Abstract
The aim of the present study was to develop a simple, sensitive, and specific approach to quantifying the SARS-CoV-2 genome in wastewater and to evaluate this approach as a means of epidemiological surveillance. Twelve wastewater samples were collected from a metropolitan area in north-eastern France during April and May 2020. In addition to the quantification of the SARS-CoV-2 genome, F-specific RNA phages of genogroup II (FRNAPH GGII), naturally present in wastewater, were used as an internal process control for the viral concentration and processing of RT-PCR inhibitors. A concentration method was required to allow the quantification of the SARS-CoV-2 genome over the longest possible period. A procedure combining ultrafiltration, phenol-chloroform-isoamyl alcohol purification, and the additional purification of the RNA extracts was chosen for the quantification of the SARS-CoV-2 genome in 100-mL wastewater samples. At the same time, the COVID-19 outbreak was evaluated through patients from the neighbouring University Hospital of Nancy, France. A regular decrease in the concentration of the SARS-CoV-2 genome from ~104 gc/L to ~102 gc/L of wastewater was observed over the eight weeks of the study, during which the population was placed under lockdown. The SARS-CoV-2 genome was even undetectable during one week in the second half of May and present but non-quantifiable in the last sample (28 May). A concordant circulation in the human community was highlighted by virological diagnosis using respiratory samples, which showed a decrease in the number of COVID-19 cases from 677 to 52 per week over the same period. The environmental surveillance of COVID-19 using a reliable viral quantification procedure to test wastewater is a key approach. The real-time detection of viral genomes can allow us to predict and monitor the circulation of SARS-CoV-2 in clinical settings and survey the entire urban human population.
Keywords: SARS-CoV-2, Wastewater, Concentration methods, Prevalence
1. Introduction
In December 2019, the first outbreak of a new coronavirus disease (COVID-19) was reported in Wuhan, China. A new coronavirus linked to Severe Acute Respiratory Syndrome (SARS-CoV-2) was identified. It displayed more than 80% sequence homology with the previous SARS-CoV, identified in 2003 (Cheung et al., 2020). By March 2020, 114 countries reported outbreaks and the WHO declared a pandemic linked to SARS-CoV-2 (WHO, 2020). France was severely affected, with more than 2,550,000 confirmed COVID-19 infections and more than 62,000 deaths by the end of 2020.
Even though many cases of COVID-19 are asymptomatic, when symptomatic it is characterized mainly by fever and respiratory symptoms, but there are also a variety of other symptoms frequently associated with the disease. These include gastrointestinal complaints, including anorexia, diarrhoea, and vomiting/nausea at frequencies of up to 27%, 12% and 10%, respectively (Cheung et al., 2020). SARS-CoV-2 is able to productively replicate in human gut enterocytes (Lamers et al., 2020) and the SARS-CoV-2 genome has been detected in the stools of both symptomatic and asymptomatic patients (Cai et al., 2020; Gao et al., 2020; Holshue et al., 2020; Tang et al., 2020; Wölfel et al., 2020; Zhang et al., 2020). Viral RNA may be detected in stools up to 10 days after viral clearance from the respiratory tract, regardless of the severity of the disease (Cheng et al., 2020). The viral concentration can be high, potentially reaching 107 genome copies (gc)/g (Wölfel et al., 2020).
As expected, very high concentrations of the SARS-CoV-2 genome have also been detected in wastewater (Ahmed et al., 2020; Hindson, 2020; Medema et al., 2020; Nemudryi et al., 2020; Randazzo et al., 2020; Wu et al., 2020a; Wurtzer et al., 2020). Concentrations may reach close to 106 gc/L (Wu et al., 2020a; Wurtzer et al., 2020). The presence of SARS-CoV-2 in wastewater indicates a potential health risk, but also a data source that can be used for epidemiological purposes (Lodder and de Roda Husman, 2020; Thompson et al., 2020). Wastewater-based epidemiology is not a new idea. It has been used to detect chemicals (Choi et al., 2020) and other viruses, such as norovirus, hepatitis A virus, poliovirus, and Aichivirus (Asghar et al., 2014; Hellmer et al., 2014; Lodder et al., 2012, 2013). Many countries (including The Netherlands, Spain, France, Australia, and Israel) now support the idea of using this type of epidemiological approach in the surveillance of human populations and to possibly launch an early warning system to predict future outbreaks (Ahmed et al., 2020; Bar Or et al., 2020; Medema et al., 2020; Wu et al., 2020b; Wurtzer et al., 2020).
Whatever its purpose — whether epidemiological or to assess public health risk — the first step is to define a method of quantifying SARS-CoV-2 in wastewater. The methodologies that have been used to date are highly diverse. The concentration methods were based on various principles, including ultrafiltration (Ahmed et al., 2020; Medema et al., 2020; Nemudryi et al., 2020; Trottier et al., 2020); protein precipitation (Kumar et al., 2020; La Rosa et al., 2020b; Wu et al., 2020a); electronegative membranes (Ahmed et al., 2020) and ultracentrifugation (Wurtzer et al., 2020). Recovery rates are currently unknown or extrapolated from those of similar viruses such as porcine epidemic diarrhoea virus (PEDV) and mengovirus (MgV) (Randazzo et al., 2020).
The aim of this study was to define the dynamics of viral concentration in wastewater during the first lockdown of the French population. This objective required a simple, specific, and sensitive approach to quantifying the SARS-CoV-2 genome in wastewater. In the present study, we took advantage of the wide circulation of SARS-CoV-2 in north-eastern France to compare two methods of virus concentration. We assessed the recovery rate of SARS-CoV-2 by virus quantification directly in four unconcentrated wastewater samples. We also included an internal process control, using F-specific RNA phages of genogroup II (FRNAPH GGII). We applied the most reliable method to investigate the temporal variations of SARS-CoV-2 genome concentrations in wastewater over 12 weeks in an area of France with 250,000 inhabitants, which has been significantly impacted by COVID-19.
2. Material and methods
2.1. Sewage samples
Samples were collected between 2 April and 28 May 2020 in the French Grand Est region. In that region, the population was under lockdown from 17 March to 10 May 2020. A total of 12 water samples (400 mL) were collected from a wastewater treatment plant (WWTP) weekly or twice weekly. The average volume of influent treated in this WWTP was ~118,000 m3/day (~250,000 inhabitants). The wastewater samples were captured at regular intervals, just after decantation, but before activated sludge treatment. The samples were stored at ˗20 °C until analysis.
2.2. Clinical samples
At the same time, 19,850 samples were examined by the laboratory of the local University Hospital to ascertain the presence of the SARS-CoV-2 genome in nasopharyngeal swabs. These samples were collected between week 10 and week 22 from suspected COVID-19 patients hospitalized in the local university hospital (60%) and from retirement home residents (40%). More than 95% of the people sampled during this period were resident in the urban area of the WWTP. The procedure used for the detection of the SARS-CoV-2 genome was based on primers and probes designed to target two RdRp (RNA-dependent RNA polymerase) gene segments (RdRp_IP2 and RdRp_IP4), in accordance with a procedure developed by the French National Reference Centre for Respiratory Viruses (Institut Pasteur, 2020).
2.3. Concentration procedures
Two concentration procedures, based on ultrafiltration and on PEG 6000 precipitation, respectively, were compared in the present study.
For the ultrafiltration procedure, a Centricon® Plus-70 centrifugal ultrafilter with a cut-off of 100 kD (Merck Millipore) was used. Before processing the samples, the ultrafilter was washed with 60 mL deionised water by centrifugation at 1500 × g for 15 min to remove the trace amounts of glycerine, in accordance with the manufacturer's recommendations. Two volumes of 50 mL wastewater were both filtered by centrifugation at 1500 × g for 15 min. After each centrifugation step, the concentrate was recovered by inverting the system and applying centrifugation (1000 × g for 2 min). The resulting concentrate's volume was around 1.5 mL. The ultrafilter was then washed with 3.5 mL of deionised water. The washing solution was added to the 1.5 mL concentrate to produce the final concentrate sample (5 mL). In order to recover the maximum amount of virus genome from the ultrafilter, two further washing steps were undertaken, each using 5 mL NucliSENS® lysis buffer (bioMérieux) for an incubation time of 5 min. The entire volume (15 mL) was then used for nucleic acid extraction. Twelve water samples were subjected to this procedure.
PEG 6000 precipitation was performed in a 250-mL centrifuge bottle containing 3 g beef extract powder, 3 g NaCl, and 0.37 g glycine for 100 mL of wastewater. After the dissolution of the beef extract powder, 20 g of PEG 6000 were added. The sample was gently stirred at 4 °C for 2 h and then maintained at 4 °C overnight. The pellet obtained after centrifugation at 4500 × g and 4 °C for 45 min was resuspended in deionised water to obtain a concentrated 5 mL sample. Ten millilitres of NucliSENS® lysis buffer were then added to the concentrate. After incubation for 10 min at room temperature, the entire volume (15 mL) was used for nucleic acid extraction. This concentration method was tested on the first four water samples of our study.
In tandem with each concentration procedure, 5 mL samples of unconcentrated water were used for viral genome extraction. They were submitted to the same procedure as the concentrated samples. Ten millilitres of NucliSENS® lysis buffer was added to the water, which was incubated at room temperature for 10 min, prior to the nucleic acid extraction.
2.4. Nucleic acid extraction
Both concentrated and unconcentrated wastewater samples reached the same volume of 15 mL after the lysis step. Phenol-chloroform-isoamyl alcohol purification was then undertaken with both concentrated and unconcentrated samples. This was realized in a 50 mL conical tube, containing 4 g of a mixture of high-vacuum silicon grease (Dow Corning®) and silicon dioxide (Sigma) (90:10 w/w), which was labelled the separation tube. The sample in lysis buffer (15 mL) was transferred to the separation tube and 15 mL phenol-chloroform-isoamyl alcohol (25:24:1 pH 7.8–8.2, Acros Organics™) were added. After 15 s of vigorous stirring by hand and centrifugation (3500 × g for 5 min), the hydrophile supernatant (15 mL) was recovered. The nucleic acid extraction was continued using 70 μL of magnetic silica beads and the NucliSENS® easyMAG™ platform (bioMérieux). The extracted nucleic acids were eluted in 100 μL of elution buffer.
A complementary step was applied to some of the samples to remove residual environmental inhibitors using OneStep PCR Inhibitor Removal kit (ZymoResearch). The RNA samples were stored at ˗80 °C until viral genome quantification. A negative control made of sterile phosphate buffered saline was included at the beginning of each nucleic acid extraction procedure.
2.5. Viral genome quantification
Viral RNA quantification was performed using real-time RT-PCR and RT-digital droplet PCR (RT-ddPCR).
For the FRNAPH GGII genome, the VTB4-Fph GII set published by Wolf et al. (2010) was used. For the SARS-CoV-2 genome, two primer sets were selected: the “RdRp_IP4” set developed by the Pasteur Institute developed by the Pasteur Institute (Institut Pasteur, 2020) for the RdRp gene and the “E” set developed by Corman et al. (2020) for the envelope protein (E) gene. The specificity of these sets for other respiratory viruses, including human coronaviruses, has been previously described (Corman et al., 2020; Etievent et al., 2020). The SARS-CoV-2 and FRNAPH GGII genomes were quantified in the same RNA extracts. For the RdRp_IP4, E, and VTB4-Fph GGII sets, quantification was performed using an RNA UltraSens™ One-Step Quantitative RT-PCR system (Applied Biosystems™). The RdRp_IP4 and E sets were applied to 5 μL of RNA in a 25-μL reaction volume with final concentrations of 0.4 μM for each primer and 0.2 μM for the probe. The VTB4-Fph GGII set was used on 2 μL of RNA in a 20-μL reaction volume with final concentrations of 1 μM for each primer and 0.3 μM for the probe. A StepOnePlus Real-Time PCR System (Applied Biosystems™) was used for these three real-time RT-PCR assays. For the RdRp_IP4 and E sets, the RT step was performed at 50 °C for 20 min and PCR amplification was performed at 95 °C for 2 min, followed by 50 cycles of 15 s at 95 °C and 30 s at 58 °C. For the VTB4-Fph GGII set the RT step was performed at 50 °C for 30 min and PCR amplification was performed at 95 °C for 2 min, followed by 45 cycles of 15 s at 95 °C and 40 s at 58 °C. Negative and positive controls were included in each experiment. Quantification was carried out using standard curve ranges. RNA extracted from patients who had tested positive for SARS-CoV-2 was quantified using ddRT-PCR with the E set, as described below. Quantified RNA was then used to obtain the standard curve for both RdRp_IP4 and E genes. The nCoV-ALL-Control plasmid (Eurofins genomic) was also used for the standard curve of the E gene. This plasmid containing ampicillin resistance gene was maintained in TOP10 chemically competent E. coli (Invitrogen) and quantified using Qubit 4 fluorometer (Invitrogen). At last, RNA extracted from GA phage suspension was used to obtain the standard curve for FRNAPH GGII. RNA extracted from GA phage suspension was quantified by ddRT-PCR using VTB4-Fph the GGII set as described below. The standard curves ranged from 1 × 10−1 to 1 × 104 gc/reaction for the RdRp_IP4 and E genes of SARS-CoV-2 and from 2.8 × 10−1 to 2.8 × 104 gc/reaction for the GA phage. The limit of detection (LoD) was 1 gc/RT-qPCR reaction for the RdRp and E genes. The limit of quantification (LoQ) ranged from 1 to 10 gc/RT-qPCR reaction for the RdRp gene and reached 1 gc/RT-qPCR reaction for the E gene. When we take the analytical volumes of the wastewater sample concentrations subjected to RT-qPCR analysis into consideration, the LoQ ranged from 2 × 102 to 2 × 103 gc/L of wastewater for the RdRp gene and reached 2 × 102 gc/L of wastewater for the E gene.
The ddRT-PCR assays were performed using the E set of SARS-CoV-2 and the VTB4-Fph GGII set for GA phage. Amplifications were carried out in a 20-μL reaction mixture containing 5 μL of RNA and 15 μL of One-Step RT-ddPCR™ Kit for Probes (Bio-Rad). The reaction mix contained 0.9 μM of each primer and 0.3 μM of the probe. The samples were placed in the droplet generator using 70 μL of generator oil to generate up to 20,000 droplets per sample. The resulting picolitre droplet emulsions (40 μL) were transferred to a Veriti 96-Well Thermal Cycler (Applied Biosystems). After amplification, the plate was transferred to the QX100TM Droplet Reader (Bio-Rad) and QuantaSoft™ Software (Bio-Rad) was used to measure the number of positive droplets per well. Droplets were designated positive or negative based on their fluorescence amplitude, using thresholding. The starting concentration of each target RNA molecule was then calculated, by modelling a Poisson distribution.
For each sample and each targeted virus (SARS-CoV-2 and FRNAPH GGII) the recovery rate was calculated as follows: recovery rate = genome copies (gc) in 5 mL unconcentrated sample × 20 × 100/gc in 100 mL of the concentrate. The high concentration of the FRNAPGH GGII genome allowed for the determination of the recovery rate in all wastewater samples. It was also possible to determine the recovery rate for SARS-CoV-2 in 5 of the 12 samples.
2.6. Estimation of RT-qPCR inhibition
The estimation of RT-qPCR inhibition in the wastewater samples was based on the concentrations of FRNAPH GGII detected. Logarithmic dilutions (1/10 and 1/100) were performed in PCR-grade water following viral RNA extraction. The RT-qPCR assay was then carried out on both undiluted and diluted RNA extracts. The percentage of inhibition was estimated for undiluted and 1/10 samples by taking the concentration obtained from the 1/100 samples as a reference, due to the high dilution of potential inhibitors.
2.7. Statistical analysis
The statistical analyses were performed using XLSTAT 2020.3.1.2 software. As normal distribution of the data and the homogeneity of variances could not be met, two non-parametric tests were used. A Kruskal-Wallis test was performed on k-independent samples (k > 3) to compare the viral genome copy values obtained using the different concentration and purification methods. A Wilcoxon signed-rank test was performed on paired samples to compare the recovery rates following the two methods and the genome concentrations of the two targeted genes. P values < 0.05 were considered statistically significant.
3. Results and discussion
3.1. Concentration method for SARS-CoV-2 quantification in wastewater
The epidemiological surveillance of SARS-CoV-2 by quantifying its genome in wastewater requires reliable methods of concentration and detection. Beginning our study in a region that was highly impacted by COVID-19 allowed us to estimate recovery rates for SARS-CoV-2.
FRNAPH are usually present in wastewater at concentrations that are relatively stable over time and around the world (Lucena et al., 2003). We obtained just under 2.5 × 107 gc/L which was high enough to allow us to evaluate both recovery rates and PCR inhibition problems. Using these bacteriophages, the presence of PCR inhibitors was detected at even a low volume (5 mL) of unconcentrated wastewater. Indeed, the PCR inhibition varied between 88% and 100% in undiluted RNA extracts (n = 5) and between 14% and 42% in 1/10 diluted RNA extracts (n = 5). Following phenol-chloroform purification, the PCR inhibitors had been completely removed from 5 mL of wastewater. In these five wastewater samples, the level of contamination by SARS-CoV-2 also enabled the quantification of the virus in 5 mL unconcentrated samples and showed that, following phenol-chloroform purification, the concentrations of SARS-CoV-2 genome detected in the undiluted samples had multiplied by between a factor of 2 and a factor of 10. The positive impact of this method on viral RNA extraction from environmental samples such as wastewater, sediments and animal stool samples has already been shown by previous studies (Miura et al., 2011; Hartard et al., 2015). The phenol-chloroform purification method was systematically applied in the following experiments. A complementary method for the removal of PCR inhibitors in RNA extracts was tested during the comparison of the concentration methods (as described below).
Two concentration procedures, based on protein precipitation using PEG 6000 and on ultrafiltration using the Centricon® 70-Plus 100 kD device, respectively, were compared. Recovery rates for both FRNAPH GGII and SARS-CoV-2 could be determined (Table 1 ) for both methods in four distinct wastewater samples collected in April 2020, since these samples tested positive for the SARS-CoV-2 genome even when only 5 mL of unconcentrated wastewater was analysed. The mean recovery rate of FRNAPH GGII reached 47.7 ± 18.5% and 39.2 ± 23.9% using ultrafiltration and protein precipitation, respectively. Because some of the results obtained for FRNAPH GGII showed residual PCR inhibition, the RNA extracts previously obtained by both methods were then purified using a PCR inhibitor removal kit. This treatment led to mean recovery rates of 47.1 ± 10.4% and 43.3 ± 20.3% for ultrafiltration and protein precipitation, respectively. This supplementary treatment therefore led to a slight increase in the mean recovery rate of protein precipitation. It also led to a decrease in the standard deviation in both concentration methods. The concentration values obtained for FRNAPH GGII using the two concentration methods, with or without the use of the PCR inhibitor removal kit, were not significantly different (p value = 0.098, Kruskall-Wallis test). For SARS-CoV-2, the mean recovery rates of the RdRp_IP4 gene reached 64.1 ± 50.2% and 32.4% ± 20.2% using ultrafiltration and protein precipitation, respectively. Additional purification of the RNA extracts led to mean recovery rates of 55.8 ± 46.9% and 23.5 ± 15.0% for ultrafiltration and protein precipitation, respectively. Thus, the mean recovery rate obtained for SARS-CoV-2 (RdRp_IP4 gene) using ultrafiltration were twice as high as those obtained using protein precipitation. Moreover, the highest recovery rate was obtained using ultrafiltration in the four wastewater samples. As observed in the case of the phage, additional purification reduced the standard deviation values. The genome concentration values were not significantly different between the two concentration methods (p value = 0.088, Kruskall-Wallis test). Amplification of the E gene was then performed on the purified RNA extracts, resulting in mean recovery rates of 64.0 ± 41.6% and 45.0 ± 44.6% for ultrafiltration and protein precipitation, respectively (Table 1). The statistical analysis of the data obtained for both the RdRp_IP4 and E genes showed significantly higher recovery rates for ultrafiltration than for protein precipitation (p value = 0.009, Wilcoxon signed-rank test).
Table 1.
Recovery rates (%) of SARS-CoV-2 and FRNAPH GGII genomes in wastewater samples using two concentration methods (protein precipitation and ultrafiltration) both alone and combined with the use of a PCR inhibitor removal kit.
| Virus/Primers set |
SARS-CoV-2/RdRp_IP4 |
SARS-CoV-2/E |
FRNAPH GGII/VTB4-Fph GGII |
|||
|---|---|---|---|---|---|---|
| Concentration method | PP | UF | PP | UF | PP | UF |
| Without PCR Inhibitor removal kit | ||||||
| Sample 1 | 56.0 | 131.0 | ND | ND | 58.2 | 49.1 |
| Sample 2 | 12.0 | 56.0 | ND | ND | 4.5 | 26.0 |
| Sample 3 | 41.8 | 60.2 | ND | ND | 43.9 | 71.1 |
| Sample 4 | 19.8 | 9.2 | ND | ND | 50.0 | 44.4 |
| Mean ± SD | 32.4 ± 20.2 | 64.1 ± 50.2 | ND | ND | 39.2 ± 23.9 | 47.7 ± 18.5 |
| With PCR Inhibitor removal kit | ||||||
| Sample 1 | 40.5 | 119.0 | 108.0 | 119.0 | 68.2 | 52.7 |
| Sample 2 | 27.0 | 61.0 | 23.0 | 45.0 | 19.1 | 37.5 |
| Sample 3 | 22.4 | 32.6 | 43.0 | 70.0 | 37.2 | 58.9 |
| Sample 4 | 4.3 | 10.6 | 6.0 | 22.0 | 50.0 | 39.4 |
| Mean ± SD | 23.5 ± 15.0 | 55.8 ± 46.9 | 45.0 ± 44.6 | 64.0 ± 41.6 | 43.3 ± 20.3 | 47.1 ± 10.4 |
PP: protein precipitation, UF: ultrafiltration, ND: not done.
Recovery rates and the inhibition of molecular detection methods applied for SARS-CoV-2 in wastewater have been poorly described. Nevertheless, our SARS-CoV-2 recovery rate was higher than those described in a recent review paper (La Rosa et al., 2020a) reporting five studies that described recovery rates for coronavirus in water matrices. But these recovery rates were all defined using artificial spiking with different types of coronaviruses (bovine enteric coronavirus, transmissible gastroenteritis virus [TGEV], murine hepatitis virus, and SARS-CoV). The recovery rate varied from 0 to 21.4% for SARS-CoV seeded in wastewater (n = 4) (Wang et al., 2005). Our higher recovery rates for the SARS-CoV-2 naturally present in the wastewater samples could be primarily explained by the choice of concentration method. Increasing PEG concentration might lead to a decrease in the standard deviation values as suggested by the recovery rate of 41.9 ± 6.5% for TGEV seeded in concentrated surface water (Bar Or et al., 2020). Two of the studies reviewed by La Rosa et al. (2020a) used adsorption/elution on glass wool with the objective of analysing large volumes (5–50 L) of surface water (Bar Or et al., 2020) and dechlorinated tap water (Abd-Elmaksoud et al., 2014). Such methods are commonly used as primary concentration methods and are followed by secondary concentration methods — PEG precipitation in the case of these two studies. However, the combination of concentration methods used on large volumes of water commonly leads to a decrease in recovery rates. By working on municipal wastewater spiked with different enveloped and non-enveloped viruses, Ye et al. (2016) have shown that ultrafiltration is a more suitable method for wastewater than ultracentrifugation or PEG precipitation because the recovery rate of ultrafiltration is 25% compared with 5% by ultracentrifugation or PEG precipitation. Since Ye et al. (2016) used culture as their detection method, the low recovery rates may be explained by the possible inactivation of coronavirus by the greater ultracentrifugal forces and the lower precipitation capacity of enveloped viruses compared with non-enveloped viruses in PEG precipitation. Wurtzer et al. (2020) showed the ability of ultracentrifugation to concentrate SARS-CoV-2 from wastewater, but the recovery rate was not given, and detection was done using RT-qPCR instead of culture methods. It is important to note that the ultrafiltration method could be used for the detection of infectious SARS-CoV-2 in wastewater, as shown with FRNAPH by Medema et al. (2020). The environmental conditions may have a negative impact on the integrity of the SARS-CoV-2 particles, with implications for detection methods and viral risk assessment; this requires further research.
The procedure combining ultrafiltration, phenol-chloroform purification, and additional purification of the RNA extracts was chosen to monitor the concentrations of the SARS-CoV-2 genome in wastewater during longitudinal wastewater sampling. Our approach based on ultrafiltration may be performed with different devices such as Centricon® (Ahmed et al., 2020; Medema et al., 2020), Corning® Spin-X® UF (Nemudryi et al., 2020) or Vivaspin® (Trottier et al., 2020) concentrators.
3.2. Quantification of the SARS-CoV-2 genome in wastewater
Our longitudinal study was performed in a French geographical area with one of the highest prevalence rates of COVID-19. The population was placed under lockdown from 17 March until 11 May 2020, but the number of cases detected at the University Hospital increased continuously until the end of March, when the wastewater sampling period started. Twelve wastewater samples, collected between 2 April and 28 May 2020, were analysed in one replicate using both the ultrafiltration procedure on 100 mL and the direct analysis of 5 mL to quantify the SARS-CoV-2 and FRNAPH GGII genomes.
The FRNAPH GGII genome could be quantified in both 5 mL and 100 mL of wastewater in all the samples (Fig. 1 ). The mean concentrations reached 2.1 × 107 ± 1.1 × 107 gc/L and 1.6 × 107 ± 1.4 × 107 gc/L in unconcentrated and concentrated samples, respectively. From these concentration values, the recovery rates of FRNAPH GGII ranged from 14.1% to 133.8%.
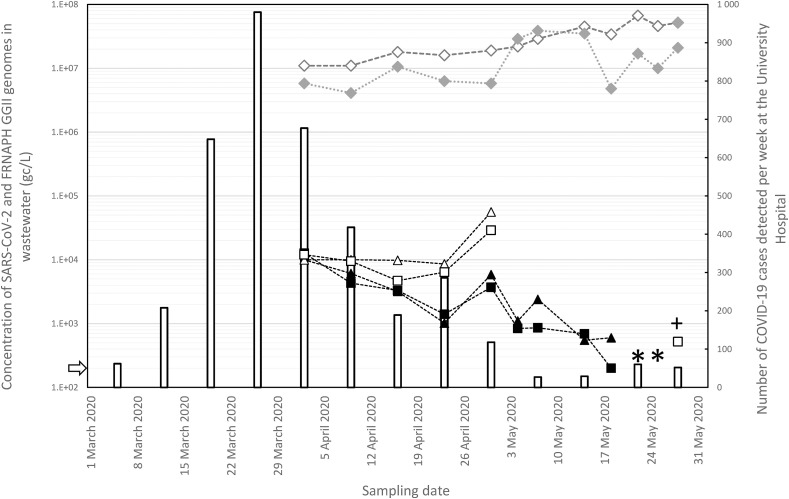
Fig. 1.
Quantitative time-course monitoring of SARS-CoV-2 and FRNAPH-GGII genomes in wastewater samples in the Nancy metropolitan area and evolution of the number of cases per week at the local University Hospital. Quantification in 100 mL of wastewater (ultrafiltration) of the SARS-CoV-2 genome targeting RdRp (full triangles) and E (full squares) genes and of the FRNAPH GGII genome (full diamonds). Quantification in 5 mL of wastewater: open forms. Negative samples of the SARS-CoV-2 genome are marked with an asterisk. The sample positive for the RdRp gene but below the LoQ is marked with a plus sign. The number of cases of COVID-19 detected at the local University Hospital is shown per week (bars). The limit of detection (LoD) for RdRp and E genes (2 × 102 gc/L) is represented by a white arrow on the left y-axis. The lines given for genome concentration values are only designed to provide a visual guide.
We propose a recovery rate of over 10% as a quality control, to validate the results. This goes beyond the current ISO standards for the molecular detection of viruses in water and food, which require recovery efficiencies of over 1% (International Organization for Standardization - ISO 2019; Lowther et al. 2019). Quantifying FRNAPH at the same time as SARS-CoV-2 also allowed for the normalisation of faecal material content, which may vary daily, depending on rainfall or variations in faecal content. In our samples, FRNAPH genomes ranged from 1.1 × 107 to 6.7 × 107 gc/L, showing that the faecal pollution was relatively stable during the period in question. No significant rainfall events were reported during the sampling period. To compare the variations in SARS-CoV-2 genome concentrations in different wastewater samples may require the normalisation of genome content. Other variables, such as temperature, pH, turbidity, total suspended solids, 5-day biological oxygen demand (BOD5), antecedent dry days, and conductivity may explain the variability of faecal indicator bacteria (FIB) (Mohammed et al., 2018; Paule-Mercado et al., 2016) and probably of other faecal pathogens.
The SARS-CoV-2 genome could be quantified in 5 mL of wastewater, without using any concentration procedures, between 2 April and 30 April (Fig. 1). By analysing 100 mL of wastewater, a decrease in the concentration of the SARS-CoV-2 genome was observed over the course of 8 weeks (Fig. 1). The SARS-CoV-2 genome was even undetectable during one week (22 and 25 May) and present but below the LoQ in the last sample (28 May). The concentrations of RdRp and E genes in the twelve samples appeared to be similar (p value = 0.496, Wilcoxon signed-rank test). Moreover, the concentration values obtained using real-time RT-qPCR and ddRT-PCR for the E gene were similar (p value = 0.734, Wilcoxon signed-rank test). This suggests that RT-PCR inhibitors were effectively removed by our protocol. Indeed, ddRT-PCR is far less influenced by inhibitors than RT-qPCR (Sun et al., 2019).
The decrease in the genome concentrations detected in wastewater may be related to the decrease in the number of COVID-19 cases observed at the Nancy University Hospital (Fig. 1). Between 2 April and 23 April, the number of cases decreased from 677/week to 286/week and the concentration values of the SARS-CoV-2 genome detected in 100 mL of wastewater decreased from 1.2 × 104 gc/L to 3.0 × 103 gc/L. During this period, the virus genome could be detected in only 5 mL of wastewater and decreased only from 1 × 104 gc/L to 7 × 103 gc/L. The analysis of only 5 mL of wastewater could be less representative of the variation in viral genome concentration. After a single data point showing an increase in genome concentration in wastewater in both 5 mL and 100 mL (30 April), the SARS-CoV-2 genome was no longer detected in 5 mL of wastewater and the concentrations dropped below 103 gc/L in the 100 mL analysis. For three of the samples during this period, either no genes were detected or only one gene was detected, and the number of cases varied between 27/week and 52/week. Nevertheless, SARS-CoV-2 was still detected in wastewater in the last sample (28 May) collected 17 days after the end of the lockdown. We observed a parallel decrease in cases in patients and genome concentration in wastewater, confirming the link between the circulation of the virus in the human population and its presence in wastewater. This clearly confirms the findings of other studies showing such a relationship (Wurtzer et al., 2020). One of the first studies on the detection of the SARS-CoV-2 genome in wastewater (Medema et al., 2020) even showed that it was possible to detect the virus in wastewater during the early stages of the COVID-19 epidemic.
4. Conclusion
We developed a method of concentrating SARS-CoV-2 from wastewater with one of the highest recovery rates described in the literature. The quantification of the SARS-CoV-2 genome was carried out in both concentrated and unconcentrated wastewater which was naturally contaminated; this gives our results a high degree of reliability. Moreover, this study provides additional data to validate proof-of-concept for a link between the outbreak in the human community and the concentrations of faecally excreted viruses in wastewater. This method can contribute to the monitoring of the epidemic and improve the management of potential viral recirculation.
Funding
This work has benefited from the financial support of the French Government's Ministry of Higher Education and Research and Innovation (Ministère de l'enseignement supérieur, de la recherche et de l'innovation [MESRI]). This study was supported by the French scientific interest group Obépine (https://www.reseau-obepine.fr) for the surveillance of SARS-CoV-2 in wastewater. All the authors' salaries were funded by either Lorraine University (France), the University Hospital of Nancy (France) or SUEZ (France).
Declaration of competing interest
None.
Acknowledgements
The authors would like to thank Sibel Berger for her contribution to the acquisition of the RT-qPCR assays for SARS-CoV-2 at LCPME. The authors would also like to thank Christophe Merlin and Hélène Guilloteau for their contributions to the plasmid replication.
References
- Abd-Elmaksoud S., Spencer S.K., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food Environ. Virol. 2014;6:253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Jochen F., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., Bassioni L. El, Akande A.O., Maamoun E. Al, Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210(Issue Suppl. l_1):S294–S303. doi: 10.1093/infdis/jiu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E. 2020. Regressing SARS-CoV-2 Sewage Measurements onto COVID-19 Burden in the Population: A Proof-of-Concept for Quantitative Environmental Surveillance. medRxiv preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Yang Z., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Liu P., Wang X., Ge Y., Xia A., Tian H., Chang H., Wang C., Li J., Wang J., Zeng M. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;ciaa198 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F., Chan P.P., Lung K., Tso E., Liu R., Ng Y., Chu M.Y., Chung T.W., Tam A.R., Yip C.C., Leung K.-H., Yim-Fong Fung A., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J., To K.K., Chan K.-H., Yuen K.-Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterol. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Thomas K.V., O'Brien J.W., Mueller J.F. In: A New Paradigm for Environmental Chemistry and Toxicology. Jiang G., Li X., editors. Springer; Singapore: 2020. Mining population exposure and community health via wastewater-based epidemiology. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etievant S., Bal A., Escuret V., Brengel-Pesce K., Bouscambert M., Cheynet V., Generenaz L., Oriol G., Destras G., Billaud G., Josset L., Frobert E., Morfin M., Gaymard A. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J. Clin. Med. 2020;9:E1871. doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.Y., Chen Y.X., Fang J.Y. 2019 Novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020;21(3):125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartard C., Rivet R., Banas S., Gantzer C. Occurrence of and sequence variation among F-specific RNA bacteriophage subgroups in feces and wastewater of urban and animal origins. Appl. Environ. Microbiol. 2015;81(8):6505–6515. doi: 10.1128/AEM.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson J. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17(5):259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M., DeBolt C., Lindquist S., Lofy K., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M., Weldon W., Biggs H., Uyeki T., Pillai S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institut Pasteur, Paris . 2020. Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2.https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2 Available online: [Google Scholar]
- Kumar M., Kumar Patel A., Shah A.V., Raval J., Rajpara N., Joshi M., Joshid C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G.J., van Schayek P., Mykytyn A.Z., Duimel H.Q., van Donsclaar E., Riesebosch S., Kuijpers H.J.H., Schippers D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;69(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Buisman A.M., Rutjes S.A., Heijne J.C., Teunis P.F., de Roda Husman A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012;78(11):3800–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Rutjes S.A., Takumi K., de Roda Husman A.M. Aichi virus in sewage and surface water, The Netherlands. Emerg. Infect. Dis. 2013;19(8):1222–1230. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther J.A., Bosch A., Butot S., Ollivier J., Mäde D., Rutjes S., Hardouin G., Lombard P., In’t Veld P., Leclercq A. Validation of EN ISO method 15216 - Part 1 – Quantification of hepatitis A virus and norovirus in food matrices. Int. J. Food Microbiol. 2019;288:82–90. doi: 10.1016/j.ijfoodmicro.2017.11.014. [DOI] [PubMed] [Google Scholar]
- Lucena F., Méndez X., Morón A., Calderón E., Campos C., Guerrero A., Cárdenas M., Gantzer C., Schwartzbrood L., Skraber S., Jofre J. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J. Appl. Microbiol. 2003;94(5):808–815. doi: 10.1046/j.1365-2672.2003.01812.x. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Miura T., Masago Y., Sano D., Omura T. Development of an effective method for recovery of viral genomic RNA from environmental silty sediments for quantitative molecular detection. Appl. Environ. Microbiol. 2011;77(12):3975–3981. doi: 10.1128/AEM.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., Hameed I.A., Razak Seidu R. Comparative predictive modelling of the occurrence of faecal indicator bacteria in a drinking water source in Norway. Sci. Total Environ. 2018;628–629:1178–1190. doi: 10.1016/j.scitotenv.2018.02.140. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Wilkinson R., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6):100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule-Mercado M.A., Ventura J.S., Memon S.A., Jahng D., Kang J.-H., Lee C.-H. Monitoring and predicting the fecal indicator bacteria concentrations from agricultural, mixed land use and urban stormwater runoff. Sci. Total Environ. 2016;550:1171–1181. doi: 10.1016/j.scitotenv.2016.01.026. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Bosch A., Myrmel M. Extended direct lysis method for virus detection on berries including droplet digital RT-PCR or real time RT-PCR with reduced influence from inhibitors. J. Virol. Methods. 2019;271:113638. doi: 10.1016/j.jviromet.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Inf. Disp. J. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184:116181. doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020;10:100157. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Xia W.J., Zhu X.M., Gu C.Q., Yin J., Wei W., Yao W., Liu C., Li J.F., Ou G.R., Wang M.N., Fang T.Y., Wang G.J., Qiu Y.H., Wu H.H., Chao F.H., Li J.W. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital. J. Virol. Methods. 2005;130(1–2):156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Greening G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010;76(5):1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. In: Gilbert Jack A., editor. Vol. 5. 2020. SARS-CoV-2 Titers in Wastewater are Higher than Expected from Clinically Confirmed Cases; pp. e00614–e00620. (mSystems). (4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. 2020. SARS-CoV-2 Titers in Wastewater Foreshadow Dynamics and Clinical Presentation of New COVID-19 Cases. medRxiv preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25(50):2000776. doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization) https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Last accessed 23 March 2020.