Abstract
Atypical teratoid/rhabdoid tumors (ATRTs) are very aggressive childhood malignancies of the central nervous system. The underlying genetic cause are inactivating bi-allelic mutations in SMARCB1 or (rarely) in SMARCA4. ATRT-SMARCA4 have been associated with a higher frequency of germline mutations, younger age, and an inferior prognosis in comparison to SMARCB1 mutated cases. Based on their DNA methylation profiles and transcriptomics, SMARCB1 mutated ATRTs have been divided into three distinct molecular subgroups: ATRT-TYR, ATRT-SHH, and ATRT-MYC. These subgroups differ in terms of age at diagnosis, tumor location, type of SMARCB1 alterations, and overall survival. ATRT-SMARCA4 are, however, less well understood, and it remains unknown, whether they belong to one of the described ATRT subgroups. Here, we examined 14 ATRT-SMARCA4 by global DNA methylation analyses. We show that they form a separate group segregating from SMARCB1 mutated ATRTs and from other SMARCA4-deficient tumors like small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) or SMARCA4 mutated extra-cranial malignant rhabdoid tumors. In contrast, medulloblastoma (MB) samples with heterozygous SMARCA4 mutations do not group separately, but with established MB subgroups. RNA sequencing of ATRT-SMARCA4 confirmed the clustering results based on DNA methylation profiling and displayed an absence of typical signature genes upregulated in SMARCB1 deleted ATRT. In summary, our results suggest that, in line with previous clinical observations, ATRT-SMARCA4 should be regarded as a distinct molecular subgroup.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00401-020-02250-7.
Keywords: SMARCA4, BRG1, ATRT, Rhabdoid, DNA methylation, RNA sequencing
Introduction
Malignant rhabdoid tumors (MRTs) are highly aggressive malignancies usually affecting young children and infants. They may occur in any part of the body, but the majority (66%) is detected in the central nervous system (CNS), where they are called atypical teratoid/rhabdoid tumors (ATRT) [9]. With an incidence of 1.4 per million in Germany [48], ATRT is a rare tumor entity even in pediatric oncology. Still, it is the most common embryonal CNS tumor in children younger than 12 months [36, 48]. Independent of tumor location, loss of function mutations in components of the SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex are a characteristic feature and represent the sole recurrent genetic alteration in all MRTs [16, 17, 29]. In the vast majority of MRTs, pathogenic variants (hereafter “mutations”) affect the SMARCB1 gene. In rare cases (about 0.5–2% of ATRT [12, 22]), SMARCA4 is mutated instead [17, 42, 43]. Since these mutations result in loss of the respective protein, loss of staining for either SMARCB1 or SMARCA4 by immunohistochemistry is used as a diagnostic tool to ensure the diagnosis of an ATRT [30]. About one-third of patients with SMARCB1-deficient MRTs carry germline mutations within the SMARCB1 gene [3, 7, 20]. Although based on very small numbers, it appears as if patients with an ATRT-SMARCA4 are even more often carriers of germline mutations. Furthermore, the latter group of patients is younger and, as their survival is even shorter, their tumors seem to be even more aggressive [17, 20]. However, due to the small number of patients suffering from an ATRT-SMARCA4, accompanied by the lack of suitable in vitro and in vivo models, knowledge regarding biological mechanisms involved in tumor development is limited. Besides MRTs, a role of SMARCA4 has been described for a variety of tumor entities including non-small cell lung cancer or thoracic sarcomas [28, 39]. In small cell carcinoma of the ovary, hypercalcemic type (SCCOHT), loss of the SMARCA4 protein has been proposed to be the main driving event in tumorigenesis [46]. In other tumor types, such as Burkitt lymphoma or medulloblastoma (MB), heterozygous missense mutations in SMARCA4 have been identified, but their role in tumor development remains elusive [23, 26, 38, 41].
Recently, DNA methylation profiling has become an attractive asset in the diagnostics of CNS tumors [4]. Based on global DNA methylation and/or gene expression, ATRTs can be divided into three distinct subgroups [15, 22, 47], designated as ATRT-TYR, ATRT-SHH, and ATRT-MYC [19]. The subgroups differ in the expression of distinct genes, the activation of specific signaling pathways, and in clinical parameters. For instance, ATRT-TYR, especially when older than 12 months, have been associated with a slightly better prognosis compared to the other subgroups or younger patients [12]. In addition to the subgrouping of ATRTs, extra-cranial MRTs (eMRTs) can also be further divided into subgroups based on molecular characteristics [5, 6]. Of note, DNA methylation profiles of eMRTs share many characteristics with ATRT-MYC.
Although these studies helped to increase the knowledge of the disease, they were predominantly based on SMARCB1 mutated ATRTs and eMRTs. Hitherto, three SMARCA4-deficient cases were included in one of the studies [22] and clustered to ATRT-SHH. However, it remains unknown, if ATRT-SMARCA4 belong to this subgroup in general, if they are divided into the same subgroups as SMARCB1-deficient ones, or, if they make up their own subgroup. To address these questions, we examined DNA methylation profiles and RNA sequencing data. Our results suggest that ATRT-SMARCA4 should be regarded as a separate molecular ATRT subgroup distinct from ATRT-MYC, ATRT-SHH, and ATRT-TYR.
Materials and methods
Tumor samples
We used published and unpublished data sets for all analyses presented here (Supplementary Table 1, online resource). Clinical data for previously unreported cases are summarized in Table 1 as well as in Supplementary Tables 2 and 3, online resource. DNA methylation or gene expression data from published data sets are available at the gene expression omnibus (GEO), accession numbers GSE70678, GSE90496, and GSE123601 and/or in the original publications [4–6, 8, 22, 34, 47]. Diagnosis of the respective tumor entity was assured by a (neuro) pathologist. To characterize the individual SMARCA4 mutations, i.e., predict the respective amino acid change, coding impact, and clinical significance/pathogenicity, we used the search engine VarSome [25]. Predicted pathogenicity was based on ClinVar [27] and The American College of Medical Genetics and Genomics (ACMG) classification [40]. Survival of SMARCB1 and SMARCA4 mutated ATRTs was compared by performing a log-rank (Mantel-Cox) test using the Prism Software Version 7 (GraphPad Software, Inc. San Diego, USA).
Table 1.
Overview of ATRT-SMARCA4 included in this study
| Case No | Tumor location | Sex | Age at diagnosis [months] | Overall survival [months] | Dead/ alive | Brain Tumor Classifier Result (score) | SMARCA4 mutation in the tumor | Predicted AA change | Coding impact | Clinical significance (ClinVar) | Clinical significance (ACMG) |
SMARCA4 germline mutation |
DNA methylation data | RNA seq data | SMARC4 protein (IHC) | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | supra | Female | 3 | 1 | Dead | No matcha | c.3407delG | p.G1136Afs*4 | Frameshift | n/a | Pathogenic | Yes | √ | √ | Lost | Patient #8 [17] |
| 02 | supra | Male | 11 | n/a | N/a | No matcha | c.1666C > Tb | p.Q556* | Nonsense | Pathogenic | Pathogenic | n/a | √ | √ | Lost | |
| 03 | supra | Female | 11 | 2 | Dead | No matcha | c.352C > T | p.Q118* | Nonsense | n/a | Pathogenic | Yes | √ | √ | Lost | |
| 04 | supra | Male | 3 | n/a | Dead | ATRT-SHH (0.99) | Breakage within the SMARCA4 region (FISH) | - | n/a | n/a | n/a | n/a | √ | x | Lost | |
| 05 | infra | Female | 10 | 2 | Dead | ATRT-SHH (0.99) | c.3565C > T | p.R1189* | Nonsense | Pathogenic | Pathogenic | Yesc | √ | x | Lost |
Patient #1 [17] Patient III2 [43] |
| 06 | infra | Female | 7 | 1 | Dead | ATRT-SHH (0.98) | c.4038_4075del38 | p.I1347* | Nonsense | n/a | Pathogenic | Yes | √ | x | Lost | Patient #4 [17] |
| 07 |
supra, Multiple |
Male | 22 | 3 | Dead | No matcha | Heterozygous SMARCA4 deletion (FISH) | - | n/a | n/a | n/a | no | √ | √ | Lost | Patient #5 [17] |
| 08 | infra | Male | 17 | 28 | Dead | No matcha | c.2335G > A | p.D779N | Missense | Uncertain significance | Uncertain significance | Yes | √ | x | Retained | Patient #9 [17] |
| 09 | supra, metastatic | Male | 3 | 1 | Dead | ATRT-SHH (0.94) | c.2920delC | p.F975Sfs*44 | Frameshift | Pathogenic | Pathogenic | Yes | √ | x | Lost | |
| 10 | infra | Female | 46 | 21 | Dead | ATRT-SHH (0.92) | c.3574C > T | p.R1192C | Missense | Uncertain significance | Uncertain significance | no | √ | √ | Retained | [32] |
| 11 | supra | Male | 1 | n/a | Dead | No matcha | n/a | n/a | n/a | n/a | n/a | n/a | √ | x | Lost | |
| 12 | infra | Male | 13 | 22 | Alive | No matcha | c.1757_1760del | p.K586Rfs*26 | Framehsift | Pathogenic | Pathogenic | n/a | √ | √ | Lost | |
| 13 | supra | Male | 4 | 9 | Alive | ATRT-SHH (0.96) | c.3168 + 5G > A | Intron | None | n/a | Uncertain significance | n/a | √ | √ | Lost | |
| 14 | supra | Male | 3 | 3 | Dead | ATRT-SHH (0.99) | c.3277C > T | p.R1093* | Nonsense | Likely Pathogenic | Pathogenic | n/a | √ | √ | Lost |
aInterpreted by the brain tumor classifier (www.molecularneuropathology.org)
bInferred from RNA Seq data
cInferred since the same SMARCA4 mutation found in the patient’s tumor was found in her sister’s and father’s germline; infra infratentorial, n/a data not available, supra supratentorial, √ analysis done, x analysis not done
DNA methylation profiling
For DNA isolation from FFPE tissue, 10 × 10 µm sections were cut and DNA isolated using the ReliaPrep™ FFPE gDNA Miniprep System (Promega) according to manufacturer’s instructions. About 100–500 ng DNA was used for bisulfite conversion by the EZ DNA Methylation Kit (Zymo Research). Afterwards, the DNA Clean & Concentrator-5 (Zymo Research) and the Infinium HD FFPE DNA Restore Kit (Illumina) were employed to clean and restore the converted DNA. Finally, either the HumanMethylation450 BeadChip array or the Infinium MethylationEPIC BeadChip Kit (both Illumina) were used to quantify the methylation status of 450,000 or 850,000 CpG sites, respectively, on an iScan device (Illumina).
RNA sequencing
For RNA isolation from FFPE tissue, 10 × 10 µm sections were cut and RNA isolated using the Maxwell® RSC RNA FFPE Kit (Promega). The RNA integrity was analyzed with the RNA 6000 Nano Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). From total RNA, the ribosomal RNA was depleted with the help of the RiboCop rRNA Depletion Kit (Lexogen) followed by RNA sequencing library generation using the CORALL Total RNA-Seq Library Prep Kit (Lexogen). Concentrations of all samples were measured with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and fragment lengths distribution of the final libraries was analyzed with the DNA High Sensitivity Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). All samples were normalized to 2 nM and pooled at equimolar concentrations. The library pool was sequenced on the NextSeq500 (Illumina) with 1 × 75 bp, with 24.5–35.1 million reads per sample.
Bioinformatics
IDAT files were processed as previously described [45]. In detail, raw data files were loaded into R using the minfi package (v.1.32.0). Since we included data derived from EPIC and 450 K arrays, we used single-sample normalization method (ssNoob) [10] for normalization of all samples. Furthermore, we excluded probes targeting the sex chromosomes. For t-distributed stochastic neighbor embedding (t-SNE) analysis, the Rtsne (v.0.15) package was employed. Probes were selected by standard deviation > 0.25, resulting in 40,426 probes for Fig. 1a and 14,772 probes for Fig. 1b. Perplexity was set to 28 and 19 for Fig. 1a, b, respectively. The heatmap were build based on the 1000 most variable probes by standard deviation with R package pheatmap (v1.0.12, https://CRAN.R-project.org/package=pheatmap), using the clustering method “ward.D2”. Global DNA methylation levels were calculated as described in [22] and significant differences determined by a Wilcoxon Rank Sum Test. Genome-wide chromosomal losses and gains as well as gains and losses at the SMARCA4 locus were calculated as published previously [6, 37].
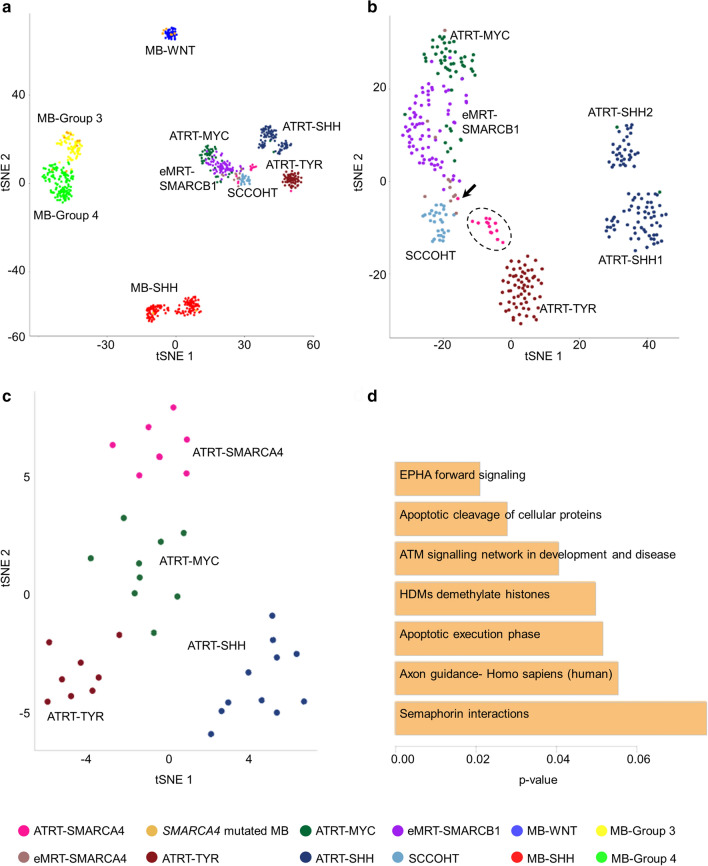
Fig. 1.
Unsupervised t-SNE analysis based on the 40,426 most variant probes of DNA methylation profiles of ATRT-SMARCA4 (n = 14, pink), eMRT-SMARCA4 (n = 11, brown), MBs with SMARCA4 mutation (n = 18, ocher), SMARCB1 mutated ATRTs [n = 62 ATRT-TYR (dark red), n = 93 ATRT-SHH (blue), n = 53 ATRT-MYC (dark green)], eMRT-SMARCB1 (n = 74, purple), SMARCA4 wild-type MBs [n = 37 MB-WNT (dark blue), n = 135 MB-SHH (red), n = 75 MB-Group 3 (yellow), n = 138 MB-Group 4 (green)] and SCCOHTs (n = 28, light blue) (a). Unsupervised t-SNE analysis based on the 14,772 most variant probes of DNA methylation profiles of the same samples but without MB samples (b). Unsupervised t-SNE analysis based on the 1000 most variable genes across all tumor samples, i.e., n = 8 ATRT-SMARCA4 (pink), n = 8 ATRT-TYR (red), n = 11 ATRT-SHH (blue) and n = 9 ATRT-MYC (green), Of note, ATRT-SMARCA4 cases 10 and 13 displayed strong similarities, as their data points almost overlap on the t-SNE plot (c). Significantly enriched pathways in ATRT-SMARCA4 are sorted according to p-values and depicted in a bar chart (d). Arrow indicates case 7
Fastq files from single end or paired end sequencing were aligned to hg19 human genome using STAR aligner (v2.5.2a). Read counts were obtained from the STAR generated BAM files with featureCounts (v2.0.0) using hg19 gene annotation (Ensembl release 87). The count matrix was used to obtain normalized reads via variance stabilizing transformation applied through DESeq2 R package (v1.26.0) and then batch corrected using removeBatchEffect function of R package limma (v3.42.2). Batch corrections was performed between sequencing data generated from FFPE samples (the SMARCA4-deficient samples) and the data derived from FF samples (SMARCB1 samples).
For t-SNE analysis, we used the packages R-tsne and employed only the 500, 1000, and 2000 most variable genes throughout all samples. Perplexity was set to 30 and theta to 0.5.
For differential expression analysis, we used the DESeq2 R package and performed correction for multiple testing using the Benjamini Hochberg method. For boxplots of meaningful, differentially expressed genes, we performed ANOVA analysis with post-hoc contrast testing using the built in aov function of R. For the genes MITF, MYC, and GLI2—upregulated only in one of the ATRT subgroups—we used a t.test of ATRT-SMARC4 versus the subgroups ATRT-TYR, ATRT-MYC, and ATRT-SHH. For pathway enrichment analysis, we employed the ConensusPathDB tool (http://cpdb.molgen.mpg.de/) using the “enrichment analysis” option and default datasets to be tested. Visualization was performed using the ClueGo tool (version 2.5.7) from Cytoscape using default parameters.
Data availability
The DNA methylation and gene expression data of this study have been deposited in NCBI’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession numbers GEO: GSE161692 and GSE160748, respectively.
Results
ATRT-SMARCA4 do not group with the known ATRT subtypes based on DNA methylation pattern
In this study, we included 8 unpublished and six previously reported ATRT samples with a verified SMARCA4 alteration [17, 32, 43]. All clinical data including genetics and survival are depicted in Table 1 and Supplementary Fig. 1, online resource. We performed global DNA methylation analysis of all 14 tumor samples and used the Heidelberg Brain Tumor classifier (v11b4) [4] to validate, whether they matched to one of the established ATRT subgroups. Fifty percent of the samples (7/14) were classified (score > 0.9) as an ATRT-SHH, 21% (3/14) showed the best, but not a specific match with ATRTs (score = 0.58–0.87), whereas 29% (4/14) did not match to any methylation class at all. Aiming to better characterize our tumor samples in an unsupervised way, we performed t-SNE. To this end, we included previously published and unpublished DNA methylation profiles of other SWI/SNF-deficient tumors, i.e., SCCOHTs and eMRTs (Supplementary Table 1–2, online resource [5, 6, 8]). The latter comprised samples with either a SMARCA4 or a SMARCB1 mutation. Furthermore, we included MBs with heterozygous SMARCA4 missense mutations as well as SMARCA4 wild-type MBs and further ATRTs (Supplementary Table 3, online resource [4, 34]).
We recognized that MBs with SMARCA4 mutations grouped with their annotated MB subgroups (Fig. 1a). ATRT-SMARCA4 located close to the SCCOHTs, ATRTs, and eMRTs without grouping distinctly to one tumor entity. We then removed all MB samples and recalculated the t-SNE (Fig. 1b) and additionally performed unsupervised hierarchical clustering (Supplementary Fig. 2, online resource). Thereby, we recognized that ATRT-SMARCA4 arranged as a separate class, whereas the eMRTs with SMARCA4 deficiency were located in between the SCCOHTs and the SMARCA4 wild-type eMRTs. Case 7 (arrow in Fig. 1b) grouped with SMARCA4-deficient eMRTs instead of ATRTs. However, this patient did not only harbor multiple supratentorial lesions, but also tumor masses at the left cardiac ventricle and in the right proximal femur. Therefore, we cannot rule out that case 7 is in fact the metastasis of an eMRT. Finally, ATRT-SMARCA4 had the lowest methylation levels of all ATRTs (Supplementary Fig. 3). Taken together, DNA methylation patterns of ATRT-SMARCA4 indicate that these tumors cannot be included in the established classification of SMARCB1 mutated ATRTs.
The transcriptome of ATRT-SMARCA4 is distinct from SMARCB1 altered ATRTs and suggests enhanced Ephrin signaling as a possible tumor driver
Next, we performed RNA sequencing of 8 ATRT-SMARCA4 (Table 1) and compared them to SMARCB1 mutated ATRT by t-SNE analysis (Fig. 1c). We recognized four distinct subgroups in the t-SNE plot corresponding to ATRT-TYR, ATRT-SHH, ATRT-MYC, and ATRT-SMARCA4. As expected, SMARCA4 expression levels were reduced, whereas SMARCB1 expression levels were higher in SMARCA4-deficient tumor samples compared to all other ATRT subgroups (Supplementary Fig. 4a,b, online resource). SMARCA2, which is supposed to be post-transcriptionally downregulated in SCCOHTs [24], displayed lower expression values as well (Supplementary Fig. 4c, online resource). Besides these SWI/SNF-related genes, the SMARCA4-deficient samples featured high expression values of EPHA5, ROCK1, and FGF10 as well as low expression levels of GLI2, MITF, MYC, and DMRT2 (Supplementary Fig. 4d-j, online resource). Concordantly, functional analysis using ConsensusPathDB [18] identified EPHA forward signaling including the genes EPHA5 and ROCK1 as the most significant enriched pathway (Fig. 1d). As EPHA5 is known to be significantly upregulated in SCCOHT as well [1], Ephrin signaling might be one of the tumor-promoting pathways in tumors driven by SMARCA4 deficiency. Finally, network analysis suggested further processes including neuronal function associated terms such as Neuroactive ligand-receptor interaction and Membrane depolarization to be altered in ATRT-SMARCA4 (Supplementary Fig. 5, online resource). Overall, comparing the transcriptomes of SMARCA4 and SMARCB1 mutated ATRTs sustains the notion that the former forms a separate subgroup marked by Ephrin signaling rather than fitting into the established ATRT subclasses.
SMARCA4 deficiency is not the result of large chromosomal deletions
The ATRT subgroups ATRT-TYR, ATRT-SHH, and ATRT-MYC are not only different in regard to their DNA methylation profiles and their transcriptomes, but also harbor different kinds of tumor-driving SMARCB1 alterations [19]. To examine the genetic basis of SMARCA4 deficiency in our cohort, we investigated chromosomal gains and losses (Supplementary Fig. 6, online resource) and alterations specifically at the SMARCA4 locus located on chr19p13.2 (Supplementary Fig. 7, online resource). As expected, the only chromosomal alterations in SMARCB1-deficient ATRTs were losses on chromosome 22q, harboring SMARCB1. Deletions of chromosomal regions of 4q and 6q were detected in SMARCB1 mutated eMRTs. Neither ATRTs nor eMRTs nor SCCOHTs displayed large chromosomal alterations, including chromosomal arm 19p. The SMARCA4 locus revealed no deletions in any of the examined tumor samples.
Discussion
In this study, we collected 14 samples of ATRT-SMARCA4 aiming at the molecular characterization of these tumors. Based on clinical data, our cohort fits well with all previously published cases [17], as the patients were very young infants, who died shortly after diagnosis. Since we were particularly interested in whether these tumors belong to one or more of the three known ATRT subgroups (ATRT-TYR, ATRT-SHH, ATRT-MYC), we performed DNA methylation and RNA sequencing analysis to examine them on a molecular level.
First, only half of our cases were recognized as ATRTs by the Heidelberg Brain Tumor classifier. The ones that were recognized matched all to ATRT-SHH. This was in line with the previously published observation that 3 SMARCA4 mutant cases matched to ATRT-SHH in an unsupervised hierarchical clustering analysis of DNA methylation data [22]. Still, 50% of our cases could not be diagnosed using the Heidelberger classifier. This uncovers a problem in the diagnostics of these tumors, as the classifier has become an import neuropathological tool, in particular for rare and aggressive brain tumors. In t-SNE analyses, ATRT-SMARCA4 grouped overall together with SMARCB1-deficient RTs and SCCOHTs, but did not locate specifically within one of these tumor subclasses. Furthermore, they clustered apart from the established ATRT subgroups and formed a separate subcluster within the group of SMARCA4-deficient tumors. In contrast, MBs carrying heterozygous SMARCA4 missense mutations, clearly grouped together with one of the four MB subgroups on t-SNE. This finding suggests that in ATRTs the SMARCA4 alterations, which are either homozygous nonsense or homozygous missense mutations, have a major impact on the tumor’s methylome, whereas the heterozygous missense mutations in MB might be less important in this regard. SCCOHTs and eMRTs, especially those with SMARCA4 deficiency, showed high similarities in our analyses, even though not being randomly intermingled. Our results are, therefore, in line with previously published studies discussing whether SCCOHTs shall be regarded as a subgroup of RTs [8, 11].
ATRTs and eMRTs with SMARCA4 deficiency as well as SMARCB1 mutated eMRTs and ATRT-MYC were hypomethylated and differed significantly from the hypermethylated ATRT-SHH and ATRT-TYR. This finding is in agreement with other studies that have previously investigated global methylation levels of SMARCB1-deficient RTs [6, 19]. Furthermore, SCCOHTs also displayed the lowest methylation levels, indicating that SMARCA4-deficient tumors demonstrate a rather hypomethylated phenotype. This may promote tumor development and progression by changing the global heterochromatin structure and activating proto-oncogenes or germline specific genes [31, 49]. Furthermore, global hypomethylation is associated with a poor prognosis in cancer entities, such as myeloma or type I ovarian cancers [35, 44] and increases during the malignant transformation of meningioma [13]. This might be associated with the even more aggressive behavior of ATRT-SMARCA4 compared to SMARCB1 mutated ones.
We identified possible signature genes for ATRT-SMARCA4, as EPHA5, ROCK1, and FGF10 were up- and DMRT2 significantly downregulated in these tumors. The most significant enriched pathway was Ephrin forward signaling that is important for embryonic CNS development and also known to be altered in different cancer entities such as glioma [33]. As several pharmacological tools for the inhibition of Ephrin signaling including those targeting EPHA5 have been tested for other diseases, this might be a good starting point for future studies on ATRT-SMARCA4 [14, 21, 50].
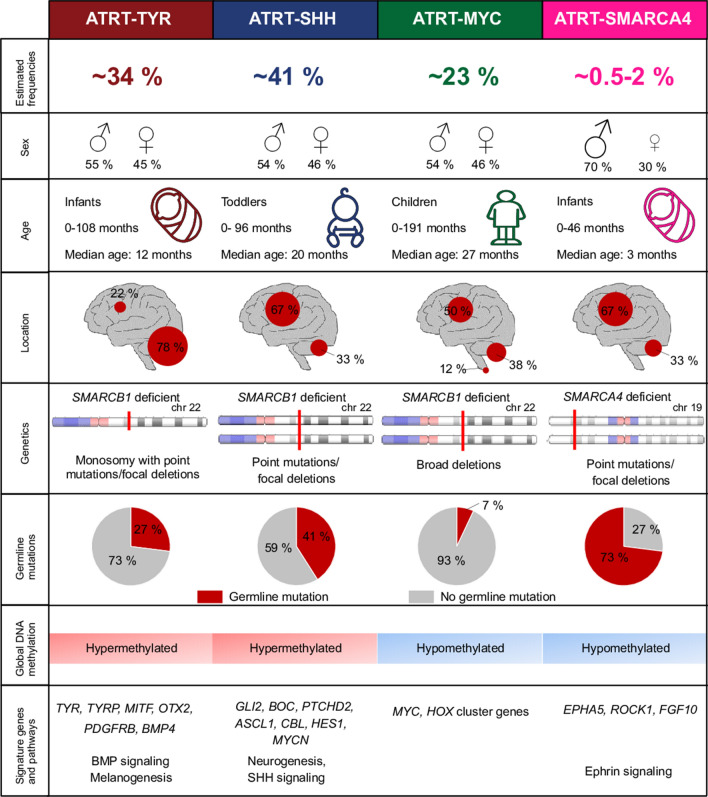
Overall, based on the data presented here, we propose that ATRT-SMARCA4 should be regarded as a separate ATRT subgroup. As reported previously [17], they affect young children, who are often carriers of germline mutations. Additionally, the male predominance is even more pronounced and their global DNA methylation patterns as well as their global transcriptomics differ from the SMARCB1 mutated ATRT subgroups. The data are summarized in Fig. 2, which gives an overview on ATRT subgroups including the here described new findings. In future studies, these findings might be used to adjust therapeutic options in the clinic and consequently improve the survival of patients.
Fig. 2.
Proposed model for including ATRT-SMARCA4 in the subgroup classification of ATRTs. Note that frequencies for each subgroup are based on published datasets and represent only a rough estimation. Estimated frequencies of SMARCB1-deficient ATRT subgroups (n = 321), their male to female ratios (n = 82 for ATRT-TYR, n = 105 for ATRT-SHH, n = 56 for ATRT-MYC), age (n = 62 for ATRT-TYR, n = 72 for ATRT-SHH, n = 43 for ATRT-MYC), and locations (n = 68 for ATRT-TYR, n = 91 for ATRT-SHH, n = 48 for ATRT-MYC) as well as information regarding genetics, signature genes and pathways is based on the study by Ho et al. [19]. Frequencies of germline mutations was taken from Frühwald et al. [12]. Frequencies of ATRT-SMARCA4 is estimated based on studies published by Johann et al. [22] and Frühwald et al. [12]. Information concerning the sex ratio (n = 19), age (n = 19), location (n = 19), and germline mutations (n = 10) of ATRT-SMARCA4 are taken from the study presented here and published reports [2, 17]. Genetics, global DNA methylation levels as well as signature genes and pathways of ATRT-SMARCA4 are based on the here generated results. Design is inspired by the model proposed by Ho et al. [19]
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Anne Reichstein and Florian Oyen (University Medical Center Hamburg-Eppendorf) for excellent technical support. This study was supported by the Deutsche Krebshilfe, the Wilhelm Sander Stiftung, and the Hamburger Krebsgesellschaft. U.S. receives generous funding from the Fördergemeinschaft Kinderkrebszentrum Hamburg. MH is supported by DFG (HA 3060/8-1) and IZKF Münster (Ha3/017/20).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dörthe Holdhof and Pascal D. Johann contributed equally to this work.
References
- 1.Auguste A, et al. Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) beyond SMARCA4 Mutations: a comprehensive genomic analysis. Cells. 2020 doi: 10.3390/cells9061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bookhout C, Bouldin TW, Ellison DW. Atypical teratoid/rhabdoid tumor with retained INI1 (SMARCB1) expression and loss of BRG1 (SMARCA4) Neuropathology. 2018;38(3):305–308. doi: 10.1111/neup.12452. [DOI] [PubMed] [Google Scholar]
- 3.Bourdeaut F, et al. Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res. 2011;17(1):31–38. doi: 10.1158/1078-0432.Ccr-10-1795. [DOI] [PubMed] [Google Scholar]
- 4.Capper D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun HE, et al. Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell. 2016;29(3):394–406. doi: 10.1016/j.ccell.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun HE, et al. Identification and analyses of extra-cranial and cranial rhabdoid tumor molecular subgroups reveal tumors with cytotoxic T cell infiltration. Cell Rep. 2019;29(8):2338–2354.e7. doi: 10.1016/j.celrep.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton KW, et al. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer. 2011;56(1):7–15. doi: 10.1002/pbc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahiminiya S, et al. Molecular analyses reveal close similarities between small cell carcinoma of the ovary, hypercalcemic type and atypical teratoid/rhabdoid tumor. Oncotarget. 2016;7(2):1732–1740. doi: 10.18632/oncotarget.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finetti MA, et al. Translational genomics of malignant rhabdoid tumours: current impact and future possibilities. Semin Cancer Biol. 2020;61:30–41. doi: 10.1016/j.semcancer.2019.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Fortin JP, Triche TJ, Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulkes WD, et al. No small surprise—small cell carcinoma of the ovary, hypercalcaemic type, is a malignant rhabdoid tumour. J Pathol. 2014;233(3):209–214. doi: 10.1002/path.4362. [DOI] [PubMed] [Google Scholar]
- 12.Frühwald MC, et al. Age and DNA methylation subgroup as potential independent risk factors for treatment stratification in children with atypical teratoid/rhabdoid tumors. Neuro Oncol. 2020;22(7):1006–1017. doi: 10.1093/neuonc/noz244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, et al. DNA methylation in the malignant transformation of meningiomas. PLoS ONE. 2013;8(1):e54114. doi: 10.1371/journal.pone.0054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giorgio C, et al. Ephrin or not? Six tough questions on Eph targeting. Expert Opin Ther Targets. 2020;24(5):403–415. doi: 10.1080/14728222.2020.1745187. [DOI] [PubMed] [Google Scholar]
- 15.Han ZY, et al. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun. 2016;7:10421. doi: 10.1038/ncomms10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasselblatt M, et al. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52(2):185–190. doi: 10.1002/gcc.22018. [DOI] [PubMed] [Google Scholar]
- 17.Hasselblatt M, et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol. 2014;128(3):453–456. doi: 10.1007/s00401-014-1323-x. [DOI] [PubMed] [Google Scholar]
- 18.Herwig R, et al. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11(10):1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 19.Ho B, et al. Molecular subgrouping of Atypical Teratoid / Rhabdoid Tumors (ATRT)—a reinvestigation and current consensus. Neuro Oncol. 2020 doi: 10.1093/neuonc/noz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holsten T, et al. Germline variants in SMARCB1 and other members of the BAF chromatin-remodeling complex across human disease entities: a meta-analysis. Eur J Hum Genet. 2018;26(8):1083–1093. doi: 10.1038/s41431-018-0143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain R, et al. Pharmacological inhibition of Eph receptors enhances glucose-stimulated insulin secretion from mouse and human pancreatic islets. Diabetologia. 2013;56(6):1350–1355. doi: 10.1007/s00125-013-2877-1. [DOI] [PubMed] [Google Scholar]
- 22.Johann PD, et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29(3):379–393. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2016;238(3):389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopanos C, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35(11):1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kretzmer H, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landrum MJ, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–d844. doi: 10.1093/nar/gkz972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Loarer F, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47(10):1200–1205. doi: 10.1038/ng.3399. [DOI] [PubMed] [Google Scholar]
- 29.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–2988. doi: 10.1172/jci64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis D et al. (2016) WHO classification of tumours of the central nervous system. Revised 4th edition. ed., France: International Agency for Research on Cancer
- 31.Madakashira BP, Sadler KC. DNA methylation, nuclear organization, and cancer. Front Genet. 2017;8:76. doi: 10.3389/fgene.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masliah-Planchon J, et al. SMARCA4-mutated atypical teratoid/rhabdoid tumor with retained BRG1 expression. Pediatr Blood Cancer. 2016;63(3):568–569. doi: 10.1002/pbc.25772. [DOI] [PubMed] [Google Scholar]
- 33.Nakada M, Hayashi Y, Hamada J. Role of Eph/ephrin tyrosine kinase in malignant glioma. Neuro Oncol. 2011;13(11):1163–1170. doi: 10.1093/neuonc/nor102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Notaro S, et al. Evaluation of folate receptor 1 (FOLR1) mRNA expression, its specific promoter methylation and global DNA hypomethylation in type I and type II ovarian cancers. BMC Cancer. 2016;16:589. doi: 10.1186/s12885-016-2637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostrom QT, et al. Alex’s lemonade stand foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2015;16(Suppl 10):x1–x36. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaff E, et al. Molecular subgrouping of primary pineal parenchymal tumors reveals distinct subtypes correlated with clinical parameters and genetic alterations. Acta Neuropathol. 2020;139(2):243–257. doi: 10.1007/s00401-019-02101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reisman DN, et al. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- 40.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders J, et al. Loss of BRG1 (SMARCA4) immunoexpression in a pediatric non-central nervous system tumor cohort. Pediatr Dev Pathol. 2019 doi: 10.1177/1093526619869154. [DOI] [PubMed] [Google Scholar]
- 43.Schneppenheim R, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86(2):279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sive JI, et al. Global hypomethylation in myeloma is associated with poor prognosis. Br J Haematol. 2016;172(3):473–475. doi: 10.1111/bjh.13506. [DOI] [PubMed] [Google Scholar]
- 45.Thomas C, et al. Desmoplastic myxoid tumor, SMARCB1-mutant: clinical, histopathological and molecular characterization of a pineal region tumor encountered in adolescents and adults. Acta Neuropathol. 2020;139(2):277–286. doi: 10.1007/s00401-019-02094-w. [DOI] [PubMed] [Google Scholar]
- 46.Tischkowitz M, et al. Small-cell carcinoma of the ovary, hypercalcemic type-genetics, new treatment targets, and current management guidelines. Clin Cancer Res. 2020;26(15):3908–3917. doi: 10.1158/1078-0432.Ccr-19-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torchia J, et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell. 2016;30(6):891–908. doi: 10.1016/j.ccell.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tulla M, et al. Incidence, trends, and survival of children with embryonal tumors. Pediatrics. 2015;136(3):e623–e632. doi: 10.1542/peds.2015-0224. [DOI] [PubMed] [Google Scholar]
- 49.Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017;396:130–137. doi: 10.1016/j.canlet.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, et al. Identification and therapeutic intervention of coactivated anaplastic lymphoma kinase, fibroblast growth factor receptor 2, and Ephrin Type-A receptor 5 Kinases in hepatocellular carcinoma. Hepatology. 2019;69(2):573–586. doi: 10.1002/hep.29792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA methylation and gene expression data of this study have been deposited in NCBI’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession numbers GEO: GSE161692 and GSE160748, respectively.