coronavirus (TCoV), IBV and marine coronaviruses in group III; and SARS-CoV, MERS-CoV and SARS-CoV-2 in a distinct group IV (Fig. 2). The following discusses current preclinical small molecules, such as drugs, antibodies, and RNA interference techniques against the NPs of domestic animal CoVs.
Fig. 2.
Phylogenetic analysis of NP of coronaviruses. Full length NP sequences were retrieved from the GenBank database and the Maximum Likelihood method was performed to infer the evolutionary history. The bootstrap consensus tree from 500 replicates was shown and the branches were labelled with the percentage of replicate trees in the bootstrap test. PRCV, porcine respiratory coronavirus; SeCoV, swine enteric coronavirus; TGEV, transmissible gastroenteritis virus; CCoV, canine coronavirus; FECV, feline enteric coronavirus; FIPV, feline infectious peritonitis virus; PEDV, porcine epidemic diarrhea virus; HCoV-229E, human coronavirus 229E; HCoV-NL63, human coronavirus NL63; HCoV-HKU1, human coronavirus HKU1; MHV, mouse hepatitis virus; ECoV, Equine coronavirus; BCoV, bovine coronavirus; virus; HCoV-OC43, human coronavirus OC43; PDCoV, porcine deltacoronavirus; IBV, infectious bronchitis virus; TCoV, turkey coronavirus; BdCoV, bottlenose dolphin coronavirus; Whale-CoV, whale coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
5.1. Porcine coronaviruses
Porcine coronaviruses can be classified into the genera Alphacoronavirus, Betacoronavirus and Deltacoronavirus in the family Coronaviridae. Porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV), belonging to the Alphacoronavirus genus, are two major pathogens that cause gastroenteritis in pigs, with especially severe cases in neonatal piglets [52]. An emerging porcine deltacoronavirus (PDCoV) reported in 2009 has pathological lesions that are clinically indistinguishable from those of PEDV and TGEV. The high evolution rate of CoVs has led to many variants — a non-pathogenic porcine respiratory coronavirus (PRCV) descended from TGEV has specific mutations in its spike gene. Co-circulation of porcine CoVs has been described worldwide, a circumstance that eventually generated a novel pathogenic recombinant variant containing PEDV SP on a TGEV backbone (swine enteric coronavirus, SeCoV) [53]. To date, there are neither specific treatments nor antivirals available for porcine coronaviruses, making it difficult to control the rapid emergence and spread of these viruses.
The classical PEDV has caused epidemics in Europe since the 1970s, and the outbreaks in China in 2010s were caused by the emergence of highly virulent PEDV strains. Commercial PEDV vaccines derived from classical PEDV are not effective against highly virulent strains. Deejai et al., developed a virtual screening system based on the three-dimensional structure of PEDV NPs. The results demonstrated that 1,286 compounds in the FDA-approved drug database interacted with the RNA-binding region of the NP [54]. Three of the top-scoring compounds, trichlormethiazide, D-(+) biotin, and glutathione reduced free acid, presented an inhibitory effect on the binding between NPs and nucleic acids in vitro, indicating that these compounds interfere with the RNA-binding activity of NPs. The inhibitory activity of both trichlormethiazide and D-(+) biotin against PEDV occurred at 0.094 mg/mL with significant cell death observed above this concentration. Although glutathione reduced free acid had a minor cytotoxic effect on Vero cells, inhibition of PEDV replication required a higher concentration (1.5 mg/ml). According to the NP-compound docking result, a potential mechanism shared among these three compounds is via interacting with His74 of NPs by H-bonds or ammonium ions [54].
In addition to chemical compounds, another study revealed that RNA interference targeting PEDV NPs could be effective in inhibiting virus replication. Shi et al., analyzed 25 sequences of PEDV NPs from different countries and designed short hairpin RNAs (shRNAs) against regions conserved between classical and emerging PEDV [55]. Cell viability was not affected by transfection of plasmids expressing the shRNAs. Two shRNAs targeting the NTD and one shRNA targeting the CTD of NPs showed dose-dependent inhibitory effects on both NP expression and PEDV replication in swine intestine epithelial cells. Particularly, all of the designed shRNAs had comparable antiviral function against the classical CV777 strain and the emerging LNCT2 strain [55].
During virus infection, innate immune pathways are quickly stimulated, and specific antiviral proteins can be upregulated to counter pathogens at the first line. Bone marrow stromal cell antigen 2 (BST2; tetherin; CD317; HM1.24), an IFN-inducible antiviral protein, has been shown to inhibit the release of budding virions from cells infected with either DNA or RNA viruses, including PEDV [56]. Upregulation of BST2 was identified in Vero and porcine kidney cells, which is correlated to the inhibition of PEDV replication. Multiple mechanisms through which BST2 targets the NP of PEDV have been reported. BST2 physically interacted with the full-length NP at its glycosyl-phosphatidylinositol domain; therefore, the integrity of NP is important to the stability of the BST2-NP complex. Furthermore, the overexpression of BST2 significantly reduced the production of PEDV NP by enhancing protein degradation via autophagy [56].
Several monoclonal antibodies against the PEDV NP have been utilized to develop methods for early detection and characterization of NP epitopes. The specificity of these antibodies has been investigated and, based on the results of NP cross-reactivity between PEDV and TGEV, precise diagnostic assays for differentiating various swine coronavirus infections with similar clinical symptoms have been established [57], [58]. However, these monoclonal antibodies have not been tested for their therapeutic or prophylactic potential in a coronavirus infection. Small molecule inhibitors, such as benzothiazolium and cardenolides, had anti-TGEV activity in swine testicle cells [59], [60]. TGEV NPs were used as a read-out in the in vitro assay, and dose-dependent effects of the compounds were related to NP expression. Both NP expression and virus replication were negatively correlated with the concentration of the compounds in cells [59]. Nonetheless, more detailed experiments are required to confirm whether these compounds directly influence the function of TGEV NPs.
5.2. Avian coronaviruses
Gammacoronaviruses are mainly associated with avian and marine hosts. However, the avian CoV, IBV, is one of the major causes of highly contagious respiratory diseases in domestic fowl. Turkey coronavirus (TCoV) also belongs to this genus, but there is relatively limited antiviral information regarding it. Two different groups of small molecules have been found to interact with the RNA-binding domain of IBV NP protein. Eucalyptol, also named 1, 8-cineole, is an achiral aromatic component found in many plants, such as Eucalyptus and Salvia leaves. Its anti-inflammatory and apoptosis-inducing effects have been well-documented. The maximum noncytotoxic concentration of eucalyptol on Vero cells has been determined to be 3.90 mM, with an IC50 of 0.61 mM against IBV infection [61]. Eucalyptol has different inhibitory effects at various stages of IBV infection. The highest percent of inhibition, 82.61%, occurred when cells were treated with eucalyptol during the replication period of IBV infection. When the virus was pretreated with eucalyptol prior to infection, replication was inhibited by 61.68%. However, treating cells with eucalyptol prior to infection or during the adsorption period only provided about 10% and 5% inhibition of virus replication, respectively. An in silico NP and eucalyptol model demonstrated that the H-bond formed between the oxygen atom in the core of eucalyptol and Tyr92 at the N-terminus of the IBV NP. Protein spatial analysis revealed that eucalyptol strongly associated with 5 active residues in the RNA-binding domain of IBV NPs (Tyr92, Pro134, Phe137, Asp138 and Tyr140). These residues are fully or partially conserved in various IBV strains, indicating that eucalyptol may have cross-strain anti-IBV activity. (−)-Pinenes also directly target the RNA-binding domain of IBV NPs, and are found in two isoforms as the major component of pine tree oil, or turpentine [62]. Its antimicrobial activities have been reported. Both the cyclohexene ring of (−)-α-pinene and (−)-β-pinene bind strongly to residues Tyr92 and Pro134 of IBV NPs. (−)-α-pinene had slightly higher bioactivity with the IBV NP compared to (−)-β-pinene, as the interaction energy of the former was −36.83 kcal mol−1 and −35.59 kcal mol−1 for the latter. Consistently, IC50 of (−)-α-pinene against IBV replication was lower (0.98 mM) than that of (−)-β-pinene (1.32 mM).
In addition to interfering with the function of the RNA-binding domain of IBV NPs, Scott et al., found that a hammerhead ribozyme can specifically cleave NP mRNA of IBV [63]. Hammerhead ribozymes, the catalytic RNA, constitute of a catalytic domain that cleaves target RNA and the flanking sequences are responsible for the specificity of the ribozyme. As revealed by both gel electrophoresis and quantitative polymerase chain reaction, the RNA of IBV NPs was specifically cleaved by a hammerhead ribozyme that leads to decline of NP RNA level. In addition to IBV, hammerhead ribozyme strategy has successfully restricted the mouse coronavirus MHV and other viruses in different viral families by targeting various genes, including Arenaviridae (to cleave the S genomic RNA of lymphocytic choriomeningitis virus) [64], Birnaviridae (RNA polymerase gene of infectious bursal disease virus) [65], Paramyxoviridae (nucleocapsid mRNA of mumps virus) [66], Picornaviridae (5′ untranslated region of hepatitis C virus) [67] and Retroviridae (5′ untranslated region of HIV-1) [68].
5.3. Feline coronaviruses
Feline coronaviruses are classified as Alphacoronavirus; other members of this subgroup include CCoV and TGEV. Currently, two feline coronaviruses are circulating worldwide: feline enteric coronavirus (FECV) and FIPV [69]. FECV is prevalent in domestic cats and usually causes mild or subclinical symptoms. In contrast, FIPV, derived from parental FECV with specific mutations, is highly virulent and causes fatal peritonitis in cats of any age, but especially between the ages of 4 to 16 months [69]. To date, there is no effective treatment for FIPV, and mortality is extremely high. Several groups have identified small molecules with anti-FIPV potential, such as small interfering RNA (siRNA) which inhibits FIPV infection in vitro using the FIPV nucleocapsid gene or protein as a read-out; nevertheless, whether these drugs directly target with NPs has been approved yet. According to two different studies, siRNA targeting the FIPV NP gene is an efficient strategy to reduce the copy number of genomic and messenger FIPV RNA, viral protein expression, and production of viral progeny [70], [71]. The potency of siRNA specific to the NP gene is comparable to that of siRNA targeting a 5′ leader sequence essential to controlling coronavirus gene expression. Importantly, siRNA is effective against FIPV even at the lowest tested concentration (5 nM), and can be used to inhibit FIPV replication within a broad range of multiplicity of infection [70]. Combinations of siRNAs specific to different regions of the FIPV genome resulted in more effective viral inhibition [71]. The selection of these siRNA was based on homology between various FIPV strains, suggesting the possibility of a cross-strain inhibitory effect.
At present, RNA interference strategy has been shown to inhibit PEDV, IBV and FIPV infection. However, it is the only approach to restrict FIPV replication in cells. It is worthy of noting that all of validations were performed in cells or by in vitro system. Further investigation of safety and effect of indicated compounds are required in animal models. A brief summary of inhibitors against domestic animal CoVs was listed in Table 2.
Table 2.
Therapeutics against NPs of domestic animal CoVs.
| Compounds | Virus | Binding sites | Mechanism | Assay system | Dose | Reference |
|---|---|---|---|---|---|---|
| Trichlormethiazide | PEDV | His74, Tyr76, Arg113 | Interference with RNA-binding activity of NP | Cell based infection | 0.094 mg/ mL | [54] |
| D-(+) biotin | PEDV | Ser 17, His74 | Interference with RNA-binding activity of NP | Cell based infection | 0.094 mg/ mL | [54] |
| Glutathione reduced free acid | PEDV | Tyr19, Ser17, His74, Arg113 | Interference with RNA-binding activity of NP | Cell based infection | 1.5 mg/ mL | [54] |
| shRNA-N307 | PEDV | NP gene sequence: 307–327 | Inhibitory effect on NP production | Cell based infection | 1–4 μg/ 5x104 cells | [55] |
| shRNA-N463 | PEDV | NP gene sequence: 463–483 | Inhibitory effect on NP production | Cell based infection | 1–4 μg/ 5x104 cells | [55] |
| shRNA-N1071 | PEDV | NP gene: 1071–1091 | Inhibitory effect on NP production | Cell based infection | 1–4 μg/ 5x104 cells | [55] |
| BST2 | PEDV | Full length NP | Interaction with NP NTD | Cell based infection | N/A | [56] |
| Eucalyptol | IBV | Tyr92, Pro134, Phe137, Asp138, Tyr140 | Interference with RNA-binding activity of NP | Cell based infection | 3.90 mM | [61] |
| (−)-α-pinene | IBV | Ala33, Ser34, Gln37, Tyr92, Pro134, Phe137, Asp138, Gln139, Gly147, Pro149 | Interaction with NP RBD | Cell based infection | 7.88 mM | [62] |
| (−)-β-pinene | IBV | Ala33, Ser34, Gln37, Tyr92, Pro134, Phe137, Asp138, Gln139, Gly147, Pro149 | Interaction with NP RBD | Cell based infection | 6.09 mM | [62] |
| Hammerhead ribozyme | IBV | NP mRNA | Cleavage of NP mRNA | In vitro assays | 0.5–10 μM | [63] |
| siRNA NP (N1) | FIPV | FIPV genome (27112–27130) | Reduction of viral genomic RNA | Cell based infection | 5 nM | [70] |
| siRNA NP (N2) | FIPV | FIPV genome (27885–27837) | Reduction of viral genomic RNA | Cell based infection | 100 nM | [70] |
| siRNA-N | FIPV | FIPV genome (27507–27531) | Reduction of viral genomic RNA | Cell based infection | 100 nM | [71] |
6. Concluding remarks
As NPs share several conserved residues and structures across a large variety of coronavirus strains, they are a promising target for drug discovery with the potential to develop cross-strain activity. Most studies focused on finding compounds that interact with RNA-binding domain at the N-terminus, which specifically suppresses the efficiency of coronavirus genome replication and virion assembly. Virtual screening of high resolution structural information of native interacting interfaces against multiple drug libraries is the main method of discovering novel therapeutic agents. Alternatively, potential compound candidates can be selected according to structure-based stabilization of non-native protein–protein interactions. As such, specific ligands may enhance or inhibit the interaction between NP molecules, leading to abnormal oligomerization and therefore, anti-coronavirus activity. Although these data are all preclinical and more detailed experiments and analysis of NP-compound structures are required, this review could be a basis for developing effective treatments for coronavirus infection.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The project was partly supported by the Ministry of Science and Technology, Taiwan (MOST-108-2321-B-005-010-MY3).
References
- 1.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donnelly C.A., Ghani A.C., Leung G.M., Hedley A.J., Fraser C., Riley S. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed A.E. The predictors of 3- and 30-day mortality in 660 MERS-CoV patients. BMC Infect Dis. 2017;17:615. doi: 10.1186/s12879-017-2712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C., Group C.T.T. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Kehtan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., Group C.S. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2020 doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J., Tao H., Yan Y., Huang S.Y., Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12 doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Oliveira O.V., Rocha G.B., Paluch A.S., Costa L.T. Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1772885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahedi H.M., Ahmad S., Abbasi S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y., Liu M.o., Zhao W., Zhang J., Zhang X., Wang K.e. Isolation of virus from a SARS patient and genome-wide analysis of genetic mutations related to pathogenesis and epidemiology from 47 SARS-CoV isolates. Virus Genes. 2005;30(1):93–102. doi: 10.1007/s11262-004-4586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutta N.K., Mazumdar K., Gordy J.T., Dutch R.E. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol. 2020;94(13) doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol. 2002;76(10):4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu I.M., Gustafson C.L., Diao J., Burgner J.W., 2nd, Li Z., Zhang J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J Biol Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem Biophys Res Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 23.Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macneughton M.R., Davies H.A. Ribonucleoprotein-like structures from coronavirus particles. J Gen Virol. 1978;39:545–549. doi: 10.1099/0022-1317-39-3-545. [DOI] [PubMed] [Google Scholar]
- 25.Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J Virol. 2005;79(2):1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies H.A., Dourmashkin R.R., Macnaughton M.R. Ribonucleoprotein of avian infectious bronchitis virus. J Gen Virol. 1981;53:67–74. doi: 10.1099/0022-1317-53-1-67. [DOI] [PubMed] [Google Scholar]
- 27.Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J Virol. 2011;85(2):675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surjit M., Liu B., Chow V.T.K., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281(16):10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C.-Y., Chang C.-k., Chang Y.-W., Sue S.-C., Bai H.-I., Riang L. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368(4):1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T.K., Lal S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol. 2005;79(17):11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.-K., Hsu Y.-L., Chang Y.-H., Chao F.-A., Wu M.-C., Huang Y.-S. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J Virol. 2009;83(5):2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S.Y., Liu C.L., Chang Y.M., Zhao J., Perlman S., Hou M.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 38.Chen I.-J., Yuann J.-M., Chang Y.-M., Lin S.-Y., Zhao J., Perlman S. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim Biophys Acta. 2013;1834(6):1054–1062. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., Leibowitz J.L. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J Mol Biol. 2009;394(3):544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo H., Chen J., Chen K., Shen X., Jiang H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 42.Luo H., Ye F., Sun T., Yue L., Peng S., Chen J. In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys Chem. 2004;112:15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81(8):3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C.K., Jeyachandran S., Hu N.J., Liu C.L., Lin S.Y., Wang Y.S. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol Biosyst. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- 45.Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int J Nanomed. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson M.P., McCormick T.G., Nance C.L., Shearer W.T. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: potential for HIV-1 therapy. J Allergy Clin Immunol. 2006;118:1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Hamza A., Zhan C.-G. How can (-)-epigallocatechin gallate from green tea prevent HIV-1 infection? Mechanistic insights from computational modeling and the implication for rational design of anti-HIV-1 entry inhibitors. J Phys Chem B. 2006;110(6):2910–2917. doi: 10.1021/jp0550762. [DOI] [PubMed] [Google Scholar]
- 48.Lin S.M., Lin S.C., Hsu J.N., Chang C.K., Chien C.M., Wang Y.S. Structure-based stabilization of non-native protein-protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J Med Chem. 2020;63:3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 49.White K.M., Abreu P., Jr., Wang H., De Jesus P.D., Manicassamy B., Garcia-Sastre A. Broad spectrum inhibitor of influenza A and B viruses targeting the viral nucleoprotein. ACS Infect Dis. 2018;4:146–157. doi: 10.1021/acsinfecdis.7b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarma P., Shekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D.P., Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain) J Biomol Struct Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1753580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yadav R., Imran M., Dhamija P., Suchal K., Handu S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1778536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deejai N., Roshorm Y.M., Kubera A. Antiviral compounds against nucleocapsid protein of porcine epidemic diarrhea virus. Anim Biotechnol. 2017;28:120–130. doi: 10.1080/10495398.2016.1232268. [DOI] [PubMed] [Google Scholar]
- 55.Shi D.a., Wang X., Shi H., Zhang J., Han Y., Chen J. Significant Interference with porcine epidemic diarrhea virus pandemic and classical strain replication in small-intestine epithelial cells Using an shRNA expression vector. Vaccines (Basel) 2019;7(4):173. doi: 10.3390/vaccines7040173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong N., Shan T., Wang H., Jiao Y., Zuo Y., Li L. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. 2020;16(10):1737–1752. doi: 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W., Chen W., Huang J., Jin L., Zhou Y., Chen J. Generation, identification, and functional analysis of monoclonal antibodies against porcine epidemic diarrhea virus nucleocapsid. Appl Microbiol Biotechnol. 2019;103:3705–3714. doi: 10.1007/s00253-019-09702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., Fang L., Zhan J., Shi X., Liu Q., Lu Q. Identification and characterization of linear B cell epitopes on the nucleocapsid protein of porcine epidemic diarrhea virus using monoclonal antibodies. Virus Res. 2020;281:197912. doi: 10.1016/j.virusres.2020.197912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang C.W., Chang H.Y., Hsu H.Y., Lee Y.Z., Chang H.S., Chen I.S. Identification of anti-viral activity of the cardenolides, Na(+)/K(+)-ATPase inhibitors, against porcine transmissible gastroenteritis virus. Toxicol Appl Pharmacol. 2017;332:129–137. doi: 10.1016/j.taap.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang C.W., Yang Y.N., Liang P.H., Chen C.M., Chen W.L., Chang H.Y. Novel small-molecule inhibitors of transmissible gastroenteritis virus. Antimicrob Agents Chemother. 2007;51:3924–3931. doi: 10.1128/AAC.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z., Wu N., Fu Y., Yang G., Wang W., Zu Y. Anti-infectious bronchitis virus (IBV) activity of 1,8-cineole: effect on nucleocapsid (N) protein. J Biomol Struct Dyn. 2010;28:323–330. doi: 10.1080/07391102.2010.10507362. [DOI] [PubMed] [Google Scholar]
- 62.Yang Z., Wu N., Zu Y., Fu Y. Comparative anti-infectious bronchitis virus (IBV) activity of (-)-pinene: effect on nucleocapsid (N) protein. Molecules. 2011;16(2):1044–1054. doi: 10.3390/molecules16021044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Callison S.A., Hilt D.A., Jackwood M.W. In vitro analysis of a hammerhead ribozyme targeted to infectious bronchitis virus nucleocapsid mRNA. Avian Dis. 2005;49:159–163. doi: 10.1637/7259-081104R. [DOI] [PubMed] [Google Scholar]
- 64.Xing Z., Whitton J.L. An anti-lymphocytic choriomeningitis virus ribozyme expressed in tissue culture cells diminishes viral RNA levels and leads to a reduction in infectious virus yield. J Virol. 1993;67:1840–1847. doi: 10.1128/jvi.67.4.1840-1847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akin A., Lin T.L., Wu C.C. A ribozyme targeted to RNA polymerase gene of infectious bursal disease virus effectively cleaves and inhibits expression of the viral gene product. Acta Virol. 1999;43:341–347. [PubMed] [Google Scholar]
- 66.Albuquerque-Silva J., Milican F., Bollen A., Houard S. Ribozyme-mediated decrease in mumps virus nucleocapsid mRNA level and progeny in infected vero cells. Antisense Nucleic Acid Drug Dev. 1999;9:279–288. doi: 10.1089/oli.1.1999.9.279. [DOI] [PubMed] [Google Scholar]
- 67.Macejak D.G., Jensen K.L., Jamison S.F., Domenico K., Roberts E.C., Chaudhary N. Inhibition of hepatitis C virus (HCV)-RNA-dependent translation and replication of a chimeric HCV poliovirus using synthetic stabilized ribozymes. Hepatology. 2000;31(3):769–776. doi: 10.1002/hep.510310331. [DOI] [PubMed] [Google Scholar]
- 68.Hotchkiss G., Maijgren-Steffensson C., Ahrlund-Richter L. Efficacy and mode of action of hammerhead and hairpin ribozymes against various HIV-1 target sites. Mol Ther. 2004;10:172–180. doi: 10.1016/j.ymthe.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 69.Pedersen N.C. An update on feline infectious peritonitis: diagnostics and therapeutics. Vet J. 2014;201(2):133–141. doi: 10.1016/j.tvjl.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDonagh P., Sheehy P.A., Norris J.M. In vitro inhibition of feline coronavirus replication by small interfering RNAs. Vet Microbiol. 2011;150:220–229. doi: 10.1016/j.vetmic.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anis E.A., Wilkes R.P., Kania S.A., Legendre A.M., Kennedy M.A. Effect of small interfering RNAs on in vitro replication and gene expression of feline coronavirus. Am J Vet Res. 2014;75:828–834. doi: 10.2460/ajvr.75.9.828. [DOI] [PubMed] [Google Scholar]