Abstract
Background
Circular RNAs (circRNAs) have been implicated in the progression and chemoresistance development of hepatocellular carcinoma (HCC). However, the precise parts of circ_0031242 in HCC chemoresistance are still not fully understood.
Methods
The levels of circ_0031242, miR-924 and POU class 3 homeobox 2 (POU3F2) were detected by quantitative real-time polymerase chain reaction (qRT-PCR) assay or Western blot analysis. IC50 value for cisplatin (DDP) and cell viability were measured by the cell counting kit-8 (CCK-8) assay. Cell migration, invasion and apoptosis were assessed by transwell assay and flow cytometry, respectively. Targeted correlations among circ_0031242, miR-924 and POU3F2 were verified by the dual-luciferase reporter and RNA immunoprecipitation (RIP) assays.
Results
Our data revealed that circ_0031242 was associated with HCC resistance to DDP. The silencing of circ_0031242 diminished DDP resistance, suppressed cell viability, migration, invasion and promoted apoptosis of DDP-resistant HCC cells (Huh7-R and SNU-387-R) in vitro, as well as enhanced DDP sensitivity in vivo. Mechanistically, circ_0031242 directly interacted with miR-924 by binding to miR-924. Moreover, miR-924 was a downstream effector of circ_0031242 function. POU3F2 was a direct target of miR-924, and miR-924 overexpression regulated DDP-resistant HCC cell progression and DDP resistance by down-regulating POU3F2. Furthermore, circ_0031242 modulated POU3F2 expression through sponging miR-924.
Conclusion
Our findings identified that circ_0031242 functioned as an important regulator in DDP-resistant HCC cell progression and DDP resistance through the miR-924/POU3F2 axis, illuminating circ_0031242 as a potential therapeutic target for the chemoresistant HCC.
Keywords: HCC, chemoresistance, circ_0031242, miR-924, POU3F2
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide.1 Significant improvement has recently been achieved in the prevention, detection and therapy of HCC.2 However, the development of chemoresistance has become a major problem in successful HCC treatment.3 A clearer understanding of the molecular basis of HCC chemoresistance will provide a novel opportunity to develop better therapeutic interventions.
Circular RNAs (circRNAs) are covalently closed biomolecules that have been implicated in normal development and human diseases.4 Some circRNAs have been recently identified as important regulators in HCC chemoresistance by functioning as sponges of microRNAs (miRNAs).5 Li et al showed that circRNA F-box protein 11 (circFBXO11) enhanced oxaliplatin resistance of HCC by sequestering miR-605.6 Luo et al demonstrated that circ_101505 regulated HCC cell resistance to cisplatin (DDP) by working as a miR-103 sponge.7 As for circRNA protein arginine methyltransferase 5 (circ_PRMT5), it was reported to enhance the malignant progression of gastric cancer and non-small cell lung cancer.8,9 Moreover, circ_PRMT5 (also called circ_0031242) was overexpressed in HCC and functioned as a potential driver in HCC tumorigenesis by sponging miR-188-5p.10 Nevertheless, the critical, detailed roles of circ_0031242 in HCC chemoresistance are still not fully understood.
Emerging evidence has demonstrated the crucial involvement of miRNAs in the chemoresistance of HCC.11,12 Previous work identified miR-924 as a tumor suppressor in HCC.13 The study also highlighted the involvement of miR-924 in the competing endogenous RNA (ceRNA) network. A previous report showed that POU class 3 homeobox 2 (POU3F2) was overexpressed in HCC tissues and served as a tumor driver of HCC.14 Of interest, computational algorithms (CircInteractome and TargetScan) predicted two putative complementary regions among circ_0031242, miR-924 and POU3F2, hinting a potential ceRNA regulatory network in HCC chemoresistance.
Here, we sought to identify the precise parts of circ_0031242 in HCC chemoresistance. Aiming to this goal, we established two DDP-resistant HCC cell lines. Focusing on the molecular basis mediated by circ_0031242, we further investigated whether the miR-924/POU3F2 axis was involved in circ_0031242-mediated regulation in DDP-resistant HCC cell progression and drug resistance.
Materials and Methods
Clinical Specimens and Cells
The subjects of the human study were 30 primary patients with HCC and 30 recurrent HCC patients after treatment with DDP-based chemotherapy at the Third Affiliated Hospital of Shandong First Medical University. The Ethics Committee of the Third Affiliated Hospital of Shandong First Medical University approved the study protocol and all subjects gave informed consent. Tumor specimens were obtained from all subjects and 30 adjacent noncancerous hepatic tissues were collected from random 30 patients. All clinical specimens were stored at −80°C to measure circ_0031242, miR-924 and POU3F2 expression using quantitative real-time polymerase chain reaction (qRT-PCR) assay and Western blot analysis. This study was carried out in accordance with the Declaration of Helsinki.
Human THLE-2 hepatocyte cells, Huh7 and SNU-387 hepatoma cells (Bnbio, Beijing, China) were cultivated at 37°C in 5% CO2. Bronchial epithelial cell growth medium (BEGM, Lonza, Basel, Switzerland) was used for the cultivation of THLE-2 cells, and Dulbecco’s modified eagles medium (DMEM, Lonza) was used for Huh7 and SNU-387 cells. The culture media was also supplemented with 10% fetal calf serum (FCS, Lonza) and 1% antibiotics (penicillin/streptomycin, Lonza).
The two DDP-resistant HCC cells (Huh7-R and SNU-387-R) were generated by exposing the corresponding parental cells to gradually increasing concentrations of DDP (Selleck, Shanghai, China) over 10 months as reported15 with minor modifications. An additional 2 µg/mL of DDP was added in the culture media to maintain the resistance phenotype. The level of DDP resistance was evaluated by the cell counting kit-8 (CCK-8) assay.
Cell Transfection
Chemical siRNA specific to circ_0031242 (si-circ_0031242, 5ʹ-AAUUCCAAGGUGCAAUAGCGG-3ʹ) or the nontarget siRNA (si-NC, 5ʹ-GGUACACCCUCCAUGGUAAUU-3ʹ), miR-924 mimic (5ʹ-CGUUCUGUAGUGUUCUGAGA-3ʹ) or a scrambled nontarget sequence (miRNA NC mimic, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), and the inhibitor of miR-924 (miR-924 inhibitor, 5ʹ-UCUCAGAACACUACAGAACG-3ʹ) or its negative control sequence (inhibitor NC, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) were obtained from Ribobio (Guangzhou, China). The sequence of POU3F2 (Accession: NM_005604.4) was amplified by PCR and cloned into pcDNA3.1(+) plasmid (Bnbio) with BamH I and Xho I restriction sites to create POU3F2 overexpressing plasmid (pc-POU3F2), with a nontarget plasmid as the negative control (pc-NC). After reaching ~50% confluence, Huh7-R and SNU-387-R cells were transiently transfected with 50 nM of the indicated oligonucleotide and 100 ng of plasmid using Lipofectamine 3000 reagent (Invitrogen, Bleiswijk, the Netherlands). Forty-eight hours post-transfection, the cells were harvested for further analysis.
qRT-PCR
Total RNA was isolated from tissue specimens and cultured cells by using a modified Trizol co-purification technique.16 The levels of circ_0031242 and POU3F2 mRNA were quantified using PrimeScriptTM One-Step TB Green® RT-PCR Kit (TaKaRa, Dalian, China) in a 20 μL of reaction containing 100 ng of total RNA with primers specific for circ_0031242: 5ʹ-TACCATTGGCCTCTAGCCCT-3ʹ (forward) and 5ʹ-CAAGGGGAATCACAGCCCAT-3ʹ (reverse), or POU3F2: 5ʹ-TGGGATTTACCCAAGCGGAC-3ʹ (forward) and 5ʹ-CGCCTGCAAAGGTCAAAACA-3ʹ (reverse). Quantification of miR-924 was carried out using miRCURY LNATM Universal RT miRNA PCR (Exiqon, Denmark) with primer for miR-924: 5ʹ-TCTTCTCTGTTTTGGCCA-3ʹ (forward) and 5ʹ-GAACATGTCTGCGTATCTC-3ʹ (reverse). Each reaction was individually run on a 480 II Light Cycler (Roche, Basel, Switzerland) in triplicate. Results were calculated using the 2−ΔΔCt method and normalized based on the expression of β-actin: 5ʹ-CTCGCCTTTGCCGATCC-3ʹ (forward) and 5ʹ-GGGGTACTTCAGGGTGAGGA-3ʹ (reverse), or U6: 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ (forward) and 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ (reverse). Reactions were incubated for 10 min at 95°C, and then for 40 cycles at 94°C for 15 s, and 60°C for 60 s.
Measurement of IC50 Value of DDP and Cell Viability
The IC50 values of DDP and cell viability were determined by the colorimetric CCK-8 assay.17 For IC50 value measurement, transfected cells (1 × 104 per well) were plated in 96-well plates and exposed to various concentrations (0.5, 1, 2, 4, 8, 16, 32, 64 and 128 µg/mL) of DDP. For cell viability detection, transfected cells were plated in 96-well plates at 5 × 103 per well. After 24 h, CCK-8 solution was added in each well based on the protocols of manufacturers (Abcam, Cambridge, UK). The number of viable cells was proportional to the absorbance which could be measured by a microplate reader at 460 nm.
Transwell Migration and Invasion Assay
Transwell inserts for 24-well plates (8.0 µm-pore membrane, Corning, Tewksbury, MA, USA) were used for transwell migration assays. To measure cell invasion, the transwell inserts were precoated with Matrigel (BD Biosciences, Oxford, UK). Transfected cells in serum-free media were plated onto the top of the insert membranes at 2.5 × 104 cells for migration assays and 5 × 104 cells for invasion assays. 500 µL of culture medium containing 10% FCS was added into the bottom of the lower chamber as a chemoattractant. The wells were fixed with 70% ethanol and stained with 0.5% crystal violet after 24 h incubation at 37°C. Underneath a microscope (Leica, Wetzlar, Germany) at 100× magnification, the number of cells in five random fields was counted to get an average of cells that had traversed the membrane insert.
Determination of Cell Apoptosis
Cell apoptosis was determined by flow cytometry on an ECSCalibur (BD Biosciences). Transfected cells (2.5 × 105) were double-stained with 5 µL of fluorescein isothiocyanate (FITC)-labeled Annexin V and 5 µL of propidium iodide (PI, 50 µg/mL) as per manufacturing recommendation (Pharmingen, San Diego, CA, USA). A total of 10,000 events were analyzed per sample by the Cell Quest software (BD Biosciences) and results were expressed as a percentage of total cell number.
Western Blot
Western blot was carried out using standard protocols as reported.18 Total protein isolated with ice-cold RIPA lysis buffer (EpiZyme, Shanghai, China) were fractionated on 4–12% Bis-Tris gels (Invitrogen), blotted on polyvinylidene fluoride membranes (Corning) and then blocked with 5% non-fat milk. Antibodies against P-glycoprotein (P-gp, ab235954), multidrug resistance-associated protein 1 (MRP1, ab225885), POU3F2 (ab137469), Bcl-2 (ab182858), Bax (ab32503), β-catenin (ab32572), c-myc (ab32072) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, ab22555) were used as primary antibody as per the directions of manufacturers (Abcam). Horseradish peroxidase-conjugated anti-rabbit IgG antibody (ab97057) was used as the secondary antibody. Immunoreactive proteins were visualized by Enhanced Chemiluminescence (Millipore, Shanghai, China) and analyzed using Multi-Gauge software (Fujifilm, Tokyo, Japan).
Generation of Stable circ_0031242 Knockdown Cell Line
Lentiviruses encoding shRNA-circ_0031242 (sh-circ_0031242) or a nontarget shRNA (sh-NC) were obtained from Ribobio. Huh7-R cells were infected with the lentiviruses in media containing 8 µg/mL polybrene. The cells with positive transduction were selected by puromycin (1 µg/mL) more than 3 days.
Animal Studies
In this study, 4-6-week-old female BALB/c nude mice were purchased from Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) and divided into 4 groups (n = 6 per group): PBS, DDP, DDP+sh-NC or DDP+sh-circ_0031242. About 2 × 106 sh-NC-infected, sh-circ_0031242-transduced or un-transduced Huh7-R cells were subcutaneously implanted into the left flank of 4-6-week-old female BALB/c nude mice (6 mice per group, Vital River Laboratory, Beijing, China). After one week of implantation, DDP administration (3 mg/kg) was conducted every 4 days by intraperitoneal injection. Tumor growth was periodically assessed by calculating tumor volume as follows: volume = length × width2/2. In the end, tumor tissues were harvested from all sacrificial mice.
Dual-Luciferase Reporter Assay
The miRNAs that potentially bind to circ_0031242 were searched using the CircInteractome online tool (https://circinteractome.nia.nih.gov/mirna_target_sites.html), and the putative targets of miR-924 were predicted by the TargetScan software (http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg). The fragments of circ_0031242 and POU3F2 3ʹUTR containing the miR-924-pairing region or miss-matched miR-924-binding sites were individually inserted into the pmirGLO plasmid (Promega, Mannheim, Germany). Huh7-R and SNU-387-R cells were seeded at 1 × 105 cells per well into 24-well plates 24 h before transfection with 200 ng of each reporter construct and 50 nM of miR-924 mimic or negative control using Lipofectamine 3000 reagent. The ratio of Renilla/firefly luciferase was determined with the Promega Dual-luciferase assay using 20 µL of cell lysates.
RNA Immunoprecipitation (RIP) Assay
RIP assays were done on Huh7-R and SNU-387-R cells with anti-Argonaute2 (anti-Ago2) or anti-IgG antibody as recommended by the manufacturers (Millipore). Cell lysates were incubated with magnetic beads-coupled the above antibodies at 4°C for 4–6 h. Total RNA from the beads was isolated to quantify the enrichment levels of circ_0031242 and miR-924 by qRT-PCR as above.
Statistical Analysis
Data were analyzed for quantitative variables using a Student’s t-test (two-tailed) or analysis of variance (ANOVA) followed by Tukey-Kramer post hoc test. The Mann–Whitney U-test was used to analyze the differences of gene expression in clinical specimens. Pearson’s rank correlation was used to test the correlations among circ_0031242, miR-924 and POU3F2 in DDP-resistant HCC tissues. All analyses were conducted using the GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). Data were presented as the mean ± standard deviation with at least three times of independent experiments and were interpreted statistically significant as P less than 0.05.
Results
Overexpression of circ_0031242 Was Associated with DDP Resistance in HCC
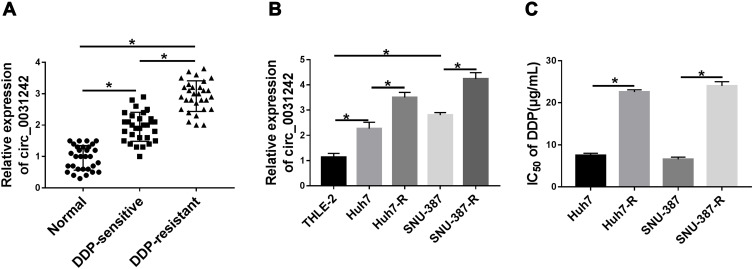
To examine whether circ_0031242 was involved in DDP resistance of HCC, we firstly determined its expression pattern in the tumor samples from primary patients (DDP-sensitive) and recurrent patients after treatment with DDP-based chemotherapy (DDP-resistant). As demonstrated by qRT-PCR, the expression of circ_0031242 (mature sequence and structural representation were provided in Supplement Figure 1) was remarkably increased in HCC tissues compared to the normal controls (Figure 1A). Moreover, recurrent HCC patients had a higher circ_0031242 level than primary patients (Figure 1A). Moreover, circ_0031242 was dramatically up-regulated in HCC cell lines compared with the THLE-2 cells, and it was higher in DDP-resistant HCC cells than that in their parental cells (Figure 1B). Additionally, by contrast, the IC50 value of DDP was significantly elevated in DDP-resistant HCC cells (Huh7-R and SNU-387-R, Figure 1C).
Figure 1.
Circ_0031242 was overexpressed and associated with DDP resistance in HCC. The expression of circ_0031242 by qRT-PCR in 30 healthy hepatic tissues, HCC tissues from 30 primary patients (DDP-sensitive) and 30 recurrent patients after treatment with DDP-based chemotherapy (DDP-resistant) (A), THLE-2, Huh7, SNU-387, Huh7-R and SNU-387-R cells (B). (C) The IC50 value of DDP by CCK-8 assay in Huh7, SNU-387, Huh7-R and SNU-387-R cells. *P < 0.05 by a two-tailed Student’s t-test or ANOVA followed by Tukey-Kramer post hoc test.
Silencing of circ_0031242 Mitigated DDP Resistance, Suppressed Cell Viability, Migration, Invasion, and Enhanced Apoptosis in Huh7-R and SNU-387-R Cells
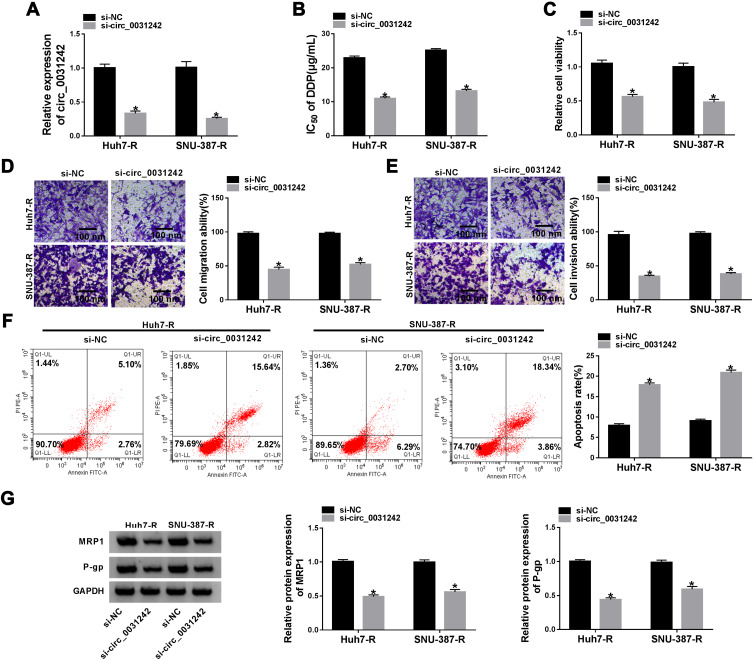
To determine the biological effect of circ_0031242 on the malignant behaviors of DDP-resistant HCC cells, we performed “phenocopy” silencing by siRNA that direct targets circ_0031242. Transient transfection of si-circ_0031242, but not the scrambled sequence control, prominently down-regulated the expression of circ_0031242 in both Huh7-R and SNU-387-R cell lines (Figure 2A). In contrast to the si-NC control, circ_0031242 knockdown led to a significant decrease in the IC50 values of DDP (Figure 2B). Functional experiments revealed that the silencing of circ_0031242 caused a prominent repression in cell viability (Figure 2C), migration (Figure 2D), invasion (Figure 2E), as well as a striking enhancement in cell apoptosis (Figure 2F). Moreover, in comparison to the negative control, circ_0031242 silencing resulted in reduced levels of multidrug resistance-associated proteins MRP1 and P-gp in the two DDP-resistant HCC cell lines (Figure 2G). Additionally, the silencing of circ_0031242 significantly reduced anti-apoptotic protein Bcl-2 expression and elevated pro-apoptotic Bax protein level in both Huh7-R and SNU-387-R cell lines (Supplement Figure 2A and B).
Figure 2.
Circ_0031242 knockdown declined DDP resistance, cell viability, migration, invasion and accelerated apoptosis in Huh7-R and SNU-387-R cells. Huh7-R and SNU-387-R cells were transfected with si-NC or si-circ_0031242. (A) Circ_0031242 expression by qRT-PCR in transfected cells. (B) The IC50 value of DDP in transfected cells by CCK-8 assay. (C) Cell viability by CCK-8 assay. (D and E) Cell migration and invasion by transwell assay. (F) Cell apoptosis by flow cytometry. (G) The levels of MRP1 and P-gp in transfected cells by Western blot. *P < 0.05 by a two-tailed Student’s t-test.
Silencing of circ_0031242 Enhanced DDP Sensitivity in vivo
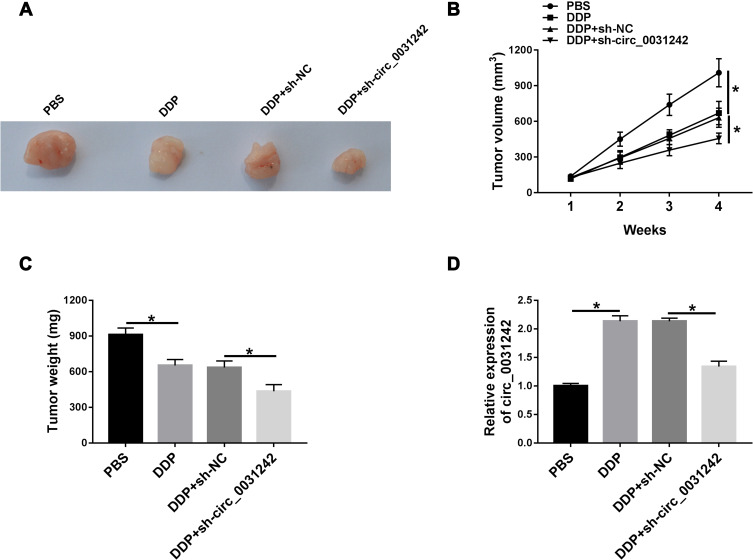
We then elucidate whether circ_0031243 could modulate DDP sensitivity in vivo. Strikingly, DDP administration led to a diminishment of tumor growth compared with control group (Figure 3A–C). Moreover, simultaneous sh-circ_0031242 transduction and DDP treatment caused a more significant repression in tumor growth (Figure 3A–C). Additionally, DDP treatment resulted in increased expression of circ_0031242, and this effect was abolished by sh-circ_0031242 in the xenograft tissues (Figure 3D).
Figure 3.
Circ_0031242 knockdown enhanced DDP resistance of Huh7-R cells in vivo. Representative pictures after 4 weeks implantation (A) and growth curves (B) of the xenograft tumors formed by sh-NC-infected, sh-circ_0031242-transduced or un-transduced Huh7-R cells under DDP or PBS treatment. Tumor average weight (C) and circ_0031242 level by qRT-PCR (D) of xenograft tumors formed by sh-NC-infected, sh-circ_0031242-transduced or untransduced Huh7-R cells under DDP or PBS treatment, on 4 weeks after implantation. *P < 0.05 by ANOVA followed by Tukey-Kramer post hoc test.
Circ_0031242 Directly Interacted with miR-924
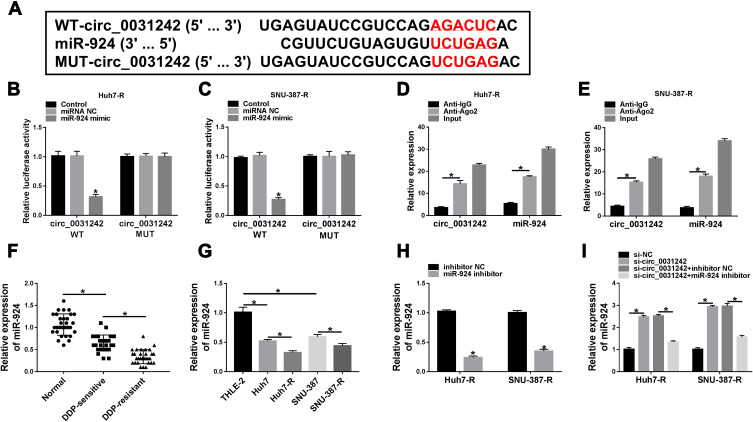
In order to further understand the role of circ_0031242, we performed a detailed analysis for its targeted miRNAs using the CircInteractome online web (https://circinteractome.nia.nih.gov/mirna_target_sites.html). Among these candidates, we selected 6 miRNAs that were associated with HCC tumorigenesis, and our data showed that miR-924 was the most significantly up-regulated miRNA in si-circ_0031242-transfected DDP-resistant cells (Supplement Figure 3A and B). CircInteractome online tool predicted that circ_0031242 harbored a target sequence (AGACUC) for miR-924 (Figure 4A). When we cloned the circ_0031242 segment containing the putative miR-924-pairing sites into a luciferase plasmid, the cotransfection of the luciferase reporter (circ_0031242 WT) and miR-924 mimic into the two DDP-resistant HCC cell lines produced lower luciferase activity than cells cotransfected with miRNA NC control (Figure 4B and C). However, the mutation of the target sequence (circ_0031242 MUT) dramatically abolished the repressive impact of miR-924 (Figure 4B and C). RIP experiments showed that the enrichment levels of circ_0031242 and miR-924 were synchronously elevated by anti-Ago2 antibody (Figure 4D and E), demonstrating their potential endogenous interaction. Of interest, miR-924 level was lower in HCC group than that in normal control group (Figure 4F and G). Moreover, in contrast to the DDP-sensitive counterparts, miR-924 expression was prominently decreased in DDP-resistant tissues and cells (Figure 4F and G). Additionally, a strong inverse correlation between miR-924 and circ_0031242 level was found in DDP-resistant tissues (Supplement Figure 4A). We then evaluated whether circ_0031242 influenced miR-924 expression in the two DDP-resistant cell lines. The transfection efficiency of miR-924 inhibitor was gauged by qRT-PCR (Figure 4H). In comparison to their counterparts, the transfection of si-circ_0031242 led to a striking up-regulation in miR-924 expression, and this effect was dramatically reversed by miR-924 inhibitor (Figure 4I).
Figure 4.
Circ_0031242 directly interacted with miR-924 through pairing to miR-924. (A) Schematic of the putative miR-924-pairing sites within circ_0031242 identified by CircInteractome online tool and the mutation in the target region. (B and C) Relative luciferase activity in Huh7-R and SNU-387-R cells cotransfected with circ_0031242 wild-type (circ_0031242 WT) or mutant-type (circ_0031242 MUT) luciferase reporter constructs and miR-924 mimic or miRNA NC mimic. (D and E) The enrichment levels of circ_0031242 and miR-924 in cell lysates incubated with anti-Ago2 or anti-IgG antibody. The expression of miR-924 by qRT-PCR in 30 healthy hepatic tissues, HCC tissues from 30 primary patients (DDP-sensitive) and 30 recurrent patients after treatment with DDP-based chemotherapy (DDP-resistant) (F), THLE-2, Huh7, SNU-387, Huh7-R and SNU-387-R cells (G), Huh7-R and SNU-387-R cells transfected with inhibitor NC or miR-924 inhibitor (H), Huh7-R and SNU-387-R cells transfected with si-NC, si-circ_0031242, si-circ_0031242+inhibitor NC or si-circ_0031242+miR-924 inhibitor (I). *P < 0.05 by a two-tailed Student’s t-test or ANOVA followed by Tukey-Kramer post hoc test.
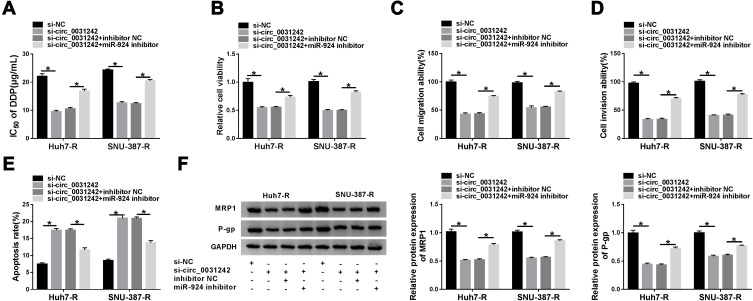
Regulatory Impact of circ_0031242 Silencing on Cell DDP Resistance, Viability, Migration, Invasion, and Apoptosis Was Reversed by miR-924 Down-Regulation
We then asked whether miR-924 was involved in the regulation of circ_0031242 knockdown on DDP-resistant HCC cell behaviors. To address this, Huh7-R and SNU-387-R cells were cotransfected with si-circ_0031242 and miR-924 inhibitor. By contrast, si-circ_0031242-mediated IC50 decrease was remarkably abated by miR-924 knockdown (Figure 5A). Moreover, circ_0031242 silencing-mediated viability repression (Figure 5B), migration inhibition (Figure 5C), invasion reduction (Figure 5D) and apoptosis enhancement (Figure 5E) were dramatically abolished by the down-regulation of miR-924. Additionally, the reduced impact of circ_0031242 silencing on MRP1 and P-gp expression was strikingly abrogated by miR-924 inhibitor (Figure 5F).
Figure 5.
The regulatory impact of circ_0031242 silencing on DDP-resistant HCC cell behaviors was abated by miR-924 down-regulation. Huh7-R and SNU-387-R cells were transfected with si-NC, si-circ_0031242, si-circ_0031242+inhibitor NC or si-circ_0031242+miR-924 inhibitor, followed by the measurement of IC50 value of DDP by CCK-8 assay (A), cell viability by CCK-8 assay (B), cell migration and invasion by transwell assay (C and D), cell apoptosis by flow cytometry (E), MRP1 and P-gp levels by Western blot (F). *P < 0.05 by ANOVA followed by Tukey-Kramer post hoc test.
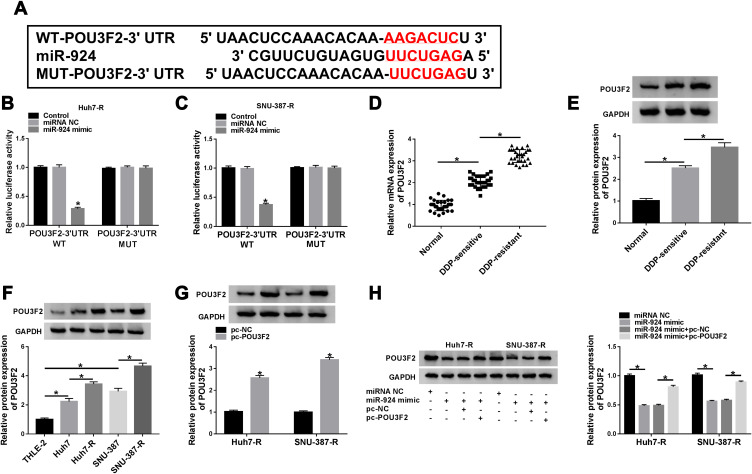
POU3F2 Was Directly Targeted by miR-924
To identify the downstream targets of miR-924, we used the software TargetScan. Among these candidates, we selected 6 genes that were involved in HCC carcinogenesis. Our data showed that POU3F2 mRNA expression was the most significantly down-regulated in Huh7-R and SNU-387-R cells transfected with miR-924 mimic (Supplement Figure 3C and D). To identify the direct relationship between miR-924 and POU3F2, we generated the POU3F2 3ʹUTR wild-type (POU3F2-3ʹUTR WT) or mutant-type luciferase reporter (POU3F2-3ʹUTR MUT) (Figure 6A) and tested them in dual-luciferase reporter assays. Transient transfection of POU3F2-3ʹUTR WT in the presence of miR-924 mimic caused a prominent reduction of luciferase activity, and this effect was completely abrogated by the mutation of the miR-924-pairing sequence (Figure 6B and C). POU3F2 was overexpressed in HCC tissues and cells compared with the normal controls, and a more distinct up-regulation of POU3F2 was found in DDP-resistant HCC tissues and cells (Figure 6D–F). In addition, in DDP-resistant HCC tissues, POU3F2 mRNA expression was inversely correlated with miR-924 level and positively correlated with circ_0031242 expression (Supplement Figure 4B and C). Western blot data revealed that POU3F2 overexpressing plasmid (pc-POU3F2) induced a significant up-regulation of POU3F2 protein level in both Huh7-R and SNU-387-R cell lines (Figure 6G). More importantly, the transfection of miR-924 mimic resulted in decreased POU3F2 expression, and this effect was remarkably abolished by pc-POU3F2 (Figure 6H).
Figure 6.
POU3F2 was directly targeted by miR-924. (A) Schematic of the miR-924-pairing sequence within the 3ʹUTR of POU3F2 mRNA and mutated the miR-924-pairing sites. (B and C) Relative luciferase activity in Huh7-R and SNU-387-R cells cotransfected with POU3F2 3ʹUTR wild-type (POU3F2-3ʹUTR WT) or mutant-type (POU3F2-3ʹUTR MUT) luciferase reporter construct and miR-924 mimic or miRNA NC mimic. POU3F2 mRNA and protein levels by qRT-PCR and Western blot in healthy hepatic tissues, HCC tissues from primary patients (DDP-sensitive) and recurrent patients after treatment with DDP-based chemotherapy (DDP-resistant) (D and E), THLE-2, Huh7, SNU-387, Huh7-R and SNU-387-R cells (F), Huh7-R and SNU-387-R cells transfected with pc-POU3F2 or pc-NC (G), Huh7-R and SNU-387-R cells transfected with miRNA NC mimic, miR-924 mimic, miR-924 mimic+pc-NC or miR-924 mimic+pc-POU3F2 (H). *P < 0.05 by a two-tailed Student’s t-test or ANOVA followed by Tukey-Kramer post hoc test.
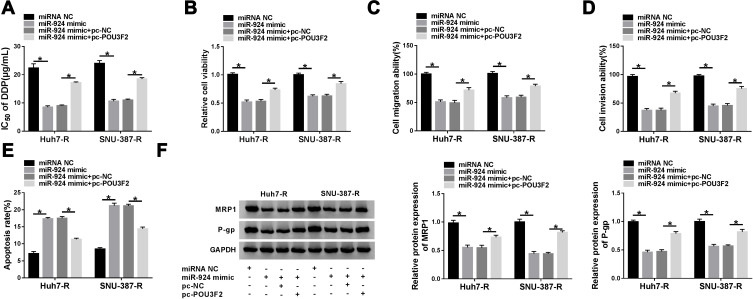
POU3F2 Was a Functional Target of miR-924 in Regulating Cell DDP Resistance, Viability, Migration, Invasion and Apoptosis
We next examined whether miR-924 regulated DDP-resistant HCC cell behaviors by POU3F2. By contrast, the increased miR-924 level led to a striking reduction in IC50 values of DDP (Figure 7A), and a significant suppression in cell viability (Figure 7B), migration (Figure 7C), invasion (Figure 7D), as well as a remarkable promotion in cell apoptosis (Figure 7E) in both Huh7-R and SNU-387-R cell lines. Furthermore, miR-924 overexpression caused a prominent decrease in the levels of MRP1 and P-gp in the two HCC cell lines (Figure 7F). Nevertheless, these effects of miR-924 overexpression were dramatically abrogated by POU3F2 up-regulation (Figure 7A–F).
Figure 7.
The increased miR-924 level regulated DDP-resistant HCC cell behaviors by down-regulating POU3F2. Huh7-R and SNU-387-R cells were transfected with miRNA NC mimic, miR-924 mimic, miR-924 mimic+pc-NC or miR-924 mimic+pc-POU3F2 (POU3F2 overexpressing plasmid). (A) The IC50 value of DDP in transfected cells by CCK-8 assay. (B) Cell viability by CCK-8 assay. (C and D) Cell migration and invasion by transwell assay. (E) Cell apoptosis by flow cytometry. (F) The levels of MRP1 and P-gp in transfected cells by Western blot. *P < 0.05 by ANOVA followed by Tukey-Kramer post hoc test.
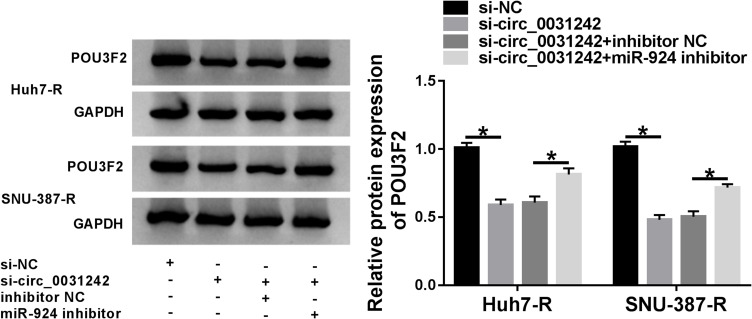
Circ_0031242 Regulated POU3F2 Expression by Working as a Sponge of miR-924
Subsequently, we undertook to examine whether, if so, how circ_0031242 influenced POU3F2 expression. As expected, in contrast to their counterparts, the transfection of si-circ_0031242 led to a significant reduction of POU3F2 protein expression in both Huh7-R and SNU-387-R cell lines, and this effect was remarkably reversed by miR-924 inhibitor (Figure 8).
Figure 8.
Circ_0031242 regulated POU3F2 expression by sponging miR-924. POU3F2 protein expression by Western blot in Huh7-R and SNU-387-R cells transfected with si-NC, si-circ_0031242, si-circ_0031242+inhibitor NC or si-circ_0031242+miR-924 inhibitor. *P < 0.05 by ANOVA followed by Tukey-Kramer post hoc test.
Discussion
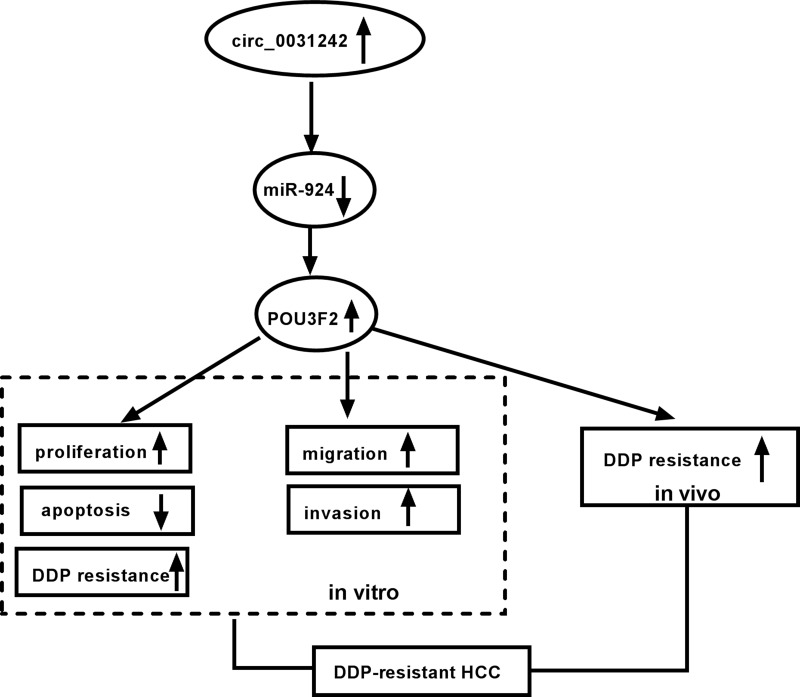
Chemoresistance is a major problem in successful HCC therapy.19 Aberrant expression of circRNAs has been reported to impact HCC carcinogenesis and chemoresistance development.5,20,21 In this study, our results showed that circ_0031242 expression was associated with HCC resistance to DDP. We showed that the silencing of circ_0031242 diminished cell DDP resistance, repressed cell progression in vitro and enhanced DDP sensitivity in vivo. More importantly, we provided a molecular explanation, at least in part, for the regulation of circ_0031242 in HCC DDP resistance (Figure 9).
Figure 9.
The schematic model of the circ_0031242/miR-924/POU3F2 axis on DDP-resistant HCC cell progression and drug resistance. In DDP-resistant HCC tissues and cells, circ_0031242 was up-regulated, resulting in decreased miR-924. Then, the reduced miR-924 level elevated POU3F2 expression. Finally, the increased expression of POU3F2 enhanced DDP resistance, promoted cell viability, migration, invasion, as well as repressed apoptosis in DDP-resistant HCC cells, thereby leading to DDP-resistant HCC progression.
The ceRNA hypothesis suggests that circRNAs modulate gene expression by sequestering miRNAs, reducing their repression to target mRNAs.22,23 Here, we first identified that circ_0031242 directly targeted miR-924. MiR-924 could regulate the pathogenesis of inflammatory bowel disease by targeting α-defensin 5.24 MiR-924 was also found to be overexpressed in leiomyoma progenitor cells.25 Moreover, Fan et al identified that miR-924-mediated ceRNA network influenced the tumorigenesis of HBV-related HCC.13 Our results demonstrated the modulation of circ_0031242 on DDP-resistant HCC cell progression and DDP resistance through miR-924.
POU3F2, a POU-homeodomain transcription factor, has been shown as a crucial modulator in various cancers, such as melanoma, colon cancer and prostate cancer.26–28 Moreover, recent work reported that POU3F2 was up-regulated in HCC tissues and the increased expression of POU3F2 predicted a poor prognosis in these patients.14 The study focusing on the involvement of POU3F2 in HCC aggressive progression also argued for a ceRNA regulatory network.14 Here, we were first to identify that miR-924 directly targeted POU3F2. Moreover, we first showed that the increased level of miR-924 modulated DDP-resistant HCC cell behaviors by down-regulating POU3F2. More importantly, we pointed out the role of circ_0031242 as a miR-924 sponge to control POU3F2 expression. MRP1 and P-gp, two members of ATP-binding cassette transporter proteins, exert important functions in the development of multidrug resistance in human cancer.29,30 Our results showed the regulation of circ_0031242 on the expression of MRP1 and P-gp by the miR-924/POU3F2 axis, and a future challenging is to identify how the novel network regulates their expression in DDP-resistant HCC cells. Additionally, the Wnt/β-catenin signaling is one of the cancer-related pathways and promotes the tumorigenesis of various cancers, including HCC.31 Moreover, the Wnt/β-catenin signaling was discovered to be implicated in the DDP resistance development of HCC.32 Our data also demonstrated that circ_0031242 regulated the Wnt/β-catenin/c-myc signaling by the miR-924/POU3F2 axis in DDP-resistant HCC cells (Supplement Figure 5). However, the direct evidence between the Wnt/β-catenin/c-myc pathway and circ_0031242-mediated regulation in DDP-resistant HCC cell progression and drug resistance is lack, which is expected to be explored in further work.
In summary, the findings in this paper identified the precise parts of circ_0031242 in HCC chemoresistance, with the ability to regulate DDP-resistant HCC cell progression and DDP resistance by targeting the miR-924/POU3F2 axis. These findings provided a strong rationale for developing circ_0031242 as a potential therapeutic target for the chemoresistant HCC.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, Gao M, Li Z, et al. p53 and P-glycoprotein influence chemoresistance in hepatocellular carcinoma. Front Biosci (Elite Ed). 2018;10:461–468. [DOI] [PubMed] [Google Scholar]
- 4.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 5.Wu MY, Tang YP, Liu JJ, Liang R, Luo XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer. 2020;11(10):2993–3001. doi: 10.7150/jca.39854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Qin X, Wu R, et al. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med. 2020;24(9):5152–5161. doi: 10.1111/jcmm.15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Y, Fu Y, Huang R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discov. 2019;5(1):121. doi: 10.1038/s41420-019-0202-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi: 10.1016/j.gene.2019.144099 [DOI] [PubMed] [Google Scholar]
- 9.Du W, Li D, Guo X, et al. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif Cells Nanomed Biotechnol. 2019;47(1):4120–4130. doi: 10.1080/21691401.2019.1671857 [DOI] [PubMed] [Google Scholar]
- 10.Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19(3):269–279. doi: 10.1016/j.aohep.2020.01.002 [DOI] [PubMed] [Google Scholar]
- 11.Ding B, Lou W, Xu L, Fan W. Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci Rep. 2018;38(5):BSR20180915. doi: 10.1042/BSR20180915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espelt MV, Bacigalupo ML, Carabias P, Troncoso MF. MicroRNAs contribute to ATP-binding cassette transporter- and autophagy-mediated chemoresistance in hepatocellular carcinoma. World J Hepatol. 2019;11(4):344–358. doi: 10.4254/wjh.v11.i4.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan H, Lv P, Mu T, et al. LncRNA n335586/miR-924/CKMT1A axis contributes to cell migration and invasion in hepatocellular carcinoma cells. Cancer Lett. 2018;429:89–99. doi: 10.1016/j.canlet.2018.05.010 [DOI] [PubMed] [Google Scholar]
- 14.Ding S, Jin Y, Hao Q, Kang Y, Ma R. LncRNA BCYRN1/miR-490-3p/POU3F2, served as a ceRNA network, is connected with worse survival rate of hepatocellular carcinoma patients and promotes tumor cell growth and metastasis. Cancer Cell Int. 2020;20(1):6. doi: 10.1186/s12935-019-1081-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuo L, Liu J, Wang B, Gao M, Huang A. Differential miRNA expression profiles in hepatocellular carcinoma cells and drug-resistant sublines. Oncol Rep. 2013;29(2):555–562. doi: 10.3892/or.2012.2155 [DOI] [PubMed] [Google Scholar]
- 16.Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106(8):947–952. doi: 10.1002/jso.23174 [DOI] [PubMed] [Google Scholar]
- 17.Präbst K, Engelhardt H, Ringgeler S, Hübner H. Basic colorimetric proliferation assays: MTT, WST, and resazurin. Methods Mol Biol. 2017;1601:1–17. [DOI] [PubMed] [Google Scholar]
- 18.Stevens JJ, Graham B, Dugo E, et al. Arsenic trioxide induces apoptosis via specific signaling pathways in HT-29 colon cancer cells. J Cancer Sci Ther. 2017;9(1):298–306. doi: 10.4172/1948-5956.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler SM, Hosseini K, Hussein UK, et al. Hepatocellular carcinoma and nuclear paraspeckles: induction in chemoresistance and prediction for poor survival. Cell Physiol Biochem. 2019;52(4):787–801. [DOI] [PubMed] [Google Scholar]
- 20.Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. doi: 10.1016/j.lfs.2019.116660 [DOI] [PubMed] [Google Scholar]
- 21.Zhou S, Wei J, Wang Y, Liu X. Cisplatin resistance-associated circRNA_101237 serves as a prognostic biomarker in hepatocellular carcinoma. Exp Ther Med. 2020;19(4):2733–2740. doi: 10.3892/etm.2020.8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi: 10.1136/jmedgenet-2015-103334 [DOI] [PubMed] [Google Scholar]
- 23.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 24.Miles DR, Shen J, Chuang AY, et al. Alpha-defensin 5 expression is regulated by microRNAs in the Caco-2 intestinal epithelial cell line. J Inflamm Bowel Dis Disord. 2016;1(1):105. [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzarini R, Caffarini M, Delli Carpini G, et al. From 2646 to 15: differentially regulated microRNAs between progenitors from normal myometrium and leiomyoma. Am J Obstet Gynecol. 2020;222(6):596.e1–596.e9. doi: 10.1016/j.ajog.2019.12.016 [DOI] [PubMed] [Google Scholar]
- 26.Kobi D, Steunou AL, Dembélé D, et al. Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell Melanoma Res. 2010;23(3):404–418. doi: 10.1111/j.1755-148X.2010.00697.x [DOI] [PubMed] [Google Scholar]
- 27.Chen H-Y, Islam A, Yuan T-M, et al. Regulation of tNOX expression through the ROS-p53-POU3F2 axis contributes to cellular responses against oxaliplatin in human colon cancer cells. J Exp Clin Cancer Res. 2018;37(1):161. doi: 10.1186/s13046-018-0837-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop JL, Thaper D, Vahid S, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263 [DOI] [PubMed] [Google Scholar]
- 29.Robey RW, Pluchino KM, Hall MD, et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18(7):452–464. doi: 10.1038/s41568-018-0005-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Jaitak V. Natural products as multidrug resistance modulators in cancer. Eur J Med Chem. 2019;176:268–291. doi: 10.1016/j.ejmech.2019.05.027 [DOI] [PubMed] [Google Scholar]
- 31.Khalaf AM, Fuentes D, Morshid AI, et al. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma. 2018;5:61–73. doi: 10.2147/JHC.S156701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Liu S, Li M, Huang P, Li X. circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco Targets Ther. 2019;12:9539–9549. doi: 10.2147/OTT.S229507 [DOI] [PMC free article] [PubMed] [Google Scholar]