Abstract
The challenge to avoid or reduce cardiopulmonary bypass-related injuries in cardiovascular surgery remains a major issue. Remote ischemic preconditioning (RIPC) remains a promising strategy whose clinical applications appear to be significantly more realistic and extensive as compared with other conservative or surgical strategies. However, considering its underlying mechanism(s) are still unclear, novel ideas and methods must be explored to enhance its potential in clinical applications. Long noncoding RNAs (LncRNAs) are a kind of RNAs that have been implicated in the occurrence and development of cardiovascular diseases. The differently expressed LncRNAs and their biological effects during RIPC have not been explored previously. In this study, mouse and human LncRNA microarrays were used to investigate the expression signatures of LncRNAs and mRNAs in the myocardial tissue after RIPC. Therafter, homology comparisons were used to screen homologous genes from differentially expressed LncRNAs. Competing endogenous RNA (ceRNA) mechanism analysis were employed to find the matching relationship among homologous LncRNA, mRNA and microRNA. 554 differentially expressed mouse LncRNAs (281 up-regulated/273 down-regulated) and 1392 differentially expresssed human LncRNAs (635 up-regulated/757 down-regulated) were selected for further analysis. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to quantify these LncRNAs, homology comparison and ceRNA mechanism analysis provided a pair of homologous LncRNAs (ENST00000574727 & ENSMUST00000123752) for further research investigation. Overall, in this study, a number of differentially expressed LncRNAs were identified which may play an important role the regulation of both inflammation and cell proliferation. The findings may thus unveil the mystery of RIPC and discover a novel protective mechanism for the mitigation of cardiovascular ischemia-reperfusion disease.
Keywords: LncRNA, remote ischemic preconditioning (RIPC), myocardial ischemia-reperfusion injury (MIRI), microarray, ceRNA mechanism
Introduction
The potential strategies to avoid or reduce the injury of important organs caused by cardiopulmonary bypass remains a major focus of clinical and basic research in the field of cardiovascular surgery. Available, ischemic preconditioning has been possibly considered to be the best strategy of endogenous protection so far. As the implementation of direct ischemic preconditioning is greatly limited in clinical applications, researchers in recent years have turned their attention to the more realistic one, remote ischemic preconditioning (RIPC).
RIPC, is a low-risk, non-invasive, economical treatment [1], it can protect heart, brain, liver, kidney and other organs after ischemia-reperfusion (I/R) [2] and its protective effects have been validated in many animal experimental studies and clinical practice [3]. However, the underlying molecular mechanism(s) of the functions of RIPC remain unclear, and its procedure is substantially complex, involving many factors, including the expression of a wide variety of molecules and even genes. With the deepening of the understanding of mechanisms(s) regulating RIPC, the research in this area has significantly increasing in recent years.
RIPC primarily achieves its protective effects through three different steps: signal generation, signal transmission to the target organ, and target organs response to the signal [4]. In the past five years, RIPC has been extensively used in the conservative treatment of heart disease, however, the results have been found to be predominantly disappointing, mainly due to a lack of a thorough understanding of its pathogenesis which has resulted in the paucity of strong interventional procedures.
LncRNA is a kind of RNA molecules with a length of more than 200 bp. Like circRNA and tiRNA, its importance has been ignored for a long time, though it discovered much ealier. A number of previous studies have shown that LncRNA can play an important role in the occurrence and development of cardiovascular diseases by regulating the proliferation, apoptosis and autophagy in various kinds of cells [5]. The role of LncRNAs in the regulation of organ ischemia-reperfusion injury (IRI) has also been gradually confirmed in prior studies. A few articles have reported that LncRNA can alleviate MIRI by regulating apoptosis [6]. LncRNA can also be used as a molecular sponge for adsorbing microRNA and an epigenetic regulator that can modulate MIRI [7]. Interestingly, lncRNA microarray analysis has revealed that significant changes in the expression profile of LncRNAs in myocardium may occur during the early stages of reperfusion and the genes regulated by these LncRNAS have been found to be related to the control of immune response, spermine metabolism, chemotactic activity and chemokine receptor binding which have been also closely related to several MIRI-related signal pathways. In addition, lncRNA may also play an important role in myocardial ischemia-reperfusion through modulating gene co-expression network [8]. The regulatory effect of LncRNAs on MIRI is gradually being recognized, but their mechanisms remain largely unknown. In addition, the potential roles that LncRNAs might play in RIPC and their underlying regulatory mechanism(s) remain unexplored. The possibility of using LncRNAs as potential therapeutic targets has also not been investigated before. In view of the above lacuna in the field, in this study, LncRNA microarray has been used to analyze the changes of myocardial LncRNAs expression during the early stage after RIPC in both mice and humans. In addition, using bioinformatics and homology analysis, various differentially expressed LncRNAs have been identified and screened out. These highly modulated LncRNAs are further verified by qRT-PCR to confirm potential candidate LncRNAs and thus build a strong foundation for further functional research, mechanism related studies and clinical research in the future.
LncRNAs contain significant information and can potentially involve multiple molecular mechanism(s) than microRNA, but little is known about the various regulatory network of gene expression mediated by LncRNA and their potential mechanisms. Therefore, a detailed exploration about the potential mechanism(s) of RIPC from the perspective of gene regulation may enhance our overall understanding of RIPC procedure. Thus, this study aimed at unraveling the underlying mechanism of RIPC related LncRNAs may have substantial clinical implications and practical significance for discovering novel intervention strategies as well as exploring new therapeutic targets for MIRI.
Materials and methods
Surgical management and grouping of patients
24 children patients with non-cyanotic congenital heart disease were included from the Department of Cardiothoracic surgery, the second affiliated Hospital of Wenzhou Medical University and Yuying Children’s Hospital. Exclusion criteria used were as following: 1) patients with various severe complications such as severe liver disease, kidney disease or lung disease that can significantly affect the peripheral vascular disease of the upper limb; 2) patients or their families that refused to participate in the clinical trial. Withdrawal criteria used were as following: patients with anesthesia, surgical accidents and other adverse events during the experimental observation who were forced to stop the trial. All the expenses for the treatment of the patients were paid by the Children’s Heart Center of the second affiliated Hospital of Wenzhou Medical University, which did not increase the additional financial burden of the subjects enrolled in this study. This study was approved by the Ethics Committee of the Department of Cardiothoracic surgery of the second affiliated Hospital of Wenzhou Medical University and the Ethics Committee of Yuying Children’s Hospital.
In addition, self-control study on pre and post RIPC was performed in these patients; the cardiac tissues obtained before and after RIPC were randomized into control group and RIPC group respectively. After anesthesia and thoracotomy, a small amount of right atrial tissue was taken from above the purse-string suture area of the right atrium intubation. Thereafter, the cuff was placed on patients’ upper arm, inflated to 200 mmHg to block the blood flow for 5 minutes then cuff was relaxed to restore perfusion for 5 minutes., This procedure was repeated 4 times, and a small amount of right atrial tissue was used again. After the specimen was collected, the operation was initiated. Echocardiography and electrocardiogram were used to detect heart rhythm, cardiac correction and recovery of cardiac function during the operation or at least 3 days after operation.
Grouping and treatment of experimental animals
C57BL/6 mice, 6-8 weeks old, weighing 22-26 g were provided by Vital River Laboratory Animal Technology Co., Ltd., Beijing, China. The mice were kept in a quiet room with 12/12 h cycle light/dark at a constant temperature of 20~22°C and provided with adequate water and food supply. All the surgical procedures and postoperative care were carried out in accordance with the guidelines for the Care and use of Experimental Animals issued by the Chinese Academy of Health. All the operations involving mice were approved by the Animal Research Committee of Wenzhou Medical University (wydw2016-0334). Forty C57BL/6 mice were randomly divided into various groups as following: 1) Sham group (n=10): only undergo dissociation of the left femoral artery and thoracotomy; 2) MIRI group (n=10): ligation and reperfusion of the left anterior descending branch were given on the basis of the separation of the left femoral artery and thoracotomy; 3) RIPC+MIRI group (n=10): separating left femoral artery for RIPC, left anterior descending branch was ligated and reperfused by thoracotomy; 4) RIPC group (n=10): for separating left femoral artery for RIPC, and the heart was obtained by thoracotomy.
RIPC in C57BL/6 mice
After endotracheal intubation and anesthetization with isoflurane, the limbs of mice were fixed with tape, a small incision was made along the left groin, and the femoral artery was removed. The left femoral artery was gently clamped with a microvascular clamp for 5 minutes, thereafter the vascular clamp was relaxed, and perfusion was restored for 5 minutes. The same cycle was repeated for four more times.
MIRI mice model
After isoflurane induced anesthesia, the mice were endotracheal intubated and adjusted to the right lateral position. Thoracotomy was performed on the third or fourth intercostal on the left side to expose the pericardium. The pericardium was thereafter opened with microscopic forceps and the left anterior descending branch of coronary artery was liagted with 6-0 silk thread to induce myocardial ischemia. After 30 minutes of myocardial ischemia, ligation was made loose so that the heart can be reperfused. Meanwhile, a small hose was indwelled in chest to extract residual air in the chest, and sutured layer by layer rapidly to chest cavity. Then mice were resuscitated by 100% oxygen ventilation and transferred to a warm pad. Thereafter, 180 minutes later, the re-anesthetized mice were killed, and the samples needed for the experiment were obtained. The occlusion and reperfusion of the left anterior descending branch could be judged by the significant changes in the myocardial color.
Total RNA purification
Total RNA was extracted from the collected myocardial tissue with the Trizol reagent (15596026; Ambion, Cambridge, UK). Thereafter, RNA content was quantified with 260/280 UV spectrophotometry. The isolated high-purity total RNA after quantification was subjected to LncRNA microarray analysis or qRT-PCR testing.
LncRNA microarray analysis
Arraystar human and mouse LncRNA microarrays V3.0 provided by KangChen Bio-tech Co. Ltd. (Shanghai, China) were used to detect the various RNA samples. RNA labeling and array hybridization was conducted as previously described [9,10]. The differential expression levels of LncRNAs and mRNAs between the RIPC group and the Sham group were thereafter compared. In addition, among the various detected up-regulated RNAs, adjusted P value <0.05 and either whose FC value (fold change >2 & Chip intensity >500 or FC value >3 & Chip intensity >50 were considered as differentially expressed. However, as for down-regulated RNAs, adjusted P value <0.05 and either whose FC value >2 & Chip intensity >1000 or FC value >3 & Chip intensity >50 were defined as differentially expressed.
GO and KEGG pathway analysis
KangChen Bio-tech Co. Ltd. (Shanghai, China) was commissioned to conduct GO and KEGG pathway analysis to systematically analyze the differentially expressed genes and enrich important GO terms and KEGG pathways (P<0.05) [11]. The significance of the P value was evaluated by FDR (false discovery rate) and recommended FDR value was considered less than 0.05.
Western blot analysis for tissues
The total protein was extracted from mice’s myocardial tissue (5 mg) and the protein concentration was determined with Enhanced BCA Protein Assay Kit (Beyotime Biotechnology). The following primary antibodies were used in this study: 1:1000 diluted Rabbit anti-Bax (50599-2-Ig; Proteintech Group, Inc), Bcl2 (26593-1-AP; Proteintech Group, Inc), Caspase3 (66470-2-Ig; Proteintech Group, Inc), cleaved-Caspase3 (WL01992; Wanlei Biotechnology Co., Ltd.), TNF-α (17590-1-AP; Proteintech Group, Inc), IL-6 (WL02841; Wanlei Biotechnology Co., Ltd.) primary antibody. The next day, 1:3000 diluted Goat Anti-Rabbit antibody (SA00001-2; Proteintech Group, Inc) was added, and ECL was used for coloration, scanning and analyzing the results with the Gel imaging system. After quantification, the ratio of each protein to GAPDH was used for comparison and analysis.
Myocardial infarct size measurement
The myocardial infarct size was measured through Evans Blue (E2129; Sigma, St. Louis, MO)/TTC (2,3,5-triphenyltetrazolium chloride, T8877; Sigma) double staining method. The viable tissues were stained red and white by TTC was defined as area at risk (AAR). Non-ischemic myocardial tissues were stained deep blue by Evans Blue. Infarct area (INF) appeared to be pale after staining. The percent of infarcted area over the area at risk (INF/AAR ratio, IAR%) was thereafter calculated as described previously [12].
Hematoxylin-eosin staining
The hearts of mice were fixed in 4% paraformaldehyde. After 24 h, fixed hearts were embedded with paraffin and the embedded hearts were sliced (5 μm thick). The sections are thereafter stained with hematoxylin and eosin according to the standard protocol and analyzed by light microscopy.
Plasma cTnI, TNF-α concentration
The serum levels of TNF-α, and cTnI were measured using a mouse Enzyme-Linked ImmunoSorbent Assay kit (Boyun Biotech Co., Ltd, Shanghai) in accordance with the manufacturer’s instructions.
qRT-PCR analysis
The total RNA was extracted from myocardial tissues using the Trizol reagent (15596026; Ambion, Cambridge, UK), RNA content was thereafter quantified by 260/280 UV spectrophotometry. The same volume of RNA solution was used to reverse transcriptase using the RT-PCR kit (FSQ-101; TOYOBO, Kita-ku, Osaka, Japan). The expressions of various LncRNAs were quantified by SYBR Green (170-8882AP; Bio-Rad, Hercules, CA) in a two-step, real-time RT-PCR using CFX96 Touch Real-Time PCR Detection System. The expression of LncRNA was normalized to GAPDH mRNA content and was calculated using the comparative methods.
Genes homology analysis & CeRNA mechanism analysis
Genes Homology Analysis and CeRNA Mechanism Analysis were performed by KangChen Bio-tech Co. Ltd. (Shanghai, China). The various detected up-regulated homologous LncRNAs, whose chip intensity >500, FC value >1.0, P<0.05 and E-value <0.05 were selected. The detected down-regulated homologous LncRNAs, whose chip intensity >50, FC value >1.0, P<0.05 and E-value <0.05 were selected. Finally, the ceRNA mechanism analysis of stable differentially expressed homologous LncRNAs was performed.
Statistical analysis
All data in this article were expressed as mean ± SEM. The comparisons between groups were assessed by t-test. All statistical analyses were performed using Graph Pad Prism Software (Version 8.0, La Jolla, CA). P<0.05 was considered as statistically significant.
Results
RIPC attenuated myocardial apoptosis and inflammation after MIRI in mice
The levels of serum TNF-α and cTnI were measured by enzyme linked immunosorbent assay (Elisa) kit. Moreover, upon comparison with the sham group, RIPC group did not display a significantly elevated levels of TNF-α and cTnI in the serum. However, in MIRI group and RIPC+MIRI group, serum TNF-α and cTnI levels were significantly increased whereas the levels of these cytokines in RIPC+MIRI group were significantly lower than that in MIRI group (Figure 1A). The results showed that MIRI was accompanied by an increase of troponin and inflammatory factors which clearly indicated myocardial injury and inflammatory state and effective RIPC operation could inhibit these events and thereby protect the myocardium. In addition, western blot data showed that cleaved Caspase-3, Bax, TNF-α and IL-6 were up-regulated, while Bcl-2 was down-regulated (Figure 1C, 1D). These observations suggested that local myocardium was in the state of inflammatory microenvironment and myocardial apoptosis, while RIPC significantly inhibited the expression of various pro-apoptotic as well as pro-inflammatory factors and significantly alleviated myocardial damage.
Figure 1.
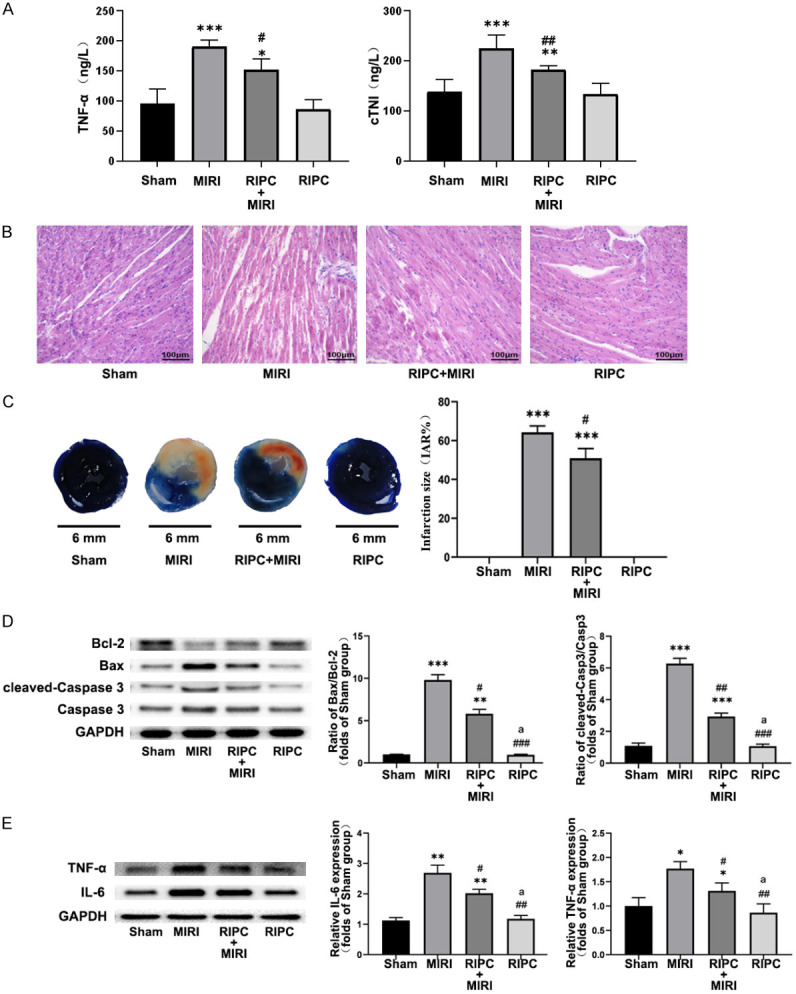
The verification of the protective effects of ischemic preconditioning in vivo. ELISA results of TNF-α and cTNI indicate that RIPC attenuated myocardial injury after ishemia/reperfusion (A), meanwhile representative western blot data showed that RIPC can reduce apoptosis and inflammation after MIRI (D, E), RIPC also attenuated harmful myocardial structural changes (B) and myocardial ischemic infarction (C). The comparison of the mean values between the two groups was performed using the independent-sample t test. All data has been presented as mean ± Standard Error of Mean (SEM). “*” means compared to Sham group, “#” means compared to MIRI group, “a” means compared to RIPC+MIRI group. *P<0.05, **P<0.01, ***P<0.0001.
RIPC reversed structural changes in the myocardium after MIRI in mice
HE staining clearly showed that MIRI caused local swelling, myocardial necrosis, myocardial fiber disorder and rupture, accompanied by substantial inflammatory cell infiltration (Figure 1B). In contrast, these pathological changes were greatly found to be attenuated in the RIPC group, where the structure and morphology of cardiomyocytes were relatively normal, with mild swelling, partial rupture of myocardial fibers, and a small fraction of inflammatory cell infiltration in interstitium. The myocardial fibers in the sham and RIPC groups were found to be normal and the stripes were generally clear, and no myocardial remodeling, necrosis or neutrophil infiltration was observed.
RIPC significantly reduced the infarct size after MIRI
After TTC-Evans blue staining, upon comparison with sham operation and RIPC groups, IAR% increased remarkably in MIRI group and RIPC+MIRI group while IAR% in RIPC+MIRI group was relatively smaller than MIRI group to a certain extent (Figure 1C). The results indicated that the infarct size predominantly increased during MIRI, and RIPC could protect the myocardium and thereby significantly reduce the infarct size.
Microarray hybridization data of mouse and human
Arraystar mouse LncRNA microarray v3.0 and arraystar human LncRNA microarray v3.0 were specially designed for the global profiling of mouse & human LncRNAs and protein-coding transcripts. Thereafter, LncRNAs were carefully selected from authoritative public transcriptome databases (including UCSC known genes, Gencode, Refseq, etc.) and literature review of papers with high impact factors. Heat maps of hierarchical clustering results were designed to depict various differentially-regulated lncRNA and mRNAs between the two groups. It was deciphered from the results of scatterplot that expression of log-2 ratio and distribution of lncRNAs and mRNAs in RIPC group and nc group was almost similar in both the mouse and human (Figure 2).
Figure 2.
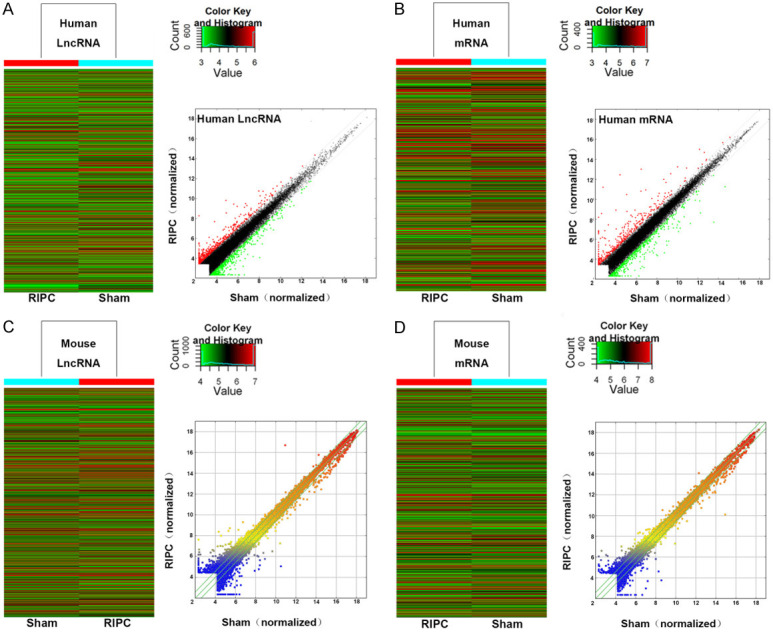
LncRNA and mRNA expression profile comparison between the RIPC group and the sham group in both the human and mouse. The hierarchical clustering of all target value human lncRNAs (A) and mRNAs (B). The scatterplot was used for assessing human lncRNA (A) and mRNA (B) expression variation between the RIPC group and the sham group. The hierarchical clustering of all the target value mouse lncRNAs (C) and mRNAs (D). The scatterplot was used for assessing mouse lncRNA (C) and mRNA (D) expression variation between the RIPC group and the sham group. Green lines inside the scatterplot picture represent the fold change lines (the default FC value was given 2.0). LncRNAs and mRNAs above the top green line were detected as up-regulated and below the bottom green line were detected as down-regulated, both of them were indicated as FC value >2.0 in expression. For example, “red dots” stand for higher relative expression levels (A), and “green dots” indicate low relative expression levels (B).
The expression levels of differentially regulated lncRNAs and mRNAs of human and mouse
For analysis purpose, presupposed >2-fold differential expression was set as screening condition. Overall, 1392 differentially regulated LncRNAs were selected from human LncRNA microarray, including 635 up-regulated and 757 down-regulated ones. 554 differentially regulated LncRNAs from mouse lncRNA microarray, including 281 up-regulated and 273 down-regulated ones have been shown in Table 1. Moreover, analysis of the differential expression profile of mRNAs, as well as top10 differentially expressed LncRNAs and mRNA after RIPC in both human and mouse have been listed in Table 2.
Table 1.
The number of various differentially expressed LncRNAs
| LncRNAs | Spacies | FC2-5 | FC5-10 | FC≥10 | Total |
|---|---|---|---|---|---|
| Up | Mouse | 247 | 24 | 10 | 281 |
| Down | Mouse | 239 | 29 | 5 | 273 |
| Up | Human | 617 | 13 | 5 | 635 |
| Down | Human | 731 | 21 | 5 | 757 |
Table 2.
Top10 differentially expressed LncRNAs and mRNA after RIPC in the human and mouse
| Sepuence name | Gene symbol | FC |
|---|---|---|
| Top10 Up-regulated human LncRNAs | ||
| ENST00000507926 | CTD-2306M5.1 | 61.2128103 |
| NR_028308 | BRE-AS1 | 30.9319247 |
| ENST00000564531 | RP11-1100L3.8 | 14.7943499 |
| uc001jpa.1 | BC022313 | 14.3805754 |
| NR_026860 | LINC00473 | 12.6209272 |
| TCONS_00019322 | XLOC_009167 | 9.7478118 |
| ENST00000457058 | RP11-109I13.2 | 9.2358625 |
| ENST00000417650 | RP11-439L18.1 | 8.5209731 |
| ENST00000508443 | CTB-47B11.3 | 7.243086 |
| TCONS_00017313 | XLOC_008149 | 7.0343197 |
| Top10 Up-regulated human mRNAs | ||
| ENST00000338488 | NR4A3 | 350.7761867 |
| NM_005252 | FOS | 264.2330283 |
| NM_173200 | NR4A3 | 92.7188392 |
| NM_001114171 | FOSB | 62.3703608 |
| NM_006732 | FOSB | 58.6511585 |
| NM_002089 | CXCL2 | 42.6043755 |
| ENST00000307407 | IL8 | 29.8900665 |
| ENST00000294829 | FAM71A | 22.987126 |
| NM_000450 | SELE | 21.876733 |
| NM_001174090 | SLC4A11 | 20.5018814 |
| Top10 Down-regulated human LncRNAs | ||
| uc001ukd.1 | AX747752 | 19.4261082 |
| uc001tdk.2 | KRT19P2 | 15.7030871 |
| ENST00000519609 | RP11-32D16.1 | 13.8072969 |
| ENST00000552167 | RP11-359M6.1 | 13.1343691 |
| ENST00000431401 | RP1-290I10.5 | 10.7059655 |
| uc003cmy.1 | DQ592230 | 9.8065073 |
| NR_024089 | LINC00162 | 9.4575664 |
| ENST00000464537 | RP11-420J11.2 | 8.9155883 |
| ENST00000581210 | KRT16P1 | 8.2424453 |
| chrX:136212300-136224525+ | chrX:136212300-136224525 | 7.8726809 |
| Top10 Down-regulated human mRNAs | ||
| NM_001083537 | FAM86B1 | 57.6879463 |
| NM_005823 | MSLN | 17.0867096 |
| NM_016352 | CPA4 | 15.2902808 |
| NM_002276 | KRT19 | 13.1581887 |
| NM_001126102 | HP | 13.1148645 |
| NM_005143 | HP | 12.4122693 |
| ENST00000322165 | HSD17B6 | 12.0160265 |
| NM_005556 | KRT7 | 11.9192372 |
| NM_013404 | MSLN | 11.5713044 |
| NM_017625 | ITLN1 | 10.5037021 |
| Top10 Up-regulated mouse LncRNAs | ||
| ENSMUST00000136102 | Plekhm2 | 54.2768466 |
| NR_045285 | Gm5083 | 39.6142257 |
| BG068257 | mouselincRNA1118 | 19.6325586 |
| ENSMUST00000153836 | Asxl3 | 16.6523791 |
| BF180946 | mouselincRNA0005 | 16.1865307 |
| TCONS_00032441 | XLOC_024323 | 14.4913109 |
| ENSMUST00000175707 | Vmn1r-ps142 | 12.7990311 |
| TCONS_00004283 | XLOC_003982 | 11.4696996 |
| mouselincRNA0499- | mouselincRNA0499 | 11.1981243 |
| uc007tsn.1 | Tcrd | 10.8516461 |
| Top10 Up-regulated mouse mRNAs | ||
| NM_009341 | Tcp10b | 16.1522272 |
| NM_011325 | Scnn1b | 13.3807131 |
| NM_153524 | Mrgpra4 | 9.0141778 |
| NM_146334 | Olfr1330 | 8.9121771 |
| NM_001081449 | Vmn2r54 | 7.8662994 |
| NM_011456 | Serpinb9e | 7.587836 |
| NM_001099301 | Obp2b | 7.2571405 |
| NM_001159422 | Ccdc58 | 6.7617881 |
| NM_146789 | Olfr1230 | 5.6599084 |
| NM_001005748 | Phactr1 | 5.4333252 |
| Top10 Down-regulated mouse LncRNAs | ||
| ENSMUST00000173563 | Gm2302 | 43.4009886 |
| AK017761 | AK017761 | 16.2941162 |
| mouselincRNA0933- | mouselincRNA0933 | 13.2961955 |
| ENSMUST00000152439 | Gm14009 | 11.5085008 |
| TCONS_00004249 | XLOC_003927 | 11.1955942 |
| uc009qiv.2 | DQ690356 | 9.9517056 |
| ENSMUST00000152128 | Gm16080 | 9.6625515 |
| NR_045965 | 1700084F23Rik | 9.6499683 |
| ENSMUST00000157528 | Gm26348 | 9.6431601 |
| ENSMUST00000123663 | Gm13707 | 9.5857546 |
| Top10 Down-regulated mouse mRNAs | ||
| NM_177855 | Med12l | 28.4145113 |
| NM_001163028 | Bcmo1 | 27.8440333 |
| NM_147038 | Olfr1416 | 19.690464 |
| NM_010199 | Fgf12 | 16.3330758 |
| NM_008081 | B4galnt2 | 12.6905967 |
| NM_010990 | Olfr48 | 12.6787376 |
| NM_001166375 | March1 | 11.8556742 |
| NM_053015 | Mlph | 11.3830897 |
| NM_001081153 | Unc13c | 8.8654521 |
| NM_009411 | Tpbpa | 7.2739333 |
GO and KEGG analyses of differentially regulated mRNAs of human and mouse
Thereafter, GO analysis (Biological processes, cellular components, molecular functions) was performed to analyze the major potentially relevant functions of the closest coding genes according to the GO database that provided the essential functional classifications for the National Center for Biotechnology Information (NCBI). In this study, GO analysis initially reflected the functional significance of differentially expressed (up-regulated and down-regulated) mRNA in myocardium which experienced RIPC in both the human and mouse. In the human and mouse groups, the most enriched GO terms (top 10) of 3 structured networks have been shown in Figure 3, and the top 10 enriched pathways have been shown in Figure 4. Moreover, the pathway analysis indicated that majority of up-regulated mRNAs were involved in the regulation of MAPK and TNF signaling pathways, while most down-regulated mRNAs were involved in the modulation of NF-κB signaling pathway and apoptosis in humans. Moreover, the pathway analysis of mice tissues revealed that significantly down-regulated mRNAs participated in cAMP signaling pathway, inflammation-mediated TRP pathway and so forth, whereas the up-regulated mRNAs related signaling pathways did not appear to be closely related to myocardial ischemia-reperfusion injury.
Figure 3.
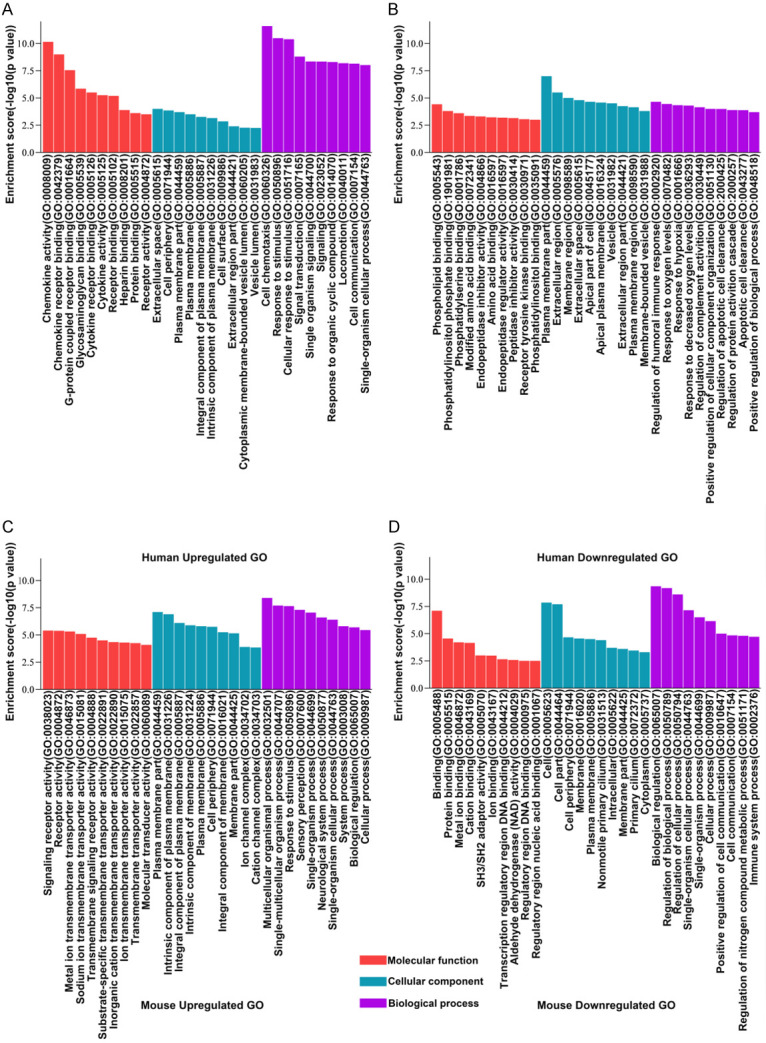
Gene Ontology (GO) analysis for functional classification of the differentially expressed LncRNAs. The GO categories contained 3 different domains: biological process (BP), molecular function (MF), and cellular component (CC). The up-regulated GO of human (A), the down-regulated GO of human (B), the up-regulated GO of mouse (C), the down-regulated GO of mouse (D) were arranged sequentially. P value denotes the significance of GO term enrichment in the differentially expressed protein-coding RNAs list. The lower the P value was, more significant was the GO term (p value ≤ 0.05 is recommended).
Figure 4.
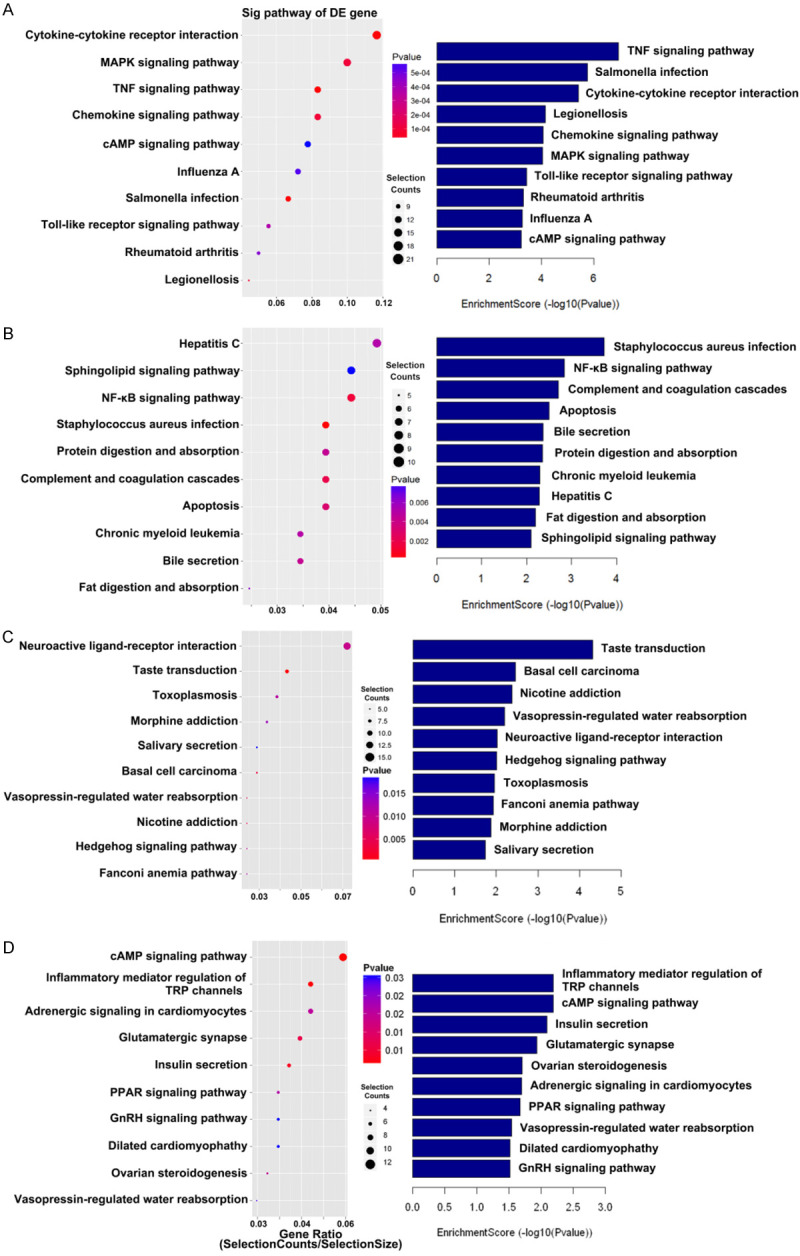
Pathway enrichment analysis. A, B. The up-regulated gene and down-regulated gene pathway of the human. C, D. The up-regulated gene and down-regulated gene pathway of the mouse. This figure shows the top 10 significant pathways related to both up-regulated and down-regulated genes. The P value (Fisher P value) denoted the significance of the pathway correlated to the various conditions. The lower the p value was, the more significant the pathway (the recommended P value cut-off was 0.05). The size of selection counts point indicated the number of corresponding genes. The larger the point was, the more genes involved in this pathway.
Homology analysis between the human LncRNAs & mouse LncRNAs
Furthermore, among the preliminary screened mouse and human LncRNAs, homology analysis was performed to select homologous LncRNAs. We noticed that there were 5 pairs of up-regulated homologous LncRNAs, and, 6 pairs of down-regulated homologous LncRNAs. Homologous genes are highly conserved during the process of biological evolution, may exert important biological functions, and can act as an important target for disease treatment. Therefore, it was hypothesized that the selected 11 pairs of homologous LncRNAs might have a significant potential for further study. The 11 different pairs of homologous LncRNAs have been listed in Table 3.
Table 3.
The list of detected homologous LncRNAs
| No. | Sequence name | Regulation | FC | Spacies |
|---|---|---|---|---|
| 1 | NR_026778 | Up | 2.7203589 | human |
| ENSMUST00000154437 | Up | 1.0567851 | mouse | |
| 2 | Uc010rog.2 | Up | 2.1087689 | human |
| NR_003513 | Up | 1.0640434 | mouse | |
| 3 | ENST00000446406 | Up | 2.2950602 | human |
| NR_001592 | Up | 1.3577539 | mouse | |
| 4 | ENST00000527314 | Up | 1.5297628 | human |
| ENSMUST00000163913 | Up | 9.7726336 | mouse | |
| 5 | NR_037719 | Up | 3.1086571 | human |
| NR_033133 | Up | 5.9252023 | mouse | |
| 6 | ENST00000565841 | Down | 3.9193896 | human |
| AK050947 | Down | 8.2915752 | mouse | |
| 7 | NR_073081 | Down | 2.5207779 | human |
| AK158081 | Down | 1.2406486 | mouse | |
| 8 | ENST00000509649 | Down | 2.3379888 | human |
| NR_002928 | Down | 1.1492311 | mouse | |
| 9 | uc001tdk.2 | Down | 15.7030871 | human |
| ENSMUST00000126460 | Down | 1.0307194 | mouse | |
| 10 | ENST00000574727 | Down | 7.2125214 | human |
| ENSMUST00000123752 | Down | 1.1493588 | mouse | |
| 11 | NR_037845 | Down | 1.0515052 | human |
| AK133331 | Down | 4.1421506 | mouse |
Verification of various differentially expressed LncRNAs in microarray by qRT-PCR
The removal of mitochondrial LncRNAs and those which cannot be designed with suitable primers, including detected differentially expressed 11 pairs of homologous LncRNAs, and another 17 LncRNAs which displayed a more stable expression but lower FC. It was found that ENSMUST00000163913, ENST00000565841, ENSMUST00000126460 could not be effectively designed with suitable primers. Finally, only 36 LncRNAs were left for qRT-PCR verification (their sequence details and primers have been indicated in Table 4). According to the Genebank sequence, each primer sequence was designed and synthesized by Shanghai Zhili Biology Co., Ltd. The relative expression of top 9 differentially expressed LncRNAs after qRT-PCR has been listed in Figure 5A, and the microarray consistency was found to be reliable.
Table 4.
The list of primers of selected LncRNAs
| Sequence name | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| uc010rog.2 | TGACTGACTATGACTGCTGAG | TGCTATCTTCCTGTTGCCATT |
| ENST00000566457 | TCTGCTCGGCTCTGTCAT | GGCTCTTGGTTATGGTTCTTG |
| ENST00000509649 | GCACATTTGACTTGGAAGCA | CGCCCAGCCAACATTCTT |
| ENST00000527314 | TAGGAGATCGGGACCAGCTT | GTGCGGACAACCTCAGCTAC |
| NR_037845 | GTGGCGTGATGTCTGCTTA | GAGGCTGAAGTAGGCGAATC |
| ENST00000574727 | CCTGTTCTGTGTATTGCTTCC | CTGACTCTCCTTGCTACCATT |
| ENSMUST00000123752 | AAGACTGCTCTGCCTGGT | GCTCTGAAGACATCCGTTGA |
| NR_040512 | GCAGGAATATGGAGCACAGT | ACTTCAGGAGACAGTAACAGAC |
| ENSMUST00000173563 | TCAGAGTGATTCAGACAGTGTT | AAGGTCCAGCCAGCAGAT |
| ENSMUST00000154036 | CTGGCTGAACTGGAACTTAGA | CCAAGGAACCTGACATCTTCT |
| AK133331 | TGGTGCTTATCTAACTGGATGT | TGCTGTGGTCTCGTCTTAC |
| ENSMUST00000148722 | GTGTCTCCTGTACTGACTTGT | CTTCGCCATCCATCCTGAA |
| TCONS_00003974 | TCATGGTTCTTCAGGCTGTATA | ATGCTGGCTCCTCTTCTCT |
| NR_001592 | GAGACTCAAAGCACCCGTGA | AGATGGACGACAGGTGGGTA |
| ENST00000446406 | GGAGCACCTTGGACATCT | CCTACTCCACACTCCTCAC |
| NR_033133 | CCAACAGCCATCTCCGTAG | AGACAGACACAGAAACAAAGAC |
| NR_037719 | GCTCACTGACACCGTTTG | TCATTGGCTTCCAGGACTG |
| ENSMUST00000130038 | CCTTACTCCCTGCCCTTCT | ATCCACTTGTCTCTGCTTCTC |
| ENSMUST00000134981 | TGCTGCTTCCTGTTGTGT | TGTTCTTCCTTCGTCTATATGG |
| NR_046186 | ACTTACCCTTGAAGAGGAAATG | CTCATCAGTGCTATCATCATCA |
| TCONS_00023846 | GATGTAATTGTATGCGACCTG | AACCTCTCAATCCCAGATTT |
| TCONS_00019322 | AGCAGAAGCAACAGGTCAA | ACGGAGTAAGCAAGTCACAA |
| ENSMUST00000136102 | AGCAGGTGCAGTTGAGTG | GGAACTTGTTATGTAGCAGAGG |
| ENSMUST00000128411 | AGAAGTGTTATTGCCACCAGAT | TGCTGACCAGGATAGACCAA |
| AK050947 | TTGGTATGAAGGTGTGAGAGG | TGGAGGTCTGTAGCACTGTA |
| NR 073081 | AGGACTGGCACAAGTTCTG | TTCTCAGCGAAGTACACCTT |
| AK158081 | TGAGAGGCAGGCAAGGAA | TAGCGGTGGAGTGGTCAA |
| uc001tdk.2 | AGCCACTACTACAGGACCAT | GCCAGTTGAGCATTGTTGAT |
| ENSMUST00000145313 | AATGCTGTCTCACCTACCAT | CAGAGTAGATGCTTGAAGGAG |
| ENSMUST00000146263 | CTGTGGCTGAAGAGTCTCC | CAATTCTGGTGATGATGTCTGA |
| ENST00000420279 | AACCACATGAAGCAACTTAGC | TCGGTAGACTCCAAGCAGAA |
| NR_026778 | GGCGGCAAGACAGACATT | CCTTCTAGGTATCACCTCATCC |
| ENSMUST00000154437 | AAAGCCCTTCAAGCTCCTAG | TCCAAGTCCAGTCACAATAGAT |
| NR_003513 | GGCAGAGCAGCAGTTGAT | GACCTCCACTACGCACCTA |
| NR_002928 | GCTGTCACCATTGCTTGTT | TCCTAGAACTGCCAACCATC |
| AK133331 | TGGTGCTTATCTAACTGGATGT | TGCTGTGGTCTCGTCTTAC |
Figure 5.
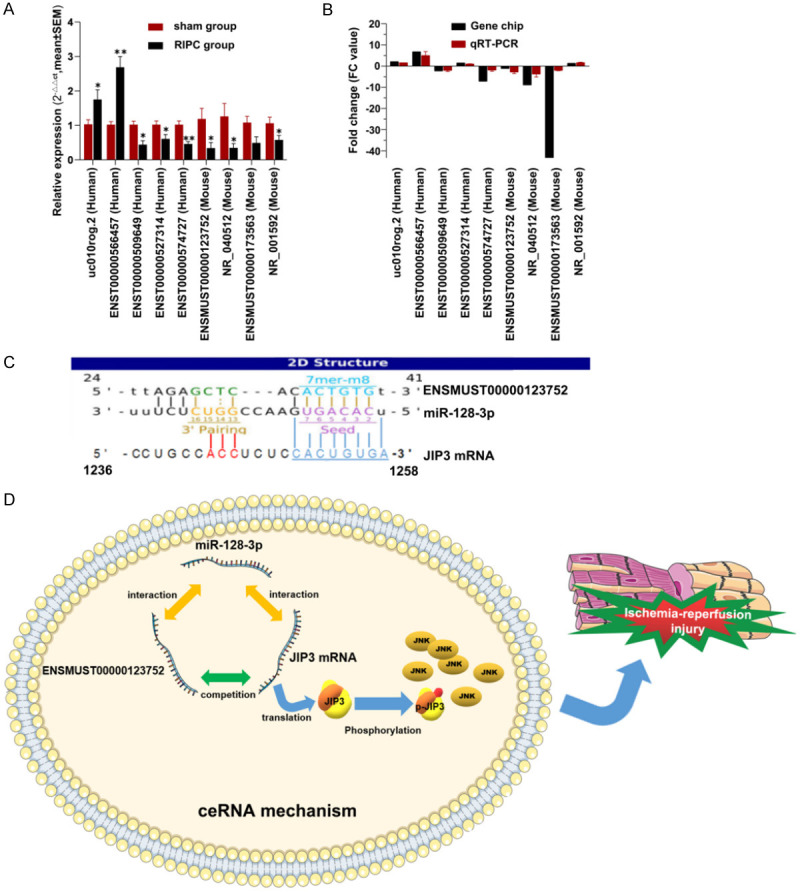
qRT-PCR validation of differential expressed lncRNAs. 8 LncRNAs were confirmed by qRT-PCR analysis to display significant changes between RIPC group and sham group (A). The data was expressed as the mean ± SEM (*P<0.05, **P<0.01). (B) qRT-PCR patterns of 9 lncRNAs were found to be consistent with that of the microarray data. The competing binding site was predicted by ceRNA mechanism analysis (C) and interaction has been shown in conception map (D).
CeRNA mechanism analysis of the homologous LncRNA ENSMUST00000123752
In order to facilitate the follow-up exploration and verification, ceRNA mechanism analysis was performed for the binding site prediction of mouse LncRNA (LncRNA ENSMUST00000123752), and it was named as LncRNA ENSMUST00000123752 ischemic preconditioning-related LncRNA1 (IPCRL1), and the corresponding homologous LncRNA ENST00000574727 was named as ischemic preconditioning-related LncRNA2 (IPCRL2). There was a complementary binding site found between IPCRL1 and miR-128-3p. Interestingly, it was noted that the predicted target protein JNK-interacting protein3 (JIP3, Mapk8ip3), and its mRNA could bind to the same binding site (Figure 5C). Additionally, CeRNA mechanism was shown in endogenous competing relation conception map (Figure 5D).
Discussion
MIRI remains a major problem in clinical practice and it is the basic pathophysiological process that occurs during the treatment of cardiopulmonary resuscitation, thrombolysis of myocardial infarction, cardiopulmonary bypass and so on. Therefore, finding safe and effective treatment modalities and drugs to alleviate MIRI remains one of the primary focuses of research in the field of cardiovascular medicine. When myocardial tissue has been subjected to non-fatal ischemia-reperfusion, it can often show adaptive compensatory responses with defensive and protective significances, which are called ischemic preconditioning. In recent years, myocardial ischemic preconditioning has become one of the most effective strategies for both the prevention and treatment of MIRI. However, RIPC has a more realistic and extensive application space as the implementation of direct ischemic preconditioning has been greatly limited in clinical practice. The protective effect of RIPC has been confirmed in several animal experiments and clinical studies and it has been found to be beneficial in protecting heart, brain, liver, kidney and other organs after ischemia-reperfusion. We found that after MIRI, myocardial tissue swelling, myocardial necrosis, myocardial fiber disorder and rupture was observed.
Besides protein-coding genes, non-coding RNAs have attracted significant attention in recent years and can regulate various biological processes. According to the findings of the Human Genome Project, the number of total protein-coding genes in humans accounts for less than 2% of the entire human genome sequence, and the remaining 90% of the non-coding sequences can be transcribed, resulting in a large number of non-coding RNAs. Moreover, as a result of the rapid development of high-throughput RNA sequencing technology, a large number of novel non-coding RNAs have been discovered in recent years. The most famous non-coding RNAs identified is microRNA (miRNA), which has been implicated to play a key role in regulating a variety of biological and pathological processes [13,14]. Yu et al. have reported previously that silencing LncRNA AK139328 can significantly up-regulate the expression of miR-204-3p and thereafter effectively inhibit cardiomyocyte autophagy, thus attenuating MIRI in diabetic mice [15]. On the contrary, Zhu et al. have reported that LncRNA AK139128 can mediate autophagy and apoptosis of cardiomyocytes after MIRI by targeting miR-499/FOXO4 axis [16]. Moreover, Liang et al. have revealed that the LncRNA ROR/miR-124-3p/TRAF6 axis can regulate both apoptosis and inflammation of human cardiomyocytes induced by hypoxia/reoxygenation [17]. We hypothesized in this study that differentially expressed LncRNAs after RIPC may also play an important role in MIRI, hence we used high-throughput LncRNA microarray to screen and potentially identify the differentially expressed LncRNAs in mouse and human myocardium in the early stage after RIPC.
In addition, without existing databases and limited reference information being available to reveal the functional annotations of various identified LncRNAs, a correlation between LncRNA and mRNA was constructed, and the potential functions of various differentially expressed mRNA was systematically deciphered through GO annotation and pathway analysis.
We observed that in human’s pathway analysis, MAPK signaling pathway, cAMP signaling pathway, Toll-like receptor signaling pathway and TNF signaling pathway were most significantly enriched in up-regulated genes, while NF-κB signaling and apoptosis pathways were mostly enriched in the down-regulated genes. For example, for NF-κB signaling pathway, mRNAs of various activating factors of this pathway such as TRADD, TNF R1, RANKL, LTB, LTA and crucial kinase IKKβ were markedly observed to be down-regulated. However, for apoptosis pathway p53 mRNA was found to be down-regulated, which in addition, to down-regulated TRADD, as well as TNF R1 can also be effectively coupled with apoptosis pathway. Meanwhile, the four pathways enriched in up-regulated genes appeared to be activated; however, this result cannot be explained by only studying the activation status of these pathways. In fact, among the various up-regulated genes in these different pathways, several product proteins of them belonged to anti-injury factors, and their up-regulation can be related to the antagonism of damage-related signaling pathways to some extent such as HSP72 in MAPK pathway, Hip1 at the terminal end of cAMP signaling pathway. Hip1 also plays an important role in Hedgehog signaling pathway and can improve cardiac angiogenesis, promote cardiac repairment and thereby enhance cardiac functions after myocardial infarction. The activation of Epac2-Rap1 signaling cascade in cAMP pathway is generally considered to exert adverse effects but prior studies have indicated that the existence of Epac2-Rap1 signal can significantly reduce the production of mitochondrial ROS and attenuate the susceptibility to myocardial arrhythmia [18]. In addition, numerous studies have shown that inhibition of cAMP-PKA pathway can significantly suppress apoptosis. Wu et al. found that the inhibition of miR-200a can attenuate the cAMP/PKA signal pathway by up-regulating the expression of DRD2 in PD rats, thus inhibiting the apoptosis of striatum [19]. Moreover, Xu et al. have reported that MIRI can increase DRG cAMP, and an intrathecal injection of selective cAMP-PKA inhibitors could effectively reduce myocardial injury [20].
Interestingly, the activation of these different signaling pathways reflect that the human’s heart in the early stage of RIPC may be in possible state where it can be co-expressing both pro-injury and anti-injury related factors simultaneously. However, because of the particularity of sample collections, we cannot completely rule out the possible influence of patients’ inherent cardiac pathological state on production of various pro-injury factors.
We noticed that in mouse’s pathway analysis, Hedgehog signaling pathway was predominantly enriched in up-regulated genes, while cAMP signaling pathway, dilated cardiomyopathy related pathway, adrenergic signaling in cardiomyocytes and inflammatory mediator regulation of TRP channels were mostly enriched in the down-regulated genes.
Interestingly, the majority of significant signaling pathways affected above are well known for their close relationship with apoptosis, inflammation and oxidative stress which are the most commonly observed pathological processes in MIRI. The remaining signaling pathways were directly or indirectly involved in maintenance of cardiac structure, as well as regulation of myocardial angiogenesis, myocardial regeneration and repairment. However, although the regulatory relationship and specific mechanisms between these differentially expressed LncRNAs and various signaling pathways cannot be determined at present, but these enriched signaling pathways have been found to be inextricably linked with MIRI, hence the differentially expressed LncRNA after RIPC may exhibit potential beneficial functions, thus establishing the significance of our findings. We further deepened the association through the homologous genes in these differentially expressed LncRNAs and identified the potential clinically meaningful targets and pathways for various LncRNAs, so as to clearly establish a complete chain of evidence related to RIPC-LncRNAs-biological targets-signaling pathways.
In fact, after homology analysis and qRT-PCR verification, a pair of human and mouse homologous LncRNAs (ENST00000574727 & ENSMUST00000123752) were identified for our further research object. ENST00000574727, IPCRL2, human LncRNA, also known as ALOX12P2-003 or ALOX12E, is located at chromosome 17 forward strand, with a length of 4326bp. ENSMUST00000123752, IPCRL1, mouse LncRNA, also known as Alox12e-002, Alox12-ps1; Alox12-ps2, is a 793 bp LncRNA, located at Chromosome 11 reverse strand. The functions of these LncRNAS have not been discovered and confirmed previously. We observed that demonstrated a similar down-regulation trend as that of JNK mRNA. Hence, their potential functions were deciphered through a ceRNA mechanism analysis and subsequently matching the complementary binding sites. Finally, a possible target for IPCRL1 through ceRNA mechanism analysis was established. This ceRNA mechanism analysis showed that there were several complementary matched sequences between the 3’-UTR of JIP3 (Mapk8ip3) mRNA and miR-128-3p seed site which could also complementarily bind to IPCRL1. This suggested that there may possibly be a competitive adsorption of miR-128-3p between IPCRL1 and JIP3 mRNA. Therefore, a novel hypothesis related to RIPC was established and it was hypothesized that through the LncRNA/miR-128-3p/JIP3 mRNA ceRNA mediated mechanisms, the robust activation of JNK pathway could be effectively inhibited thereby leading to the reduction of MIRI. However, further experimental demonstration in gene knockout animals, are required to validate these observations.
In recent years, miR-128-3p has been reported to be closely associated with myocardial infarction and myocardial ischemia-reperfusion injury. For instance, Liu et al. found that Selenomethionine could attenuate LPS-induced myocardial inflammation and oxidative stress through modulation of miR-128-3p/p38MAPK/NF-κB pathway, also clarified the negative regulation relationship between miR-128-3p and p38 MAPK [21]. On the contrary, Ma et al. discovered that the fat-1 transgenic myocardium had a better cardiac function, a smaller fibrotic area, and fewer apoptotic cardiomyocytes than the wild type myocardium after myocardial infarction. These findings indicated that the upregulation of 9 different miRNAs, including miR-128-3p, could be closely related to the enhancement of myocardial anti-apoptotic ability [22]. In addition, Chen et al. found that the inhibition of miR-128-3p by Tongxinluo can effectively protect the human cardiomyocytes from IRI via upregulation of p70s6k1/p-p70s6k1 cascade [23]. At the same time, other studies have proposed that miR-128-3p may be closely related to the MAPK signaling pathway [21,24,25], For example, JNK pathway can function as an important part of MAPK pathway. A few studies have confirmed that that miR-128-3p can negatively regulate the phosphorylation and activation of JNK pathway, for example, Xie et al. reported that miR-128-3p can negatively control the JNK/MAPK pathway by repressing multiple factors required for the phosphorylation of the JNK/MAPK pathway and thereby further promote cells from undifferentiated myoblasts to differentiate into skeletal muscle cells [26]. Moreover, in the differential expression profile of various mRNA, the down-regulation of JNK3 was also observed. Moreover, a series of prior high-quality studies related to the ceRNA mechanism of LncRNAs, have suggested that miR-128-3p can play a role of bridge connection [27-29]. It appears to be a similar case, as JIP family proteins have become an important research hotspot for heart diseases and injury, which has been mainly based on the irreplaceable role of JIP in JNK activation. More interestingly, different members belonging to JIP family, sucha s JIP1 and JIP3 actually have diametrically opposite effects. For example, He et al. concluded that the TNFα-activated JNK activity in H9c2 cells could be completely abolished by JIP-1 and when JIP-1 was over-expressed in cardiomyocytes, cell death was found to be significantly reduced after simulated myocardial infarction [31]. Xu et al. suggested that the depletion of endogenous JIP1 by siRNA treatment could significantly increase the IR-induced JNK activity, whereas siRNA-mediated depletion of endogenous JIP3 effectively inhibited JNK activity [32]. Ma et al. reported that JIP3-sliencing can significantly reduce myocardial hypertrophy by inactivating JNK [33]. Yin et al. used JIP3 siRNA to reduce the expression of JIP3 and found that the levels of MA2K7/p-JNK/cleaved caspase-3 decreased substantially [34] and the activation of JNK pathway was effectively suppressed. Overall findings suggest that, JIP3, as the direct substrate of JNK [30], can play an important role in the activation of JNK pathway.
Moreover, the result of the study of microRNA, which is another kind of non-coding RNA, which can control diverse biological functions and anti-microRNA therapy, has been successfully and safely used in the phase II clinical trials of hepatitis [35]. Thus based on our data and understanding, an in-depth study of the role of different LncRNAs in RIPC and elucidation of their potential mechanisms of action could lead to a rapid development of targeted therapy for cardiovascular diseases undergoing MIRI which could be significantly useful in clinical settings.
In summary, a number of differentially expressed LncRNAs were identified and validated, which may be closely related to MIRI. Additionally, several LncRNAs with stable expression and significant FC, including homologous LncRNAs IPCRL1 and IPCRL2 were selected and validated in this study. The data also suggested that IPCRL1 may mediate MIRI by targeting JIP3 activation through a ceRNA regulation mechanism. However, the current established hypothesis has to be further verified by several more experiments. Overall, by continuously exploring the functions and mechanisms of these LncRNAs, the mystery of RIPC could be unraveled and a novel protective mechanism of cardiovascular ischemia-reperfusion disease could be identified in future.
Acknowledgements
This work was supported by the Science and Technology Project of Medical and Health of Zhejiang Province (2017RC021), the Special Project for Significant New Drug Research and Development in the Major National Science and Technology Projects of China (2020ZX09201002), Key Discipline Program of Pediatric Surgery of Health Bureau of Zhejiang Province (No. 11-ZC27), Research Center for diagnosis and treatment of cardiac and vascular diaease of Zhejiang, P. R.China (JBZX-202001) and the National Natural Science Foundation of China (No. 82070274).
Disclosure of conflict of interest
None.
Abbreviations
- RIPC
Remote ischemic preconditioning
- LncRNA
Long noncoding RNA
- MIRI
Myocardial ischemia-reperfusion injury
- I/R
ischemia-reperfusion
- IRI
ischemia-reperfusion injury
- ELISA
enzyme linked immunosorbent assay
- TNF-α
tumor necrosis factor-α
- cTNI
cardiac troponin I
- qRT-PCR
Quantitative real-time polymerase chain reaction
- CeRNA
Competing endogenous RNA
- FC
fold change
- AAR
Area at risk
- INF
Infarct area
- HE
hematoxylin-eosin
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NCBI
National Center for Biotechnology Information
- FDR
false discovery rate
- IPCRL1
Ischemic preconditioning-related LncRNA1
- IPCRL2
Ischemic preconditioning-related LncRNA2
- MAPK
mitogen-activated protein kinase
- JIP3
JNK-interacting protein3
- TRP
Transient Receptor Potential
References
- 1.Hausenloy DJ, Yellon DM. Ischaemic conditioning and reperfusion injury. Nat Rev Cardiol. 2016;13:193–209. doi: 10.1038/nrcardio.2016.5. [DOI] [PubMed] [Google Scholar]
- 2.Zhao W, Meng R, Ma C, Hou B, Jiao L, Zhu F, Wu W, Shi J, Duan Y, Zhang R, Zhang J, Sun Y, Zhang H, Ling F, Wang Y, Feng W, Ding Y, Ovbiagele B, Ji X. Safety and efficacy of remote ischemic preconditioning in patients with severe carotid artery stenosis prior to carotid artery stenting: a proof-of-concept, randomized controlled trial. Circulation. 2017;135:1325. doi: 10.1161/CIRCULATIONAHA.116.024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skyschally A, Gent S, Amanakis G, Schulte C, Kleinbongard P, Heusch G. Across-species transfer of protection by remote ischemic preconditioning with species-specific myocardial signal transduction by RISK and SAFE pathways. Circ Res. 2015;117:279. doi: 10.1161/CIRCRESAHA.117.306878. [DOI] [PubMed] [Google Scholar]
- 4.Rassaf T, Totzeck M, Hendgen-Cotta UB, Shiva S, Heusch G, Kelm M. Circulating nitrite contributes to cardioprotection by remote ischemic preconditioning. Circ Res. 2014;114:1601–1610. doi: 10.1161/CIRCRESAHA.114.303822. [DOI] [PubMed] [Google Scholar]
- 5.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, De Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 6.Wang K, Liu F, Liu CY, An T, Zhang J, Zhou LY, Wang M, Dong YH, Li N, Gao JN, Zhao YF, Li PF. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23:1394–1405. doi: 10.1038/cdd.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon RA, Jaé N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Li G, Lu H, Li W, Li X, Liu H, Li X, Li T, Yu B. Expression profiling and ontology analysis of long noncoding RNAs in post-ischemic heart and their implied roles in ischemia/reperfusion injury. Gene. 2014;543:15–21. doi: 10.1016/j.gene.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Wang J, Xue Y, Fang A, Wu S, Huang K, Tao L, Wang J, Shen Y, Wang J, Pan L, Li L, Ji K. Microarray analysis reveals a potential role of lncRNA expression in 3,4-benzopyrene/angiotensin II-activated macrophage in abdominal aortic aneurysm. Stem Cells Int. 2017;2017:9495739. doi: 10.1155/2017/9495739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H, Tian H, Shi R, Mao X. lncRNA expression signatures in periodontitis revealed by microarray: the potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem. 2015;116:640–647. doi: 10.1002/jcb.25015. [DOI] [PubMed] [Google Scholar]
- 11.Zheng B, Liu H, Wang R, Xu S, Liu Y, Wang K, Hou X, Shen C, Wu J, Chen X, Wu P, Zhang G, Ji Z, Wang H, Xiao Y, Han J, Shi H, Zhao S. Expression signatures of long non-coding RNAs in early brain injury following experimental subarachnoid hemorrhage. Mol Med Rep. 2015;12:967–973. doi: 10.3892/mmr.2015.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S, Tao L, Wang J, Xu Z, Wang J, Xue Y, Huang K, Lin J, Li L, Ji K. Amifostine pretreatment attenuates myocardial ischemia/reperfusion injury by inhibiting apoptosis and oxidative stress. Oxid Med Cell Longev. 2017;2017:4130824. doi: 10.1155/2017/4130824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 14.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 15.Yu S, Dong B, Fang Z, Hu X, Tang L, Zhou S. Knockdown of lncRNA AK139328 alleviates myocardial ischaemia/reperfusion injury in diabetic mice via modulating miR-204-3p and inhibiting autophagy. J Cell Mol Med. 2018;22:4886–4898. doi: 10.1111/jcmm.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Z, Zhao C. LncRNA AK139128 promotes cardiomyocyte autophagy and apoptosis in myocardial hypoxia-reoxygenation injury. Life Sci. 2019;2019:116705. doi: 10.1016/j.lfs.2019.116705. [DOI] [PubMed] [Google Scholar]
- 17.Liang Y, Liu Q, Xu G, Zhang J, Chen Y, Hua F, Deng C, Hu Y. The lncRNA ROR/miR-124-3p/TRAF6 axis regulated the ischaemia reperfusion injury-induced inflammatory response in human cardiac myocytes. J Bioenerg Biomembr. 2019;51:381–392. doi: 10.1007/s10863-019-09812-9. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Kirton HM, Al-Owais M, Thireau J, Richard S, Peers C, Steele DS. Epac2-Rap1 signaling regulates reactive oxygen species production and susceptibility to cardiac arrhythmias. Antioxid Redox Signal. 2017;27:117–132. doi: 10.1089/ars.2015.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D, Wang S, Wen X, Han X, Wang Y, Shen M, Fan S, Zhuang J, Zhang Z, Shan Q, Li M, Hu B, Sun C, Lu J, Zheng Y. Inhibition of microRNA-200a upregulates the expression of striatal dopamine receptor D2 to repress apoptosis of striatum via the cAMP/PKA signaling pathway in rats with Parkinson’s disease. Cell Physiol Biochem. 2018;51:1600–1615. doi: 10.1159/000495649. [DOI] [PubMed] [Google Scholar]
- 20.Xu S, Xu Y, Cheng X, Huang C, Pan Y, Jin S, Xiong W, Zhang L, He S, Zhang Y. Inhibition of DRG-TRPV1 upregulation in myocardial ischemia contributes to exogenous cardioprotection. J Mol Cell Cardiol. 2020;135:175–184. doi: 10.1016/j.yjmcc.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Wang S, Zhang Q, Li X, Xu S. Selenomethionine alleviates LPS-induced chicken myocardial inflammation by regulating the miR-128-3p-p38 MAPK axis and oxidative stress. Metallomics. 2020;12:54–64. doi: 10.1039/c9mt00216b. [DOI] [PubMed] [Google Scholar]
- 22.Ma H, Chen P, Sang C, Huang D, Geng Q, Wang L. Modulation of apoptosis-related microRNAs following myocardial infarction in fat-1 transgenic mice vs wild-type mice. J Cell Mol Med. 2018;22:5698–5707. doi: 10.1111/jcmm.13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Xu C, Zhang J, Li Q, Cui H, Li X, Chang L, Tang R, Xu J, Tian X, Huang P, Xu J, Jin C, Yang Y. Inhibition of miR-128-3p by Tongxinluo protects human cardiomyocytes from ischemia/reperfusion injury via upregulation of p70s6k1/p-p70s6k1. Front Pharmacol. 2017;8:775. doi: 10.3389/fphar.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Tang M. Integrative analysis of mRNAs, miRNAs and lncRNAs in urban particulate matter SRM 1648a-treated EA. hy926 human endothelial cells. Chemosphere. 2019;233:711–723. doi: 10.1016/j.chemosphere.2019.05.294. [DOI] [PubMed] [Google Scholar]
- 25.Woo I, Christenson LK, Gunewardena S, Ingles SA, Thomas S, Ahmady A, Chung K, Bendikson K, Paulson R, McGinnis LK. Micro-RNAs involved in cellular proliferation have altered expression profiles in granulosa of young women with diminished ovarian reserve. J Assist Reprod Genet. 2018;35:1777–1786. doi: 10.1007/s10815-018-1239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie S, Li J, Chen H, Tan Y, Liu S, Zhang Y, Xu H, Yang J, Liu S, Zheng L, Huang M, Guo Y, Zhang Q, Zhou H, Qu L. Inhibition of the JNK/MAPK signaling pathway by myogenesis-associated miRNAs is required for skeletal muscle development. Cell Death Differ. 2018;25:1581–1597. doi: 10.1038/s41418-018-0063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Zhang R, Wei G, Dai S, Zhang X, Yang W, Li X, Bai C. Long non-coding RNA maternally expressed 3 increases the expression of neuron-specific genes by targeting miR-128-3p in all-trans retinoic acid-induced neurogenic differentiation from amniotic epithelial cells. Front Cell Dev Biol. 2019;7:342. doi: 10.3389/fcell.2019.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q, Meng Q, Qi M, Li F, Liu B. Shear-sensitive lncRNA AF131217.1 inhibits inflammation in huvecs via regulation of KLF4. Hypertension. 2019;73:e25–e34. doi: 10.1161/HYPERTENSIONAHA.118.12476. [DOI] [PubMed] [Google Scholar]
- 29.Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of miR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15:1139–1157. doi: 10.1007/s13311-018-0649-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zeke A, Misheva M, Reményi A, Bogoyevitch MA. JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol Mol Biol Rev. 2016;80:793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He H, Li H, Lin A, Gottlieb RA. Activation of the JNK pathway is important for cardiomyocyte death in response to simulated ischemia. Cell Death Differ. 1999;6:987–991. doi: 10.1038/sj.cdd.4400572. [DOI] [PubMed] [Google Scholar]
- 32.Xu B, Zhou Y, O K, Choy PC, Pierce GN, Siow YL. Regulation of stress-associated scaffold proteins JIP1 and JIP3 on the c-Jun NH2-terminal kinase in ischemia-reperfusion. Can J Physiol Pharmacol. 2010;88:1084–1092. doi: 10.1139/y10-088. [DOI] [PubMed] [Google Scholar]
- 33.Ma Q, Liu Y, Chen L. JIP3 deficiency attenuates cardiac hypertrophy by suppression of JNK pathway. Biochem Biophys Res Commun. 2018;503:1–7. doi: 10.1016/j.bbrc.2018.03.208. [DOI] [PubMed] [Google Scholar]
- 34.Yin C, Huang G, Sun X, Guo Z, Zhang J. DLK silencing attenuated neuron apoptosis through JIP3/MA2K7/JNK pathway in early brain injury after SAH in rats. Neurobiol Dis. 2017;103:133–143. doi: 10.1016/j.nbd.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]


